Abstract
Cryopreserved human hepatocytes were used to investigate the role of arylamine N-acetyltransferase 2 (NAT2; EC 2.3.1.5) polymorphism on the N-acetylation of isoniazid (INH). NAT2 genotype was determined by Taqman allelic discrimination assay and INH N-acetylation was measured by high performance liquid chromatography. INH N-acetylation rates in vitro exhibited a robust and highly significant (P<0.005) NAT2 phenotype-dependent metabolism. N-acetylation rates in situ were INH concentration- and time-dependent. Following incubation for 24 h with 12.5 or 100 µmol/L INH, acetyl-INH concentrations varied significantly (P = 0.0023 and P = 0.0002) across cryopreserved human hepatocytes samples from rapid, intermediate, and slow acetylators, respectively. The clear association between NAT2 genotype and phenotype supports use of NAT2 genotype to guide INH dosing strategies in the treatment and prevention of tuberculosis.
KEY WORDS: Isoniazid, N-Acetyltransferase 2, Acetylation polymorphism, Human hepatocytes, Genotype, Phenotype
Graphical abstract
Isoniazid N-acetylation rates in vitro and multiple concentrations in situ exhibited robust and highly significant N-acetyltransferase 2 (NAT2) genotype-dependent metabolism in cryopreserved human hepatocytes. The clear association between NAT2 genotype and phenotype supports use of NAT2 genotype to guide isoniazid dosing strategies in the treatment and prevention of tuberculosis.
1. Introduction
Arylamine N-acetyltransferases (E.C. 2.3.1.5) catalyze the N-acetylation of numerous arylamine and hydrazine drugs. N-Acetyltransferase 2 (NAT2) is subject to a genetic polymorphism in human populations and was identified following administration of isoniazid (INH) for the treatment of tuberculosis1. As recently reviewed, numerous single nucleotide polymorphisms in the coding exon the NAT2 gene, inherited as NAT2 haplotypes and genotypes, confer rapid, intermediate, and slow acetylator phenotypes that modify the metabolism of arylamine and hydrazine drugs in human populations2. The role of NAT2 and its genetic polymorphism in the metabolism and pharmacokinetic profile of INH has been reviewed3, 4.
INH is extensively prescribed because of the high global incidence of tuberculosis with over 10 million new cases reported in 20155. Numerous studies have investigated the effect of NAT2 genetic polymorphism in INH efficacy and toxicity in populations across the world2. The extensive use of INH for the treatment and prevention of tuberculosis is compromised by INH-induced hepatotoxicity and liver failure6.
The role of the NAT2 acetylator polymorphism on INH hepatotoxicity has been extensively investigated. As reported in each of three recent reviews7, 8, 9, published reports on the role of the NAT2 acetylator polymorphism on INH hepatotoxicity have been inconsistent and controversial. INH hepatotoxicity has been investigated in the rat, mouse and rabbit, but the course and pattern of the hepatoxicity is much different from that observed in human populations and inconsistent and controversial results have been reported9, suggesting that INH hepatotoxicity in animal models is not informative for understanding INH hepatoxicity in humans.
Increased risk of INH-induced hepatoxicity initially was proposed in rapid NAT2 acetylators because of greater generation of hydrazine metabolites10 supported by several clinical studies reporting higher incidence of INH hepatotoxicity in rapid NAT2 acetylators10, 11, 12. Recent meta-analyses of the role of NAT2 polymorphism on INH-induced hepatotoxicity have reported a higher frequency of INH-induced hepatotoxicity in NAT2 slow acetylators but no difference in risk between rapid and intermediate NAT2 acetylators7, 8, 13. A very recent review of INH metabolism and hepatotoxicity concludes that the role of NAT2 acetylator polymorphism in INH hepatotoxicity remains controversial and poorly understood9.
As previously reviewed14, while investigations of drug metabolism and toxicity often are carried out in animal models, interspecies differences may preclude accurate prediction of drug metabolic profiles in humans. Although primary cultures of human hepatocytes can produce a metabolic profile similar to that found in vivo, the availability of primary cultures of human hepatocytes is limited. Cryopreservation techniques have been developed yielding a high percentage of viable and plateable hepatocytes15, 16, 17 to facilitate investigations using annotated cryopreserved human hepatocytes for use by laboratories across the world.
