Abstract
Epidemiological studies point to race as a determining factor in cancer susceptibility. In US registries recording cancer incidence and survival by race (distinguishing “Black versus White”), individuals of African ancestry have a globally increased risk of malignancies compared to Caucasians and Asian Americans. Differences in socioeconomic status and health care access play a key role. However, the lesser disease susceptibility of Hispanic populations with comparable life-styles and socioeconomic status as African Americans, (“Hispanic paradox”) points to the concomitant importance of genetic determinants. Here, we overview the molecular basis of racial disparity in cancer susceptibility ranging from genetic polymorphisms and cancer-driver gene mutations to obesity, chronic inflammation and immune responses. We discuss implications for race-adapted cancer screening programs and clinical trials to reduce disparities in cancer burden.
Keywords: Racial disparities, Racial differences, Cancer, African, Asian
Epidemiological evidence for racial differences in cancer susceptibility and survival
Race refers to a population with common genetic and phenotypic features that separates them from other populations. Ethnicity pertains to the different cultural, socioeconomic, religious properties, including customs, language, diet and cultural identity [1]. The association of race with political ideologies and the abuse of science to promote racism have rendered the term race itself problematic. However, since this review deals with the biological basis of disparities in cancer risk we are going to employ the term “race” rather than “ethnicity”. Currently the United States Census Bureau defines six race categories: White or Caucasian, Black or African American, Asian, American Indian or Alaska Native, Native Hawaiian or Other Pacific Islander and “some other race”. Consistent with this, Hispanics refers to people with historical or cultural relationship with Spain, regardless of race, while individuals originating from Middle East or North Africa belong to the “White” race with no further distinctioni. Epidemiological data highlight large racial disparities in incidence and survival of many cancer types [2, 3]. The most comprehensive data on racial differences are derived from US cancer registries that distinguish among races as indicated aboveii. According to these, African Americans, referred to as “Black”, have higher incidence and lesser survival of all combined malignancies relative to individuals of the “White” population (Figure 1) [4]. These differences were historically attributed to confounding socio-economical and behavioral factors, such as diet, alcohol abuse, smoking, and access to screening and treatment. However, there is evidence that the survival disparity persists after normalization for these factors and in equal access settings, such as in the US Military Health System [5, 6]. In this context, while differences in incidence of specific cancer types such as head and neck squamous cell carcinoma (HNSCC) disappeared or were even reversed over the last 20 years, the survival gap between “Black and White” people remained, independent of stage at diagnosis and treatment (Figure 2 A, B) [2]. Interestingly, survival differences between Black and White patients in this non sex-related cancer type are limited to the male population, pointing to the interplay between racial and sex determinants of cancer susceptibility. As we previously reviewed, females have a generally lower cancer risk than males [7]. However, the importance of race-related determinants of cancer susceptibility goes beyond sex as indicated by the greater incidence and/or lower survival of Black versus White patients even in sex-specific cancers, such as prostate, breast and cervix (Figure 2 C–F).
Figure 1. Racial differences in cancer incidence and survival.
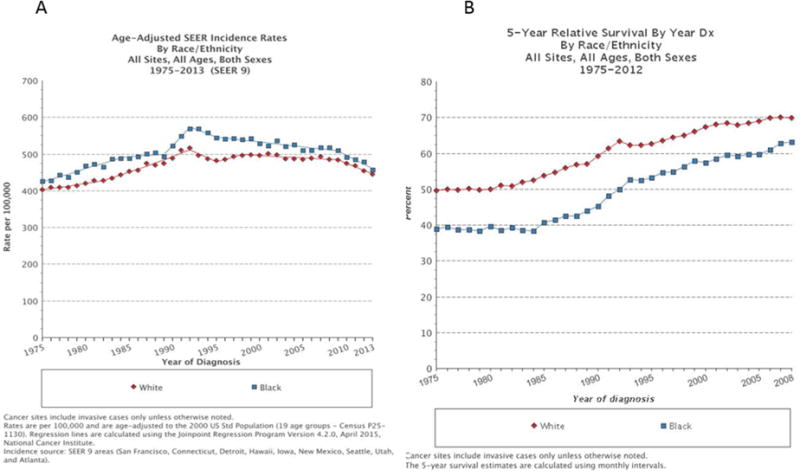
Incidence (A) and 5-year survival (B) of all cancer registered cancer sites for White and Black, all ages and both sexes, confounded for the years 1975–2013 and 1975–2012, respectively.
Data retrieved from SEER 2012ii
Figure 2. Persistence of survival disparity between Black and White over time.
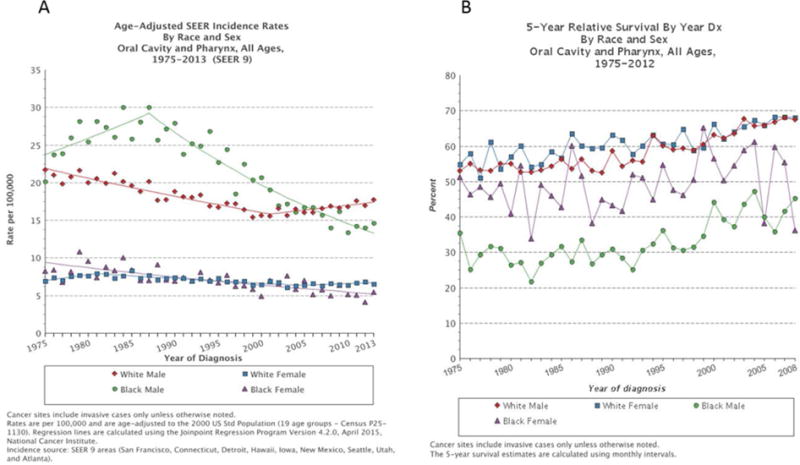
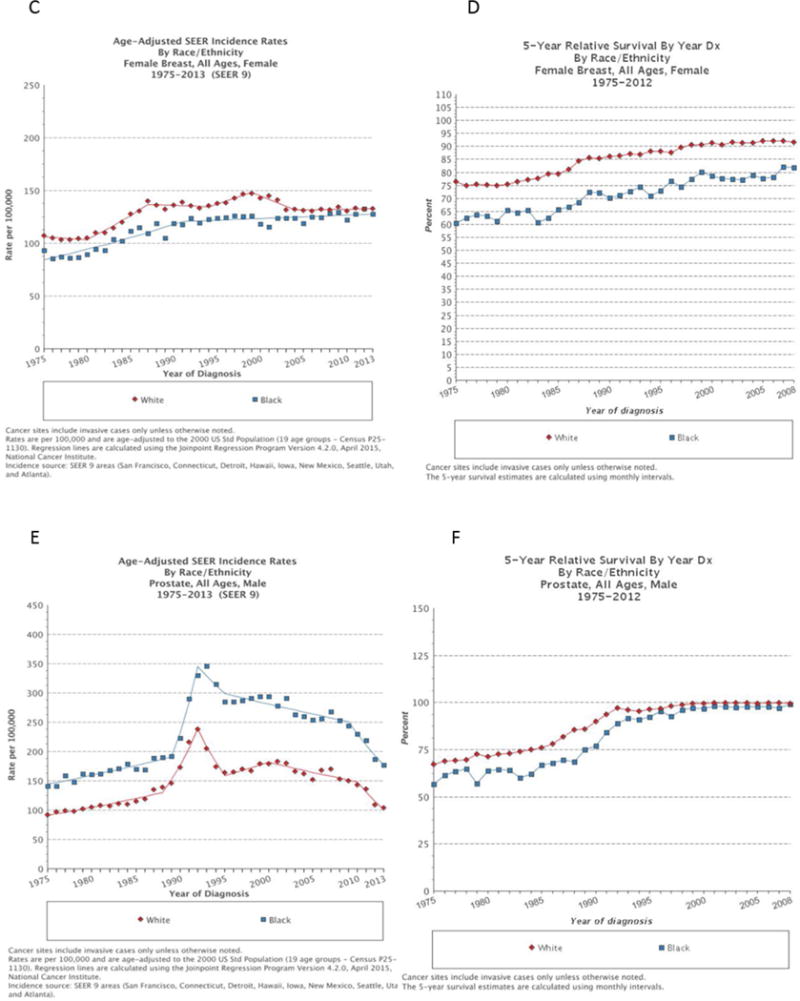
Despite a significant decline in incidence of head and neck squamous cell carcinoma (HNSCC) in Black men compared to White men (A) over the time period of 1975–2013, with currently even lower incidence rates among Black men, the survival disparity remains (B). In Black women the incidence (C) and survival (D) of breast cancer are both lower compared to White women. In prostate cancer, the higher incidence (E) in Black men persists, while the survival (F) differences diminish over time.
