Abstract
Cancer cells need to escape immune surveillance for successful tumor growth. Loss of MHC class I has been described as a major immune evasion strategy in many cancers. MHC class I transactivator (CITA), NLRC5, has found to be a key transcription coactivator of MHC class I genes. Recent genetic studies revealed that NLRC5 is a major target for cancer immune evasion mechanism. The reduced expression or activity of NLRC5 caused by promoter methylation, copy number loss, or somatic mutations is associated with defective MHC class I expression, impaired cytotoxic T cell activation and poor patient prognosis. Here we review the role of NLRC5 in cancer immune evasion and the future prospects for cancer research.
Keywords: MHC class I, NLRC5, CITA, cancer, immune evasion
NLRC5/CITA and cancer
In 1909 Paul Ehrlich first proposed the idea that the human immune system usually surveys and suppresses cancerous cells [1]. In the 1950–60’s, Macfarlane Burnet and Lewis Tomas advocated the theory of cancer immune surveillance where the immune system can recognize cancer cells through tumor-specific antigens [2–5]. Since then, many studies revealed that cancer cells develop various strategies in order to escape from the immune system [6, 7]. Impaired function of the Major Histocompatibility Complex (MHC) class I antigen presentation pathway has been known as a major factor for cancer immune evasion [8–10]. The discovery of CITA, known as NLRC5 [nucleotide-binding domain and leucine-rich repeats containing (NLR) family, caspase activation and recruitment domain (CARD) domain containing 5], a key co-transactivator of the genes involved in MHC class I pathway, including HLA-A, HLA-B, HLA-C, β2-microglobulin (B2M), immunoproteasome components (LMP)2, LMP7 and transporter associated with antigen processing 1 (TAP1) [11, 12], pointed towards the idea that NLRC5 may play an important role in immune evasion of cancers. The idea is supported by the recent mouse and human studies showing that NLRC5 plays a pivotal role in tumor immunity by regulating the expression of MHC class I genes [13, 14]. Here we discuss the role of NLRC5 in cancer immune evasion and its future potential for diagnosis and treatments.
NLRC5: a key regulator of MHC class I gene expression
MHC class I
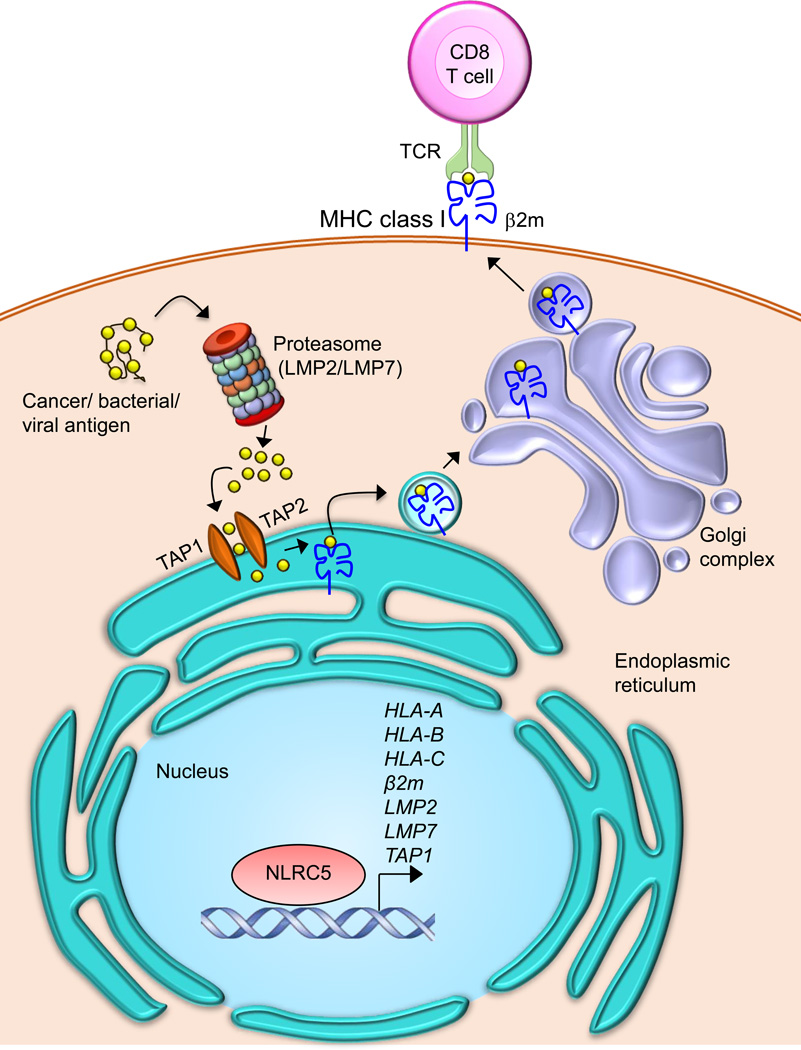
MHC class I molecules are composed of a polymorphic α chain and a non-polymorphic invariant β chain (B2M) linked by a non-covalent interaction. MHC class I genes are categorized into classical (MHC class Ia) (HLA-A, -B, -C, see Glossary) or non-classical (MHC class Ib) (HLA-E, -F, -G, see Glossary) genes [15, 16]. The antigenic peptides are produced from antigens in the cytoplasm, such as from cancer or bacterial antigens through proteasomal digestion by the immunoproteasome complex (Figure 1). These degraded peptides are then transported to the endoplasmic reticulum (ER) through TAP1 and TAP2. MHC class I molecules are loaded with the antigenic peptides in the lumen of the ER with the aid of the peptide loading complex (PLC) [17]. These peptide loaded MHC class I molecules are exported to the cell surface and presented to CD8+ T cells to elicit the antigen-specific CD8+ T cell activation (Figure 1).
Figure 1. NLRC5-dependent MHC class I antigen presentation pathways.
MHC class I molecules present cancer antigens that are degraded in the cytosol by the immunoproteasome to CD8+ T cells. The degraded antigenic peptides are transported into the endoplasmic reticulum via TAP1/TAP2, where peptides are loaded onto the MHC class I complex, which is composed of a heavy chain and β2-microglobulin (β2m). Peptide loaded MHC class I complexes are transported to the cell surface through the Golgi and are presented to CD8+ T cells. NLRC5 is involved in the transactivation of MHC class I and class I related genes, which include HLA-A, HLA-B, HLA-C, β 2m, LMP2, LMP7 and TAP1. TAP- transporter associated with antigen processing; TCR- T-cell receptor; LMP2/LMP7- large multifunctional peptidase 2 and 7.
