 The flavin-dependent monooxygenase AzmF catalyses posttranslational oxime formation during biosynthesis of the azolemycin complex of novel ribosomal peptide natural products.
The flavin-dependent monooxygenase AzmF catalyses posttranslational oxime formation during biosynthesis of the azolemycin complex of novel ribosomal peptide natural products.
Abstract
Streptomycetes are a rich source of bioactive specialized metabolites, including several examples of the rapidly growing class of ribosomally-biosynthesized and post-translationally-modified peptide (RiPP) natural products. Here we report the discovery from Streptomyces sp. FXJ1.264 of azolemycins A–D, a complex of novel linear azole-containing peptides incorporating a unique oxime functional group. Bioinformatics analysis of the Streptomyces sp. FXJ1.264 draft genome sequence identified a cluster of genes that was hypothesized to be responsible for elaboration of the azolemycins from a ribosomally-biosynthesized precursor. Inactivation of genes within this cluster abolished azolemycin production, consistent with this hypothesis. Moreover, mutants lacking the azmE and azmF genes accumulated azolemycin derivatives lacking the O-methyl groups and an amino group in place of the N-terminal oxime (as well as proteolysed derivatives), respectively. Thus AzmE, a putative S-adenosyl methionine-dependent methyl transferase, is responsible for late-stage O-methylation reactions in azolemycin biosynthesis and AzmF, a putative flavin-dependent monooxygenase, catalyzes oxidation of the N-terminal amino group in an azolemycin precursor to the corresponding oxime. To the best of our knowledge, oxime formation is a hitherto unknown posttranslational modification in RiPP biosynthesis.
Introduction
Despite remarkable advances in the science of chemical synthesis, bioactive natural products continue to provide useful starting points for the development of new drugs and agrochemicals.1 Many natural products are peptides or peptide derivatives which can be divided into two major classes, nonribosomal peptides (NRPs) and ribosomally-biosynthesized and post-translationally-modified peptides (RiPPs), based on their differing biosynthetic origins.2,3 NRPs are assembled by modular multienzyme assembly lines and are widely used in the clinic as antibiotics (e.g. vancomycin,4 penicillins/cephalosporins,5 and daptomycin6), immunosuppressants (e.g. cyclosporine7) and anticancer agents (e.g. bleomycin8). The advent of high throughput genome sequencing in the 21st century coupled with the development of in silico sequence mining tools has revealed RiPPs as a second major class of peptide natural products, endowed with a high degree of structural diversity and a wide variety of bioactivities.3 In comparison with NRP biosynthetic gene clusters, those responsible for RiPP biosynthesis are relatively small and are more amenable to genetic manipulation for the creation of novel derivatives.9–12
Linear azol(in)e-containing peptides (LAPs) typically incorporate several (methyl)oxazol(in)es and/or thiazol(in)es and represent an important family of RiPPs exemplified by microcin B17 and goadsporin.13,14 The enzymology underlying posttranslational azol(in)e formation has generated considerable interest in this family of RiPPs over the past two decades.15–17 However, relatively little is known about the biosynthesis of LAPs in actinobacteria,18 which are considered to be prolific producers of RiPPs.3
Here we report the discovery of azolemycins A–D, a novel complex of LAPs containing a unique oxime functional group, from Streptomyces sp. FXJ1.264. This strain, isolated from red soil collected in the Jiangxi Province of southeast China, has already been shown to produce two known classes of bioactive polyketide metabolites, the etheromycins and the anthrabenzoxocinones.19,20 We also report identification of the azolemycin biosynthetic gene cluster, and show that genes predicted to encode a flavin-dependent monoxygenase and an S-adenosyl methionine (SAM)-dependent methyl transferase are responsible for oxime formation and late stage methylation, respectively, in azolemycin biosynthesis.
Results and discussion
Isolation and structure elucidation of the azolemycins
UHPLC-MS analysis of ethanol extracts from small-scale cultures of Streptomyces sp. FXJ1.264 grown on GYM agar identified two potentially interesting metabolites with m/z = 670.3 and 684.3. Dried ethanol extracts from large-scale (25 L) agar cultures were fractionated by silica gel column chromatography eluting with chloroform/methanol. Fractions containing the metabolites of interest were combined and subjected to Sephadex LH-20 column chromatography, eluting with methanol, to remove lipophilic impurities. Separation of the resulting mixture of metabolites by semi-preparative HPLC yielded one pair of compounds with the molecular formula C31H39N7O8S and another pair of compounds with the molecular formula C32H41N7O8S (see ESI†). Database searches using these molecular formulae showed that the compounds are novel. The purified isomers were found to readily interconvert upon prolonged storage, or in acidic solvents such as chloroform.