Additional epidemiological data show a better outcome in cancer patients with Hispanic versus African American background, in spite of comparable socioeconomic status, a phenomenon known as the “Hispanic paradox” [8]. Besides genetic factors, differences in life styles, specifically diet, have been proposed as a possible explanation for this observation [9]. Asian Americans have also a generally lower cancer risk than all other racial groups [3] and even in this case there is an interplay between genetic differences and a variety of behavioral/environmental risk factors. This review investigates possible molecular determinants of disparity in cancer susceptibility among different races, focusing in particular on African Americans relative to Caucasians and Asians, given the wealth of available information on these three races. We summarize the evidence of racial determinants of different susceptibility to various cancer types, ranging from genetic polymorphisms, epigenetic alterations and cancer-driver gene mutations, to immune/inflammatory responses and obesity, and point to future directions of investigation on as yet poorly explored areas, like cancer initiating cells and tumor microenvironment.
Genetic diversity
Racial differences in a wide variety of phenotypes and susceptibility to diseases can be attributed in part to genomic diversity. Comparative studies show greater genetic diversity and lower levels of linkage disequilibrium in African populations relative to all other Non-African populations [10]. This has been attributed to the origin of Hominids in Africa >900,000 years ago, with internal migrations and the bottleneck of migrating populations towards the rest of the world about 100’000 years ago (“Out of Africa” hypothesis) [11]. In addition, some interbreeding occurred after the spreading of Homo sapiens into the Euro-Asian continent with Neanderthals, resulting in the presence of 1.5–4% of Neanderthal DNA in the genome of modern Eurasians [12].
Neanderthal alleles have been linked with higher risk for sun-induced skin precancerous lesions (actinic keratosis) in population of Caucasian ancestry compared to Africans who develop rarely sun-induced skin cancers. A mutation in the melanocortin 1 receptor (MC1R) that reduces receptor activity and is associated with pale skin color and red hair in individuals of European ancestry has been identified in Neanderthal DNA [13]. Molecular studies revealed that MC1R is implicated in DNA repair and cell survival pathways, which helps explaining the increased melanoma risk of individuals harboring the non-functional “red hair color MC1R” variant [14].
Pigmentation is obviously protective against UV-induced cancers of the skin and can explain the different spectrum of skin tumors arising as a consequence of immunosuppression in organ transplant patients of Caucasian versus African ancestry. In fact, in a large South-African study and data comparison with transplant centers worldwide, the incidence of skin tumors was similar between “White” and “Non-White” (black and mixed-race) patients, while the tumor type was significantly different: squamous cell cancer (SCCs) and basal cell cancer of the skin being the most common malignancies in the “White” population, while Kaposi sarcoma was much more frequent in Black [15]. Four epidemiological forms of Kaposi sarcoma are known: classic (sporadic); African (endemic); AIDS-associated (epidemic); and immunosuppression-associated (iatrogenic). While human herpesvirus-8 infection appears to be the triggering agent in all cases, other determinants of cancer development are likely involved, which may differ among races. These include putative cancer cells of origin, either lymphatic or vascular endothelial progenitors, their microenvironment and, as considered in detail further below, the immune system [17]. Countering the protective role of pigmentation, other, as yet unknown, determinants render Africans more susceptible to aggressive cancer development, even in the skin [18]. In fact, although African American have a much lower incidence of UV-induced cutaneous SCCs, they develop SCCs at sites of wound healing that are much more aggressive than in Caucasians [19]. In addition, African Albinos are disproportionately affected by skin SCCs compared to the general African population [20].
Genetic polymorphisms and cancer gene mutations
Recent advances in genome-wide association studies (GWAS) have provided exciting novel insights into the genetic basis of complex common diseases like cancer. Although many genetic variants have been linked to predisposition to specific cancer types, the association of most identified variants results only in a marginally increased risk of cancer development. This unaccounted basis of genetic predisposition has been called “genetic dark matter”, in the sense that genetic susceptibility appears like a certainty, while its molecular basis cannot, as yet, be explained [21].
There are many reported single nucleotide polymorphisms (SNPs) [22] and copy number variations (CNVs) [23, 24] associated with racial diversity, potentially affecting non coding RNAs [25], epigenetic regulation [26, 27] and/or posttranslational modifications [28]. However, their biological and clinical significance in most cases is unknown. A notable exception in relation to cancer susceptibility is the TP53 P72R polymorphism, the main P/P allele being preferentially found in African Americans with colon cancer [29] and Asians with gastric cancer [30]. It has been proposed that the TP53 P/P-related cancer susceptibility is due to a faster accumulation of mutations and a larger pool of putative cancer-initiating cells. In fact, the TP53 P72R allele induces transcription of the tumor suppressor gene PRDM1B (BLIMP-1) that can promote stem cell commitment to differentiation, favoring elimination of cells with DNA-damage induced p53 activation [31].
While the significance of nucleotide differences with possibly subtle regulatory function is difficult to assess, there are also differences in incidence of cancer-driver mutations among races (Table 1). One well-documented example are EGFR mutations, which are significantly more common in Asian lung cancer patients (32–57%) than in those of other races, with important consequences for targeted therapies [32, 33].
Table 1.
Differences in clinicopathological characteristics and driver gene mutations in selected cancer types
| Cancer type | African American | Caucasian | Asian | Clinical implication | References |
|---|---|---|---|---|---|
| Lung cancer | |||||
| KRAS mutation | 17% | 26–31.6% | 10.4–11% | Mutation significantly more frequent in smokers. | [32, 101] |
| EGFR mutation | 3–19% | 3–20% | 32–57% | Mutation significantly more frequent in women and non-smokers. Predicts response to EGFR tyrosine kinase inhibitors. |
[32, 101–105] |
| ALK rearrangement (EML4-ALK fusion) | 4% | 5.6% | 4.9–67% | Associated with younger age, never smoking, advanced clinical stage. Predicts response to ALK inhibitors. |
[32, 105–108] |
| MET mutation | 0–1% | 2.2–19% | 13–14.3% | More frequent in males, smokers. | [101, 102, 109] |
| Melanoma | |||||
| BRAF mutation | 8% | 21% | 24–25.5% | BRAF or NRAS mutation mutually exclusive, more frequently associated with ulceration and poor survival. | [34, 110–112] |
| N RAS mutation | 12% | 22% | 7.2% | [34, 112] | |
| GNAQ mutation GNA11 mutation |
45–49% 32% |
18% 20% |
Present at all stages of uveal melanoma GNAQ and GNA11 mutations are mutually exclusive. |
[110, 113, 114] | |
| Prostate cancer | |||||
| Anterior localization | 49.2% | 20% | Associated with lower androgen receptor signaling. | [115] | |
| ERG rearrangement TMPRSS-ERG fusion | 27.6–31.3% | 37.4–50% | 7.5–15.9% | No correlation with clinicopathological features besides race | [35–37] |
| PTEN deletion | 6.9% | 19.8–42.3% | 14.3% | Associated with higher Gleason score, androgen independence and worse prognosis. May predict response to PI3K inhibitor. |
[36, 37] |
| SPINK1 overexpression | 23.8% | 8.2% | SPINK1 overexpression and ERG rearrangements mutually exclusive. Associated with aggressive disease. | [36] | |
| Breast cancer | |||||
| Triple negative tumors | 19.5–48.1% | 9.2–14.5% | 9% | Associated with poor survival. | [40, 41, 116] |
| Basal like tumors -Premenopausal -Postmenopausal |
39% 14% |
16% 16% |
Significantly more TP53 mutations in basal like vs luminal A tumors (44 vs 15%). | [117] | |
| Luminal A tumors -Premenopausal -Postmenopausal |
36% 59% |
51% 58% |
[117] | ||
| Luminal B tumors -Premenopausal -Postmenopausal |
9% 16% |
18% 16% |
[117] | ||
| HER2 expression | 7–19.5% | 6–13% | 8.5–20% | No differences between pre-and postmenopausal status. Predicts response to anti- HER2 antibodies. |
[40, 41, 116–120] |
| BRCA1 mutation All ages <35 years |
1.3% 16.7% |
2.2% 7.2% |
0.5% 2.4% |
8.3 % in Ashkenazi Jewish breast cancer patients of all ages, 66.7% in patients <35 years. Associated with poor survival. |
[121, 122] |
| TP53 mutation | 42.9% | 27 6% | TP53 mutations associated with poorer survival for African Americans but not for Caucasians. | [123, 124] | |
| PIK3CA mutation | 20% | 33.9% | May predict response to PI3K inhibitors. | [123] | |
| Colorectal cancer | |||||
| Proximal localization Cancer stage III, IV Lymphocytic infiltration |
49% 52% 29% |
34% 37% 12% |
Proximal (right-sided) tumors associated with worse outcome. | [125, 126] | |
| KRAS mutation | 23–44.1% | 15–34.9% | 27.8–37.9% | Associated with poor prognosis. Predicts lack of response to EGFR-antibodies. |
[125,127–129] |
| BRAF mutation | 4–6.4% | 7–13.9% | 4.5–6% | Associated with poor prognosis. | [111, 125, 127, 128] |
| Microsatellite instability (MSI) | 9% | 9% | 9% | MSI associated with favorable outcome. Negative predictor marker for 5-FU based therapy in stage II and III patients. |
[125, 128–131] |
| Gastric cancer | |||||
| HER2 expression | 23.6% | 23.9% | FIER2 expression higher in intestinal vs diffuse-type (31.8 vs 6.1 %), and gastroesophageal junction cancer vs gastric tumors (32.2 vs 21.4%). Predicts response to anti-FlER2 antibodies |
[120, 132] | |
| GIST | |||||
| c-KIT mutation | 78–79% -exon 11: 67–83% -exon 9: 11–14% | 90.7% -exon 11: 74% -exon 9: 7.3% | Exon 11 mutations predict better outcome after imatinib treatment. | [133–136] | |
| PDGFRA mutation | 6–12%- exon 18: 9% | 16%- exon 18: 8% | KIT and PDGFRA mutations mutually exclusive. | [133, 136] | |
| HNSCC | |||||
| HPV infection | 4–25% | 34–71.1% | 51.3% | Associated with better outcome. | [137–139] |
| Papillary Thyroid cancer | |||||
| BRAF mutation | 48% | 75.4–80% | Associated with aggressive clinicopathological features. | [111, 140–142] | |
| TERT promoter mutation | 4.7–25.5% | 4.4–11.3% | Associated with poor prognosis. | [142, 143] | |
| Glioblastoma | |||||
| IDH1 and 2 mutation | 10% | 16,1% | Associated with better outcome | [144, 145] | |
| TERT promoter mutation | 73.3% | TERT promoter mutation alone associated with poor prognosis, occurring together with IDH1 mutation associated with prolonged survival. | [144] | ||
| MGMT promoter méthylation | 36% | Associated with better survival. | [146] | ||
In melanoma, the frequency and type of mutation in the main driver oncogenes is also race-dependent. BRAF and NRAS mutations are found in about 30–60% and 30% of Caucasian patients, but only in 8% and 12%, respectively, of those of African ancestry [34]. BRAF mutations are mainly present in melanomas of intermittently sun-exposed skin, such as the trunk, while the occurrence in melanomas of non-exposed or chronically sun exposed skin such as the face and extremities is low [34]. These differences are consistent with a different, sun-independent pathogenesis of melanoma in black skin [34]. In prostate cancer, TMPRSS-ERG fusion is more common in Caucasian men (50%) than African (31%) or Asian (16%) [35]. Similarly, PTEN deletion, leading to increased PI3K activity, is rarely found in African (7%) and Asian (14%) prostate cancer patients but present in 20–40% of Caucasian prostate cancer samples [36, 37]. Given the lower survival of African Americans, the surprisingly lower incidence of PTEN mutations in tumors of these patients points to the possibility of alternative pathways being of greater importance.
