Abstract
Dengue is a severe mosquito-borne viral infection causing half a million deaths annually. Dengue virus NS2B/NS3 protease is a validated target for anti-dengue drug design. A series of hitherto unreported 3,5-bis(arylidene)-4-piperidones analogues 4a—4j were synthesized and screened in silico against DENV2 NS2B/NS3 protease to elucidate their binding mechanism and orientation around the active sites. Results were validated through an in vitro DENV2 NS2B/NS3 protease assay using a fluorogenic Boc-Gly-Arg-Arg-AMC substrate. Nitro derivatives of 3,5-bis(arylidene)-4-piperidones (4e and 4j) emerged as promising lead molecules for novel protease inhibitors with an IC50 of 15.22 and 16.23 µmol/L, respectively, compared to the standard, panduratin A, having IC50 of 57.28 µmol/L.
KEY WORDS: Dengue virus; NS2B/NS3 protease; Piperidone; α,β-Unsaturated ketone; Protease inhibitors
Graphical abstract
A series of hitherto unreported 3,5-bis(arylidene)-4-piperidones analogues 4a—4j were synthesized and screened in silico against DENV2 NS2B/NS3 protease to elucidate their binding mechanism and orientation around the active sites. Compounds 4e and 4j emerged as promising lead molecules for novel protease inhibitors with an IC50 of 15.22 and 16.23 µmol/L, respectively, compared to the standard, panduratin A, having IC50 of 57.28 µmol/L.
1. Introduction
Dengue virus (DENV) is a dreadful arboviral pathogen responsible for the tropical epidemic dengue fever (DF) causing high rates of global morbidity and mortality1. According to the World Health Organization (WHO), around 3.9 billion people are currently under high risk of dengue fever infection2. DENV infections have now become endemic in more than half of the world and recently an increased number of uncontrolled outbreaks with huge socio-economic implications have been reported3. DENVs exist as four closely related antigenic DENV, 1—4 serotypes, but the cross-immunity amongst each other after recovery is only partial and successive infection by different serotypes may worsen the severity due to an “antigen-dependent enhancement” effect (ADE). This ADE effect makes vaccine development against DENVs extremely difficult4. Recently, Sanofi obtained first approval for a long-anticipated tetravalent vaccine Dengvaxia® against dengue fever, but its efficacy against the different DENVs is still unclear5. Currently, there is no other vaccine or effective anti-viral therapy available in the market for the prevention or treatment of dengue fever. Therefore, there is a pressing need for development of new anti-dengue agents that are effective against all serotypes (Table 1).
Table 1.
DENV2 NS2B/NS3 protease inhibition activities of 3,5-bis(arylidene)-4-piperidones (4a—4j).
 |
||||
| Compd. | Ar | Ar׳ | Binding free energy (kcal/mol) | IC50 (µmol/L)a |
| 4a | 2-CH3Ph | 4-FPh | −10.00 | NDb |
| 4b | 2-ClPh | 4-FPh | −10.07 | ND |
| 4c | 2,4-Cl2Ph | 4-FPh | −10.39 | ND |
| 4d | 4-FPh | 4-FPh | −9.49 | ND |
| 4e | 4-NO2Ph | 4-FPh | −11.36 | 15.22±1.10 |
| 4f | 2-CH3Ph | 4-OCH3Ph | −10.53 | ND |
| 4g | 2-ClPh | 4-OCH3Ph | −10.25 | ND |
| 4h | 2,4-Cl2Ph | 4-OCH3Ph | −10.11 | ND |
| 4i | 4-FPh | 4-OCH3Ph | −9.81 | ND |
| 4j | 4-NO2Ph | 4-OCH3Ph | −11.09 | 16.23±1.30 |
| Panduratin A | – | – | −10.10 | 57.28±1.30 |
—Not applicable.
Values are indicated as means±standard deviations from 3 independent experiments performed in triplicate.
ND, Not determined as <10% inhibition at 50 µmol/L.
The dengue virus genome is a single-stranded RNA encoding three structural proteins viz., capsid C, membrane M, and the envelope E along with the non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 which are processed by trypsin-like NS2B-NS3 protease6). The catalytic triad (His51, Asp75, and Ser135) is within the NS3 protease domain, but a region of NS2B is also required for catalytic activity7. Protease complex NS2B/NS3 is essential for viral replication and therefore receives considerable attention as a therapeutic target for the development of novel dengue inhibitors8.