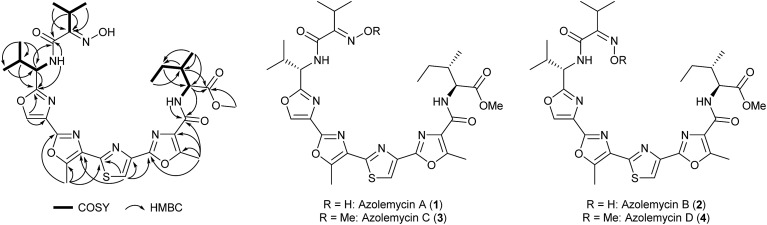
Using 1- and 2-D NMR spectroscopy, the planar structure was established for the more abundant lower molecular weight isomer (Fig. 1 and ESI†). This compound was named azolemycin A. The NMR data for the less abundant lower molecular weight isomer, azolemycin B, are very similar to those for azolemycin A. The biggest discrepancy is in the chemical shift of the signal due to the methine proton in the isopropyl group of the oxime (3.47 for azolemycin A compared with 3.05 for azolemycin B). We thus hypothesized that azolemycins A and B differ only in the configuration of the oxime. To test this hypothesis, azolemycins A and B were hydrolyzed under mildly acidic conditions and the products of the reactions were analyzed by high resolution UHPLC-MS, which showed they had identical retention times and molecular formulae, corresponding to the ketone resulting from hydrolytic cleavage of the oxime (see ESI†). To assign the configuration of the oximes in azolemycin A and B, 1H NMR chemical shifts for the E and Z isomers of the oxime derived from condensation of the N-methyl amide of α-ketoisovaleric acid with hydroxylamine were calculated, using methodology recently reported by Tantillo and coworkers.21 The methine proton in the model oxime with the E-configuration was predicted to have a chemical shift of 3.61 ppm, whereas the corresponding proton in the Z-configured oxime was predicted to have a chemical shift of 3.09 ppm (See ESI†). Thus the configurations of the oximes in azolemycins A and B were assigned as E and Z, respectively.
Fig. 1. Key correlations observed in COSY and HMBC NMR spectra of azolemycin A, and the structures of azolemycins A–D deduced from HRMS, NMR spectroscopy and degradation studies.
To determine the configurations of the stereogenic centers in azolemycin A, it was hydrolyzed in strong acid and the valine and isoleucine produced were derivatized using Marfey's reagent.22 LC-MS comparisons with Marfey's derivatives of l- and d-valine established the S-configuration for the stereocenter bearing the isopropyl group. The α-carbon in the isoleucine residue was also shown to have the S-configuration by comparison with Marfey's derivatives of l-isoleucine, l-allo-isoleucine, d-isoleucine and d-allo-isoleucine. However, the configuration of the β-carbon could not be established on the basis of such comparisons. Thus we compared the NMR data for the α-proton of the isoleucine residue with literature values. In CDCl3, the α-proton in the methyl ester of N-benzoyl-allo-isoleucine is reported to have a chemical shift of 4.90 ppm and coupling constants of 4.3 and 9.0 Hz, whereas the corresponding proton in the methyl ester of N-benzyol-isoleucine has a chemical shift of 4.70 ppm and coupling constants of 5.2 and 8.6 Hz.23 The α-proton of the isoleucine residue in azolemycin A has a chemical shift of 4.73 ppm and coupling constants of 5.4 and 8.8 Hz (also in CDCl3). This indicates that the isoleucine residue in azolemycin A does not possess the allo relative configuration, leading us to assign the S-configuration to the β-carbon. This assignment was further confirmed by comparison of the 1H NMR spectra of azolemycin A with a chemically-synthesized mimic of its 5-aryl-3-methyl-oxazole-2-carboxyl-l-isoleucine methyl ester moiety (see ESI†). Thus structures 1 and 2 (Fig. 1) were assigned to azolemycins A and B, respectively. The structure of azolemycin A has been independently confirmed by total synthesis, details of which will be reported elsewhere.24
HRMS, NMR data and chemical shift calculations for the isomeric higher molecular weight azolemycin congeners indicated that they are derivatives of azolemycins A and B in which the oxime has been O-methylated (see ESI†). These compounds were named azolemycins C (3) and D (4), respectively (Fig. 1).
