Abstract
Objective
According to the Developmental Origin of Health and Disease (DOHaD) concept, maternal obesity and accelerated growth in neonates predispose offspring to white adipose tissue (WAT) accumulation. In rodents, adipogenesis mainly develops during lactation. The mechanisms underlying the phenomenon known as developmental programming remain elusive. We previously reported that adult rat offspring from high-fat diet-fed dams (called HF) exhibited hypertrophic adipocyte, hyperleptinemia and increased leptin mRNA levels in a depot-specific manner. We hypothesized that leptin upregulation occurs via epigenetic malprogramming, which takes place early during development of WAT.
Methods
As a first step, we identified in silico two potential enhancers located upstream and downstream of the leptin transcription start site that exhibit strong dynamic epigenomic remodeling during adipocyte differentiation. We then focused on epigenetic modifications (methylation, hydroxymethylation, and histone modifications) of the promoter and the two potential enhancers regulating leptin gene expression in perirenal (pWAT) and inguinal (iWAT) fat pads of HF offspring during lactation (postnatal days 12 (PND12) and 21 (PND21)) and in adulthood.
Results
PND12 is an active period for epigenomic remodeling in both deposits especially in the upstream enhancer, consistent with leptin gene induction during adipogenesis. Unlike iWAT, some of these epigenetic marks were still observable in pWAT of weaned HF offspring. Retained marks were only visible in pWAT of 9-month-old HF rats that showed a persistent “expandable” phenotype.
Conclusions
Consistent with the DOHaD hypothesis, persistent epigenetic remodeling occurs at regulatory regions especially within intergenic sequences, linked to higher leptin gene expression in adult HF offspring in a depot-specific manner.
Keywords: Perinatal programming, Adipose tissue, Epigenetic mechanisms, Developmental origin of health and disease, Gene expression, Fat expansion
Highlights
-
•
The white adipose tissue is an important target of developmental programming.
-
•
Higher leptin gene expression occurs in offspring from obese dams in a depot-specific manner.
-
•
Leptin upregulation occurs via epigenetic malprogramming during development of adipose tissue.
-
•
Persistent genomic epigenetic remodeling occurs in adipose tissue of offspring from obese dams.
-
•
Intergenic regions were more affected than the leptin promoter region in offspring of obese dams.
1. Introduction
According to the Developmental Origin of Health and Disease (DOHaD) concept [1], maternal obesity and accelerated growth in neonates predispose offspring to obesity and metabolic pathologies [2]. Thus, white adipose tissue (WAT) is an important target of developmental programming [3], [4]. In mammals, WAT has a dual role in storing lipid and in controlling energy homeostasis through secretion of leptin [5]. In rodents, adipose tissue growth and adipogenesis mainly develop during lactation [4], [6], [7]. Adipogenesis occurs throughout the life time suggesting that WAT remains expandable throughout life. During critical developmental time windows of fetal and neonatal life, adipocyte stem cells are plastic and highly sensitive to maternal factors [8]. Genetics and nutritional and hormonal factors influence the relative contributions of hyperplasia (cell number increase) and hypertrophy (cell size increase) to the growth of WAT [9], [10], [11]. However, little is known about the cellular and molecular mechanisms underlying the phenomenon known as developmental programming [4], [12]. Among them, epigenetic modifications are likely to play an important role [4], [12], [13]. Epigenetics can be defined as somatically heritable states of gene expression resulting from changes in chromatin structure without alterations in the DNA sequence, including DNA and histone modifications and chromatin remodeling [13]. DNA methylation occurs at cytosines, mainly in the CpG dinucleotide, to form 5-methylcytosine (5 mC) and serves to establish long-term gene silencing [14]. 5-hydroxymethylcytosine (5 hmC) is another important cytosine modification catalyzed by the enzymes of the TET family [15]. Although its role is not fully elucidated, 5 hmC is enriched in active transcriptional regulatory regions. In particular, 5 hmC is enzymatically produced in WAT, where it plays a regulatory role in adipogenesis [16], [17], [18], [19]. Histone modifications influence chromatin structure and, hence, the ability of critical DNA-associated regulatory proteins that control transcription to gain access to the DNA. In particular, elevated H3K4me1/H3K27ac and lower H3K9me3 marks are linked to promoter and enhancer activation during adipocyte differentiation [16], [20]. Epigenetic marks may serve as a memory of exposure, in early life, to inappropriate environments [21]. These persistent marks may ultimately induce long-term changes in gene expression [4], [12]. Few studies have reported that maternal nutritional manipulations modulate gene expression profile in WAT's offspring via epigenetic mechanisms [22], [23], [24], [25], [26]. However, the concept of continued editing of early-life epigenetic markings or memories during adult life is based on evidence from limited experimental studies, especially in WAT programming [24], [26].
We previously showed that adult rat offspring from high-fat diet-fed dams (called HF) exhibited hypertrophic adipocyte, hyperleptinemia, and increased leptin mRNA levels in a depot-specific manner [27]. Depot-specific differences are closely associated with differential cardiometabolic risks [10], [28]. We hypothesized that leptin upregulation occurs via epigenetic malprogramming, which takes place early during development of WAT. Epigenetic modifications of the leptin promoter play a role in leptin expression during adipocyte differentiation and in obesity-related leptin upregulation. In addition to the promoter sequence, intergenic regions that have yet to be determined are likely to regulate leptin gene expression [25], [29], [30], [31], [32], [33], [34]. Here, we identified two potential enhancers (i.e., upstream and downstream) using bioinformatics analysis. We showed, for the first time, that maternal obesity differently affects epigenetic remodeling of the promoter and the two enhancers linked to higher leptin gene expression during visceral perirenal (pWAT) and subcutaneous inguinal (iWAT) deposits development in HF neonates. We also reported that retained active marks that are correlated with persistent increased leptin mRNA levels occur in a depot-specific manner in adult obesity-prone HF offspring.