MHC class I promoter
MHC class I molecules are expressed in all nucleated cells and are induced by type I (IFN-α/β) and type II (IFN-γ) interferons [18]. The MHC class I promoter encompasses conserved cis regulatory elements of enhancer A, the interferon stimulated response element (ISRE), and the SXY module, which includes W/S, X1, X2, and Y boxes [19]. The bidirectional promoter shared by TAP1 and LMP2 genes also contains a SXY module [20]. Constitutive and inducible transactivation of MHC class I genes are dependent on these cis-regulatory elements. Enhancer A contains one or two nuclear factor-κB (NF-κB) binding sites, whereas interferon regulatory factor 1 (IRF1) binds to ISRE [21]. While X1 box in the SXY module is bound by the regulatory factor X (RFX) complex, X2 box is bound by CREB or ATF1, and the Y box is bound by the NFY complex [19, 22]. These constitutively expressed proteins are assembled on the SXY module of the MHC class I promoter as a multi-protein/DNA complex termed enhanceosome [23–25]. NLRC5 binds to this multiprotein complex to form the CITA enhanceosome (see Text Box 1 for further details) and in turn transactivates MHC class I and related genes [26–28].
Box 1. CITA enhaceosome.
NLRC5 induces the transcriptional activation of MHC class I and related genes by binding within the SXY module of MHC class I promoters to form the CITA enhanceosome. The RFX trimeric complex is recruited to the X1 box and CREB/ATF1 is recruited to the X2 box. NLRC5 is recruited to the SXY module with the aid of RFX trimeric complex. Generation of this DNA/transcription factor complex on the MHC class I promoter drives the transactivation of MHC class I genes. It is therefore termed as MHC class I transactivator (CITA) enhanceosome.
NLRC5 as MHC Class I Transactivator
NLRC5 belongs to NLR protein family and holds a tripartite structure similar to other NLR proteins. NLRC5 consists of an atypical N-terminal CARD, centrally located nucleotide-binding domain (NBD), and long C-terminal leucine-rich repeats (LRRs) [27, 29–31]. NLRC5 is expressed in various human and mouse tissues, with high expression in hematopoietic cells [12, 29, 31–33]. Although NLRC5 expression is observed in non-hematopoietic tissues at various degrees, NLRC5 contribution to MHC expression in these tissues requires accurate exclusion of blood and tissue resident hematopoietic cells, which remains to be performed [26, 31]. Despite lacking a DNA-binding domain, NLRC5 can specifically associate with other DNA-binding proteins that are recruited to the SXY module of the MHC class I promoter to form the CITA enhanceosome (see Text Box 1 for further details) [12, 20, 26, 27, 32–36]. NLRC5 is involved in the trans-activation of the expression of genes involved in MHC class I antigen presentation pathway including classical MHC class Ia, non-classical MHC class Ib, B2M, LMP2, LMP7, and TAP1 [13, 20, 26, 27, 32].
NLRC5-mediated transactivation of MHC class I requires functional W/S, X1 and X2 motifs on the MHC class I promoter [35]. Particularly, NLRC5-dependent MHC class I gene expression requires the conserved consensus motif “TAACCTG” in the W/S box [20, 35]. NLRC5 interacts with the ankyrin repeats of RFXB (also known as RFXANK) protein in the RFX trimeric complex, which is bound to the X1 box [27]. RFXB lacks a DNA binding domain and hence binds through RFX5 and RFXAP, which possess the binding domain [37]. It has been shown that NLRC5 cooperates with CREB/ATF1 for the transactivation of MHC class I genes [35, 38]. However, NFY binding to the Y box or interaction of NLRC5 with NFY is yet to be determined [12, 27, 33, 36]. NLRC5 cooperates with chromatin remodeling enzymes such as histone acetyltransferases (e.g., GCN5, PCAF) that aid the NLRC5-dependent MHC class I promoter activation through epigenetic regulation [12, 27, 34]. Similar to MHC class I, NLRC5 expression is also induced by IFN-γ as well as less potently by type I IFNs through the activation of STAT1 [31, 39].
In vivo role of NLRC5 in class I-mediated immune responses
Several studies using Nlrc5-deficient mice demonstrated the critical in vivo role of NLRC5 in MHC class I-dependent antigen presentation. The dependence of MHC class I expression on NLRC5 was confirmed by observation of impaired expression of classical and non-classical MHC class-I genes (H2Db, H2Kb, H2-M3, H2-Qa1, and Tla) and genes involved in MHC class I antigen presentation pathways such as β2m, Tap1, Lmp2 in the thymus and spleen of Nlrc5-deficient mice [31, 32, 34, 40]. IFN-γ could not rescue the impaired expression of MHC class I genes, suggesting that NLRC5 is important for both constitutive and inducible gene expression [32]. Furthermore, non-lymphoid organs such as the kidney and intestine also show an impaired MHC class I expression [32]. In contrast, Nlrc5 deficiency has no effect on the expression of MHC class II transactivator (CIITA) or MHC class II genes (H2-Aα), indicative of a strict specificity of NLRC5 for the activation of MHC class I [31, 32, 34]. Mice deficient in Nlrc5 showed severe reduction in the surface expression of MHC class I (H2-D, H2-K) in CD4+ and CD8+ T cells, a significant defect in B cells, an intermediate reduction in macrophages, and a moderate defect in the bone-marrow derived dendritic cells (DCs) [31, 32, 34, 41]. The moderate decrease of expression of MHC class I molecules on the surface of DCs points towards the presence of compensatory mechanism to rescue the MHC class I deficiency in these antigen-presenting cells (see Outstanding Questions). This fact indicates that regulation of MHC class I expression by NLRC5 is cell type dependent.
Outstanding Questions Box.
What would determine the residual MHC class I expression if NLRC5 is not functional? Do residual levels of MHC class I support cancer immune surveillance?
Can the expression or methylation level of NLRC5 be a predictive biomarker for the efficacy of immunotherapies?
Would NLRC5-targeted immune editing therapy, which would augment NLRC5 expression by reducing methylation of NLRC5 promoter or increase the activity of NLRC5 using NLRC5 activating agents, be an efficient cancer therapy?
Would NLRC5-targeted therapy improve the efficacy of immunotherapies, such as cancer vaccine, checkpoint inhibitors, adoptive T cell therapy and chimeric antigen receptor (CAR)-modified T cell therapy?
The role of NLRC5 in CD8+ T cell activation was also elucidated using Nlrc5-deficient mice. While mice deficient in Ciita show drastic reduction in CD4+ T cells, mice deficient in Nlrc5 exhibit only a mild decrease in CD8+ T cells in peripheral lymphoid organs [31, 32]. The impaired proliferation and cytotoxic activity of CD8+ T cells co-cultured with Nlrc5-deficient antigen presenting cells in the presence of a specific antigenic peptide demonstrated a critical role for NLRC5 in MHC class I-dependent CD8+ T cell activation [31, 32].