Recently, comparison of the mutational landscape of colorectal tumors from Caucasian and African American patients identified two genes, ephrin type A receptor 6 (EPHA6) and folliculin (FLCN) as cancer driver genes exclusively in African Americans [38]. Overall, these differences in cancer driver mutations can contribute significantly to survival disparity among races and are of relevance in this era of targeted therapies (Table 1).
Epigenetic differences and the transciptome
At the epigenetic level, racial differences in DNA methylation were identified in healthy as well as cancer tissue, in line with the hypothesis that methylation changes are an early predisposing event occurring years before overt cancer development [39]. On the basis of differentially methylated CpG sites, Caucasian Americans, African Americans and Han Chinese-American could also be correctly clustered according to their geographical origins and associated with their distinct phenotypic features, drug metabolism and disease susceptibility [26].
Most epigenetic studies have been focusing on differences between White and Black in breast cancer, with cancer incidence being higher and survival lower in the Black population. Triple negative breast cancer (ER-, PR-, HER2-), probably the most aggressive form of breast cancer, is significantly more frequent in women of African ancestry (20–50%) compared to other races (9–15%) [40, 41], contributing to the racial survival disparities. The reason for this greater incidence is not clear, but could be related to differences in gene expression at various levels as discussed here below. Hypermethylation of TWIST, Cyclin D2, RAR-B and RASSF1A genes, implicated in cell proliferation and differentiation is more common in African American premenopausal breast cancer patients than in Caucasians [42]. These findings are difficult to reconcile with the putative tumor promoting function of the TWIST and Cyclin D2 genes. However, it is possible to speculate that the loss of the tumor suppressor gene RASSF1A and the gene encoding retinoic acid receptor beta (RAR-B) promote more undifferentiated breast tumors in African American women.
Also, gene variants of miRNA processing genes, such as AGO4, and SNPs in miRNAs regulating breast carcinogenesis are associated with differences in cancer susceptibility in African American compared to Caucasians [43].
At the transcriptome level, global gene expression analysis of breast tumors from African American and Caucasian patients matched by pathological characteristics revealed diverse molecular profiles. In several studies, two genes, CRYBB2 (crystallin beta B2) and PSPHL (L-3-phosphoserine phosphatase homolog), which have been connected, respectively, with cataract formation and pterygia, a pathological deposition of extracellular matrix of the eye connective tissue, were reported to be highly expressed in tumor epithelium from African American individuals and could be used to correctly cluster specimens according to race [44, 45]. The function of these genes in this context remains enigmatic and possible linkage to other genes of greater significance for cancer development remains to be evaluated.
Limited information is available on gene expression and/or epigenetic differences in prostate cancer of White versus Black patients and even less data exists in other major cancer types such as squamous cell cancer of various organs. In a small study, higher promoter methylation for the genes encoding SNRPN, involved in pre-mRNA processing, MST1R, a tyrosine kinase related to c-MET and ABCG5, a member of the ABC transporter superfamily, was detected in African American prostate cancer samples compared to those from Caucasians [46]. Likewise, the gene motor neuron and pancreas homeobox1 (MNX1) was shown to be upregulated to a higher extent in prostate cancer tissue from African American compared to those from European American men, [47] with the suggestion that it contributes to carcinogenesis through AKT activation and increased lipid synthesis.
Obesity and chronic inflammation
Obesity
The association between metabolic disorders such as obesity and increased incidence and mortality of postmenopausal breast, endometrial, colon, esophagus and kidney cancer has been well documented over the last 20 years [48]. It has been estimated that 14% of cancer-related deaths in men and 20% in women are caused by obesity [49]. The prevalence of obesity in African Americans and Hispanics is significantly higher than in Caucasians and Asians [50]. Black individuals have a lower maximal capacity of aerobic metabolism and greater percentage of fast contracting (type II) skeletal muscle fibers, which, together with a reduced energy consumption, predisposes them to obesity and other metabolic disorders [51]. While African Americans are particularly susceptible to obesity-related cancers, Hispanics seem to be relatively unaffected [48]. As mentioned before, this could be related to genetic but also behavioral differences, specifically diet. Even within the black population, the importance of diet is illustrated by a recent study on colon cancer risk, linking a high-fat and low-fiber western-style versus low-fat and high-fiber, African-style diets to metabolome and microbiota composition [52].
Various mechanisms have been proposed to explain how obesity promotes cancer. One possible mechanism is through insulin signaling pathways. African Americans present higher levels of insulin and after administration of glucose, the increase in serum insulin in African Americans is two to three times higher than in Caucasians [48]. Interestingly, this different insulin response is independent of any differences in body fat distribution and composition or physical activity and is already evident in children [53]. Hyperinsulinemia is caused by both increased β-cells secretion and decreased hepatic clearance in African Americans. In parallel with differences in insulin blood levels, African Americans and Hispanics are more insulin resistant than Caucasians, even after accounting for differences in body mass index (BMI) [54]. Related to the above, insulin-like growth factor 1 (IGF-1) is an important autocrine-paracrine stimulatory factor for adipose tissue growth. IGF-1 levels normally decrease with increasing BMI in Hispanics and Asian, while this decline is diminished in Caucasian and absent in African American [55], pointing to possible cancer-promoting effects of persistently elevated IGF-1 levels. Insulin resistance also increases bioavailability of IGF-1 through decreased synthesis of IGF-binding proteins (IGFBP-1 and IGFBP-2) [48]. Insulin and IGF-1 inhibit the expression of sex-hormone binding globulin (SHBG) at the same time as they stimulate secretion of female sex hormones, which, in breast and endometrial tissue, can promote cellular proliferation and inhibit apoptosis, both of which could also contribute for differences in cancer risk [56]. These complex racial variations in insulin/IGF-1 signaling in obese individuals could help explain the “Hispanic paradox”, to which we referred above [8].
Chronic Inflammation
The obesity-related cancer risk is also linked to chronic inflammation. Many conditions can trigger chronic inflammation and increase cancer susceptibility. About 15–20% of cancer related deaths are thought to be due to underlying infections and associated inflammatory responses [57].
Significant disparities between Black and White populations have been described in susceptibility and response to HIV infection [58] [59]. This disparity was also reported in a large US military cohort with equal access to health care and similar duration of HIV infection [60]. Also, in chronic hepatitis C virus (HCV) infections, racial differences in response to treatment have been reported [61] [62] [63].
Serum levels of the inflammatory proteins CRP and IL6 are higher in African American compared to various other races and this disparity persists after adjustment for BMI [64]. The G174C polymorphism in the IL6 gene with higher frequencies of the 174G allele in Non-Caucasians including African, African-American and Mexican (0.87–1.0) compared to Caucasians (0.54–0.62) leads to significantly higher IL-6 serum levels and has been proposed to contribute to racial differences in prevalence and survival of various chronic diseases including cancer [65].