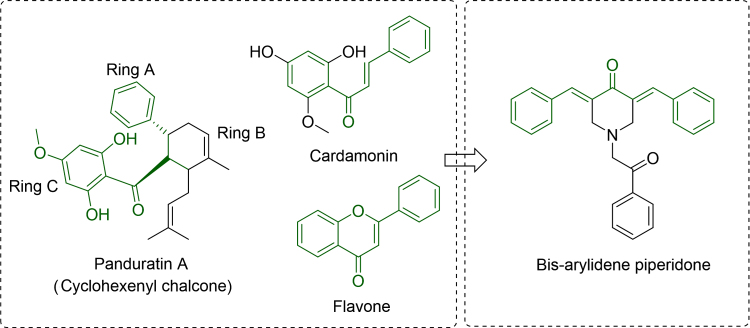
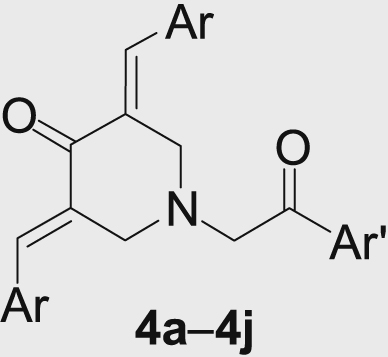
Previous studies have shown that the α,β-unsaturated ketone analogues isolated from Boesenbergia rotunda were potent inhibitors of DEN2 serine protease. Among these cyclohexenyl derivatives, 4-hydroxypanduratin A (Ki 21 µmol/L) and panduratin A (Ki 25 µmol/L) emerged as lead molecules for antidengue agents9. Subsequently, several 4-hydroxypanduratin analogues with potential dengue inhibitory activity have also been reported10. Moreover, 3,5-bis(arylidene)-4-piperidones, which are considered as a structurally distinct class of α,β-unsaturated ketones, possess marked inhibitory activities against viruses11. 3,5-Bis(arylidene)-4-piperidone derivatives constitute an important class of therapeutic agents exhibiting anticancer12, antioxidant13 and anticholinesterase14 properties as well. Prompted by the above findings and in continuation of our interest in the biological activities of 3,5-bis(arylidene)-4-piperidone15, we thought it worthwhile to incorporate the bioactive heterocycle piperidone moiety into α,β-unsaturated ketone mimics with the hope that the piperidone residue may serve as both the donor and acceptor in hydrogen-bonding interactions to improve binding affinity and also to verify its importance for the DEN2 serine protease inhibitory activity (Fig. 1).
Figure 1.
3,5-Bis(arylidene)-4-piperidones analogues as dengue protease inhibitors.
2. Results and discussion
2.1. Chemistry
The target compounds 3,5-bis(arylidene)-1-(2-oxo-2-arylethyl)piperidin-4-ones 4a—4j as depicted in Scheme 1 were obtained by the Claisen–Schmidt condensations of 4-piperidone (1) with different aromatic aldehydes in the presence of HCl and acetic acid followed by reaction with substituted phenacyl bromide16. The yields of titled compounds ranged from 68% to 87% after recrystallization with ethanol. The purity of the compounds was checked by TLC and elemental analyses. Both analytical and spectral data (NMR and IR) of all the synthesized compounds were in full agreement with the proposed structures. The IR spectrum of compound 4j showed C=O and C--N stretching vibrations at 1670 and 1232 cm−1, respectively. The 1H NMR spectral data of compound 4j showed three upfielded singlets at δ 3.85, 4.07 and 4.11 ppm due to OCH3, COCH2 and pipridine-methylene (CH2) protons. The appearance of aromatic protons and disappearance of the NH signal of the piperidine moiety further confirmed the formation of the target compounds. The 13C NMR spectral data of the compound showed three peaks in the aliphatic range of δ 54.23, 55.42 and 61.98 ppm due to pipridine-methylene (CH2), methoxy and oxoethyl carbons, respectively, whereas two carbonyl carbons were observed downfield at δ 183.19 and 194.27. The other peaks of carbon were observed at δ 113.86, 123.67, 128.43, 129.77, 130.29, 134.94, 135.85, 136.49, 148.36 and 161.81, confirming the presence of 28 carbons in the compound.
Scheme 1.
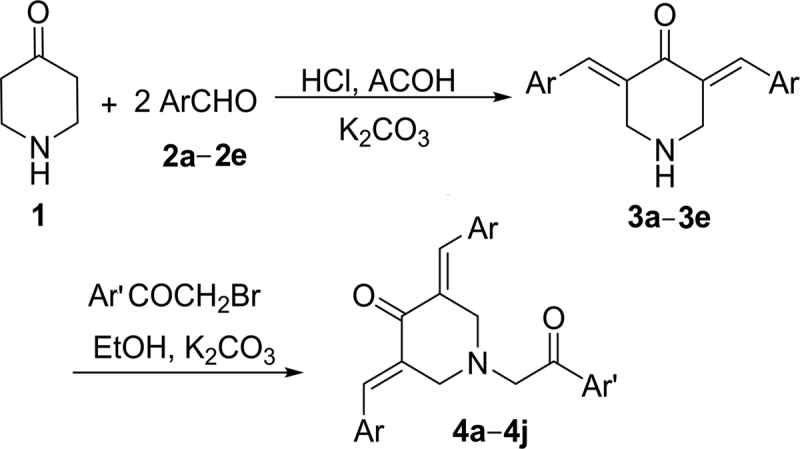
Synthesis of 3,5-bis(arylidene)-4-piperidones (4a—4j).