Moreover, the role of NLRC5 in host protection against intracellular pathogens was demonstrated in infection studies. Nlrc5-deficient mice infected with Listeria monocytogenes showed an impaired induction of antigen-specific CD8+ T cell response and had higher bacterial loads in the spleen and liver, highlighting the role of NLRC5-dependent CD8+ T cell response in pathogen clearance [32, 40]. The role played by NLRC5 in the regulation of IFN-γ production through MHC class I-mediated CD8+ T cell activation, and the role of IFN-γ in the upregulation of NLRC5 expression suggest the existence of positive feedback loop for MHC class I-dependent immune responses. The IFN-γ-NLRC5-MHC class I axis of immune system is essential for a CD8+ T cell response and the efficient killing of intracellular pathogens.
NLRC5-mediated MHC class I expression in cancer
Most cancer cells are normally eliminated by the host immune surveillance system. Anti-tumor immune responses consist of multi-step processes, starting with the generation of cancer-specific antigenic peptides till the elimination of unwanted tumor cells [6, 7]. Antigens produced from cancer cells are captured by DCs for processing at the tumor site. These DCs migrate to regional lymph nodes and present the captured antigens to T cells, which provoke the response against cancer-specific antigens. The activated effector T cells, in turn traffic and infiltrate into the tumor site. The T cell receptor (TCR) on the cytotoxic T cells recognize the antigen-MHC class I complex on the surface of target cancer cell. Eventually, the cytotoxic T cells release the cytotoxic granules, such as perforin and granzymes to kill the target cancer cells [6, 7, 42].
Recent studies using both mouse models and human cancer patient samples revealed that NLRC5 is critical for anti-cancer immunity through the activation of cytotoxic CD8+ T cells. Using mouse models, Rodriguez et al. showed that NLRC5 elicits antitumor immunity by enhancing processing and presentation of tumor antigens to CD8+ T cells [13]. Mouse melanoma cell lines stably transfected with NLRC5 exhibited high expression level of MHC class I genes, LMP2, LMP7 and TAP1 [13]. These cells were able to present melanoma antigens, particularly when co-transfected with the co-stimulatory molecule CD80, and elicited CD8+ T cell activation efficiently. Upon subcutaneous implantation, melanoma cells showed markedly reduced tumor growth in C57BL/6 host but not in Rag-1−/− mice [13]. These findings indicate that NLRC5 is crucial for CD8+ T cell-mediated antitumor immunity via MHC class I-dependent antigen presentation. In humans, Yoshihama et al. showed that NLRC5 is also essential for MHC class I-dependent CD8+ T cell-mediated tumor immunity by analyzing large datasets (~8000 samples) from human cancer patients [14]. Analysis of the gene expression across 21 solid cancer types revealed that the level of NLRC5 expression is strongly correlated with the expression of MHC class I and related genes, such as HLA-A, HLA-B, HLA-C, B2M, LMP2, LMP7 and TAP1. [14]. Furthermore, the expression level of NLRC5 is also highly correlated with that of CD8A, PRF1 (perforin-1) and GZMA (granzyme A), which are associated with cytotoxic T cell activity in tumors [43]. Interestingly, NLRC5 expression has no significant correlation with the level of CD56, indicating a non-significant role in the recruitment of NK cells to tumors. Moreover, the expression level of CIITA has no correlation with the expression of MHC class I genes. These studies show that NLRC5 expression in cancer cells is specifically important for MHC class I genes and is crucial for the recruitment and activation of CD8+ T cells in human cancers.
NLRC5-targeted immune evasion in cancer
Cancer cells utilize multiple strategies to evade the host immune system [44–46], which include the recruitment of regulatory immune cell subsets or the induction of anergy in activated T cells [47], increased resistance to cytotoxic T-cell killing [48, 49], reduced recognition of tumor-associated antigens by effector T cells [42] and suppression of effector T cell function through CTLA4, PD-1 and/or PD-L1/−L2 [50–52]. Moreover, impaired MHC class I-mediated antigen presentation has been recognized as a major immune evasion mechanism in various cancers [8, 9, 53–56]. The high frequency of loss of MHC class I has been reported in 92% of cervical cancers [57], 71% of breast cancers [58], 64% of non-small cell lung cancers [59], 67% of esophageal squamous cell carcinomas [60] and in others [61–65]. Various molecular mechanisms reported account for the loss of MHC class I, including loss of heterozygosity in HLA-A, -B, -C or B2M genes [66, 67]; somatic mutations in HLA, B2M, TAP1/2 or LMPs [67–71]; HLA gene methylation [72, 73]; post-translational changes in TAP1 [71]; and defective JAK-STAT pathway at the IFN-γ receptor signaling [74]. However, the predominant molecular mechanism for HLA loss seems to be transcriptional [53], but has been undefined for many years.
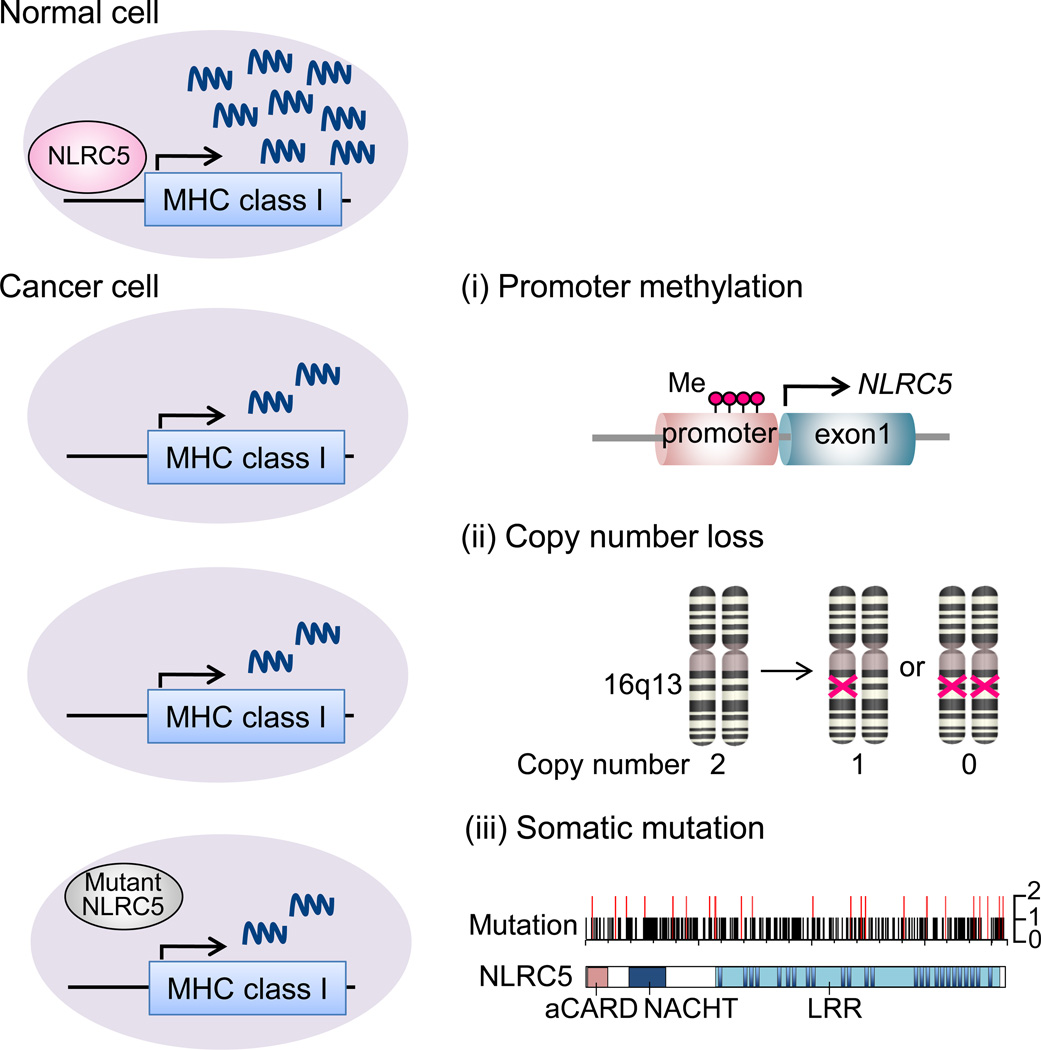
The discovery of NLRC5 as an MHC class I transactivator provided the opportunity to seek this transcriptional molecular mechanism. It is reported that NLRC5 is specifically reduced in cancer compared to corresponding normal tissues depending on the cancer type [14, 75]. This indicates that targeting NLRC5 is an immune evasion strategy in cancer. Further analysis identified three underlying molecular mechanisms targeting NLRC5 [14] (Figure 2).