Chronic inflammation results in an imbalance of circulating adipose tissue cytokines or “adipokines”, such as leptin and adiponectin, with levels of the latter decreasing with increasing BMI [66]. While there are conflicting data on leptin, high adiponectin levels are consistently correlated with reduced breast cancer risk [67]. Mechanistically, this could be linked to activation by adiponectin receptors of the antitumorigenic peroxisome proliferator-activated receptor gamma (PPARy) pathway and downstream increase of BRCA1 expression [68]. Importantly, in both Caucasians and African Americans, a SNP rs1501299 in the adiponectin gene, associated with adiponectin serum levels, also correlates with increased breast cancer risk [69].
Immune system
While racial differences in immune related functions are well established, their relationship to cancer susceptibility remains mostly to be investigated. In fact, tumor immunity is a complex phenomenon and a strong inflammatory response against pathogens or increased activity of the immune system as in autoimmune disorders does not necessarily confer protection against cancer development. On the contrary, various studies in different populations have shown an increased cancer risk -especially lymphomas - in individuals suffering from systemic inflammatory autoimmune diseases such lupus erythematodes and rheumatoid arthritis [73] [74] s[75].
Innate immunity
Susceptibility to acute infections as wells as chronic diseases including cancer is determined by genetic variations in the immune system. Genes of the immune system are subject to constant evolutionary pressure, which can vary depending on environmental conditions and relocation of populations. Individuals of African ancestry show inherent differences in their immune system relative to other races, possibly due to selective pressure in response to endemic infectious diseases in Africa [70].
The number of granzyme B secreting cytotoxic cells has been reported to be significantly lower in African American patients compared to Caucasians, suggesting that the functional activity of inflammatory cells is not identical across races [76]. Two other studies analyzed the response of cultured monocytes and macrophages to bacterial [71] and viral infections [72] in individuals of African and European ancestry. There were significant differences in gene expression before and after infection, African Americans showing stronger inflammatory responses and faster bacterial clearance [71]. Strikingly, many of the genes whose expression was altered in response to infection showed sequences that were very similar between Europeans and Neanderthals, but not Africans, suggesting that a contribution of the Neanderthal genome lead to acquisition of regulatory variants associated with reduced inflammatory responses in Euro-Asian genomes [72].
Several immunity related genes, such as the Toll-like receptor (TLR) TLR1/TLR6/TLR19 gene cluster, [77] the caspase-12 gene (CASP12), involved in cytokine production upon bacterial lipopolysaccharide stimulation, evolved under strong selective pressure and show racial variation [78].
Various studies have correlated SNPs in TLR genes to alterations in susceptibility to various infectious or inflammatory diseases, which in turn might affect cancer development [79], as described above.
Adaptive immunity
The Th1 immune response (characterized by IL2 and IFNγ secretion) results in cytotoxic CD8 cells with anti-viral and anti-tumor activity [80], while Th2-type immunity (characterized by IL4, IL5, IL9, IL10 and IL13 production, and eosinophil and basophil activation) seems to have evolved in response to parasitic infections. In many cancers, the fraction of Th1 cells is significantly decreased, while the proportion of Th2 cells is increased [80], which, for colorectal cancer, can be of prognostic value [81]. Polymorphisms present in the West African and Asian populations are linked with an increased Th2 response, with lesser acute inflammation [82], but more persistent chronic inflammation and/or suppression of antitumor immunity [80].
Individuals of African ancestry express higher levels of IL2RA, which encodes the IL2-receptor CD25, that is key for proliferation of regulatory T cells (Treg). Underlying this difference, the low-expression variant rs12251836 is common in European, and rare in African and Asian individuals [83].
Genetic variants of adaptive immune response related genes such as IL4R (interleukin 4 receptor), IL15 (interleukin 15), LTA (lymphotoxine alpha) and INFGR2 (interferon gamma receptor 2) have also been associated with enhanced cancer risk in patients of African ancestry [84].
Epigenetically, DNA methylation profiling of naïve CD4 T cells revealed hypomethylation of various genes related to apoptosis and autoimmune disorders in healthy African Americans, which may account for their greater susceptibility to these diseases than European-Americans [85].
Translational implications for cancer prevention and treatment
In the era of precision medicine, race needs to be recognized as a risk factor independent of environmental dynamics for incidence and mortality of specific cancer types and screening and treatment modalities should be adapted in order to diminish the racial survival disparity.
For instance, there is a significant difference in age-specific increase of colorectal cancer, the incidence beginning to increase at 43 years in African Americans compared to 47 years in European Americans, with a 20% higher stage-adjusted mortality. As a result, some associations recommend to start screening of average-risk African Americans at 45, rather than at 50 years of age as recommended for individuals of all races by other major agencies such as the American Cancer Society and the U.S. Preventive Services Task Force [86]. Rather than screening at an earlier age, it has been suggested that similar beneficial effects would be obtained with a 5–10% increase of total number of African American individuals that undergo the test [87].
Whether earlier initiation of screening or improving the adherence to the existing screening programs is more efficient in decreasing the cancer burden is also a question for other cancer types. In fact, African American race is considered a risk factor for prostate cancer and separate screening guidelines are proposed [88]. Similarly, genetic screening of Jewish women starting at age 25 years of age for BRCA mutations has been advised in order to identify women at high risk for developing breast and ovarian cancers in this population with relatively high BRCA mutation prevalence [89]. Given the higher mortality of breast cancer in African Americans and the higher incidence of gastric cancer in Asian individuals population-based specific screening recommendations should be considered.
In concert with variations in cellular metabolism, polymorphisms for genes encoding detoxifying and drug metabolizing enzymes such as cytochrome P450 and transporters such as P-glycoprotein are present with variable prevalence in different races. Such differences can have multiple effects at the level of conversion of various toxic compounds into DNA damaging carcinogens, steroid hormone processing and pharmacokinetics of various drugs. For instance there is emerging evidence for racial disparities in the incidence of adverse events after chemotherapy, probably as a consequence of differences in body fat composition and distribution and differential function of drug metabolizing enzymes.
It is therefore important to investigate the presence of clinically relevant genetic polymorphisms affecting various drug metabolizing enzymes in order to establish the appropriate drug doses across races and optimize therapy regimens (“pharmacoethnicity”) [90].
In clinical trials, independent of the discipline, there is often a selection bias in terms of age (younger), gender (males) and race with more European/Caucasian patients being included [91], resulting in underrepresentation of African American, Hispanic and Asian patients (<10% of clinical trial participants are minorities) [92]. In addition to improving access to novel therapies, participation of racial groups in clinical trials is critical to reach robust conclusions about the risks and benefits of drugs and specific interventions in these populations.
Racial minorities could benefit from focused accrual, taking into account their different cultural and religious background. The patient navigation model, where a lay individual is trained to provide support and education for patients enrolled in trials, has been successfully applied to improve health care access and increase participation and retention in clinical trials [92].
The complex interplay between genetic factors, obesity, chronic inflammation and environmental influences on racial disparity in cancer susceptibility and survival is illustrated in Figure 3, Key Figure.
Figure 3. Key Figure. Biological basis of disparity in cancer susceptibility among different races.
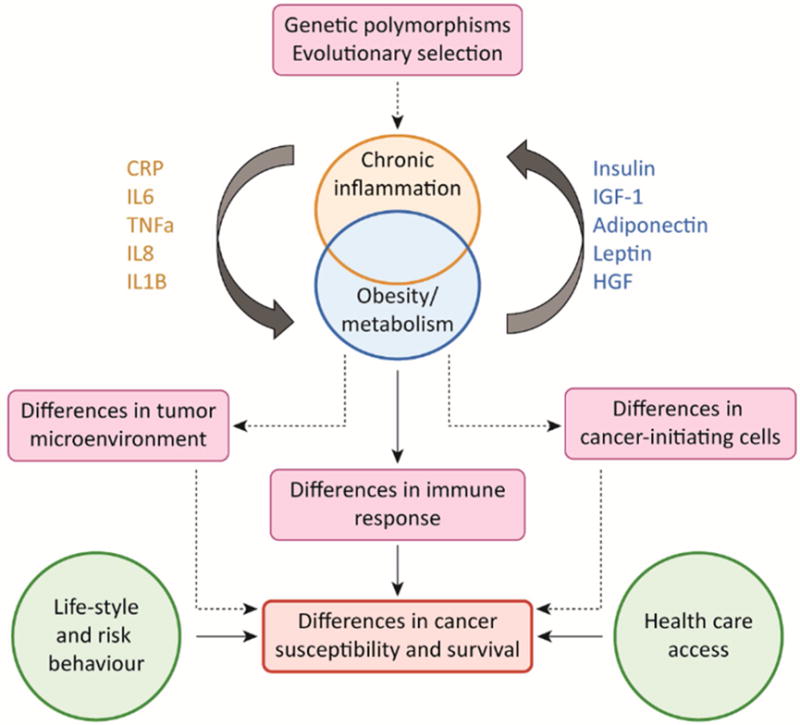
There is evidence for evolutionary selection of polymorphisms in the Th2 immune response in African populations. The prevalence of obesity in African Americans and Hispanics is significantly higher than in Caucasians and Asians. A major driver of obesity-related cancer is chronic inflammation, which is associated with an increased release of inflammatory cytokines such as TNFα, IL6, IL8, IL1-ß which results in imbalance of circulating adipose tissue cytokines or “adipokines”, such as leptin, adiponectin and hepatocyte growth factor (HGF). The racial differences in metabolism and inflammatory response contribute to differences in the immune system, how they affect cancer initiating cells and the tumor microenvironment needs to be explored. The disparity in cancer susceptibility and survival is presumably a consequence of biological and environmental factors, such as differences in life-style, risk behavior and health care access.