2. Materials and methods
2.1. Animals
Animal use accreditation by the French Ministry of Agriculture (No. 04860) has been granted to our laboratory for experimentation with rats. Experiments were conducted in accordance with the principles of laboratory animal care (European Communities Council Directive of 1986, 86/609/EEC). One-month-old virgin female Wistar rats were purchased from Charles River Laboratories (L'Arbresle, France) and housed in individual cages in a humidity-controlled room with a 12:12-h light–dark cycle. Food and water were available ad libitum. After two weeks of acclimatization on a control (C) diet (3.85 kcal/g with 10% of total calories as fat consisting of soybean oil (5.6%) and lard (4.4%), 70% as carbohydrate and 20% as protein; D12450J, Research Diets, New Brunswick, NJ, USA), female rats were fed either a high-fat (HF) diet (5.24 kcal/g with 60% of total calories as fat consisting of soybean oil (5.6%) and lard (54.4%), 20% as carbohydrate and 20% as protein; D12492B, Research Diets, New Brunswick, NJ, USA) or a C diet for 16 weeks (n = 30 per group). After mating with a male rat fed a C diet, 22-week-old pregnant females were transferred into individual cages with free access to water and to their respective diets (C or HF diet) throughout gestation and lactation. At parturition, pups were weighed and sexed. Litter size was adjusted to 8 pups per dam (four males and four females). During lactation, body weight of pups was assessed on postnatal days (PND) 12 and 21. Only male offspring have been used for further analysis. After weaning, male offspring from C or HF dams were housed individually with free access to water and C diet. Body weight of animals was measured weekly until 9 months of age. Male offspring were sacrificed at 3 different stages: PND12, PND21, and 9 months of age (n = 10 from C or HF dams). To obviate any litter effects, animals used for each experiment were randomly chosen in different litters, and only a limited number of animals (n = 1 to 2) was used from each litter. The protocol has been previously described (Figure S1) [27].
2.2. Plasma and tissue collections
12-day-old and 21-day-old neonates were separated from their dams for 6 h and 9-month-old rats were fasted 16 h before they were sacrificed by decapitation (between 9 and 10 a.m.). Trunk blood samples were collected into pre-chilled tubes containing EDTA (20 μl of a 5% solution) and centrifuged at 4000 g for 10 min at 4 °C. Plasma was stored at −20 °C. Fat pads were weighed, frozen in liquid nitrogen, and stored at −80 °C. For histology experiments, animals were fixed by intracardiac perfusion using buffered 4% paraformaldehyde solution.
2.3. Quantification of plasma leptin levels
Serum leptin concentrations were determined using a murine ELISA kit (Bertin Pharma, Cayman medical, USA) [27]. The assay sensitivity was 0.04 ng/ml and the intra-and inter-assay coefficients of variation were 5.4% and 7.3%.
2.4. Morphometric analysis of adipose tissue
Fat pad mass as well as cell-size distributions were measured in pWAT and iWAT as previously described [27]. For histology experiments, tissues were post-fixed for 24 h in buffered 4% paraformaldehyde solution and embedded in paraffin. Fixed tissues were then cut into serial 10 μm sections, mounted on gelatine-coated slides, and stained with hematoxylin of Groat and phloxin (2%), according to standard laboratory protocols. Sections were examined using light microscopy (Leica DM IRE2), and photomicrographs were captured at 20× magnification. The surface of adipocytes was evaluated in ten randomly selected fields of vision of a minimum of 500 adipocytes using Image J software (NIH, USA). Total cell number is a direct measure reflecting hyperplasia. The number of cells was estimated as previously described [11], [27].
2.5. Pyrosequencing analysis
Genomic DNA was extracted from frozen WAT using a DNA extraction kit (DNeasy blood and tissue kit, Qiagen, Courtaboeuf, France) and modified with sodium bisulfite using the EpiTect Fast Bisulfite Conversion kit (Qiagen, Courtaboeuf, France) according to the manufacturer's protocol. The percentage of cytosine methylation was determined by pyrosequencing bisulfite converted DNA using PyroMark Q24 (Qiagen, Courtaboeuf, France). Pyrosequencing primers were designed to amplify leptin CpG sites (Table S1). PCR was performed with 20 μl final reaction volumes with 1.5 μl of bisulfite DNA (10 ng), 10 μl of QuantiTect EurobioGreen PCR Mix Hi-ROX (Eurobio, Les Ulis, France), 7.7 μl of H2O, and 0.4 μl of each primer set (10 mM). Sequencing reactions used 10 μl of PCR product and 20 μl of 0.375 μM sequencing primer (Qiagen, Courtaboeuf, France). Pyrosequencing assays were validated with a DNA methylation scale (0%, 5%, 25%, 50%, 75% and 100%) (EpigenDX, Hopkinton, USA). Each assay included a bisulfite conversion check to verify full conversion of the DNA.
2.6. (Hydroxy)methylated DNA immunoprecipitation
(Hydroxy)methylated DNA immunoprecipitation assays were performed as previously described [16]. Genomic DNA was sonicated using a Bioruptor (Diagenode, Liège, Belgique) to produce fragments ranging in size from 200 to 500 bp. 10 μg of fragmented, heat-denatured DNA (10 min at 95 °C) were incubated overnight at 4 °C with either a mouse monoclonal antibody directed against 5-methylcytosine methylcytosine (MeDIP) (Mab-081-100) or a rabbit polyclonal antibody directed against 5-hydroxymethylcytosine (HMeDIP) (CS-HMC-100), respectively (Diagenode, Liège, Belgique). Reactions were performed in a final volume of 500 μl of IP buffer (10 Mm sodium phosphate, pH 7.0, 140 mM NaCl, 0.05% Triton X-100). Immunocomplexes were precipitated by incubation with 40 μl of BSA-coated protein A/G magnetic beads (Dynabeads, Thermo Fischer Scientific) for 2 h at 4 °C. Beads were subsequently washed three times with 1 ml of IP buffer and finally incubated with proteinase K (280 μg/ml) overnight at 55 °C in a buffer containing 50 mM Tris–HCl (pH8.0), 10 mM EDTA, and 0.5% SDS. (Hydroxy)methylated DNA was recovered by phenol-chloroform extraction followed by ethanol precipitation.
2.7. Chromatin immunoprecipitation
ChIP assays were performed as previously described [16] with antibodies against H3K4me1, H3K27ac, and H3K9me3 (Abcam, Cambridge, UK, ab8895, ab4729 and ab1773) and IgG (Cell Signaling Technology, Danvers, USA, #2729) as a negative control.
2.8. RNA isolation and real-time PCR
Leptin mRNA levels were determined by RT-qPCR using SyberGreen-based chemistry as previously described [27]. Cyclophilin A (PpiA) and Ribosomal Protein Lateral Stalk Subunit P1 (RPLP1) were used as reference genes for RT-qPCR. DNA (hydroxy)methylation, H3K4me1, H3K27ac, and H3K9me3 levels were analyzed using immunoprecipitated and input DNA as control. The primers are listed (Table S1).