2.2. Molecular docking studies
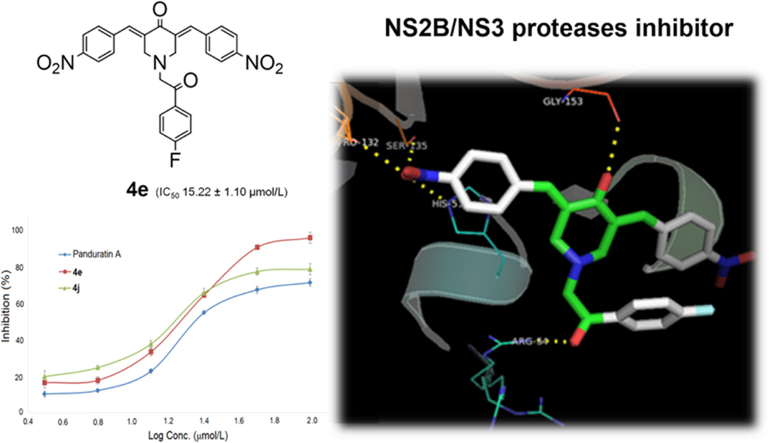
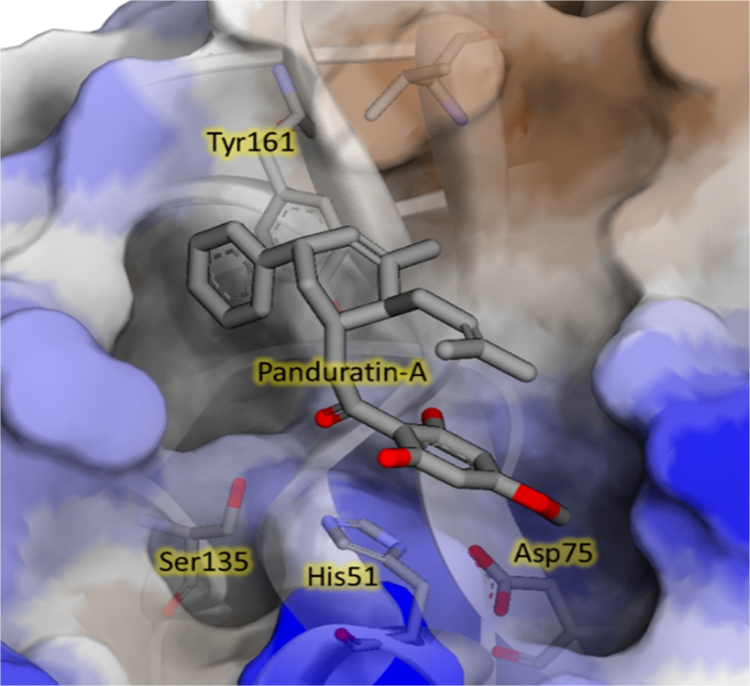
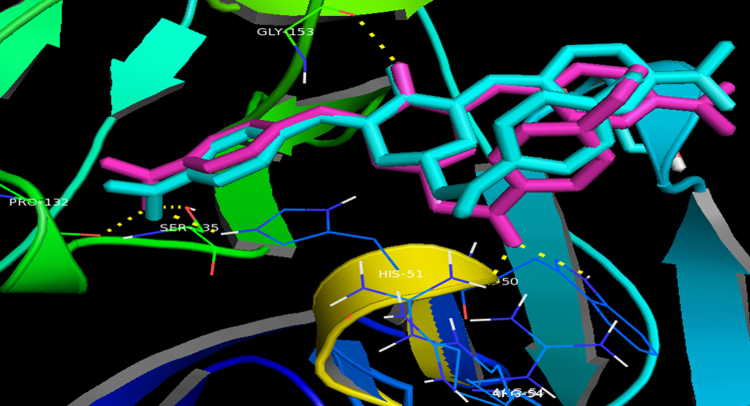
In our effort to identify novel potent NS2B/NS3 serine protease inhibitors with drug-like properties, we delved more deeply into the molecular interactions of the reference ligand panduratin A with the serine protease. AutoDock 4.2 with a Lamarkian genetic algorithm-implemented program suite was employed to identify appropriate binding modes and conformation of the ligand molecules. The crystal structure of dengue virus NS2B/NS3 protease (PDB code:2FOM, resolution 1.5 Å) was retrieved from the protein data bank (PDB) for molecular modelling studies17. The allosteric pocket proximal to the catalytic triad comprising His51, Asp75 and Ser135 residues in the NS3 protein was identified as the active site18. The carbonyl group of panduratin A projects into the oxyanion binding hole and forms hydrogen bonds with the amino hydrogens of Ser153 and Gly 151 adjacent to the catalytic site. The phenyl ring A positioned near the hydrophobic pocket S1 formed a π-π stacking interaction with Tyr161 whereas the trisubstituted phenyl moiety C aligned parallel to the pentacyclodiazo side-chain of His51 forming a CH-π interaction (Fig. 2). These binding interactions are consistent with previous reports10, 19 and gave insight into structure optimization in a further study. Based on these observations, 3,5-bis(arylidene)-4-piperidones (4a—4j) were designed as promising NS2B/NS3 protease inhibitors. Molecular docking studies of the designed molecules 4a—4j also revealed that they fit into the active site and formed hydrogen bonds with the catalytic triad. The binding free energy of compounds 4a—4j were in the range of −9.49 to −11.36 kcal/mol, indicating sufficient affinity between ligands and protein. Among these, nitro derivatives 4e and 4j were observed to have lowest docked energy of −11.36 and −11.09 kcal/mol, respectively. The docked conformations of the two ligands 4e and 4j bound to the active site of DEN2 NS2B/NS3 serine protease are shown in Fig. 3.
Figure 2.
Binding mode of panduratin A at the active site of NS2B/NS3 serine protease (PDB code: 2FOM).
Figure 3.