Identification and characterization of the azolemycin biosynthetic gene cluster
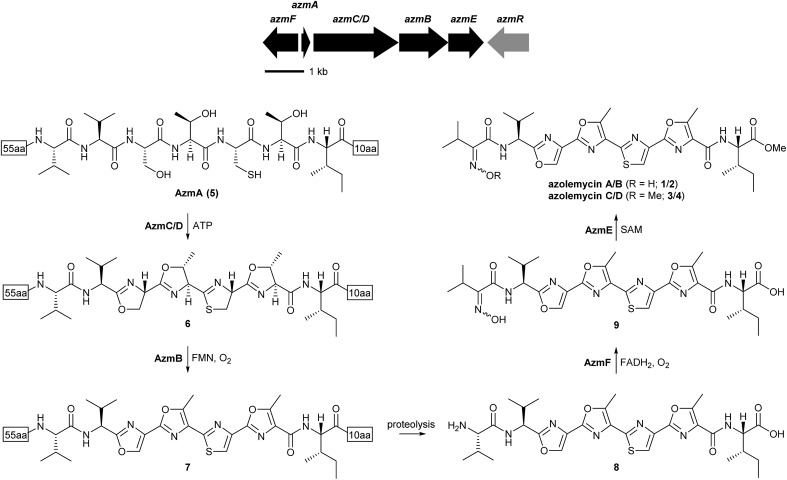
Inspection of the structure of the azolemycins suggests that they are elaborated from a peptide with the sequence VVSTCTI. Genomic DNA was extracted from Streptomyces sp. FXJ1.264 and subjected to Roche 454 FLX sequencing, resulting in a total of 9 436 877 bp of genome sequence distributed across 254 contigs (≥500 bp) with an average size of 37 153 bp. To investigate whether the peptide precursor of the azolemycins is ribosomally biosynthesized, the draft genome sequence of Streptomyces sp. FXJ1.264 was searched for coding sequences (CDSs) corresponding to the putative precursor. This resulted in identification of a 219 bp CDS, named azmA, which encodes a protein containing the putative precursor peptide flanked by 55 N-terminal residues and 10 C-terminal residues. Comparative sequence analyses of the proteins encoded by the CDSs flanking azmA indicated that they are likely involved in maturation of the putative azolemycin precursor peptide (Fig. 2 and ESI†). The azmC/D gene encodes a protein with sequence similarity to ATP-dependent cyclases known to catalyze the conversion of Ser, Thr and Cys residues to the corresponding azolines in the biosynthesis of other LAPs,16 and azmB encodes a homologue of flavin-dependent dehydrogenases that convert the azolines to the corresponding azoles.17 Thus, AzmC/D is proposed to catalyze four cyclodehydration reactions in AzmA (5) to yield 6, which undergoes AzmB-catalyzed oxidation to yield 7 (Fig. 2). Proteolytic cleavage of the N-terminal leader peptide and the C-terminal decapeptide in 7 would yield 8. Interestingly, the protease(s) responsible for this do not appear to be encoded by genes within the putative azolemycin biosynthetic gene cluster, as is the case for other azol(in)e-containing RiPPs produced by Streptomyces species, such as goadsporin and berninamycin.25,26 The azmE and azmF genes encode a putative S-adenosyl methionine (SAM)-dependent methyl transferase and a flavin-dependent monooxygenase, respectively. AzmF is predicted to oxidise the valine amino group in 8 resulting in the corresponding oxime 9 (Fig. 2). AzmE is hypothesized to catalyze methylation of the isoleucine carboxyl group in 9 to give azolemycins A and B (1 and 2). AzmE may also catalyze O-methylation of the oxime group in 1 and 2 to yield azolemycins C and D (3 and 4). The azmR gene encodes a putative LysR family transcriptional regulator, and likely plays a role in the regulation of azolemycin biosynthesis. The CDSs downstream of azmF and upstream of azmR encode proteins with no obvious role in azolemycin biosynthesis.