2.9. Bioinformatics identification of two potential enhancers regulating leptin gene expression
Public functional genomic data [16], [20] were downloaded from Gene Expression Omnibus or the UCSC Genome Browser. The identification procedure has been detailed in Figures S3,S4. CisMapper model was used to predict the involvement of identified genomic areas in the transcriptional regulation of the leptin gene [35]. Genomic sequences were aligned between mouse and rat by the EMBOSS needle [36].
2.10. Statistical analysis
All data are expressed as means ± standard error of the mean (SEM). Statistical analysis was carried out using GraphPad Prism6. A direct comparison between a pair of groups was made using an unpaired Student's t test or a two-way analysis of variance (ANOVA) for repeated measures followed by a Bonferroni post hoc test, where appropriate. Pearson test was used to assess correlations. P values <0.05 was considered statistically significant.
3. Results
3.1. Offspring from obese dams are predisposed to adiposity in a depot-specific manner
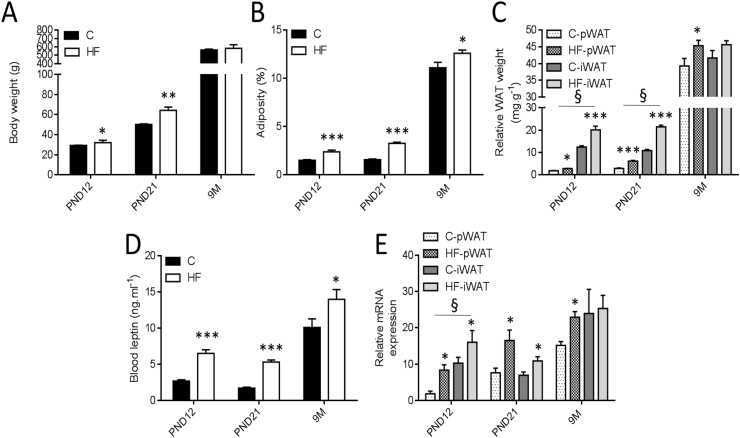
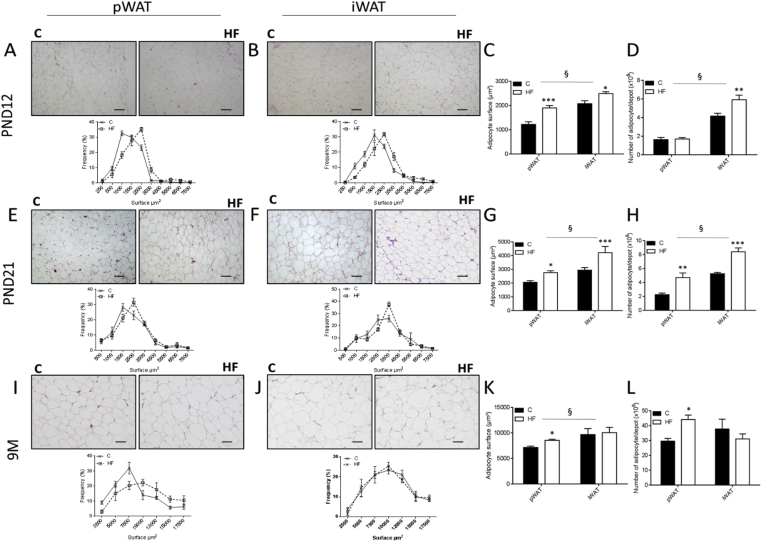
At PND12 and PND21, HF rats were heavier than C animals (Figure 1A) with elevated adiposity (Figure 1B). Both pWAT and iWAT deposits of HF offspring were larger when normalized to body weight (Figure 1C). Both fat pads exhibited an increase in average adipocyte area (Figure 2A–C,E–G) as reflected by the shift to the left of the curve and total cell number (Figure 2D,H). At 9 months of age, both groups had comparable body weight (Figure 1A) whereas HF offspring showed higher adiposity (Figure 1B). Unlike iWAT (Figure 1C,J–L), increased adiposity was associated with higher weight of pWAT (Figure 1C) and increases in both adipocyte surface (Figure 2I,K) and cell number (Figure 2L).
Figure 1.
Adiposity and leptin mRNA levels are affected in hyperleptinemic HF offspring in a depot-specific manner. Different parameters of male offspring from dams fed a C diet (called C) or high-fat diet-fed dams (called HF) were assessed on postnatal days 12 (PND12) and 21 (PND21) and 9 months of age (9M). Body weight of C and HF male offspring was determined before sacrifice (A). Adiposity (%) is defined as the weight of whole visceral and subcutaneous fat pads relative to body weight (B). Fat index for perirenal WAT (pWAT) and inguinal WAT (iWAT) was calculated (C). Serum leptin concentrations of both groups were determined by ELISA (D). Leptin mRNA levels in pWAT and iWAT were determined by RT-qPCR (E). Relative gene expression in the C group at PND12 was set to 1. Ppia and Rplp1 were used as standard genes. All data are presented as means ± SEM. Data were analyzed using two-way ANOVA followed by Bonferroni post hoc test. * Effect of maternal obesity (*P < 0.05, **P < 0.01 and ***P < 0.001); § Effect of fat pad (§P < 0.05). n = 10 per group and per age.
Figure 2.
Adipocytes of HF offspring display hypertrophy and hyperplasia in a depot-specific manner. Representative photomicrographs of paraffin-embedded sections (scale bars = 100 μm) and percentage of adipocytes in a given size range (area in μm2) in pWAT (A, E, I) and iWAT (B, F, J) at PND12 (A–B), PND21 (E–F), and 9M (I–J). Average of adipocyte surface (C, G, K) was determined in hematoxylin-eosin-stained sections and total cell number (D, H, L) was calculated by dividing fat mass by the average diameter of adipocyte at PND12 (C, D), PND21 (G, H), 9M (K, L). All data are presented as means ± SEM. Data were analyzed using two-way ANOVA followed by Bonferroni post hoc test. * Effect of maternal obesity (*P < 0.05, **P < 0.01 and ***P < 0.001); § Effect of fat pad (§P < 0.05). n = 5 per group and per age.