Binding mode of compounds 4e (cyan) and 4j (magenta) into NS2B/NS3 serine protease active site. Compound 4e formed five hydrogen bonds (yellow dotted lines) with His51 (2.9 Å), Pro132 (2.8 Å), Ser135 (2.8 Å), Gly153 (2.5 Å) and Arg54 (3.1 Å) whereas compound 4j formed six hydrogen bonds (yellow dotted lines) with His51 (2.9 Å), Pro132 (2.8 Å), Ser135 (2.8 Å), Gly153 (2.5 Å), Arg54 (2.8 Å) and Trp50 (3.1 Å).
The most promising compound 4e was observed to fit nicely into the active site of the NS2B/NS3 protease by forming five hydrogen bond interactions with the active site residues. The para-nitro group seems to be favorable in forming hydrogen bonds with His51, Pro132 and Ser135; however no interaction was observed with the catalytic triad residue Asp75. The 4-oxo substituent positioned near the oxyanion hole formed a hydrogen bond with a conserved active site residue Gly153, whereas the carbonyl group of the aryl ethanone moiety formed an additional hydrogen bond interaction with guanidino group of Arg54, allowing the molecule to bind protein more tightly and enhancing its inhibitory potential. Lipophilicity also appears to play crucial role in 4e inhibitory activity, as the phenyl ring was oriented to the lipophilic area of the binding site and formed π-π stacking with His51. Compound 4j also binded in a mode very similar to that of compound 4e, with slight variation.
2.3. Bioassays studies
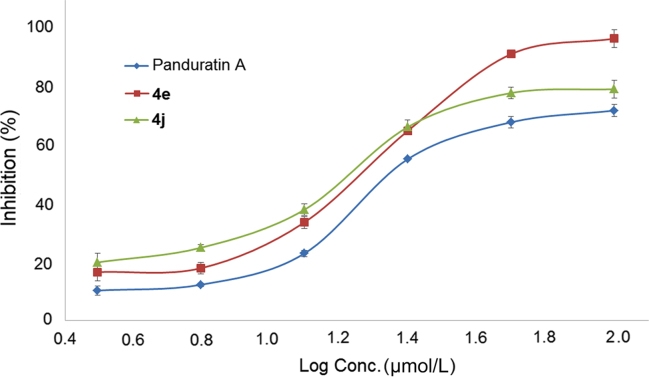
Fifty percent inhibitory concentration (IC50) values for selected compounds showing more than 10% inhibition at 50 µmol/L were determined against DENV2 NS2B/NS3 protease by using fluorogenic peptide substrate Boc-Gly-Arg-7-amino-4-methylcoumarin. Compounds 4e and 4j passed the preliminary screening and their dose-response curves are summarised in Fig. 4. Panduratin A, which was previously reported as a potent NS2B/NS3 dengue protease inhibitor was used as a positive control in this study9. The observed IC50 values for both compounds 4e and 4j were 15.22±1.10 and 16.23±1.30 µmol/L, respectively, as compared to panduratin A with an IC50 value 57.28±1.30 µmol/L. In general, it was observed that compounds having p-nitro functionality at the benzylidene moiety played an essential role in bonding with the catalytic triad due to its polar nature. The results of the in vitro study are in consistent with our docking studies and the previous findings which confirm the positive influence of the nitro group on the phenyl ring20, 21. The overall results indicate strong inhibitory activities of 3,5-bis(arylidene)-4-piperidones against NS2B-NS3 dengue protease.
Figure 4.
Dose response curves of some selected compounds on NS2B/NS3 proteases.
3. Conclusions
Based on previous findings that 3,5-bis(arylidene)-4-piperidones are endowed with strong antiviral efficacy, efforts were undertaken to design and synthesize piperidone-incorporated α,β-unsaturated ketones as novel dengue inhibitors. Molecular docking studies revealed significant molecular interactions between 3,5-bis(arylidene)-4-piperidones and the NS2B/NS3 dengue protease catalytic triad indicating their promising potential for further optimization as lead molecules. The most active inhibitors were also screened in vitro to assess their potential DENV NS2B/NS3 protease inhibitory effects and the results strongly correlated with the in silico findings.
4. Material and methods
All the chemicals and solvents were purchased from Sigma–Aldrich (USA) and QReC and were used without further purification. The synthesized compounds were purified by crystallization using ethanol and the melting points measured in open capillary tubes using a digital auto melting point apparatus by Stuart Scientific SMPI and used uncorrected. All compounds were characterized by FT-IR spectrometer using KBr pallets, 1H NMR (500 MHz, Bruker) and 13C NMR spectroscopy (125 MHz, Bruker) in CDCl3; the coupling constant (J) was reported in Hz and chemical shifts were reported in ppm (δ).
4.1. General procedure for the synthesis of 3,5-bis(arylidene)-1-(2-(aryl)-2-oxoethyl)piperidin-4-one derivatives (4a—4j)
To a stirred solution of 3,5-di(arylidene)piperidin-4-ones (0.01 mol) and K2CO3 (0.02 mol) in absolute ethanol (15 mL), substituted phenacyl bromide (0.02 mol) dissolved in ethanol was added dropwise. The completion of the reaction was monitored by TLC. The solid mass thus obtained was filtered, washed with water, and recrystallized from methanol.