Fig. 2. Organization of the azolemycin biosynthetic gene cluster and proposed pathway for azolemycin biosynthesis in Streptomyces sp. FXJ1.264. It is likely that AzmF produces exclusively the E-oxime and that azolemycin B arises from facile isomerization of the oxime group in azolemycin A. Azolemycin C and D appear to arise from AzmE-catalysed O-methylation of the oxime group in azolemycins A and B, respectively.
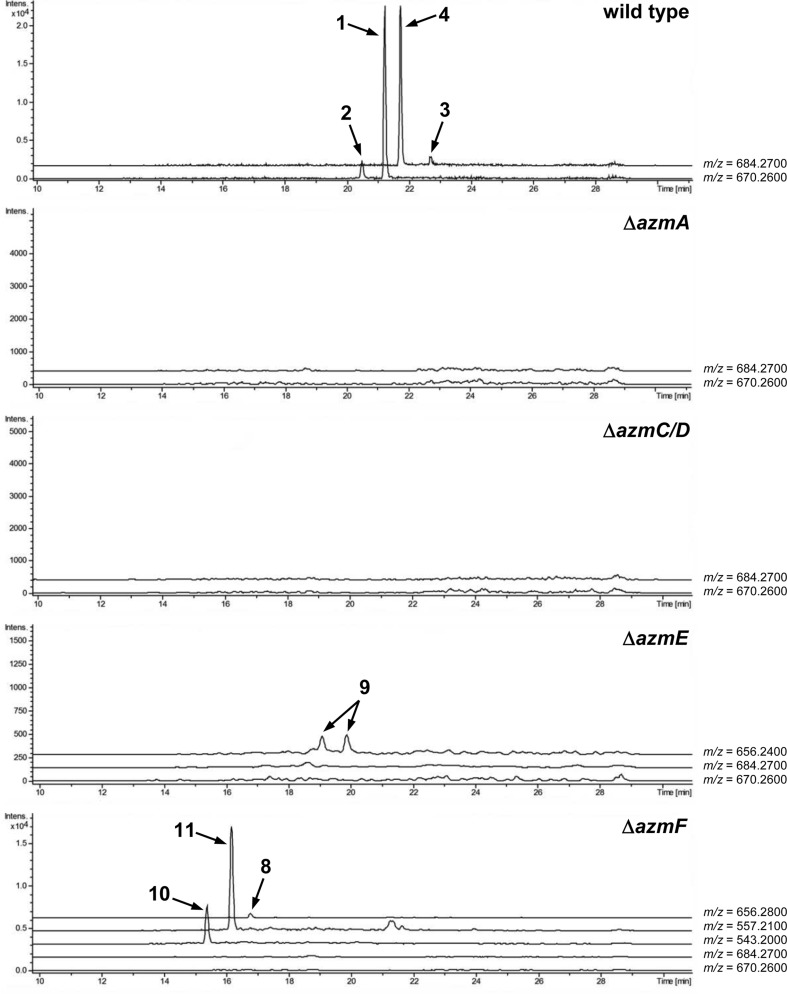
To verify that azmA encodes the azolemycin precursor peptide, a mutant of Streptomyces sp. FXJ1.264 containing a 180 bp in-frame deletion within this CDS was constructed (see ESI† for details). UHPLC-HRMS analysis of culture extracts confirmed that azolemycin production is abolished in this mutant (Fig. 3). The involvement of azmC/D in azolemycin biosynthesis was similarly established by creating a mutant of Streptomyces sp. FXJ1.264 in which this CDS has been replaced by the neo gene, which confers kanamycin resistance. UHPLC-HRMS analyses confirmed that culture extracts from this mutant does not contain azolemycins (Fig. 3).
Fig. 3. 600 dpi in TIF format)[QUESTION MARK]?>Extracted ion chromatograms from UPLC-HRMS analyses of culture extracts from wild type Streptomyces sp. FXJ1.264 and its ΔazmA, ΔazmC/D, ΔazmE and ΔazmF mutants. The m/z values used to generate each chromatogram are listed on the right.