Figure 2. Genetic and epigenetic changes in NLRC5 in cancer.
NLRC5 is a key regulator of MHC class I genes and loss of NLRC5 expression or activity leads to reduced expression of MHC class I and related genes, including HLA-A, -B, -C, B2M, LMP2, 7 and TAP1, in cancer cells. (i) DNA methylation of NLRC5 promoter is one of the major mechanisms that reduces NLRC5 expression. Red dots represent methylated site in the CpG island of the NLRC5 promoter. (ii) Copy number loss is another mechanism for reduced NLRC5 expression. The NLRC5 gene is located at the 16q13 locus in the human genome. Absence of both or one of the copies is defined as copy number loss. (iii) Somatic mutations in NLRC5 cause impaired activity of the NLRC5 protein. Mutations in NLRC5 found in one patient (black bar) and in at least two different patients (red bar), a total of 161 mutations are shown [14].
(i) DNA methylation of the NLRC5 gene promoter
Atypical methylation of CpG islands in promoter regions can transcriptionally suppress gene expression at the transcriptional level of genes that are unfavorable to cancer development [76]. DNA methylation at a CpG island in the NLRC5 promoter was quantified using a methylation-specific probe. Methylation of the NLRC5 promoter is negatively correlated with NLRC5 expression in many tumors [14]. Moreover, the methylation level of NLRC5 is also negatively correlated with the expression of NLRC5-dependent genes in the MHC class I pathway, including HLA-A, HLA-B, HLA-C, B2M, LMP2, LMP7 and TAP1. These findings indicate that methylation of NLRC5 gene is an important mechanism for immune evasion in many cancer types.
(ii) copy number loss in the NLRC5 gene
Alterations in gene copy number are frequently observed in cancer cells and are associated with altered gene expression levels [77, 78]. Copy number loss (copy number = 0 or 1) of the NLRC5 gene was observed in 28.6% of all cancer patients, highest frequency in ovarian cancer (72.2%), followed by breast cancer (59.9%) [14]. Cancer tissues with NLRC5 copy number loss showed reduced expression levels of NLRC5 as well as MHC class I genes [14].
(iii) somatic mutations in the NLRC5 gene
Somatic mutations are another major molecular mechanism of carcinogenesis [79]. Somatic mutations in NLRC5 were found in various cancers, such as colon cancer (8.6%) and melanoma (6.8%) [14]. Most of the mutations (58.5%) were missense and were distributed across the entire NLRC5 coding region with no obvious hot spots. Most mutant NLRC5 genes (54%) were incapable of activating the MHC class I promoter, even if forcibly expressed in cancer cells. Therefore, the majority of NLRC5 mutations in cancer cells are loss-of-function mutations, which cause an impaired MHC class I antigen presentation pathway [14].
Intriguingly, all aforementioned epigenetic and genetic alterations, including promoter methylation, copy number loss, or somatic mutations were observed in NLRC5 at significantly higher frequency than in any other MHC class I related genes, such as HLA-A, -B, -C, B2M, LMP2, LMP7 and TAP1 [14]. Perhaps, this is because cancer cells carrying alteration in NLRC5 may have higher chance of survival than carrying the alteration in other genes, though epigenetic or genetic alterations may happen randomly to any gene during tumor evolution. This suggests that NLRC5 is a major molecular target to induce immune evasion in cancer cells.
In summary, genetic and epigenetic changes in NLRC5 are frequently observed in various cancer cells and cause impaired expression or function of NLRC5, resulting in reduced expression of MHC class I and related genes. Reduced expression or activity of NLRC5 is associated with impaired recruitment and activation of CD8+ T cells, providing an immune evasion mechanism in various cancers [14].
NLRC5 as a prognostic biomarker
Considering the critical roles of NLRC5 in MHC class I-dependent immune responses and activation of CD8+ T cells in cancers, it was hypothesized that the expression level of NLRC5 might be associated with better prognosis of cancer patients [14]. Survival analysis to determine the impact of NLRC5 expression on overall survival of cancer patients using large cohorts stratified cancer patients into quartiles based on NLRC5 expression and methylation level. Analysis of 5-year survival in various cancer types revealed that the high NLRC5 expression quartile group shows significantly better survival compared with the low NLRC5 expression quartile in melanoma, rectal cancer, bladder cancer, uterine cancer, cervical cancer and head/neck cancer [14]. Kaplan-Meier survival analysis also showed that high NLRC5 expression is associated with significantly improved cumulative survival in melanoma, bladder and cervical cancers [14]. However, in certain cancer types, the NLRC5 expression and prognosis are not correlated. For example, brain tumors showed negative correlation, with poor prognosis in the cohort with high NLRC5 expression [14]. Perhaps, specific anatomy of brain might account for this. Brain tumors lead to inflammatory events by impaired blood–brain barrier and destruction of normal brain tissues [80, 81], resulting in the development of brain edema. Unlike other cancers, since brain mass is limited by the skull, brain edema is one major fatal complication of brain tumors. Thus, inflammation strongly influences the patient survival in brain tumor. Moreover, in some cancers, especially ones whose carcinogenesis is associated with inflammation such as lung or liver cancers, NLRC5 expression is not correlated with patient survival [14, 82]. Interestingly, high methylation level of NLRC5, but not of other MHC class I related genes (HLA-A, -B, -C, B2M, LMP2, LMP7, TAP1), was associated with poor survival in melanoma and bladder cancer patients, indicating that abnormal epigenetic changes in NLRC5 within cancer cells significantly impact clinical outcomes. These findings indicate that expression and methylation level of NLRC5 can be used as a prognostic biomarker to predict overall prognosis of cancer patients [14]. It would be interesting to see whether Nlrc5-deficient mice are susceptible to cancer types reported in the human study. Studies using animal models would further provide mechanistic information in NLRC5-dependent cancer immune surveillance.