Concluding Remarks
There has been a paradigm shift in the notion of carcinogenesis in the last decades. Cancer is no longer solely considered as an uncontrolled proliferation of genetically altered single cells but rather a consequence of disturbed tumor-stroma interactions, diminished immune surveillance and loss of tissue homeostasis and therefore, as a complex disease occurring in tissues rather than in cells exclusively [93].
However, the impact of race on cancer initiating cell populations and their surrounding stromal environment are only starting to be appreciated (see Outstanding Questions). Therefore, besides identification of race-specific cancer driver mutations, the exploration of variation in the stem cells compartment and the stromal composition in healthy and cancer tissues in diverse races is essential for a thorough understanding of carcinogenesis in different genetic backgrounds.
There is emerging evidence that the tissue composition in normal and tumor tissue differs between races, African ancestry being associated with a significant increase in stem cells populations in healthy breast and colon cancer, compared to Caucasians [94, 95]. It is also tempting to speculate that the different spatial distribution of stem cells might underlie well-known differences in tumor locations, as African American harboring significantly more right-sided colon [96] and anterior prostate tumors [97] than other races.
Recent studies showed significant alterations in the tumor microenvironment in patients of African ancestry, namely in the extent of angiogenesis, immune infiltrate, expression of genes related to epithelial to mesenchymal transition (EMT) and extracellular matrix formation [98, 99].
The interplay between behavioral/environmental factors and genetic determinants needs to be elucidated. Several studies have shown an impact of the microbiome on cancer risk and anticancer immunosurveillance [100]. In this context, the influence of diet-related racial differences on intestinal flora composition remains to be assessed. The importance of the stromal environment is illustrated by the success of immunotherapies, principally in tumors with a strong immune cell infiltrate, so called immunologically “hot tumors”.
Currently, immunotherapies, based on immune checkpoint inhibitors are rapidly becoming a powerful therapeutic tool for various cancer types, with very promising response rates and potentially severe immune related adverse events (irAEs) [101]. Despite the ever-increasing number of clinical trials with thousands of patients, so far subgroup analysis based on race of the enrolled patients have not been reported. Similarly, separate analysis based on sex has been missing. In our view, there is an unmet need to determine to what extent race affects efficacy and tolerance of immune checkpoint inhibitors targeting the CD80/CD86- CTLA4 and PD1-PD-L1 axis, in order to better select for patients who might benefit from such therapies.
However, while we encourage to further explore the contribution of biological factors to racial differences in cancer risk and mortality, we caution against trivializing the role of socio-environmental determinants such as health care access and health literacy. We believe that meaningful scientific and clinical research of racial factors is most likely to be successful using a multidisciplinary approach including geneticists, epidemiologists and social scientists.
Trends.
Racial disparities in cancer incidence and survival are not solely attributable to environmental factors such as health care access and risk behavior.
African populations show greater genetic diversity compared to all other Non-African populations. The interbreeding of the migrating population with Neanderthals in Euro-Asia resulted in the presence of 1.5–4% of Neanderthal DNA in the genome of modern Eurasians, contributing to the differences in disease susceptibility.
The prevalence of genetic polymorphisms and mutations shows racial differences for various cancer types.
The immune response in individuals of African ancestry diverges from the one in Caucasians, presumably due to distinct evolutionary pressure in response to infectious disease.
The higher cancer susceptibility of African American is linked to genetic predisposition to obesity and chronic inflammation.
Outstanding Questions.
How do behavioral and environmental factors interact with epigenetic and genetic determinants of cancer susceptibility?
How does cancer susceptibility relate to the greater genetic diversity of African populations?
Are there racial differences in the gut microbiota, which could contribute to differences in cancer risk and treatment response?
What is the contribution of racial differences in cancer stem cells and surrounding stroma in susceptibility to initiation and progression of the neoplastic process?
How do racial differences in innate and acquired immune responses affect personalized approaches to cancer therapy, specifically immunotherapy?
Acknowledgments
The authors thank Barbara Gilchrest of Harvard University for critical review of the manuscript and useful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wiencke JK. Impact of race/ethnicity on molecular pathways in human cancer. Nat Rev Cancer. 2004;4(1):79–84. doi: 10.1038/nrc1257. [DOI] [PubMed] [Google Scholar]
- 2.Aizer AA, et al. Lack of reduction in racial disparities in cancer-specific mortality over a 20-year period. Cancer. 2014;120(10):1532–9. doi: 10.1002/cncr.28617. [DOI] [PubMed] [Google Scholar]
- 3.Jin H, et al. Cancer incidence among Asian American populations in the United States, 2009–2011. Int J Cancer. 2016;138(9):2136–45. doi: 10.1002/ijc.29958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeSantis CE, et al. Cancer statistics for African Americans, 2016: Progress and opportunities in reducing racial disparities. CA Cancer J Clin. 2016 doi: 10.3322/caac.21340. [DOI] [PubMed] [Google Scholar]
- 5.Andaya AA, et al. Race and colon cancer survival in an equal-access health care system. Cancer Epidemiol Biomarkers Prev. 2013;22(6):1030–6. doi: 10.1158/1055-9965.EPI-13-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schreiber D, et al. Impact of race in a predominantly African-American population of patients with low/intermediate risk prostate cancer undergoing radical prostatectomy within an equal access care institution. Int Urol Nephrol. 2014;46(10):1941–6. doi: 10.1007/s11255-014-0773-3. [DOI] [PubMed] [Google Scholar]
- 7.Clocchiatti A, et al. Sexual dimorphism in cancer. Nat Rev Cancer. 2016;16(5):330–9. doi: 10.1038/nrc.2016.30. [DOI] [PubMed] [Google Scholar]
- 8.Ruiz JM, et al. Hispanic mortality paradox: a systematic review and meta-analysis of the longitudinal literature. Am J Public Health. 2013;103(3):e52–60. doi: 10.2105/AJPH.2012.301103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young RP, Hopkins RJ. A review of the Hispanic paradox: time to spill the beans? Eur Respir Rev. 2014;23(134):439–49. doi: 10.1183/09059180.00000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet. 2008;9:403–33. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 12.Simonti CN, et al. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 2016;351(6274):737–41. doi: 10.1126/science.aad2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lalueza-Fox C, et al. A melanocortin 1 receptor allele suggests varying pigmentation among Neanderthals. Science. 2007;318(5855):1453–5. doi: 10.1126/science.1147417. [DOI] [PubMed] [Google Scholar]
- 14.Cassidy PB, et al. Beyond Red Hair and Sunburns: Uncovering the Molecular Mechanisms of MC1R Signaling and Repair of UV-Induced DNA Damage. J Invest Dermatol. 2015;135(12):2918–21. doi: 10.1038/jid.2015.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moosa MR. Racial and ethnic variations in incidence and pattern of malignancies after kidney transplantation. Medicine (Baltimore) 2005;84(1):12–22. doi: 10.1097/01.md.0000152372.30370.6f. [DOI] [PubMed] [Google Scholar]
- 16.Tornesello ML, et al. MDM2 and CDKN1A gene polymorphisms and risk of Kaposi’s sarcoma in African and Caucasian patients. Biomarkers. 2011;16(1):42–50. doi: 10.3109/1354750X.2010.525664. [DOI] [PubMed] [Google Scholar]
- 17.Douglas JL, et al. Kaposi’s sarcoma: a model of both malignancy and chronic inflammation. Panminerva Med. 2007;49(3):119–38. [PubMed] [Google Scholar]
- 18.Greaves M. Was skin cancer a selective force for black pigmentation in early hominin evolution? Proc Biol Sci. 2014;281(1781):20132955. doi: 10.1098/rspb.2013.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gloster HM, Jr, Neal K. Skin cancer in skin of color. J Am Acad Dermatol. 2006;55(5):741–60. doi: 10.1016/j.jaad.2005.08.063. quiz 761–4. [DOI] [PubMed] [Google Scholar]