4.1.1. 1-(2-(4-Fluorophenyl)-2-oxoethyl)-3,5-bis(2-methylbenzylidene)piperidin-4-one (4a)
Yellow solid 73.43%; mp 130–132 °C; IR (KBr, cm−1): 1706 (C=O), 1233 (C–N), 1147 (C–F); 1H NMR (500 MHz, CDCl3) δ: 2.35 (s, 6H, CH3), 3.88 (s, 4H, CH2NCH2), 3.91 (s, 2H, COCH2), 7.04 (t, 2H, J=8.5 Hz, ArH), 7.11 (d, 2H, J=7.5 Hz, ArH), 7.18 (t, 2H, J=8.00 Hz, ArH), 7.23–7.26 (m, 4H, ArH), 7.87 (d, 2H, J=7.5 Hz, ArH), 8.00 (s, 2H, ArCH=).13C NMR (125 MHz, CDCl3) δ: 20.03 (CH3), 54.36 (CH2NCH2), 62.46 (COCH2), 115.58, 115.75, 125.65, 128.83, 129.00, 130.73, 130.80, 132.81, 134.10, 136.62, 138.19 (PhCH=), 166.84 (C–F), 186.79 (CO), 194.79 (COPh). Anal. Calcd. for C29H26FNO2: C, 79.25; H, 5.96; N, 3.19; Found: C, 79.30; H, 6.12; N, 3.11.
4.1.2. 3,5-bis(2-Chlorobenzylidene)-1-(2-(4-fluorophenyl)-2-oxoethyl)piperidin-4-one (4b)
Yellow solid 78.36%; mp 142–144 °C; IR (KBr, cm−1):1694 (C=O), 1222 (C–N), 1152 (C—F), 770 (C–Cl); 1H NMR (500 MHz, CDCl3) δ: 3.98 (s, 4H, CH2NCH2), 3.99 (s, 2H, OCH2), 7.07 (t, 2H, J=9.0 Hz, ArH), 7.20 (dd, 2H, J=7.0,1.50 Hz, ArH), 7.27–7.32 (m, 4H, ArH), 7.44 (dd, 2H, J=8.00, 2.00 Hz, ArH), 7.87–7.90 (m, 2H, ArH), 8.05 (s, 2H, ArCH=). 13C NMR (125 MHz, CDCl3) δ: 54.01 (CH2NCH2), 61.81 (COCH2), 115.64, 115.81, 126.56, 129.95, 130.11, 130.29, 132.00, 133.43, 133.96, 134.69, 135.07 (PhCH=), 166.89 (C–F), 186.43(CO), 194.91 (COPh). Anal. Calcd. for C27H20Cl2FNO2: C, 67.51; H, 4.20; N, 2.92; Found: C, 67.58; H, 4.26; N, 2.85.
4.1.3. 3,5-bis(2,4-Dichlorobenzylidene)-1-(2-(4-fluorophenyl)-2-oxoethyl)piperidin-4-one (4c)
Yellow solid 68.43%; mp 110–112 °C; IR (KBr, cm−1): 1698 (C=O), 1226 (C–N), 1104 (C—F), 832 (C–Cl); 1H NMR (500 MHz, CDCl3) δ: 3.90 (s, 4H, CH2NCH2), 3.95 (s, 2H, OCH2), 7.09 (t, 2H, J=8.5 Hz, ArH), 7.14 (d, 2H, J=8.0 Hz, ArH), 7.26 (d, 2H, J=9.0 Hz, ArH), 7.45 (d, 2H, J=2.5 Hz, ArH), 7.88–7.91 (m, 2H, ArH), 7.92 (s, 2H, s, ArCH=). 13C NMR (125 MHz, CDCl3) δ: 53.92 (CH2NCH2), 61.83 (OCH2), 115.82, 127.01, 129.92, 130.68, 130.76, 130.96, 131.87, 133.48, 134.33, 135.47, 135.86 (PhCH=), 166.99 (C–F), 186.09 (CO), 194.80 (COPh). Anal. Calcd. for C27H18Cl4FNO2: C, 59.04; H, 3.30; N, 2.55; Found: C, 59.10; H, 3.35; N, 2.78.
4.1.4. 3,5-bis(4-Fluorobenzylidene)-1-(2-(4-fluorophenyl)-2-oxoethyl)piperidin-4-one (4d)
Yellow solid 87.30%; mp 135–137 °C; IR (KBr, cm−1): 1691 (C=O), 1225 (C–N), 1158 (C–F); 1H NMR (500 MHz, CDCl3) δ: 4.02 (s, 4H, CH2NCH2), 4.05 (s, 2H, OCH2), 7.06–7.11 (q, 6H, J=8.5 Hz, ArH), 7.34–7.37 (q, 4H, J=5.5 Hz, ArH), 7.79 (s, 2H, ArCH=), 7.93–7.96 (q, 2H, J=6.0 Hz, ArH).13C NMR (125 MHz, CDCl3) δ: 54.39 (CH2NCH2), 62.45 (COCH2), 115.74, 115.91, 130.81, 130.89, 131.95, 132.39, 132.52, 135.90, 164.92 (C–F) 166.95 (C–F), 186.77 (CO), 194.82 (COPh). Anal. Calcd. for C27H20Cl3F3NO2: C, 72.48; H, 4.51; N, 3.13; Found: C, 72.55; H, 4.57; N, 3.05.