3.2. Offspring from obese dams display persistent hyperleptinemia and increased adipose tissue leptin mRNA levels in a depot-specific manner
HF offspring had higher plasma leptin concentration at PND12, PND21, and 9 months compared with C rats (Figure 1D). In pWAT, HF offspring showed increased leptin mRNA levels at the three stages whereas in iWAT, higher leptin mRNA levels were only observed at PND12 and PND21 (Figure 1E). Leptin mRNA levels and plasma concentration are correlated with average adipocyte area and total cell number in both deposits (Figure S2).
3.3. Offspring from obese dams show persistent epigenetic mark modifications in the leptin promoter in a depot-specific manner
We first assessed epigenetic mark modifications in the leptin promoter of both deposits. To do this, we analyzed single CpG sites by bisulfite pyrosequencing. This technique does not distinguish between 5 mC and 5 hmC [37]. To differentiate between the distribution and levels of 5 hmC and 5 mC marks, we then measured the enrichment of 5 hmC and 5 mC by MeDIP-qPCR and HMeDIP-qPCR.
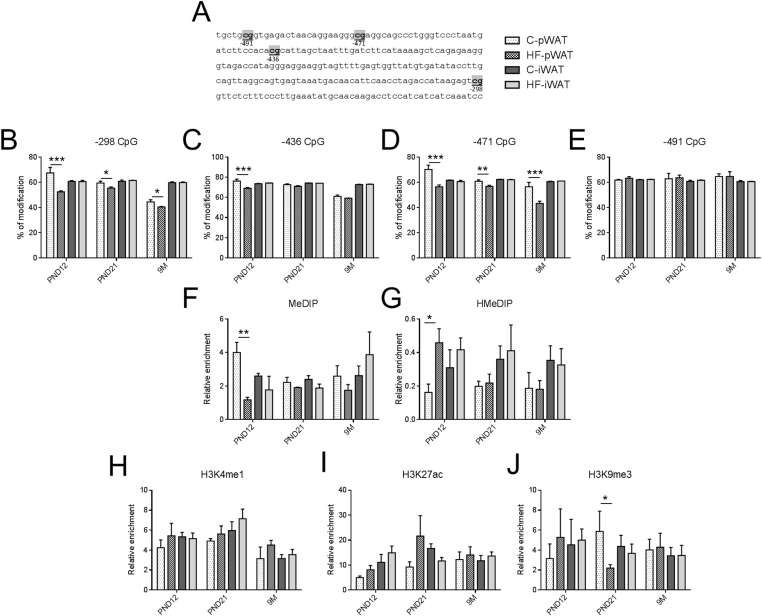
At PND12, in pWAT, three CpG (−298, −436, −471) showed lower percentage of modifications in the leptin promoter of HF offspring (Figure 3B–E). This was associated with a decrease in 5 mC (Figure 3F) and an increase in 5 hmC (Figure 3G). At PND21 and 9 months of age, HF offspring exhibited lower modifications in two CpG sites (−298, −471) (Figure 3B–E) and a reduction of H3K9me3 only at PND21 (Figure 3J) but no variation was detectable in MeDIP and HMeDIP (Figure 3F–G). No change was observed in the leptin promoter of iWAT (Figure 3).
Figure 3.
The leptin promoter exhibits persistent depletion in 5 mC in pWAT of HF offspring. Epigenetic modifications of four CpG indicated in gray (A) and located in the leptin promoter at −298 bp (B), −436 bp (C), −471 bp (D) and −491 bp (E) of the transcription start site were assessed in male C and HF offspring in two fat pads (pWAT and iWAT) at PND12, PND21 and 9 months. To discriminate the nature of DNA modifications, DNA extracted from both depots was immunoprecipitated with antibodies against DNA methylation (MeDIP) (F), DNA hydroxymethylation (HMeDIP) (G) and subject to qPCR using primers of the targeted regions. Histone modifications were measured after chromatin immunoprecipitation with antibodies against H3K4me1 (H), H3K27ac (active mark) (I) or H3K9me3 (inactive mark) (J) and qPCR using primers of targeted regions. Immunoprecipitation with normal rabbit IgG was used as a negative control. All data are presented as means ± SEM. Data were analyzed using two-way ANOVA followed by Bonferroni post hoc test. * Effect of maternal obesity (*P < 0.05, **P < 0.01 and ***P < 0.001). n = 4–6 per group.
3.4. Identification of two potential transcriptional enhancers regulating leptin gene expression
In addition to the promoter, other regions called enhancers whose sequences have yet to be established, located mainly located in intergenic regions, are likely to regulate gene expression [20]. Several studies support this notion for leptin gene expression [25], [29], [30], [31], [32], [33], [34]. In line with these findings, the epigenome of intergenic regions was more affected than promoter regions in offspring of perinatally malnourished dams [38].
Active transcriptional enhancers in adipocytes are characterized by a specific epigenetic signature which includes H3K27ac and 5 hmC [16]. Using these signatures, we have generated comparative chromatin state maps and discriminated two potential enhancers of the leptin gene activated during adipocyte differentiation (i.e., enhancers gaining H3K4me1/H3K27ac/5 hmc during differentiation) identified by ChIP-seq and HMeDIP-seq in 3T3-L1 cells [16], [20]. The two potential upstream and downstream enhancers are located 42 kb upstream and 4.4 kb downstream of the leptin transcription start site, respectively (Figures S3,S4). The involvement of the two enhancers in the transcriptional regulation of the leptin gene was validated by the CisMapper model (Table S2) [35].