- 20.Lekalakala PT, et al. Oculocutaneous Albinism and Squamous Cell Carcinoma of the Skin of the Head and Neck in Sub-Saharan Africa. J Skin Cancer. 2015;2015:167847. doi: 10.1155/2015/167847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxwell KN, Nathanson KL. Common breast cancer risk variants in the post-COGS era: a comprehensive review. Breast Cancer Res. 2013;15(6):212. doi: 10.1186/bcr3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rose AE, et al. Copy number and gene expression differences between African American and Caucasian American prostate cancer. J Transl Med. 2010;8:70. doi: 10.1186/1479-5876-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loo LW, et al. Genome-wide copy number alterations in subtypes of invasive breast cancers in young white and African American women. Breast Cancer Res Treat. 2011;127(1):297–308. doi: 10.1007/s10549-010-1297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang RS, et al. Population differences in microRNA expression and biological implications. RNA Biol. 2011;8(4):692–701. doi: 10.4161/rna.8.4.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyn H, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23(9):1363–72. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia YY, et al. Racial/ethnic disparities in human DNA methylation. Biochim Biophys Acta. 2014;1846(1):258–62. doi: 10.1016/j.bbcan.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Reimand J, et al. Evolutionary constraint and disease associations of post-translational modification sites in human genomes. PLoS Genet. 2015;11(1):e1004919. doi: 10.1371/journal.pgen.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katkoori VR, et al. Prognostic significance of p53 codon 72 polymorphism differs with race in colorectal adenocarcinoma. Clin Cancer Res. 2009;15(7):2406–16. doi: 10.1158/1078-0432.CCR-08-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, et al. Meta-analysis of the association between P53 codon 72 polymorphisms and gastric cancer. J Surg Oncol. 2013;107(4):360–6. doi: 10.1002/jso.23233. [DOI] [PubMed] [Google Scholar]
- 31.Weige CC, et al. Transcriptomes and shRNA suppressors in a TP53 allele-specific model of early-onset colon cancer in African Americans. Mol Cancer Res. 2014;12(7):1029–41. doi: 10.1158/1541-7786.MCR-13-0286-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steuer CE, et al. Role of race in oncogenic driver prevalence and outcomes in lung adenocarcinoma: Results from the Lung Cancer Mutation Consortium. Cancer. 2016;122(5):766–72. doi: 10.1002/cncr.29812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan DS, et al. Cancer Genomics: Diversity and Disparity Across Ethnicity and Geography. J Clin Oncol. 2016;34(1):91–101. doi: 10.1200/JCO.2015.62.0096. [DOI] [PubMed] [Google Scholar]
- 34.Akslen LA, et al. Mutation analysis of the EGFR-NRAS-BRAF pathway in melanomas from black Africans and other subgroups of cutaneous melanoma. Melanoma Res. 2008;18(1):29–35. doi: 10.1097/CMR.0b013e3282f32517. [DOI] [PubMed] [Google Scholar]
- 35.Magi-Galluzzi C, et al. TMPRSS2-ERG gene fusion prevalence and class are significantly different in prostate cancer of Caucasian, African-American and Japanese patients. Prostate. 2011;71(5):489–97. doi: 10.1002/pros.21265. [DOI] [PubMed] [Google Scholar]
- 36.Khani F, et al. Evidence for molecular differences in prostate cancer between African American and Caucasian men. Clin Cancer Res. 2014;20(18):4925–34. doi: 10.1158/1078-0432.CCR-13-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao X, et al. Distinct genomic alterations in prostate cancers in Chinese and Western populations suggest alternative pathways of prostate carcinogenesis. Cancer Res. 2010;70(13):5207–12. doi: 10.1158/0008-5472.CAN-09-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guda K, et al. Novel recurrently mutated genes in African American colon cancers. Proc Natl Acad Sci U S A. 2015;112(4):1149–54. doi: 10.1073/pnas.1417064112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song MA, et al. Racial differences in genome-wide methylation profiling and gene expression in breast tissues from healthy women. Epigenetics. 2015;10(12):1177–87. doi: 10.1080/15592294.2015.1121362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agboola AJ, et al. Molecular characteristics and prognostic features of breast cancer in Nigerian compared with UK women. Breast Cancer Res Treat. 2012;135(2):555–69. doi: 10.1007/s10549-012-2173-7. [DOI] [PubMed] [Google Scholar]
- 41.Clarke CA, et al. Age-specific incidence of breast cancer subtypes: understanding the black-white crossover. J Natl Cancer Inst. 2012;104(14):1094–101. doi: 10.1093/jnci/djs264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehrotra J, et al. Estrogen receptor/progesterone receptor-negative breast cancers of young African-American women have a higher frequency of methylation of multiple genes than those of Caucasian women. Clin Cancer Res. 2004;10(6):2052–7. doi: 10.1158/1078-0432.ccr-03-0514. [DOI] [PubMed] [Google Scholar]
- 43.Yao S, et al. Genetic variants in microRNAs and breast cancer risk in African American and European American women. Breast Cancer Res Treat. 2013;141(3):447–59. doi: 10.1007/s10549-013-2698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Field LA, et al. Identification of differentially expressed genes in breast tumors from African American compared with Caucasian women. Cancer. 2012;118(5):1334–44. doi: 10.1002/cncr.26405. [DOI] [PubMed] [Google Scholar]
- 45.Wallace TA, et al. Tumor immunobiological differences in prostate cancer between African-American and European-American men. Cancer Res. 2008;68(3):927–36. doi: 10.1158/0008-5472.CAN-07-2608. [DOI] [PubMed] [Google Scholar]
- 46.Devaney JM, et al. Genome-wide differentially methylated genes in prostate cancer tissues from African-American and Caucasian men. Epigenetics. 2015;10(4):319–28. doi: 10.1080/15592294.2015.1022019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L, et al. MNX1 Is Oncogenically Upregulated in African-American Prostate Cancer. Cancer Res. 2016;76(21):6290–6298. doi: 10.1158/0008-5472.CAN-16-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Speakman JR, Goran MI. Tissue-specificity and ethnic diversity in obesity-related risk of cancer may be explained by variability in insulin response and insulin signaling pathways. Obesity (Silver Spring) 2010;18(6):1071–8. doi: 10.1038/oby.2010.16. [DOI] [PubMed] [Google Scholar]
- 49.Calle EE, et al. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N Engl J Med. 2003;348(17):1625–38. doi: 10.1056/NEJMoa021423. [DOI] [PubMed] [Google Scholar]
- 50.Ogden CL, et al. Prevalence of Obesity Among Adults and Youth: United States, 2011–2014. NCHS Data Brief. 2015;(219):1–8. [PubMed] [Google Scholar]
- 51.Ceaser T, Hunter G. Black and White race differences in aerobic capacity, muscle fiber type, and their influence on metabolic processes. Sports Med. 2015;45(5):615–23. doi: 10.1007/s40279-015-0318-7. [DOI] [PubMed] [Google Scholar]
- 52.O’Keefe SJ, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ku CY, et al. Racial differences in insulin secretion and sensitivity in prepubertal children: role of physical fitness and physical activity. Obes Res. 2000;8(7):506–15. doi: 10.1038/oby.2000.63. [DOI] [PubMed] [Google Scholar]
- 54.Goran MI, et al. Insulin resistance and associated compensatory responses in african-american and Hispanic children. Diabetes Care. 2002;25(12):2184–90. doi: 10.2337/diacare.25.12.2184. [DOI] [PubMed] [Google Scholar]
- 55.Henderson KD, et al. Ethnic disparity in the relationship between obesity and plasma insulin-like growth factors: the multiethnic cohort. Cancer Epidemiol Biomarkers Prev. 2006;15(11):2298–302. doi: 10.1158/1055-9965.EPI-06-0344. [DOI] [PubMed] [Google Scholar]
- 56.Arcidiacono B, et al. Insulin resistance and cancer risk: an overview of the pathogenetic mechanisms. Exp Diabetes Res. 2012;2012:789174. doi: 10.1155/2012/789174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mantovani A, et al. Cancer-related inflammation. Nature. 2008;454(7203):436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 58.May M, et al. CD4(+) T cell count decreases by ethnicity among untreated patients with HIV infection in South Africa and Switzerland. J Infect Dis. 2009;200(11):1729–35. doi: 10.1086/648096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guillaume C, et al. African ethnicity can influence immunological responses to highly active antiretroviral therapy and immunological success at 48 months: a retrospective pilot study. Int J Infect Dis. 2013;17(12):e1259–62. doi: 10.1016/j.ijid.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 60.Weintrob AC, et al. Virologic response differences between African Americans and European Americans initiating highly active antiretroviral therapy with equal access to care. J Acquir Immune Defic Syndr. 2009;52(5):574–80. doi: 10.1097/QAI.0b013e3181b98537. [DOI] [PubMed] [Google Scholar]
- 61.Hu KQ, et al. Impact of Hispanic or Asian ethnicity on the treatment outcomes of chronic hepatitis C: results from the WIN-R trial. J Clin Gastroenterol. 2011;45(8):720–6. doi: 10.1097/MCG.0b013e31820d35e3. [DOI] [PubMed] [Google Scholar]
- 62.Brau N, et al. Black patients with chronic hepatitis C have a lower sustained viral response rate than non-Blacks with genotype 1, but the same with genotypes 2/3, and this is not explained by more frequent dose reductions of interferon and ribavirin*. J Viral Hepat. 2006;13(4):242–9. doi: 10.1111/j.1365-2893.2005.00682.x. [DOI] [PubMed] [Google Scholar]