Concluding Remarks
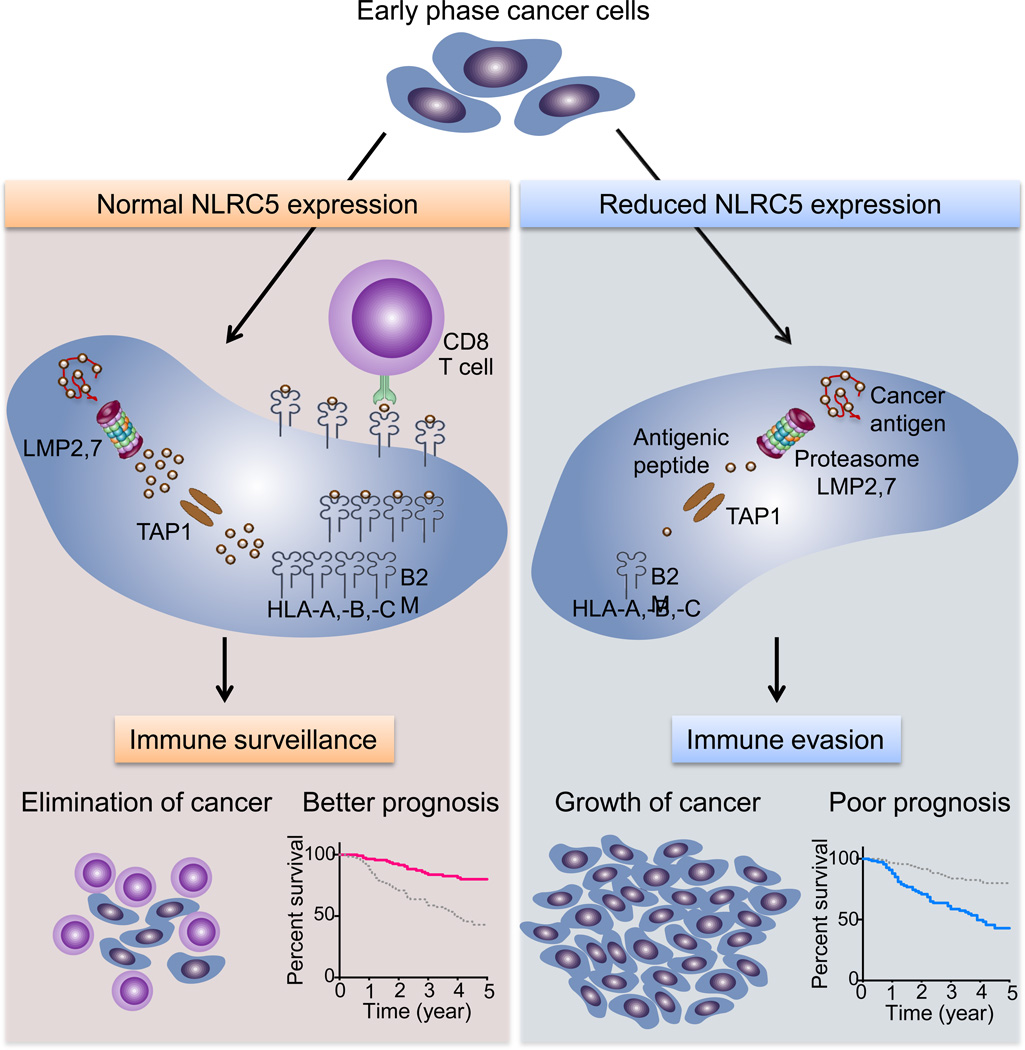
The discovery of NLRC5/CITA as a novel cancer immune evasion mechanism shed light on the understanding of how human cancers escape the immune system. Genetic and epigenetic alterations in cancer cells impact the expression and activity of NLRC5 and of MHC class I-dependent immune responses. DNA methylation of the NLRC5 promoter, copy number loss, and mutations in the NLRC5 gene result in reduced expression of MHC class I and of related genes along the antigen presentation pathway. Cancer cells with an impaired MHC class I system can escape from host immune surveillance of anti-tumor CD8+ T cells and promote efficient tumor development (Figure 3, Key Figure).
Figure 3. Key Figure. Targeting NLRC5 for immune evasion.
NLRC5-dependent MHC class I expression is essential for CD8+ T-cell-mediated anti-tumor responses. NLRC5 induces the expression of genes encoding critical components in the MHC class I pathway, which is essential for the cancer antigen presentation and recruitment/activation of cytotoxic CD8+ T cells. This anti-tumor immunity usually works effectively and cancer cells are eliminated at the early stage of development. Even if tumors are formed, patients have good prognosis. However, aberrant genetic and epigenetic changes in NLRC5 may occur during the evolution of cancer cells. Mutations, copy number loss or promoter methylation of NLRC5 cause impaired MHC class I antigen presentation due to reduced expression of HLA-A, -B, -C, B2M, LMP2, 7 and TAP1. These changes result in diminished anti-tumor CD8+ T-cell responses and a reduced infiltration in cancer tissues. Cancer cells, upon successful immune evasion, can achieve efficient tumor growth, leading to poor prognosis of the patients. Survival curves were generated for melanoma, with a high NLRC5 expression quartile group (pink line) and a low quartile group (blue line) [14].
Although the correlation between NLRC5 and patient prognosis is significant in multiple cancer types, the fact that NLRC5 is preferentially expressed in hematopoietic cells might question the causative role of NLRC5 in tumorigenesis and its potential relevance as a therapeutic target. Infiltrating cells such as TILs may contribute to the upregulation of NLRC5 or MHC-I expression in tumor tissues. On the other hand, several lines of evidence indicate that alterations in NLRC5 gene in cancer might be directly related to cancer development and that NLRC5 might be attractive target for cancer therapy. NLRC5 expression is reduced in multiple cancer types compared to corresponding normal tissues. This reduction cannot be explained by infiltration of hematopoietic cells. Furthermore, it is unlikely that aberrant epigenetic changes in NLRC5 promoter and genetic changes such as copy number loss or somatic mutations occur in infiltrated cells. Therefore, although NLRC5 expression in infiltrating cells may account for the upregulation of NLRC5 expression in some patient groups of various cancers, it is still likely that genetic and epigenetic changes in NLRC5 locus in cancer cells are associated with tumorigenesis and poor prognosis.