3.5. Offspring from obese dams show persistent epigenetic mark modifications in the upstream enhancer in a depot-specific manner
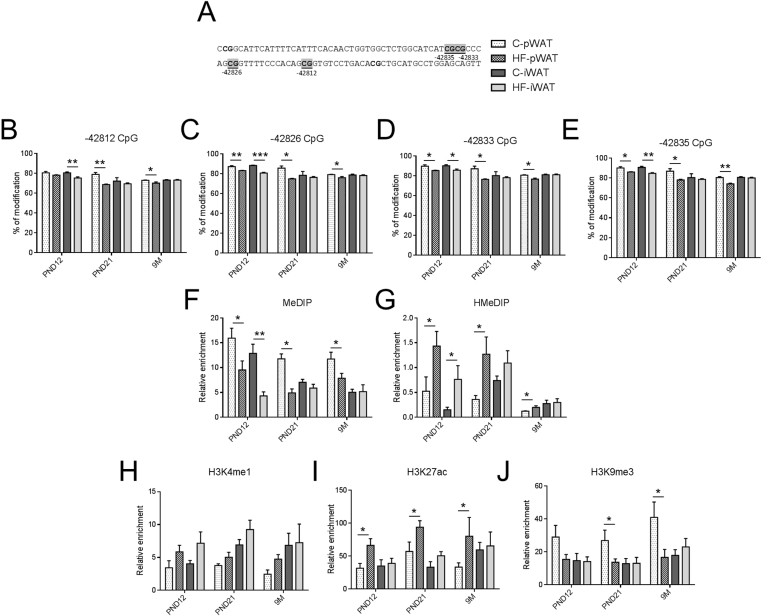
At PND12, in both fat pads, the percentage of modifications of the four CpG (−42812, −42826, −42833, −42835 bp) was lower in the upstream enhancer of HF offspring (Figure 4B–E). This was consistent with loss of 5 mC (Figure 4F) and enrichment of active marks such as 5 hmC (Figure 4G) and H3K27ac (Figure 4I). A tendency towards enrichment of active H3K4me1 (Figure 4H) and loss of inactive mark H3K9me3 (Figure 4J) were observed in the upstream enhancer of HF offspring.
Figure 4.
The upstream enhancer displays persistent depletion in 5 mC and H3K9me3 and enrichment in 5hmc and H3K4me1/H3K27ac in pWAT of HF offspring. Epigenetic modifications of four CpG indicated in gray (A) and located in the upstream enhancer at −42812 bp (B), −42826 bp (C), −42833 bp (D) and −42835 bp (E) of the transcription start site were assessed in male C and HF offspring in two fat pads (pWAT and iWAT) at PND12, PND21, and 9 months. To discriminate the nature of DNA modifications, DNA extracted from both depots was immunoprecipitated with antibodies against DNA methylation (MeDIP) (F), DNA hydroxymethylation (HMeDIP) (G) and subject to qPCR using primers of targeted regions. Histone modifications were measured after chromatin immunoprecipitation with antibodies against H3K4me1 (H), H3K27ac (active mark) (I) or H3K9me3 (inactive mark) (J) and qPCR using primers of the targeted regions. Immunoprecipitation with normal rabbit IgG was used as a negative control. All data are presented as means ± SEM. Data were analyzed using two-way ANOVA followed by Bonferroni post hoc test. * Effect of maternal obesity (*P < 0.05, **P < 0.01 and ***P < 0.001). n = 4–6 per group.
At PND21 and 9 months of age, the four CpG dinucleotides within the upstream enhancer of pWAT were similarly affected by maternal obesity. CpGs displayed a decrease in percentage of modifications (Figure 4B–E). This was correlated with lower levels of 5 mC (Figure 4F) and H3K9me3 (Figure 4J) and higher levels of 5 hmC (Figure 4G) and H3K27ac (Figure 4I). A tendency towards enrichment of active H3K4me1 (Figure 4H) was observed in the upstream enhancer of HF offspring (Figure 4H). No change was observed in the upstream enhancer of iWAT (Figure 4B–E). No persistent changes were observed in the downstream enhancer of both fat pads (Figure S5). Consistently, all these epigenetic changes were correlated with leptin mRNA levels at PND12, PND21, and 9M in both deposits.
4. Discussion
In the current study, we showed that persistent increased leptin mRNA levels in HF offspring's WAT may occur via epigenetic mechanisms which take place during the early postnatal period. In WAT, increased leptin mRNA levels are correlated with hypertrophy [39] and modified methylation of the leptin promoter. However, the underlying molecular mechanisms and the sequences regulating leptin gene expression remain unclear. In addition to the promoter sequence, additional regions that are not yet determined are expected to regulate leptin gene expression [25], [29], [30], [31], [32], [33], [34]. As a first step, we identified in silico two potential enhancers (i.e. upstream and downstream) involved in the transcriptional regulation of the leptin gene that exhibit strong dynamic epigenomic remodeling (i.e., increased H3K4me1/H3K27ac, DNA hydroxymethylation and decreased DNA methylation) during adipocyte differentiation [16], [20].
We previously showed that HF offspring had a normal birthweight and, then, exhibited a rapid weight gain during lactation, a key period of adipose tissue development [4], [27]. The accelerated postnatal growth in offspring is frequently associated with persisting adiposity throughout life [40]. HF offspring showed elevated plasma leptin concentration throughout life. Although plasma leptin is considered as a function of adipose tissue mass, plasma leptin levels and leptin gene expression are not only a result of greater adiposity [41]. In addition, further experiments are needed to determine the extent to which maternal obesity may affect central and peripheral leptin sensitivity in HF offspring. Here, we found that among iWAT and pWAT, only the latter showed a persistent “expandable” (i.e. hypertrophy, hyperplasia) phenotype with elevated leptin mRNA levels in adulthood. Contrary to subcutaneous WAT, increased visceral WAT mass is strongly associated with cardiometabolic risk and mortality [10], [28]. The varying outcomes may reflect differences in postnatal programming. One possible reason is the heterogeneity of the adipose lineage between fat depots in terms of spatiotemporal adipogenic potential, gene expression profile, growth rate, and biological properties [9], [28]. Numerous studies demonstrated that the ability of adipose precursors to differentiate during adipogenesis is dependent on the anatomic location and the local microenvironment, leading to the concept of depot-specific adipogenesis [13], [42]. Thus, subcutaneous depots have increased rates of adipose turnover and new adipocyte formation compared to other visceral fat mass [43], [44]. An emerging hypothesis is that anatomically distinct WAT depots (i.e., visceral and subcutaneous fat pads) likely represent distinct “mini-organs” [10], [28]. During lactation, adipocyte stem cells are plastic and highly sensitive to maternal factors [8]. Thus, maternal obesity and modified milk composition may influence offspring's energy and hormonal status [45], [46], thereby modifying the expression of the leptin gene via epigenetic changes in a depot-specific manner. In rodents, few studies have examined depot- and sex-specific consequences of maternal obesity in offspring's WAT and there is little agreement among them [4]. We previously demonstrated that maternal obesity programs visceral depots only in HF male offspring [27]. Given the sex-specific nature of fat depots and the sexual dimorphism in developmental programming mechanisms [47], further experiments are needed to determine whether iWAT is a target of programming in HF female offspring.