- 63.Rodriguez-Torres M, et al. Peginterferon alfa-2a and ribavirin in Latino and non-Latino whites with hepatitis C. N Engl J Med. 2009;360(3):257–67. doi: 10.1056/NEJMoa0805062. [DOI] [PubMed] [Google Scholar]
- 64.Morimoto Y, et al. Ethnic differences in serum adipokine and C-reactive protein levels: the multiethnic cohort. Int J Obes (Lond) 2014;38(11):1416–22. doi: 10.1038/ijo.2014.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berger FG. The interleukin-6 gene: a susceptibility factor that may contribute to racial and ethnic disparities in breast cancer mortality. Breast Cancer Res Treat. 2004;88(3):281–5. doi: 10.1007/s10549-004-0726-0. [DOI] [PubMed] [Google Scholar]
- 66.Li S, et al. Adiponectin levels and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2009;302(2):179–88. doi: 10.1001/jama.2009.976. [DOI] [PubMed] [Google Scholar]
- 67.Vona-Davis L, Rose DP. Adipokines as endocrine, paracrine, and autocrine factors in breast cancer risk and progression. Endocr Relat Cancer. 2007;14(2):189–206. doi: 10.1677/ERC-06-0068. [DOI] [PubMed] [Google Scholar]
- 68.Pignatelli M, et al. Enhancement of BRCA1 gene expression by the peroxisome proliferator-activated receptor gamma in the MCF-7 breast cancer cell line. Oncogene. 2003;22(35):5446–50. doi: 10.1038/sj.onc.1206824. [DOI] [PubMed] [Google Scholar]
- 69.Kaklamani VG, et al. Adiponectin pathway polymorphisms and risk of breast cancer in African Americans and Hispanics in the Women’s Health Initiative. Breast Cancer Res Treat. 2013;139(2):461–8. doi: 10.1007/s10549-013-2546-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11(1):17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- 71.Nedelec Y, et al. Genetic Ancestry and Natural Selection Drive Population Differences in Immune Responses to Pathogens. Cell. 2016;167(3):657–669 e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 72.Quach H, et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 2016;167(3):643–656 e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu KH, et al. Cancer Risk in Patients With Inflammatory Systemic Autoimmune Rheumatic Diseases: A Nationwide Population-Based Dynamic Cohort Study in Taiwan. Medicine (Baltimore) 2016;95(18):e3540. doi: 10.1097/MD.0000000000003540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Koshiol J, et al. Racial differences in chronic immune stimulatory conditions and risk of non-Hodgkin’s lymphoma in veterans from the United States. J Clin Oncol. 2011;29(4):378–85. doi: 10.1200/JCO.2010.30.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mao S, et al. Systemic lupus erythematosus and malignancies risk. J Cancer Res Clin Oncol. 2016;142(1):253–62. doi: 10.1007/s00432-015-2032-0. [DOI] [PubMed] [Google Scholar]
- 76.Basa RC, et al. Decreased Anti-Tumor Cytotoxic Immunity among Microsatellite-Stable Colon Cancers from African Americans. PLoS One. 2016;11(6):e0156660. doi: 10.1371/journal.pone.0156660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Laayouni H, et al. Convergent evolution in European and Rroma populations reveals pressure exerted by plague on Toll-like receptors. Proc Natl Acad Sci U S A. 2014;111(7):2668–73. doi: 10.1073/pnas.1317723111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saleh M, et al. Differential modulation of endotoxin responsiveness by human caspase-12 polymorphisms. Nature. 2004;429(6987):75–9. doi: 10.1038/nature02451. [DOI] [PubMed] [Google Scholar]
- 79.Trejo-de la OA, et al. Relevance of single-nucleotide polymorphisms in human TLR genes to infectious and inflammatory diseases and cancer. Genes Immun. 2014;15(4):199–209. doi: 10.1038/gene.2014.10. [DOI] [PubMed] [Google Scholar]
- 80.Tan TT, Coussens LM. Humoral immunity, inflammation and cancer. Curr Opin Immunol. 2007;19(2):209–16. doi: 10.1016/j.coi.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 81.Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–4. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 82.Hotez PJ, et al. Helminth infections: the great neglected tropical diseases. J Clin Invest. 2008;118(4):1311–21. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ye CJ, et al. Intersection of population variation and autoimmunity genetics in human T cell activation. Science. 2014;345(6202):1254665. doi: 10.1126/science.1254665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quan L, et al. Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. Int J Cancer. 2014;134(6):1408–21. doi: 10.1002/ijc.28458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Coit P, et al. Ethnicity-specific epigenetic variation in naive CD4+ T cells and the susceptibility to autoimmunity. Epigenetics Chromatin. 2015;8:49. doi: 10.1186/s13072-015-0037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paquette IM, et al. African Americans should be screened at an earlier age for colorectal cancer. Gastrointest Endosc. 2015;82(5):878–83. doi: 10.1016/j.gie.2015.03.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta S, et al. Strategies for reducing colorectal cancer among blacks. Arch Intern Med. 2012;172(2):182–4. doi: 10.1001/archinternmed.2011.594. [DOI] [PubMed] [Google Scholar]
- 88.Shenoy D, et al. Do African-American men need separate prostate cancer screening guidelines? BMC Urol. 2016;16(1):19. doi: 10.1186/s12894-016-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Metcalfe KA, et al. A comparison of the detection of BRCA mutation carriers through the provision of Jewish population-based genetic testing compared with clinic-based genetic testing. Br J Cancer. 2013;109(3):777–9. doi: 10.1038/bjc.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Phan VH, et al. Ethnic differences in drug metabolism and toxicity from chemotherapy. Expert Opin Drug Metab Toxicol. 2009;5(3):243–57. doi: 10.1517/17425250902800153. [DOI] [PubMed] [Google Scholar]
- 91.Logroscino G, et al. Current Issues in Randomized Clinical Trials of Neurodegenerative Disorders at Enrolment and Reporting: Diagnosis, Recruitment, Representativeness of Patients, Ethnicity, and Quality of Reporting. Front Neurol Neurosci. 2016;39:24–36. doi: 10.1159/000445410. [DOI] [PubMed] [Google Scholar]
- 92.Fouad MN, et al. Patient Navigation As a Model to Increase Participation of African Americans in Cancer Clinical Trials. J Oncol Pract. 2016;12(6):556–63. doi: 10.1200/JOP.2015.008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 94.Nakshatri H, et al. Ethnicity-Dependent and -Independent Heterogeneity in Healthy Normal Breast Hierarchy Impacts Tumor Characterization. Sci Rep. 2015;5:13526. doi: 10.1038/srep13526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Leavell BJ, et al. Associations between markers of colorectal cancer stem cells and adenomas among ethnic groups. Dig Dis Sci. 2012;57(9):2334–9. doi: 10.1007/s10620-012-2195-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ashktorab H, et al. A 50-year review of colorectal cancer in African Americans: implications for prevention and treatment. Dig Dis Sci. 2009;54(9):1985–90. doi: 10.1007/s10620-009-0866-5. [DOI] [PubMed] [Google Scholar]
- 97.Sundi D, et al. Pathological examination of radical prostatectomy specimens in men with very low risk disease at biopsy reveals distinct zonal distribution of cancer in black American men. J Urol. 2014;191(1):60–7. doi: 10.1016/j.juro.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kinseth MA, et al. Expression differences between African American and Caucasian prostate cancer tissue reveals that stroma is the site of aggressive changes. Int J Cancer. 2014;134(1):81–91. doi: 10.1002/ijc.28326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Martin DN, et al. Differences in the tumor microenvironment between African-American and European-American breast cancer patients. PLoS One. 2009;4(2):e4531. doi: 10.1371/journal.pone.0004531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zitvogel L, et al. Microbiome and Anticancer Immunosurveillance. Cell. 2016;165(2):276–87. doi: 10.1016/j.cell.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 101.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348(6230):56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 102.Tan DS, et al. Intertumor heterogeneity of non-small-cell lung carcinomas revealed by multiplexed mutation profiling and integrative genomics. Int J Cancer. 2014;135(5):1092–100. doi: 10.1002/ijc.28750. [DOI] [PubMed] [Google Scholar]
- 103.Krishnaswamy S, et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin Cancer Res. 2009;15(18):5714–23. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang W, et al. EGFR mutations in US Hispanic versus non-Hispanic white patients with lung adenocarcinoma. Arch Pathol Lab Med. 2014;138(4):543–5. doi: 10.5858/arpa.2013-0311-OA. [DOI] [PubMed] [Google Scholar]