NLRC5 is not only a major target of cancer immune evasion but also the potential molecule useful for diagnosis and treatments (see Outstanding Questions). NLRC5 has potential as a predictive biomarker for response to immunotherapies. Available therapies, such as cancer vaccines, checkpoint inhibitors (e.g. anti-CTLA4, anti-PD-1/PDL1), adoptive T cells, chimeric antigen receptor (CAR)-modified T cells [83] and administration of immunostimulatory cytokines (IL-2, GM-CSF)[6, 7, 42, 84] rely on the activity of T cells. Since NLRC5 expression is required for efficient cytotoxic CD8+ T cell responses, it is reasonable to assume that NLRC5 is important for the effect of these immunotherapies.
Furthermore, NLRC5 can be a potential target for novel cancer therapy. NLRC5-targeted immune editing therapy would augment NLRC5 expression or activity by reducing methylation of NLRC5 or using NLRC5 activating agents. Although NLRC5-targeted therapy would be ineffective in the case of absence of functional NLRC5 due to homozygous mutations or zero copy number, such genetic defects of NLRC5 are very rare (data not published). Therefore, it would be possible that many cancers are candidates for NLRC5-targeted therapy. Additionally, NLRC5-targeted therapy would be useful to compensate the deficiency of cancer immunotherapy. Although checkpoint inhibitors are a breakthrough cancer treatment, the efficacy is still limited [85, 86]. Even if T cells are effectively activated, when cancer cells succeed to evade immune responses, therapeutic efficacy is restricted. To compensate this, various trials combining therapeutics are ongoing, such as anti-CTLA4 (ipilimumab) and anti-PD-1 (nivolumab) [87], vaccination and checkpoint inhibitors [88, 89], or targeted therapy (BRAF and MEK inhibitors) and checkpoint inhibitors [90]. Likewise, NLRC5-targeted therapy would make a promising candidate for a combination therapy with checkpoint inhibitors.
Trends Box.
MHC class I transactivator (CITA)/NLRC5 is a key transcriptional coactivator of genes involved in MHC class I antigen presentation pathway, including HLA-A, -B, -C, β2-microglobulin, LMP2, LMP7 and TAP1.
NLRC5 is a major target of immune evasion in cancer. Genetic and epigenetic alterations such as copy number loss, DNA methylation or somatic mutations cause the reduction of MHC class I expression or activity, leading to impaired CD8+ T-cell activation.
NLRC5 is a novel biomarker for cancer patient survival. Reduced expression and increased methylation levels of NLRC5 are associated with poor patient prognosis.
Acknowledgments
This work was supported by grants from the NIH (R01DK074738), Broad Foundation (IBD-0328), National Multiple Sclerosis Society (PP1779), CSTR, TAM Genomics and CTEHR to KSK, and fellowship from Sumitomo Life Welfare and Culture Foundation to SY.
Glossary
- Classical MHC class I (MHC class Ia)
Highly polymorphic MHC class I (HLA-A, HLA-B and HLA-C in human) molecules that present endogenous antigenic peptides to CD8+ T cells
- Non-classical MHC class I (MHC class Ib)
These are less polymorphic genes compared to classical MHC class I genes. This category of MHC class I genes includes HLA-E, HLA-F, HLA-G in human. Non-classical MHC class I molecules act as ligands for activating or inhibiting NK cells
- Peptide loading complex (PLC)
This complex comprises the MHC-I α chain and β2- microglobulin, TAP1, TAP2, tapasin, calreticulin, and ERp57. PLC aids in loading of the antigenic peptides onto MHC class I
- RFX trimeric complex
The RFX trimeric complex is composed of RFX5, RFXAP, and RFXANK/RFXB. This trimeric complex binds to MHC class I promoter along with other transcription factors to transactivate MHC class I expression
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ehrlich P. Ueber den jetzigen Stand der Karzinomforschung. Ned Tijdschr Geneeskd. 1909:73–290. [Google Scholar]
- 2.Burnet M. Cancer: a biological approach. III. Viruses associated with neoplastic conditions. IV. Practical applications. British medical journal. 1957;1:841–847. doi: 10.1136/bmj.1.5023.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas L. On immunosurveillance in human cancer. The Yale journal of biology and medicine. 1982;55:329–333. [PMC free article] [PubMed] [Google Scholar]
- 4.Burnet FM. The concept of immunological surveillance. Progress in experimental tumor research. 1970;13:1–27. doi: 10.1159/000386035. [DOI] [PubMed] [Google Scholar]
- 5.Thomas L. Discussion. In: Lawrence HS, editor. Cellular and Humoral Aspects of the Hypersensitive States. New York: Hoeber-Harper; 1959. pp. 529–532. [Google Scholar]
- 6.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Motz GT, Coukos G. Deciphering and reversing tumor immune suppression. Immunity. 2013;39:61–73. doi: 10.1016/j.immuni.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrone S, Marincola FM. Loss of HLA class I antigens by melanoma cells: molecular mechanisms, functional significance and clinical relevance. Immunology today. 1995;16:487–494. doi: 10.1016/0167-5699(95)80033-6. [DOI] [PubMed] [Google Scholar]
- 9.Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;27:5869–5885. doi: 10.1038/onc.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aptsiauri N, et al. Regressing and progressing metastatic lesions: resistance to immunotherapy is predetermined by irreversible HLA class I antigen alterations. Cancer immunology, immunotherapy : CII. 2008;57:1727–1733. doi: 10.1007/s00262-008-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meissner TB, et al. NLRC5: a newly discovered MHC class I transactivator (CITA) Microbes Infect. 2012;14:477–484. doi: 10.1016/j.micinf.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kobayashi KS, van den Elsen PJ. NLRC5: a key regulator of MHC class I-dependent immune responses. Nature reviews. Immunology. 2012;12:813–820. doi: 10.1038/nri3339. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez GM, et al. NLRC5 elicits antitumor immunity by enhancing processing and presentation of tumor antigens to CD8(+) T lymphocytes. Oncoimmunology. 2016;5:e1151593. doi: 10.1080/2162402X.2016.1151593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshihama S, et al. NLRC5/MHC class I transactivator is a target for immune evasion in cancer. Proceedings of the National Academy of Sciences of the United States of America. 2016 doi: 10.1073/pnas.1602069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao GF, et al. Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8alphaalpha. The Journal of biological chemistry. 2000;275:15232–15238. doi: 10.1074/jbc.275.20.15232. [DOI] [PubMed] [Google Scholar]
- 16.Parham P, et al. Nature of polymorphism in HLA-A, -B, and -C molecules. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blum JS, et al. Pathways of antigen processing. Annual review of immunology. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van den Elsen PJ, et al. Transcriptional regulation of antigen presentation. Current opinion in immunology. 2004;16:67–75. doi: 10.1016/j.coi.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 19.Gobin SJ, et al. The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor kappa B. Journal of immunology. 1998;161:2276–2283. [PubMed] [Google Scholar]