We then decided to assess the kinetic of epigenetic changes within three regulatory regions linked to increased leptin gene expression during development of offspring's WAT and in adulthood. PND12 is an active period for epigenomic remodeling (i.e., lower DNA methylation and inactive histone modification H3K9me3 and higher DNA hydroxymethylation and active histone modification H3K4me1/H3K27ac) in both fat pads, especially in the upstream enhancer, consistent with leptin gene induction during adipogenesis. Unlike iWAT, some of these epigenetic marks were still observable in pWAT of weaned HF offspring. The global reduction of marks between lactation and weaning may reflect modified offspring's food intake from milk to solid diet. Retained active marks (i.e., the promoter and the upstream enhancer) were only observed in pWAT of adult HF offspring and were correlated with persistent increased leptin gene expression, hypertrophy and higher weight. Although additional studies are required to assess the functionality of the upstream enhancer, we speculate that this region is more active in pWAT of HF offspring resulting in long-term effects on leptin gene expression. Consistent with these findings, changes in cytosine modifications during developmental programming take place not only at promoters but also at additional regulatory regions within intergenic sequences [38]. Both enhancers display at least 80% sequence homology between mouse and rat. These two regions are partially conserved between rat and human with 68.8% sequence homology for the upstream enhancer and 56.4% sequence homology for the downstream enhancer. Recent studies comparing key selected mammals showed that enhancers are far less evolutionarily stable than are promoters across species and that sequence conservation is a rather poor predictor of functionality [48]. However, the fact that, unlike the downstream enhancer, the upstream enhancer displays similar epigenomic remodeling (enrichment in H3K27ac and H3K4me1/2/3) during human adipose stem cells differentiation suggests that the upstream enhancer is also functional and active in human adipocytes [20]. We also showed that the global 5 mC level of leptin regulatory sequences in 9-month-old HF offspring was lower than 12-day-old HF neonates, suggesting that a demethylation process occurs during development of WAT.
5 mC is the only known modification that targets the DNA itself and is usually associated with gene silencing [14]. In most cases, the extent of 5 mC of a promoter is inversely correlated with the activity of the respective gene. Generally, 5 mC physically blocks the binding of a transcription factor to its target, leading to gene silencing. The role of 5 hmC in the regulation of transcriptional activity is still not well understood. 5 hmC can be considered as a demethylation intermediate or as a stable epigenetic mark that is enriched in active genomic region [49]. The enrichment of H3K4me1/H3K27ac and the reduction of H3K9me3 are characteristic of enhancer and promoter activation [20]. These processes take place on a large scale during adipocyte differentiation [20], [50]. A potential limitation of our approach lies in the fact that we performed DNA analysis using whole WAT, which may contain different cell type abundance depending on the stage of development. However, gene expression profiling using several endothelial, hematopoietic, and immune cell markers suggests that the composition of HF offspring's WAT is not modified regardless of the stage of development.
Besides epigenetic programming mechanisms, alterations of hormone and metabolic environment during fetal and neonatal development can contribute to persistent deregulation of energy homeostasis in progeny. Although underlying mechanisms remain unclear, increased hormone levels in the fetal compartment or neonate may result in long-term fat expansion with permanent changes in mRNA and plasma hormone levels in adulthood. Over the past few decades, the adipocytokine leptin has been considered as the main programming factor of the hypothalamus adipose-axis that plays a pivotal role in the body weight set point [41]. Several studies support the notion that fat expansion with persistent changes in leptin expression and sensitivity in HF offspring may be partly due to increased neonatal leptin levels, whose origin remains open to debate (i.e. breast milk, developing WAT). First, perinatal leptin administration and amplified postnatal leptin surge have long-term detrimental effects resulting in leptin resistance, increased leptin gene expression, and higher adiposity in adulthood [46], [51]. Second, leptin displays marked neurotrophic effects on hypothalamic neuronal development [52]. Third, leptin directly activates adipogenesis by promoting differentiation of preadipocytes [53], whereas it shows antilipogenic effects on mature adipocytes [54].
5. Conclusion
Overall, our data show that editing of early-life epigenetic markings occur during development of WAT and might persist throughout life in a depot-specific manner. Consistent with the DOHaD hypothesis, it may account for fat pad differences in the long-term effects on leptin gene expression and “expandable phenotype” observed in adult obesity-prone progeny.
Author contributions
S.L., F.O., D.E., A.G., J.E., C.B. designed the study, researched data, and contributed to discussions and manuscript. L. B., L.M., A.D-C., C.P. researched data. C.L., C.G., J.L., D.V., C.J. contributed to discussion.
Funding
This study was supported by grants from the French “Heart and Arteries” Foundation (FCA13T1) and grants of the French Ministry of Education. Work at INSERM U1011 was supported by grants from the Fondation pour la Recherche Médicale (Equipe labellisée, DEQ20150331724) and “European Genomic Institute for Diabetes” (E.G.I.D., ANR-10-LABX-46).
Acknowledgements
The authors thank Barbara Deracinois for technical assistance during sacrifice.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.05.010.