- 105.Shi Y, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER) J Thorac Oncol. 2014;9(2):154–62. doi: 10.1097/JTO.0000000000000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pal SK, et al. Targeted therapies for non-small cell lung cancer: an evolving landscape. Mol Cancer Ther. 2010;9(7):1931–44. doi: 10.1158/1535-7163.MCT-10-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rodig SJ, et al. Unique clinicopathologic features characterize ALK-rearranged lung adenocarcinoma in the western population. Clin Cancer Res. 2009;15(16):5216–23. doi: 10.1158/1078-0432.CCR-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Soda M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 109.Wong DW, et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer. 2009;115(8):1723–33. doi: 10.1002/cncr.24181. [DOI] [PubMed] [Google Scholar]
- 110.Bollig-Fischer A, et al. Racial diversity of actionable mutations in non-small cell lung cancer. J Thorac Oncol. 2015;10(2):250–5. doi: 10.1097/JTO.0000000000000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu X, et al. Oncogenic GNAQ and GNA11 mutations in uveal melanoma in Chinese. PLoS One. 2014;9(10):e109699. doi: 10.1371/journal.pone.0109699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Qiu T, et al. Detection of BRAF mutation in Chinese tumor patients using a highly sensitive antibody immunohistochemistry assay. Sci Rep. 2015;5:9211. doi: 10.1038/srep09211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Si L, et al. Prevalence of BRAF V600E mutation in Chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer. 2012;48(1):94–100. doi: 10.1016/j.ejca.2011.06.056. [DOI] [PubMed] [Google Scholar]
- 114.Onken MD, et al. Oncogenic mutations in GNAQ occur early in uveal melanoma. Invest Ophthalmol Vis Sci. 2008;49(12):5230–4. doi: 10.1167/iovs.08-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Van Raamsdonk CD, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363(23):2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Faisal FA, et al. Racial Variations in Prostate Cancer Molecular Subtypes and Androgen Receptor Signaling Reflect Anatomic Tumor Location. Eur Urol. 2016;70(1):14–7. doi: 10.1016/j.eururo.2015.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tao L, et al. Breast Cancer Mortality in African-American and Non-Hispanic White Women by Molecular Subtype and Stage at Diagnosis: A Population-Based Study. Cancer Epidemiol Biomarkers Prev. 2015;24(7):1039–45. doi: 10.1158/1055-9965.EPI-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Carey LA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA. 2006;295(21):2492–502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 119.Porter PL, et al. Racial differences in the expression of cell cycle-regulatory proteins in breast carcinoma. Cancer. 2004;100(12):2533–42. doi: 10.1002/cncr.20279. [DOI] [PubMed] [Google Scholar]
- 120.Iqbal J, et al. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA. 2015;313(2):165–73. doi: 10.1001/jama.2014.17322. [DOI] [PubMed] [Google Scholar]
- 121.Yan M, et al. HER2 aberrations in cancer: implications for therapy. Cancer Treat Rev. 2014;40(6):770–80. doi: 10.1016/j.ctrv.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 122.John EM, et al. Prevalence of pathogenic BRCA1 mutation carriers in 5 US racial/ethnic groups. JAMA. 2007;298(24):2869–76. doi: 10.1001/jama.298.24.2869. [DOI] [PubMed] [Google Scholar]
- 123.Zhu Y, et al. BRCA mutations and survival in breast cancer: an updated systematic review and meta-analysis. Oncotarget. 2016 doi: 10.18632/oncotarget.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Keenan T, et al. Comparison of the Genomic Landscape Between Primary Breast Cancer in African American Versus White Women and the Association of Racial Differences With Tumor Recurrence. J Clin Oncol. 2015;33(31):3621–7. doi: 10.1200/JCO.2015.62.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shiao YH, et al. Racial disparity in the association of p53 gene alterations with breast cancer survival. Cancer Res. 1995;55(7):1485–90. [PubMed] [Google Scholar]
- 126.Xicola RM, et al. Excess of proximal microsatellite-stable colorectal cancer in African Americans from a multiethnic study. Clin Cancer Res. 2014;20(18):4962–70. doi: 10.1158/1078-0432.CCR-14-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sunakawa Y, et al. Prognostic Impact of Primary Tumor Location on Clinical Outcomes of Metastatic Colorectal Cancer Treated With Cetuximab Plus Oxaliplatin-Based Chemotherapy: A Subgroup Analysis of the JACCRO CC-05/06 Trials. Clin Colorectal Cancer. 2016 doi: 10.1016/j.clcc.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 128.Yoon HH, et al. Racial Differences in BRAF/KRAS Mutation Rates and Survival in Stage III Colon Cancer Patients. J Natl Cancer Inst. 2015;107(10) doi: 10.1093/jnci/djv186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ye JX, et al. KRAS and BRAF gene mutations and DNA mismatch repair status in Chinese colorectal carcinoma patients. World J Gastroenterol. 2015;21(5):1595–605. doi: 10.3748/wjg.v21.i5.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jeon CH, et al. Genetic alterations of APC, K-ras, p53, MSI, and MAGE in Korean colorectal cancer patients. Int J Colorectal Dis. 2008;23(1):29–35. doi: 10.1007/s00384-007-0373-0. [DOI] [PubMed] [Google Scholar]
- 131.Sussman DA, et al. Colorectal Tumors from Different Racial and Ethnic Minorities Have Similar Rates of Mismatch Repair Deficiency. Clin Gastroenterol Hepatol. 2016 doi: 10.1016/j.cgh.2016.03.037. [DOI] [PubMed] [Google Scholar]
- 132.Ribic CM, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349(3):247–57. doi: 10.1056/NEJMoa022289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van Cutsem E, et al. HER2 screening data from ToGA: targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer. 2015;18(3):476–84. doi: 10.1007/s10120-014-0402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Braconi C, et al. KIT and PDGFRalpha mutations in 104 patients with gastrointestinal stromal tumors (GISTs): a population-based study. Ann Oncol. 2008;19(4):706–10. doi: 10.1093/annonc/mdm503. [DOI] [PubMed] [Google Scholar]
- 135.Debiec-Rychter M, et al. Use of c-KIT/PDGFRA mutational analysis to predict the clinical response to imatinib in patients with advanced gastrointestinal stromal tumours entered on phase I and II studies of the EORTC Soft Tissue and Bone Sarcoma Group. Eur J Cancer. 2004;40(5):689–95. doi: 10.1016/j.ejca.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 136.Yeh CN, et al. Kinase mutations and imatinib mesylate response for 64 Taiwanese with advanced GIST: preliminary experience from Chang Gung Memorial Hospital. Ann Surg Oncol. 2007;14(3):1123–8. doi: 10.1245/s10434-006-9288-1. [DOI] [PubMed] [Google Scholar]
- 137.Li J, et al. Presence of PDGFRA and DOG1 mutations in gastrointestinal stromal tumors among Chinese population. Int J Clin Exp Pathol. 2015;8(5):5721–6. [PMC free article] [PubMed] [Google Scholar]
- 138.Jiron J, et al. Racial disparities in Human Papillomavirus (HPV) associated head and neck cancer. Am J Otolaryngol. 2014;35(2):147–53. doi: 10.1016/j.amjoto.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Settle K, et al. Racial survival disparity in head and neck cancer results from low prevalence of human papillomavirus infection in black oropharyngeal cancer patients. Cancer Prev Res (Phila) 2009;2(9):776–81. doi: 10.1158/1940-6207.CAPR-09-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Iqbal A, et al. Role of human papillomavirus infection and other factors in patients with head and neck squamous cell carcinoma. Oral Dis. 2014;20(3):288–93. doi: 10.1111/odi.12110. [DOI] [PubMed] [Google Scholar]
- 141.Brzezianska E, et al. Investigation of V600E BRAF mutation in papillary thyroid carcinoma in the Polish population. Neuro Endocrinol Lett. 2007;28(4):351–9. [PubMed] [Google Scholar]
- 142.Wang Z, et al. Clinical impact of BRAF mutation on the diagnosis and prognosis of papillary thyroid carcinoma: a systematic review and meta-analysis. Eur J Clin Invest. 2016;46(2):146–57. doi: 10.1111/eci.12577. [DOI] [PubMed] [Google Scholar]
- 143.Sun J, et al. BRAF V600E and TERT Promoter Mutations in Papillary Thyroid Carcinoma in Chinese Patients. PLoS One. 2016;11(4):e0153319. doi: 10.1371/journal.pone.0153319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu C, et al. TERT promoter Mutation and Its Association with Clinicopathological Features and Prognosis of Papillary Thyroid Cancer: A Meta-analysis. Sci Rep. 2016;6:36990. doi: 10.1038/srep36990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Killela PJ, et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget. 2014;5(6):1515–25. doi: 10.18632/oncotarget.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yan W, et al. Correlation of IDH1 mutation with clinicopathologic factors and prognosis in primary glioblastoma: a report of 118 patients from China. PLoS One. 2012;7(1):e30339. doi: 10.1371/journal.pone.0030339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kristensen LS, et al. Assessment of Quantitative and Allelic MGMT Methylation Patterns as a Prognostic Marker in Glioblastoma. J Neuropathol Exp Neurol. 2016;75(3):246–55. doi: 10.1093/jnen/nlv024. [DOI] [PMC free article] [PubMed] [Google Scholar]