- 20.Ludigs K, et al. NLRC5 exclusively transactivates MHC class I and related genes through a distinctive SXY module. PLoS Genet. 2015;11:e1005088. doi: 10.1371/journal.pgen.1005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gobin SJ, et al. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601–611. doi: 10.1016/s1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- 22.Hobart M, et al. IFN regulatory factor-1 plays a central role in the regulation of the expression of class I and II MHC genes in vivo. Journal of immunology. 1997;158:4260–4269. [PubMed] [Google Scholar]
- 23.Gobin SJ, et al. The MHC-specific enhanceosome and its role in MHC class I and beta(2)-microglobulin gene transactivation. Journal of immunology. 2001;167:5175–5184. doi: 10.4049/jimmunol.167.9.5175. [DOI] [PubMed] [Google Scholar]
- 24.Moreno CS, et al. Purified X2 binding protein (X2BP) cooperatively binds the class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. Journal of immunology. 1995;155:4313–4321. [PubMed] [Google Scholar]
- 25.Moreno CS, et al. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 26.Meissner TB, et al. NLR family member NLRC5 is a transcriptional regulator of MHC class I genes. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:13794–13799. doi: 10.1073/pnas.1008684107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meissner TB, et al. The nucleotide-binding domain of NLRC5 is critical for nuclear import and transactivation activity. Biochem Biophys Res Commun. 2012;418:786–791. doi: 10.1016/j.bbrc.2012.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neerincx A, et al. NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. Journal of immunology. 2012;188:4940–4950. doi: 10.4049/jimmunol.1103136. [DOI] [PubMed] [Google Scholar]
- 29.Benko S, et al. NLRC5 limits the activation of inflammatory pathways. Journal of immunology. 2010;185:1681–1691. doi: 10.4049/jimmunol.0903900. [DOI] [PubMed] [Google Scholar]
- 30.Gutte PG, et al. Unusual structural features revealed by the solution NMR structure of the NLRC5 caspase recruitment domain. Biochemistry. 2014;53:3106–3117. doi: 10.1021/bi500177x. [DOI] [PubMed] [Google Scholar]
- 31.Staehli F, et al. NLRC5 deficiency selectively impairs MHC class I-dependent lymphocyte killing by cytotoxic T cells. Journal of immunology. 2012;188:3820–3828. doi: 10.4049/jimmunol.1102671. [DOI] [PubMed] [Google Scholar]
- 32.Biswas A, et al. Cutting edge: impaired MHC class I expression in mice deficient for nlrc5/class I transactivator. Journal of immunology. 2012;189:516–520. doi: 10.4049/jimmunol.1200064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neerincx A, et al. NLRC5, at the Heart of Antigen Presentation. Front Immunol. 2013;4:397. doi: 10.3389/fimmu.2013.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins GR, et al. Regulation of Class I Major Histocompatibility Complex (MHC) by Nucleotide-binding Domain, Leucine-rich Repeat-containing (NLR) Proteins. The Journal of biological chemistry. 2012;287:24294–24303. doi: 10.1074/jbc.M112.364604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner TB, et al. NLRC5 Cooperates with the RFX Transcription Factor Complex To Induce MHC Class I Gene Expression. Journal of immunology. 2012;188:4951–4958. doi: 10.4049/jimmunol.1103160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neerincx A, et al. A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. The Journal of biological chemistry. 2010;285:26223–26232. doi: 10.1074/jbc.M110.109736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steimle V, et al. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 38.Raval A, et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol Cell. 2001;7:105–115. doi: 10.1016/s1097-2765(01)00159-9. [DOI] [PubMed] [Google Scholar]
- 39.Tong Y, et al. Enhanced TLR-induced NF-kappaB signaling and type I interferon responses in NLRC5 deficient mice. Cell Res. 2012;22:822–835. doi: 10.1038/cr.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao Y, et al. NLRC5 regulates MHC class I antigen presentation in host defense against intracellular pathogens. Cell Res. 2012;22:836–847. doi: 10.1038/cr.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rota G, et al. T Cell Priming by Activated Nlrc5-Deficient Dendritic Cells Is Unaffected despite Partially Reduced MHC Class I Levels. Journal of immunology. 2016;196:2939–2946. doi: 10.4049/jimmunol.1502084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellman I, et al. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rooney MS, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schreiber RD, et al. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 45.Dunn GP, et al. Cancer immunoediting: from immunosurveillance to tumor escape. Nature immunology. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 46.Zitvogel L, et al. Cancer despite immunosurveillance: immunoselection and immunosubversion. Nature reviews. Immunology. 2006;6:715–727. doi: 10.1038/nri1936. [DOI] [PubMed] [Google Scholar]
- 47.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nature reviews. Cancer. 2005;5:263–274. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 48.Medema JP, et al. Blockade of the granzyme B/perforin pathway through overexpression of the serine protease inhibitor PI-9/SPI-6 constitutes a mechanism for immune escape by tumors. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11515–11520. doi: 10.1073/pnas.201398198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ochsenbein AF. Immunological ignorance of solid tumors. Springer seminars in immunopathology. 2005;27:19–35. doi: 10.1007/s00281-004-0192-0. [DOI] [PubMed] [Google Scholar]
- 50.Uyttenhove C, et al. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nature medicine. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 51.Terness P, et al. Inhibition of allogeneic T cell proliferation by indoleamine 2,3-dioxygenase-expressing dendritic cells: mediation of suppression by tryptophan metabolites. The Journal of experimental medicine. 2002;196:447–457. doi: 10.1084/jem.20020052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Keir ME, et al. PD-1 and its ligands in tolerance and immunity. Annual review of immunology. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garrido F, et al. "Hard" and "soft" lesions underlying the HLA class I alterations in cancer cells: implications for immunotherapy. International journal of cancer. 2010;127:249–256. doi: 10.1002/ijc.25270. [DOI] [PubMed] [Google Scholar]
- 54.Seliger B. Novel insights into the molecular mechanisms of HLA class I abnormalities. Cancer immunology, immunotherapy : CII. 2012;61:249–254. doi: 10.1007/s00262-011-1153-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Leone P, et al. MHC class I antigen processing and presenting machinery: organization, function, and defects in tumor cells. Journal of the National Cancer Institute. 2013;105:1172–1187. doi: 10.1093/jnci/djt184. [DOI] [PubMed] [Google Scholar]