Conflicts of interest
The author declares that he has no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Barker D.J.P. Developmental origins of adult health and disease. Journal of Epidemiology and Community Health. 2004;58(2):114–115. doi: 10.1136/jech.58.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leddy M.A., Power M.L., Schulkin J. The impact of maternal obesity on maternal and fetal health. Reviews in Obstetrics & Gynecology. 2008;1(4):170–178. [PMC free article] [PubMed] [Google Scholar]
- 3.Lukaszewski M.-A., Eberlé D., Vieau D., Breton C. Nutritional manipulations in the perinatal period program adipose tissue in offspring. American Journal of Physiology, Endocrinology and Metabolism. 2013;305(10):E1195–E1207. doi: 10.1152/ajpendo.00231.2013. [DOI] [PubMed] [Google Scholar]
- 4.Lecoutre S., Breton C. Maternal nutritional manipulations program adipose tissue dysfunction in offspring. Frontiers in Physiology. 2015;6:158. doi: 10.3389/fphys.2015.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng Y., Scherer P.E. Adipokines as novel biomarkers and regulators of the metabolic syndrome. Annals of the New York Academy of Sciences. 2010;1212:1–19. doi: 10.1111/j.1749-6632.2010.05875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berry D.C., Stenesen D., Zeve D., Graff J.M. The developmental origins of adipose tissue. Development (Cambridge, England) 2013;140(19):3939–3949. doi: 10.1242/dev.080549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenwood M.R., Hirsch J. Postnatal development of adipocyte cellularity in the normal rat. Journal of Lipid Research. 1974;15(5):474–483. [PubMed] [Google Scholar]
- 8.Tang Q.Q., Lane M.D. Adipogenesis: from stem cell to adipocyte. Annual Review of Biochemistry. 2012;81:715–736. doi: 10.1146/annurev-biochem-052110-115718. [DOI] [PubMed] [Google Scholar]
- 9.Björntorp P., Sjöström L. Number and size of adipose tissue fat cells in relation to metabolism in human obesity. Metabolism: Clinical and Experimental. 1971;20(7):703–713. doi: 10.1016/0026-0495(71)90084-9. [DOI] [PubMed] [Google Scholar]
- 10.Pellegrinelli V., Carobbio S., Vidal-Puig A. Adipose tissue plasticity: how fat depots respond differently to pathophysiological cues. Diabetologia. 2016;59(6):1075–1088. doi: 10.1007/s00125-016-3933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lemonnier D. Effect of age, sex, and sites on the cellularity of the adipose tissue in mice and rats rendered obese by a high-fat diet. The Journal of Clinical Investigation. 1972;51(11):2907–2915. doi: 10.1172/JCI107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson M., Godfrey K.M., Lillycrop K.A., Burdge G.C., Gluckman P.D. Developmental plasticity and developmental origins of non-communicable disease: theoretical considerations and epigenetic mechanisms. Progress in Biophysics and Molecular Biology. 2011;106(1):272–280. doi: 10.1016/j.pbiomolbio.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Öst A., Pospisilik J.A. Epigenetic modulation of metabolic decisions. Current Opinion in Cell Biology. 2015;33:88–94. doi: 10.1016/j.ceb.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Portela A., Esteller M. Epigenetic modifications and human disease. Nature Biotechnology. 2010;28(10):1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 15.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oger F., Dubois-Chevalier J., Gheeraert C., Avner S., Durand E., Froguel P. Peroxisome proliferator-activated receptor α regulates genes involved in insulin/insulin-like growth factor signaling and lipid metabolism during adipogenesis through functionally distinct enhancer classes. Journal of Biological Chemistry. 2014;289(2):708–722. doi: 10.1074/jbc.M113.526996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiki K., Shinoda A., Kano F., Sato R., Shirahige K., Murata M. PPARγ-induced PARylation promotes local DNA demethylation by production of 5-hydroxymethylcytosine. Nature Communications. 2013;4:2262. doi: 10.1038/ncomms3262. [DOI] [PubMed] [Google Scholar]
- 18.Yu P., Ji L., Lee K.J., Yu M., He C., Ambati S. Subsets of visceral adipose tissue nuclei with distinct levels of 5-hydroxymethylcytosine. PLoS One. 2016;11(5):e0154949. doi: 10.1371/journal.pone.0154949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo Y., Park J.H., Weigel C., Liesenfeld D.B., Weichenhan D., Plass C. TET-mediated hydroxymethylcytosine at the Pparγ locus is required for initiation of adipogenic differentiation. International Journal of Obesity. 2017;41(4):652–659. doi: 10.1038/ijo.2017.8. [DOI] [PubMed] [Google Scholar]
- 20.Mikkelsen T.S., Xu Z., Zhang X., Wang L., Gimble J.M., Lander E.S. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Attig L., Gabory A., Junien C. Early nutrition and epigenetic programming: chasing shadows. Current Opinion in Clinical Nutrition and Metabolic Care. 2010;13(3):284–293. doi: 10.1097/MCO.0b013e328338aa61. [DOI] [PubMed] [Google Scholar]
- 22.Borengasser S.J., Zhong Y., Kang P., Lindsey F., Ronis M.J.J., Badger T.M. Maternal obesity enhances white adipose tissue differentiation and alters genome-scale DNA methylation in male rat offspring. Endocrinology. 2013;154(11):4113–4125. doi: 10.1210/en.2012-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Q.-Y., Liang J.-F., Rogers C.J., Zhao J.-X., Zhu M.-J., Du M. Maternal obesity induces epigenetic modifications to facilitate Zfp423 expression and enhance adipogenic differentiation in fetal mice. Diabetes. 2013;62(11):3727–3735. doi: 10.2337/db13-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang X., Yang Q., Fu X., Rogers C.J., Wang B., Pan H. Maternal obesity epigenetically alters visceral fat progenitor cell properties in male offspring mice. The Journal of Physiology. 2016;594(15):4453–4466. doi: 10.1113/JP272123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jousse C., Parry L., Lambert-Langlais S., Maurin A.-C., Averous J., Bruhat A. Perinatal undernutrition affects the methylation and expression of the leptin gene in adults: implication for the understanding of metabolic syndrome. FASEB Journal. 2011;25(9):3271–3278. doi: 10.1096/fj.11-181792. [DOI] [PubMed] [Google Scholar]
- 26.Masuyama H., Hiramatsu Y. Effects of a high-fat diet exposure in utero on the metabolic syndrome-like phenomenon in mouse offspring through epigenetic changes in adipocytokine gene expression. Endocrinology. 2012;153(6):2823–2830. doi: 10.1210/en.2011-2161. [DOI] [PubMed] [Google Scholar]
- 27.Lecoutre S., Deracinois B., Laborie C., Eberlé D., Guinez C., Panchenko P.E. Depot- and sex-specific effects of maternal obesity in offspring's adipose tissue. Journal of Endocrinology. 2016;230(1):39–53. doi: 10.1530/JOE-16-0037. [DOI] [PubMed] [Google Scholar]