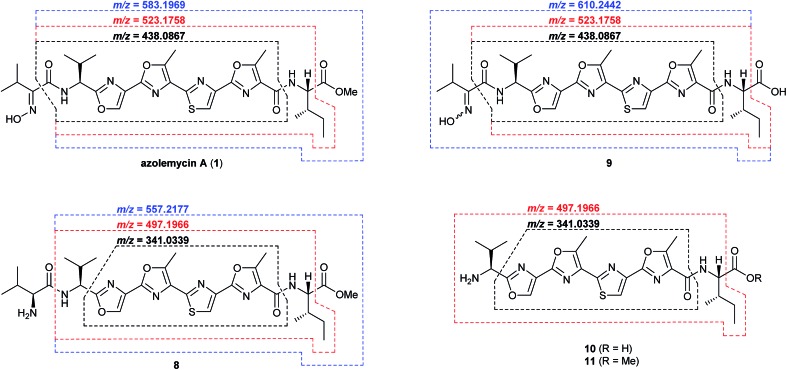
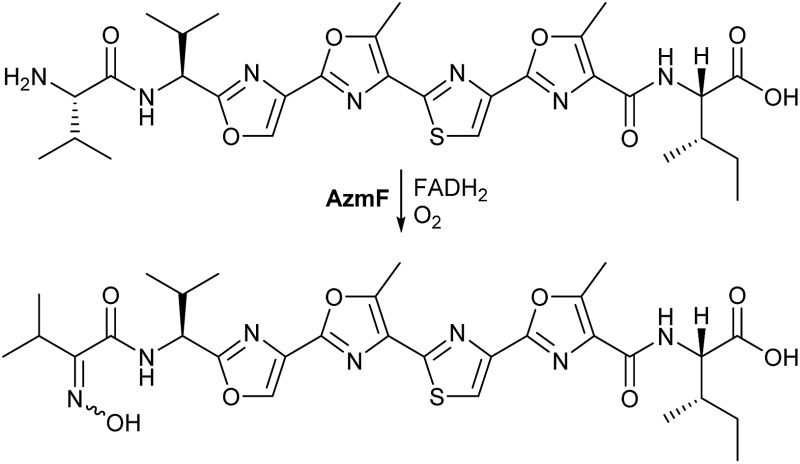
The proposed role of AzmF in azolemycin biosynthesis, as a flavin-dependent monooxygenase that catalyzes oxidation of the amino group in 8 to the corresponding oxime, was investigated by constructing a mutant of Streptomyces sp. FXJ1.264 in which azmF is replaced with the neo gene. UHPLC-HRMS analysis showed that the production of azolemycins A–D (1–4) is abolished in this mutant. However, three putative azolemycin derivatives were produced by the mutant (Fig. 3). Comparison of the MS/MS spectra for these compounds with that for azolemycin A (1) showed that they are the proposed biosynthetic intermediate 8, a derivative 10 missing the N-terminal valine residue, and a derivative 11 of azolemycins A/B (1 and 2) similarly lacking the N-terminal residue (Fig. 4 and ESI†). 10 presumably derives from 8 via proteolytic cleavage and 11 probably arises from subsequent AzmE-catalyzed methylation of 10. These data are consistent with the proposed role of AzmF in azolemycin biosynthesis and suggest that oxime formation, which is to the best of our knowledge without precedent as a posttranslational modification in RiPP biosynthesis, serves to prevent proteolytic degradation of the azolemycins (Fig. 2). In contrast to RiPPs, several NRPs containing an oxime group are known, including nocardicins A and B, caerulomycin A, althiomycin and collismycin A.21,27–29 The cytochrome P450 NocL and flavin-dependent two-component monooxygenase CrmH are responsible for late-stage oxidation of an amino group to the corresponding oxime in the biosynthesis of nocardicin and caerulomycin, respectively,30,31 and in althiomycin biosynthesis a putative di-iron-dependent oxygenase has been proposed to catalyze an analogous transformation.32,33 Finally, in collismycin biosynthesis, the putative flavin-dependent monooxygenase ClmM, which shares 46% sequence identity with AzmF, is implicated in the oxidation of a primary amine to the corresponding oxime.34
Fig. 4. Key fragment ions observed in UHPLC-HRMS/MS analyses of azolemycin A (1) in culture extracts of wild type Streptomyces sp. FXJ1.264, intermediate 9 in culture extracts of the azmE mutant, and shunt metabolites 8, 10 and 11 in the azmF mutant. Calculated m/z values based on the molecular formula of each fragment ion are shown for comparison with the measured values (see ESI† for spectra).