4.1.5. 1-(2-(4-Fluorophenyl)-2-oxoethyl)-3,5-bis(4-nitrobenzylidene)piperidin-4-one (4e)
Yellow solid 59.03%; mp 115–117 °C; IR (KBr, cm−1): 1693 (C=O), 1220 (C–N), 1150 (C–F); 1H NMR (500 MHz, CDCl3) δ: 3.42 (s, 4H, CH2NCH2), 3.98 (s, 2H,OCH2), 7.29 (t, 2H, J=9.0 Hz, ArH), 7.76–7.99 (m, 4H, ArH), 8.25 (d, 2H, ArH), 8.30 (s, 2H, ArCH=). 13C NMR (125 MHz, DMSO) δ: 54.43 (CH2NCH2), 62.27 (OCH2), 115.49, 115.64, 129.54, 130.72, 131.90, 132.11, 133.52, 134.35, 135.98 (PhCH=), 166.52 (CO), 186.72 (CO), 194.99 (COPh). Anal. Calcd. for C27H20FN3O6: C, 64.67; H, 4.02; N, 8.38; Found: C, 64.71; H, 4.08; N, 8.40.
4.1.6. 1-(2-(4-Methoxyphenyl)-2-oxoethyl)-3,5-bis(2-methylbenzylidene)piperidin-4-one (4f)
Pale yellow solid 70.00%; mp 118–120 °C; IR (KBr, cm−1): 1673 (C=O), 1257 (C–N); 1H NMR (500 MHz, CDCl3) δ: 2.35 (s, 6H, CH3), 3.84 (s, 3H, OCH3), 3.91 (s, 4H, CH2NCH2), 3.93 (s, 2H, OCH2), 6.86 (d, 2H, J=8.5 Hz, ArH), 7.22–7.27 (m, 8H, ArH), 7.84 (d, 2H, J=9.0 Hz, ArH), 8.00 (s, 2H, ArCH=); 13C NMR (125 MHz, CDCl3) δ: 20.04 (CH3), 54.42 (CH2NCH2), 55.46 (OCH3), 62.28 (COCH2), 113.69, 125.63, 128.76, 128.87, 128.91, 130.31, 130.35, 133.12, 134.19, 136.28, 138.16 (ArCH=), 163.64 (C–O), 194.88 (C=O), 207.02 (COPh). Anal. Calcd. for C30H29NO3: C, 79.80; H, 6.47; N, 3.10; Found: C, 79.88; H, 6.54; N, 3.17.
4.1.7. 3,5-bis(2-Chlorobenzylidene)-1-(2-(4-methoxyphenyl)-2-oxoethyl)piperidin-4-one (4g)
Yellow solid 85.26%; mp: 108–110 °C; IR (KBr, cm−1): 1688 (C=O), 1190 (C–N); 1H NMR (500 MHz, CDCl3) δ: 3.84 (s, 3H, OCH3), 3.90 (s, 4H, CH2NCH2), 3.95 (s, 2H, COCH2), 6.86 (d, 2H, J=9.0 Hz, ArH), 7.24 (t, 2H, J=1.5 Hz, ArH), 7.26–7.28 (m, 4H, ArH), 7.43 (d, 2H, J=2.0 Hz, ArH), 7.85 (s, 2H, J=9.0 Hz, ArH), 8.02 (s, 2H, ArCH=); 13C NMR (125 MHz, CDCl3) δ: 54.03(CH2NCH2), 55.48 (OCH3), 61.66 (COCH2), 113.74, 126.55, 128.65, 129.91, 130.06, 130.33, 130.79, 133.46, 134.15, 134.44, 135.08, 163.73 (C–O), 186.60 (C=O), 194.98 (COPh). Anal. Calcd. for C28H23Cl2NO3: C, 68.30; H, 4.71; N, 2.84; Found:C, 68.37; H, 4.73; N, 2.78.
4.1.8. 3,5-bis(2,4-Dichlorobenzylidene)-1-(2-(4-methoxyphenyl)-2-oxoethyl)piperidin-4-one (4h)
Yellow solid 77.98%; mp 156–158 °C; IR (KBr, cm−1): 1682 (C=O), 1259 (C–N); 1H NMR (500 MHz, CDCl3) δ: 3.42 (s, 4H, CH2NCH2), 3.72 (s, 2H,COCH2), 3.85 (s, 3H, OCH3), 6.88 (d, 2H, J=9.0 Hz, ArH), 7.15 (d, 2H, J=8.0 Hz, ArH), 7.25 (d, 2H, J=8.5 Hz, ArH), 7.45 (s, 2H, ArH), 7.85 (d, 2H, J=9.0 Hz, ArH), 7.92 (s, 2H, ArCH=). 13C NMR (125 MHz, CDCl3) δ: 53.94 (CH2NCH2), 55.50 (OCH3), 61.66 (COCH2), 113.81, 127.0, 128.55, 129.88, 130.30, 130.99, 131.94, 133.22, 134.52, 135.40, 135.87 (ArCH=), 163.83 (C–O), 184.15 (C=O), 194.83 (COPh). Anal. Calcd. for C28H21Cl4NO3: C, 59.92; H, 3.77; N, 2.50; Found: C, 59.96; H, 3.83; N, 2.52.