- 28.Hepler C., Gupta R.K. The expanding problem of adipose depot remodeling and postnatal adipocyte progenitor recruitment. Molecular and Cellular Endocrinology. 2017;445:95–108. doi: 10.1016/j.mce.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen W., Wang C., Xia L., Fan C., Dong H., Deckelbaum R.J. Epigenetic modification of the leptin promoter in diet-induced obese mice and the effects of N-3 polyunsaturated fatty acids. Scientific Reports. 2014;4:5282. doi: 10.1038/srep05282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melzner I., Scott V., Dorsch K., Fischer P., Wabitsch M., Brüderlein S. Leptin gene expression in human preadipocytes is switched on by maturation-induced demethylation of distinct CpGs in its proximal promoter. Journal of Biological Chemistry. 2002;277(47):45420–45427. doi: 10.1074/jbc.M208511200. [DOI] [PubMed] [Google Scholar]
- 31.Cordero P., Campion J., Milagro F.I., Goyenechea E., Steemburgo T., Javierre B.M. Leptin and TNF-alpha promoter methylation levels measured by MSP could predict the response to a low-calorie diet. Journal of Physiology and Biochemistry. 2011;67(3):463–470. doi: 10.1007/s13105-011-0084-4. [DOI] [PubMed] [Google Scholar]
- 32.Stöger R. In vivo methylation patterns of the leptin promoter in human and mouse. Epigenetics. 2006;1(4):155–162. doi: 10.4161/epi.1.4.3400. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y.-H., Dallner O.S., Birsoy K., Fayzikhodjaeva G., Friedman J.M. Nuclear Factor-Y is an adipogenic factor that regulates leptin gene expression. Molecular Metabolism. 2015;4(5):392–405. doi: 10.1016/j.molmet.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wrann C.D., Rosen E.D. New insights into adipocyte-specific leptin gene expression. Adipocyte. 2012;1(3):168–172. doi: 10.4161/adip.20574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O'Connor T., Bodén M., Bailey T.L. CisMapper: predicting regulatory interactions from transcription factor ChIP-seq data. Nucleic Acids Research. 2017;45(4):e19. doi: 10.1093/nar/gkw956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W., Cowley A., Uludag M., Gur T., McWilliam H., Squizzato S. The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Research. 2015;43(W1):W580–W584. doi: 10.1093/nar/gkv279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang Y., Pastor W.A., Shen Y., Tahiliani M., Liu D.R., Rao A. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 2010;5(1):1–9. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson R.F., Fazzari M.J., Niu H., Barzilai N., Simmons R.A., Greally J.M. Experimental intrauterine growth restriction induces alterations in DNA methylation and gene expression in pancreatic islets of rats. The Journal of Biological Chemistry. 2010;285(20):15111–15118. doi: 10.1074/jbc.M109.095133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Couillard C., Mauriège P., Imbeault P., Prud’homme D., Nadeau A., Tremblay A. Hyperleptinemia is more closely associated with adipose cell hypertrophy than with adipose tissue hyperplasia. International Journal of Obesity and Related Metabolic Disorders. 2000;24(6):782–788. doi: 10.1038/sj.ijo.0801227. [DOI] [PubMed] [Google Scholar]
- 40.Dahlhoff M., Pfister S., Blutke A., Rozman J., Klingenspor M., Deutsch M.J. Peri-conceptional obesogenic exposure induces sex-specific programming of disease susceptibilities in adult mouse offspring. Biochimica et Biophysica Acta. 2014;1842(2):304–317. doi: 10.1016/j.bbadis.2013.11.021. [DOI] [PubMed] [Google Scholar]
- 41.Breton C. The hypothalamus – adipose axis is a key target of developmental programming by maternal nutritional manipulation. Journal of Endocrinology. 2013;216(2):R19–R31. doi: 10.1530/JOE-12-0157. [DOI] [PubMed] [Google Scholar]
- 42.Jeffery E., Wing A., Holtrup B., Sebo Z., Kaplan J.L., Saavedra-Peña R. The adipose tissue microenvironment regulates depot-specific adipogenesis in obesity. Cell Metabolism. 2016;24(1):142–150. doi: 10.1016/j.cmet.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spalding K.L., Arner E., Westermark P.O., Bernard S., Buchholz B.A., Bergmann O. Dynamics of fat cell turnover in humans. Nature. 2008;453(7196):783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 44.Berry R., Rodeheffer M.S. Characterization of the adipocyte cellular lineage in vivo. Nature Cell Biology. 2013;15(3):302–308. doi: 10.1038/ncb2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun B., Purcell R.H., Terrillion C.E., Yan J., Moran T.H., Tamashiro K.L.K. Maternal high-fat diet during gestation or suckling differentially affects offspring leptin sensitivity and obesity. Diabetes. 2012;61(11):2833–2841. doi: 10.2337/db11-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kirk S.L., Samuelsson A.-M., Argenton M., Dhonye H., Kalamatianos T., Poston L. Maternal obesity induced by diet in rats permanently influences central processes regulating food intake in offspring. PloS One. 2009;4(6):e5870. doi: 10.1371/journal.pone.0005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White U.A., Tchoukalova Y.D. Sex dimorphism and depot differences in adipose tissue function. Biochimica et Biophysica Acta. 2014;1842(3):377–392. doi: 10.1016/j.bbadis.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villar D., Berthelot C., Aldridge S., Rayner T.F., Lukk M., Pignatelli M. Enhancer evolution across 20 mammalian species. Cell. 2015;160(3):554–566. doi: 10.1016/j.cell.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hahn M.A., Szabó P.E., Pfeifer G.P. 5-Hydroxymethylcytosine: a stable or transient DNA modification? Genomics. 2014;104(5):314–323. doi: 10.1016/j.ygeno.2014.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lefterova M.I., Haakonsson A.K., Lazar M.A., Mandrup S. PPARγ and the global map of adipogenesis and beyond. Trends in Endocrinology and Metabolism. 2014;25(6):293–302. doi: 10.1016/j.tem.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masuyama H., Hiramatsu Y. Additive effects of maternal high fat diet during lactation on mouse offspring. PloS One. 2014;9(3):e92805. doi: 10.1371/journal.pone.0092805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bouret S.G., Draper S.J., Simerly R.B. Trophic action of leptin on hypothalamic Neurons that regulate feeding. Science. 2004;304(5667):108–110. doi: 10.1126/science.1095004. [DOI] [PubMed] [Google Scholar]
- 53.Bol V.V., Reusens B.M., Remacle C.A. Postnatal catch-up growth after fetal protein restriction programs proliferation of rat preadipocytes. Obesity (Silver Spring, Md.) 2008;16(12):2760–2763. doi: 10.1038/oby.2008.417. [DOI] [PubMed] [Google Scholar]
- 54.Huan J.-N., Li J., Han Y., Chen K., Wu N., Zhao A.Z. Adipocyte-selective reduction of the leptin receptors induced by antisense RNA leads to increased adiposity, dyslipidemia, and insulin resistance. The Journal of Biological Chemistry. 2003;278(46):45638–45650. doi: 10.1074/jbc.M304165200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.