To explore whether AzmF homologues are involved in the biosynthesis of other LAPs, we queried the “all genomes” database of the Joint Genome Institute with the sequence of AzmF. All of the top 20 hits were from actinobacterial genomes and most of them resided in putative NRP biosynthetic gene clusters. A single hit was found within a putative LAP biosynthetic gene cluster in the genome of Actinomadura oligospora ATCC 43269. In addition to azmF, azmC/D and azmB homologues, this gene cluster contained four genes encoding putative precursor peptides (see ESI†). Experimental analysis of the metabolic product(s) will be required to establish whether the AzmF homologue encoded by this gene cluster is responsible for posttranslational oxime formation or some other type of oxidative transformation.
The function of AzmE, which belongs to the same family of SAM-dependent methyl transferases (COG3315) as the BmbA/BtmB/BotOMT/BstB enzyme responsible for posttranslational O-methylation of an aspartate side chain in bottromycin biosynthesis,10,35–37 was investigated by constructing a mutant of Streptomyces sp. FXJ1.264 in which azmE is replaced with the neo gene. UHPLC-HRMS of culture extracts showed that none of the azolemycins are produced by the mutant, but two compounds with molecular formulae corresponding to demethylated derivatives of azolemycin A (1) were identified (Fig. 3). Comparison of the MS/MS spectra of these compounds with the corresponding spectrum for azolemycin A indicated that they are the E- and Z-oxime isomers of proposed intermediate 9 in azolemycin biosynthesis (Fig. 4). O-Methylation of the carboxyl terminus is a rare posttranslational modification of RiPPs, but has recently been reported for several linear cyanobactins produced by cyanobacteria.38
Bioactivity of the azolemycins
The azolemycins were tested for antiproliferative and antimicrobial activities (see ESI†). Azolemycin C showed moderate antiproliferative effects against human colon cancer cell line HCT116 with an IC50 value of 69.2 ± 3.3 μM, with the positive control cisplatin showing an IC50 value of 22.7 ± 1.8 μM. Similarly, azolemycin B displayed modest antiproliferative activity against human bladder (T24) and cervical (HeLa) cancer cell lines with IC50 values of 88.4 ± 8.5 and 96.0 ± 3.5 μM, respectively. In comparison, cisplatin showed IC50 values of 11.5 ± 0.2 and 3.6 ± 0.2 μM, respectively, against the same cell lines. Azolemycin D showed weak antiproliferative activity against HeLa cell lines only, while azolemycin A did not exhibit any antiproliferative activity against the six cell lines tested (see ESI†). None of the four compounds showed activity against Staphylococcus aureus, Bacillus subtilis, Escherichia coli, Candida albicans or Candida pseudorugosa. These data indicate that the azolemycins probably do not function as antibiotics in the natural environment of the producing organism. The ability of the azolemycins to promote (at low concentrations) or inhibit (at high concentrations) the growth of other actinobacteria was also examined, but no activity was observed. Further experiments are required to determine whether the azolemycins possess other, more potent, biological activities.
Conclusions
Azolemycins A–D, four new LAPs containing a unique oxime functional group with modest activity against mammalian cancer cell lines, have been isolated from Streptomyces sp. FXJ1.261. The genome of Streptomyces sp. FXJ1.261 has been partially sequenced, allowing identification of the azmA gene, encoding a putative azolemycin precursor peptide. In-frame deletion of azmA abolished production of the azolemycins, as did replacement of the adjacent azmC/D gene, predicted to encode a YcaO domain-containing azoline synthetase, with a gene conferring kanamycin resistance. The azmE and azmF genes flanking azmA and azmC/D were predicted to encode a SAM-dependent methyl transferase and a flavin-dependent monoxygenase, respectively. Based on the azolemycin derivatives observed to accumulate in an azmF mutant, it was concluded that AzmF catalyzes oxidation of the amino group in the N-terminal valine residue arising from cleavage of the 55 amino acid leader peptide in AzmA to the corresponding oxime. Similarly, analysis of an azmE mutant showed that AzmE catalyses methylation of the carboxyl group in the C-terminal isoleucine residue resulting from cleavage of the C-terminal decapeptide from AzmA. In addition, AzmE appears to catalyze O-methylation of the oxime group in azolemycin A and/or B. In common with other LAP biosynthetic gene clusters in Streptomyces species, the azolemycin biosynthetic gene cluster lacks genes encoding the proteases needed to cleave the 55 amino acid leader sequence and the C-terminal decapeptide from the AzmA precursor peptide.