4.1.9. 3,5-bis(4-Fluorobenzylidene)-1-(2-(4-methoxyphenyl)-2-oxoethyl)piperidin-4-one (4i)
Light yellow solid 82.81%; mp 109–111 °C; IR (KBr, cm−1): 1682 (C=O), 1222 (C–N); 1H NMR (500 MHz, CDCl3) δ: 3.37 (s, 4H, CH2NCH2), 3.69 (s, 2H,COCH2), 3.81 (s, 3H, OCH3), 6.95 (d, 4H, J=9.0 Hz, ArH), 7.01 (t, 4H, J=9.0 Hz, ArH), 7.35–7.38 (m, 4H, ArH), 7.78 (s, 2H, ArCH=).13C NMR (125 MHz, CDCl3) δ: 54.40 (CH2NCH2), 55.50 (OCH3), 62.22 (COCH2), 113.80, 115.89, 115.89, 128.66, 130.42, 132.33, 132.40, 132.61, 132.60, 135.61 (ArCH=), 161.96 (C–F), 163.96 (C–O), 186.94 (C=O), 194.90 (COPh). Anal. Calcd. for C28H21Cl4NO3: C, 73.19; H, 5.05; N, 3.05; Found: C, 73.27; H, 5.10; N, 3.01.
4.1.10. 1-(2-(4-Methoxyphenyl)-2-oxoethyl)-3,5-bis(4-nitrobenzylidene)piperidin-4-one (4j)
Bright yellow solid 83.16%; mp 156–158 °C; IR (KBr, cm−1): 1670 (C=O), 1232 (C–N); 1H NMR (500 MHz, CDCl3) δ: 3.85 (s, 3H, OCH3), 4.07 (s, 2H, COCH2), 4.11 (s, 4H, CH2NCH2), 6.89 (d, 2H, J=9.0 Hz, ArH), 7.62 (d, 2H, J=8.5 Hz, ArH), 7.70 (d, 4H, J=7.5 Hz, ArH), 7.85 (s, 2H, ArCH=), 7.88 (d, 4H, J=9.0 Hz, ArH). 13C NMR (125 MHz, CDCl3) δ: 54.23 (CH2NCH2), 55.42 (OCH3), 61.98 (COCH2), 113.86, 123.67, 128.43, 129.77, 130.29, 134.94, 135.85, 136.49, 148.36, 161.81 (C–O), 183.19 (C=O), 194.27 (COPh). Anal. Calcd. for C28H23N3O7: C, 65.49; H, 4.51; N, 8.18; Found: C, 65.55; H, 4.60; N, 8.15.
4.2. Molecular docking studies
Molecular docking studies were performed using AutoDock 4.2 to identify appropriate binding modes and conformation of the ligand molecules. The crystal structure of dengue virus NS2B/NS3 protease (PDB code:2FOM, resolution-1.5 Å) was retrieved from the PDB and used for molecular modelling studies. The structures of all the compounds (4a—4j) were sketched using Chemdraw ultra 13.0 and converted into 3D structures using Hyperchem pro 8.0 software (www.hyper.com). Autodock tools (ADT) version 1.5.6 (www.autodock.scrips.edu) was used to prepare molecular docking. The grid box size was set to a dimension of 74×42×64 in x, y, z coordinates to cover the active site of the protease while virtual screening was performed by AutoDock 4.2.5.1. The best binding conformation was selected from the docking log (.dlg) file for each ligand and further interaction analysis was done using PyMol and Discovery Studio Visualizer 4.0.
4.3. In vitro assays
The bioassay studies were performed using a Modulus Microplate Reader with a fluorescence spectrophotometer following methods reported by Yusof et al. 22. The protease assays were carried out by preparing an enzymatic reaction mixture with a total volume of 100 µL containing standard buffer solution (Tris–HCl buffer pH 8.5), a constant amount (0.57 µmol/L) of DENV2 NS2B-NS3 solution and the synthesized compound (200 mg/L in DMSO). The mixtures were incubated at 37 °C and shaken at 200 rpm for 10 min, and then 10 mmol/L of peptide substrate which was purchased from th Peptide Institute (Japan) was added to initiate the reaction and incubation with shaking at 200 rpm (Ambient shaker incubator SI-50, Protech, Malaysia) was carried out for another 60 min. The assays were in triplicate and corrected by subtracting the absorbance of their respective blank. The selected compounds were used to generate an IC50 value in the range (6.25–200 mg/L) of sample concentration. The IC50 values were determined graphically from inhibition curves (log inhibitor concentration vs. percentage of inhibition) using graphpad module (www.graphpad.com).
Acknowledgments
The authors wish to acknowledge The Research University Team Grant (RUT/1001/PKIMIA/855006) provided by Universiti Sains Malaysia (USM).