- 56.Garrido F, et al. The urgent need to recover MHC class I in cancers for effective immunotherapy. Current opinion in immunology. 2016;39:44–51. doi: 10.1016/j.coi.2015.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vermeulen CF, et al. Frequent HLA class I loss is an early event in cervical carcinogenesis. Hum Immunol. 2005;66:1167–1173. doi: 10.1016/j.humimm.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 58.Inoue M, et al. Expression of MHC Class I on breast cancer cells correlates inversely with HER2 expression. Oncoimmunology. 2012;1:1104–1110. doi: 10.4161/onci.21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hanagiri T, et al. Clinical significance of expression of cancer/testis antigen and down-regulation of HLA class-I in patients with stage I non-small cell lung cancer. Anticancer Res. 2013;33:2123–2128. [PubMed] [Google Scholar]
- 60.Qifeng S, et al. Methylation of the promoter of human leukocyte antigen class I in human esophageal squamous cell carcinoma and its histopathological characteristics. J Thorac Cardiovasc Surg. 2011;141:808–814. doi: 10.1016/j.jtcvs.2010.04.031. [DOI] [PubMed] [Google Scholar]
- 61.Hoves S, et al. In situ analysis of the antigen-processing machinery in acute myeloid leukaemic blasts by tissue microarray. Leukemia. 2009;23:877–885. doi: 10.1038/leu.2008.391. [DOI] [PubMed] [Google Scholar]
- 62.Meissner M, et al. Defects in the human leukocyte antigen class I antigen processing machinery in head and neck squamous cell carcinoma: association with clinical outcome. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:2552–2560. doi: 10.1158/1078-0432.CCR-04-2146. [DOI] [PubMed] [Google Scholar]
- 63.Kageshita T, et al. Down-regulation of HLA class I antigen-processing molecules in malignant melanoma: association with disease progression. The American journal of pathology. 1999;154:745–754. doi: 10.1016/S0002-9440(10)65321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pandha H, et al. Loss of expression of antigen-presenting molecules in human pancreatic cancer and pancreatic cancer cell lines. Clinical and experimental immunology. 2007;148:127–135. doi: 10.1111/j.1365-2249.2006.03289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaklamanis L, et al. Loss of major histocompatibility complex-encoded transporter associated with antigen presentation (TAP) in colorectal cancer. The American journal of pathology. 1994;145:505–509. [PMC free article] [PubMed] [Google Scholar]
- 66.Maleno I, et al. Multiple mechanisms generate HLA class I altered phenotypes in laryngeal carcinomas: high frequency of HLA haplotype loss associated with loss of heterozygosity in chromosome region 6p21. Cancer immunology, immunotherapy : CII. 2002;51:389–396. doi: 10.1007/s00262-002-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paschen A, et al. Complete loss of HLA class I antigen expression on melanoma cells: a result of successive mutational events. International journal of cancer. 2003;103:759–767. doi: 10.1002/ijc.10906. [DOI] [PubMed] [Google Scholar]
- 68.Koopman LA, et al. Human leukocyte antigen class I gene mutations in cervical cancer. Journal of the National Cancer Institute. 1999;91:1669–1677. doi: 10.1093/jnci/91.19.1669. [DOI] [PubMed] [Google Scholar]
- 69.Benitez R, et al. Mutations of the beta2-microglobulin gene result in a lack of HLA class I molecules on melanoma cells of two patients immunized with MAGE peptides. Tissue antigens. 1998;52:520–529. doi: 10.1111/j.1399-0039.1998.tb03082.x. [DOI] [PubMed] [Google Scholar]
- 70.Chen HL, et al. A functionally defective allele of TAP1 results in loss of MHC class I antigen presentation in a human lung cancer. Nature genetics. 1996;13:210–213. doi: 10.1038/ng0696-210. [DOI] [PubMed] [Google Scholar]
- 71.Seliger B, et al. Immune escape of melanoma: first evidence of structural alterations in two distinct components of the MHC class I antigen processing pathway. Cancer research. 2001;61:8647–8650. [PubMed] [Google Scholar]
- 72.Serrano A, et al. Rexpression of HLA class I antigens and restoration of antigen-specific CTL response in melanoma cells following 5-aza-2’-deoxycytidine treatment. International journal of cancer. 2001;94:243–251. doi: 10.1002/ijc.1452. [DOI] [PubMed] [Google Scholar]
- 73.Nie Y, et al. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. [DOI] [PubMed] [Google Scholar]
- 74.Seliger B, et al. IFN inducibility of major histocompatibility antigens in tumors. Advances in cancer research. 2008;101:249–276. doi: 10.1016/S0065-230X(08)00407-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu R, et al. Expression profile of innate immune receptors, NLRs and AIM2, in human colorectal cancer: correlation with cancer stages and inflammasome components. Oncotarget. 2015;6:33456–33469. doi: 10.18632/oncotarget.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 77.Albertson DG, et al. Chromosome aberrations in solid tumors. Nature genetics. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 78.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–1558. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 80.Fan Z, et al. Dexamethasone alleviates tumor-associated brain damage and angiogenesis. PLoS One. 2014;9:e93264. doi: 10.1371/journal.pone.0093264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dubois LG, et al. Gliomas and the vascular fragility of the blood brain barrier. Front Cell Neurosci. 2014;8:418. doi: 10.3389/fncel.2014.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li X, et al. NLRC5 expression in tumors and its role as a negative prognostic indicator in stage III non-small-cell lung cancer patients. Oncology letters. 2015;10:1533–1540. doi: 10.3892/ol.2015.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Porter DL, et al. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. The New England journal of medicine. 2011;365:725–733. doi: 10.1056/NEJMoa1103849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 85.Weber JS, et al. Phase I/II study of ipilimumab for patients with metastatic melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:5950–5956. doi: 10.1200/JCO.2008.16.1927. [DOI] [PubMed] [Google Scholar]
- 86.Yang JC, et al. Ipilimumab (anti-CTLA4 antibody) causes regression of metastatic renal cell cancer associated with enteritis and hypophysitis. Journal of immunotherapy. 2007;30:825–830. doi: 10.1097/CJI.0b013e318156e47e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. The New England journal of medicine. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Duraiswamy J, et al. Dual blockade of PD-1 and CTLA-4 combined with tumor vaccine effectively restores T-cell rejection function in tumors. Cancer research. 2013;73:3591–3603. doi: 10.1158/0008-5472.CAN-12-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim T, et al. Combining targeted therapy and immune checkpoint inhibitors in the treatment of metastatic melanoma. Cancer biology & medicine. 2014;11:237–246. doi: 10.7497/j.issn.2095-3941.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]