Acknowledgments
We thank Prof. Xuejun Jiang and Dr Shubin Niu (Institute of Microbiology, Chinese Academy of Sciences) for their assistance with the antitumor assays. This work was supported by grants from the National Science Foundation of China (NSFC, 31170010 and 31300010) and by a BBSRC China Partnering Award (BB/L010852/1 to GLC).
Footnotes
References
- Newman D. J., Cragg G. M. J. Nat. Prod. 2012;75:311. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzer D., Finking R., Marahiel M. A. Nat. Prod. Rep. 2003;20:275. doi: 10.1039/b111145k. [DOI] [PubMed] [Google Scholar]
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., Camarero J. A., Campopiano D. J., Challis G. L., Clardy J., Cotter P. D., Craik D. J., Dawson M., Dittmann E., Donadio S., Dorrestein P. C., Entian K. D., Fischbach M. A., Garavelli J. S., Goransson U., Gruber C. W., Haft D. H., Hemscheidt T. K., Hertweck C., Hill C., Horswill A. R., Jaspars M., Kelly W. L., Klinman J. P., Kuipers O. P., Link A. J., Liu W., Marahiel M. A., Mitchell D. A., Moll G. N., Moore B. S., Muller R., Nair S. K., Nes I. F., Norris G. E., Olivera B. M., Onaka H., Patchett M. L., Piel J., Reaney M. J., Rebuffat S., Ross R. P., Sahl H. G., Schmidt E. W., Selsted M. E., Severinov K., Shen B., Sivonen K., Smith L., Stein T., Sussmuth R. D., Tagg J. R., Tang G. L., Truman A. W., Vederas J. C., Walsh C. T., Walton J. D., Wenzel S. C., Willey J. M., van der Donk W. A. Nat. Prod. Rep. 2013;30:108. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard B. K., Walsh C. T. Angew. Chem., Int. Ed. 2003;42:730. doi: 10.1002/anie.200390202. [DOI] [PubMed] [Google Scholar]
- Baldwin J. E., Abraham E. Nat. Prod. Rep. 1988;5:129. doi: 10.1039/np9880500129. [DOI] [PubMed] [Google Scholar]
- Miao V., Coeffet-LeGal M. F., Brian P., Brost R., Penn J., Whiting A., Martin S., Ford R., Parr I., Bouchard M., Silva C. J., Wrigley S. K., Baltz R. H. Microbiology (London, U. K.) 2005;151:1507. doi: 10.1099/mic.0.27757-0. [DOI] [PubMed] [Google Scholar]
- Canafax D. M., Ascher N. L. Clin. Pharm. 1983;2:515. [PubMed] [Google Scholar]
- Umezawa H., Maeda K., Takeuchi T., Okami Y. J. Antibiot. 1966;19:200. [PubMed] [Google Scholar]
- Mitchell D. A., Lee S. W., Pence M. A., Markley A. L., Limm J. D., Nizet V., Dixon J. E. J. Biol. Chem. 2009;284:13004. doi: 10.1074/jbc.M900802200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. P., Tianero M. D. B., Kwan J. C., Wyche T. P., Michel C. R., Ellis G. A., Vazquez-Rivera E., Braun D. R., Rose W. E., Schmidt E. W., Bugni T. S. Org. Lett. 2012;14:5050. doi: 10.1021/ol3022758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane C. D., Melby J. O., Molohon K. J., Susarrey A. R., Mitchell D. A. ACS Chem. Biol. 2013;8:1998. doi: 10.1021/cb4003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tianero M. D. B., Donia M. S., Young T. S., Schultz P. G., Schmidt E. W. J. Am. Chem. Soc. 2012;134:418. doi: 10.1021/ja208278k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer A., Freund S., Nicholson G., Jung G. Angew. Chem., Int. Ed. 1993;32:1336. [Google Scholar]
- Igarashi Y., Kan Y., Fujii K., Fujita T., Harada K., Naoki H., Tabata H., Onaka H., Furumai T. J. Antibiot. 2001;54:1045. doi: 10.7164/antibiotics.54.1045. [DOI] [PubMed] [Google Scholar]