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Mallhi T.H., Khan A.H., Sarriff A., Adnan A.S., Khan Y.H. Patients related diagnostic delay in dengue: an important cause of morbidity and mortality. Clin Epidemiol Glob Health. 2016;4:200–201. [Google Scholar]
- 2.Brady O.J., Gething P.W., Bhatt S., Messina J.P., Brownstein J.S., Hoen A.G. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shepard D.S., Undurraga E.A., Halasa Y.A., Stanaway J.D. The global economic burden of dengue: a systematic analysis. Lancet Inf Dis. 2016;16:935–941. doi: 10.1016/S1473-3099(16)00146-8. [DOI] [PubMed] [Google Scholar]
- 4.Bäck A.T., Lundkvist Å. Dengue viruses—an overview. Infect Ecol Epidemiol. 2013;3:19839. doi: 10.3402/iee.v3i0.19839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Recker M., Vannice K., Hombach J., Jit M., Simmons C.P. Assessing dengue vaccination impact: model challenges and future directions. Vaccine. 2016;34:4461–4465. doi: 10.1016/j.vaccine.2016.06.082. [DOI] [PubMed] [Google Scholar]
- 6.Xie X., Gayen S., Kang C., Yuan Z., Shi P.Y. Membrane topology and function of dengue virus NS2A protein. J Virol. 2013;87:4609–4622. doi: 10.1128/JVI.02424-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falgout B., Pethel M., Zhang Y.M., Lai C.J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Phong W.Y., Moreland N.J., Lim S.P., Wen D., Paradkar P.N., Vasudevan S.G. Dengue protease activity: the structural integrity and interaction of NS2B with NS3 protease and its potential as a drug target. Biosci Rep. 2011;31:399–409. doi: 10.1042/BSR20100142. [DOI] [PubMed] [Google Scholar]
- 9.Kiat T.S., Pippen R., Yusof R., Ibrahim H., Khalid N., Rahman N.A. Inhibitory activity of cyclohexenyl chalcone derivatives and flavonoids of fingerroot, Boesenbergia rotunda (L.), towards dengue-2 virus NS3 protease. Bioorg Med Chem Lett. 2006;16:3337–3340. doi: 10.1016/j.bmcl.2005.12.075. [DOI] [PubMed] [Google Scholar]
- 10.Frimayanti N., Chee C.F., Zain S.M., Rahman N.A. Design of new competitive dengue NS2B/NS3 protease inhibitors—a computational approach. Int J Mol Sci. 2011;12:1089–1100. doi: 10.3390/ijms12021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Subbagh H.I., Abu-Zaid S.M., Mahran M.A., Badria F.A., Al-Obaid A.M. Synthesis and biological evaluation of certain α,β-unsaturated ketones and their corresponding fused pyridines as antiviral and cytotoxic agents. J Med Chem. 2000;43:2915–2921. doi: 10.1021/jm000038m. [DOI] [PubMed] [Google Scholar]
- 12.Cheng D., Valente S., Castellano S., Sbardella G., Di Santo R., Costi R. Novel 3,5-bis(bromohydroxybenzylidene)piperidin-4-ones as coactivator-associated arginine methyltransferase 1 inhibitors: enzyme selectivity and cellular activity. J Med Chem. 2011;54:4928–4932. doi: 10.1021/jm200453n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kálai T., Kuppusamy M.L., Balog M., Selvendiran K., Rivera B.K., Kuppusamy P. Synthesis of N-substituted 3,5-bis(arylidene)-4-piperidones with high antitumor and antioxidant activity. J Med Chem. 2011;54:5414–5421. doi: 10.1021/jm200353f. [DOI] [PubMed] [Google Scholar]
- 14.Basiri A., Xiao M., McCarthy A., Dutta D., Byrareddy S.N., Conda-Sheridan M. Design and synthesis of new piperidone grafted acetylcholinesterase inhibitors. Bioorg Med Chem Lett. 2017;27:228–231. doi: 10.1016/j.bmcl.2016.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kia Y., Osman H., Kumar R.S., Murugaiyah V., Basiri A., Perumal S. A facile chemo-, regio- and stereoselective synthesis and cholinesterase inhibitory activity of spirooxindole-pyrrolizine-piperidine hybrids. Bioorg Med Chem Lett. 2013;23:2979–2983. doi: 10.1016/j.bmcl.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Dimmock J.R., Padmanilayam M.P., Puthucode R.N., Nazarali A.J., Motaganahalli N.L., Zello G.A. A conformational and structure-activity relationship study of cytotoxic 3,5-bis(arylidene)-4-piperidones and related N-acryloyl analogues. J Med Chem. 2001;44:586–593. doi: 10.1021/jm0002580. [DOI] [PubMed] [Google Scholar]
- 17.Erbel P., Schiering N., D׳Arcy A., Renatus M., Kroemer M., Lim S.P. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat Struct Mol Biol. 2006;13:372–373. doi: 10.1038/nsmb1073. [DOI] [PubMed] [Google Scholar]
- 18.Valle R.P., Falgout B. Mutagenesis of the NS3 protease of dengue virus type 2. J Virol. 1998;72:624–632. doi: 10.1128/jvi.72.1.624-632.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kee L.Y., Kiat T.S., Wahab H.A., Yusof R., Rahman N.A. Nonsubstrate based inhibitors of dengue virus serine protease: a molecular docking approach to study binding interactions between protease and inhibitors. Asia Pac J Mol Biol Biotechnol. 2007;15:53–59. [Google Scholar]
- 20.Tomlinson S.M., Watowich S.J. Anthracene-based inhibitors of dengue virus NS2B-NS3 protease. Antivir Res. 2011;89:127–135. doi: 10.1016/j.antiviral.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weng Z., Shao X., Graf D., Wang C., Klein C.D., Wang J. Identification of fused bicyclic derivatives of pyrrolidine and imidazolidinone as dengue virus-2 NS2B-NS3 protease inhibitors. Eur J Med Chem. 2017;125:751–759. doi: 10.1016/j.ejmech.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 22.Yusof R., Clum S., Wetzel M., Murthy H.M., Padmanabhan R. Purified NS2B/NS3 serine protease of dengue virus type 2 exhibits cofactor NS2B dependence for cleavage of substrates with dibasic amino acids in vitro. J Biol Chem. 2000;275:9963–9969. doi: 10.1074/jbc.275.14.9963. [DOI] [PubMed] [Google Scholar]