- Li Y. M., Milne J. C., Madison L. L., Kolter R., Walsh C. T. Science. 1996;274:1188. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- Dunbar K. L., Melby J. O., Mitchell D. A. Nat. Chem. Biol. 2012;8:569. doi: 10.1038/nchembio.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby J. O., Li X. P., Mitchell D. A. Biochemistry. 2014;53:413. doi: 10.1021/bi401529y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jian X. H., Pan H. X., Ning T. T., Shi Y. Y., Chen Y. S., Li Y., Zeng X. W., Xu J., Tang G. L. ACS Chem. Biol. 2012;7:646. doi: 10.1021/cb200479f. [DOI] [PubMed] [Google Scholar]
- Wang H., Liu N., Xi L., Rong X., Ruan J., Huang Y. Appl. Environ. Microbiol. 2011;77:3433. doi: 10.1128/AEM.02915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. Y., Liu N., Huang Y., Chen Y. H. Tetrahedron: Asymmetry. 2014;25:113. [Google Scholar]
- Lodewyk M. W., Soldi C., Jones P. B., Olmstead M. M., Rita J., Shaw J. T., Tantillo D. J. J. Am. Chem. Soc. 2012;134:18550. doi: 10.1021/ja3089394. [DOI] [PubMed] [Google Scholar]
- Bhushan R., Bruckner H. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2011;879:3148. doi: 10.1016/j.jchromb.2011.05.058. [DOI] [PubMed] [Google Scholar]
- Khatskevich I. E., Kalnin I. K., Karpeiskaya E. I., Klabunovskii E. I. Bull. Acad. Sci. USSR, Div. Chem. Sci. 1983;32:323. [Google Scholar]
- Anderson Z. J., Fox D. J., Org. Biomol. Chem., 2015. , , submitted for publication . [Google Scholar]
- Malcolmson S. J., Young T. S., Ruby J. G., Skewes-Cox P., Walsh C. T. Proc. Natl. Acad. Sci. U. S. A. 2013;110:8483. doi: 10.1073/pnas.1307111110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onaka H., Nakaho M., Hayashi K., Igarashi Y., Furumai T. Microbiology (London, U. K.) 2005;151:3923. doi: 10.1099/mic.0.28420-0. [DOI] [PubMed] [Google Scholar]
- Bycroft B. W., Pinchin R. J. Chem. Soc., Chem. Commun. 1975;4:121. [Google Scholar]
- Divekar P., Read G., Vining L. Can. J. Chem. 1967;45:1215. [Google Scholar]
- Shindo K., Yamagishi Y., Okada Y., Kawai H. J. Antibiot. 1994;47:1072. doi: 10.7164/antibiotics.47.1072. [DOI] [PubMed] [Google Scholar]
- Kelly W. L., Townsend C. A. J. Am. Chem. Soc. 2002;124:8186. doi: 10.1021/ja025926g. [DOI] [PubMed] [Google Scholar]
- Zhu Y. G., Zhang Q. B., Li S. M., Lin Q. H., Fu P., Zhang G. T., Zhang H. B., Shi R., Zhu W. M., Zhang C. S. J. Am. Chem. Soc. 2013;135:18750. doi: 10.1021/ja410513g. [DOI] [PubMed] [Google Scholar]
- Cortina N. S., Revermann O., Krug D., Muller R. ChemBioChem. 2011;12:1411. doi: 10.1002/cbic.201100154. [DOI] [PubMed] [Google Scholar]
- Gerc A. J., Song L., Challis G. L., Stanley-Wall N. R., Coulthurst S. J. PLoS One. 2012;7:e44673. doi: 10.1371/journal.pone.0044673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia I., Vior N. M., Gonzalez-Sabin J., Brana A. F., Rohr J., Moris F., Mendez C., Salas J. A. Chem. Biol. 2013;20:1022. doi: 10.1016/j.chembiol.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Huo L. J., Rachid S., Stadler M., Wenzel S. C., Muller R. Chem. Biol. 2012;19:1278. doi: 10.1016/j.chembiol.2012.08.013. [DOI] [PubMed] [Google Scholar]
- Gomez-Escribano J. P., Song L. J., Bibb M. J., Challis G. L. Chem. Sci. 2012;3:3522. [Google Scholar]
- Crone W. J. K., Leeper F. J., Truman A. W. Chem. Sci. 2012;3:3516. [Google Scholar]
- Leikoski N., Liu L. W., Jokela J., Wahlsten M., Gugger M., Calteau A., Permi P., Kerfeld C. A., Sivonen K., Fewer D. P. Chem. Biol. 2013;20:1033. doi: 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.