Cryopreserved human hepatocytes have been used for in vitro investigations into the metabolism of arylamine drugs and carcinogens18, 19 and exploring relationships between acetylator genotype and phenotype20. The purpose of our present study was to investigate the role of NAT2 genetic polymorphism on INH N-acetylation in cryopreserved human hepatocytes obtained from rapid, intermediate and slow acetylators.
2. Materials and methods
2.1. Source and processing of cryopreserved human hepatocytes
Cryopreserved hepatocyte samples obtained from humans were received from Bioreclamation IVT (Baltimore, MD, USA) and stored in liquid nitrogen until use. Hepatocyte samples were collected from consented donors under IRB approved protocols at their FDA licensed donor center (http://www.bioreclamationivt.com/). Hepatocytes were prepared from fresh human tissue with hepatocytes isolated and frozen within 24 h of organ removal. All hepatocytes are human transplant rejected. All hepatocytes were tested and are negative for hepatitis B and C and HIV 1 and 2. Hepatocytes were treated as containing human-derived materials as potentially infectious, as no known test methods can offer assurance that products derived from human tissues will not transmit infectious agents. Upon removal from liquid nitrogen, hepatocytes were thawed according to the manufacturer׳s instructions by warming a vial of the hepatocytes at 37 °C for 90 s and transferring to a 50 mL conical tube containing 45 mL of InVitroGRO HT medium (Bioreclamation IVT, USA). The cell suspension was centrifuged at 50 × g at room temperature for 5 min. The supernatant was discarded and cells washed once in ice-cold phosphate buffered saline (PBS) before lysing the cells in ice-cold 20 mmol/L NaPO4, 1 mmol/L dithiothreitol, 1 mmol/L EDTA, 0.2% Triton X-100, 1 mmol/L phenylmethylsulfonyl fluoride, 1 µmol/L pepstatin A, and 1 µg/mL aprotinin. The lysate was centrifuged at 15,000 × g for 20 min and the supernatant was aliquoted and stored at –70 °C. Protein concentrations in the lysates were determined using the Bio-Rad protein assay kit (Bio-Rad, Richmond, CA, USA). Hepatocyte used for all in vitro experiments were categorized at either cryosuspensions or cryoplateable hepatocytes. Hepatocytes used for in situ studies were categorized as cryoplateable and approved for studies involving drug metabolism.
2.2. NAT2 genotyping and assignment of acetylator phenotype
Prod. Type: FTPw?>Genomic DNA was isolated from pelleted cells prepared from human cryopreserved hepatocyte samples as described above by using the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer׳s instructions. The nomenclature and functional effects of single nucleotide polymorphisms present in various NAT2 haplotypes and genotypes has been reviewed2, 21. NAT2 genotypes and deduced phenotypes were determined as described previously21, 22. Controls (no DNA template) were run to ensure that there was no amplification of contaminating DNA. Individuals possessing two NAT2 alleles associated with rapid acetylation activity (NAT2*4, NAT2*12, and NAT2*13) were classified as rapid acetylators; individuals possessing one of these alleles and one allele associated with slow acetylation activity (NAT2*5, NAT2*6, NAT2*7, and NAT2*14) were classified as intermediate acetylators, and those individuals that possessed two slow acetylation alleles were classified as slow acetylators. Cryopreserved hepatocytes with rapid, intermediate and slow NAT2 acetylator genotype were selected at random for measurements of INH N-acetylation as described below. The individual NAT2 genotype, gender, and ethnicity of each individual human cryopreserved hepatocyte sample is listed together with their respective INH N-acetyltransferase enzyme activity in vitro (Table 1) and INH N-acetylation in situ (Table 2).
Table 1.
Isoniazid (INH) and N-(4-aminobenzoyl)-L-glutamic acid (ABG) N-acetyltransferase activities in vitro in human cryopreserved human hepatocyte samples.
| Sample ID | NAT2 genoytpe | Deduced NAT2 phenotype | Gender | Ethnicity | INH N-acetyltransferase activitya | ABG N-acetyltransferase activitya |
|---|---|---|---|---|---|---|
| FLA | NAT2*4/*4 | Rapid | Female | Caucasian | 0.361 | 0.494 |
| HWG | NAT2*4/*4 | Rapid | Male | Caucasian | 0.183 | 0.358 |
| GPM | NAT2*4/*4 | Rapid | Male | Hispanic | 0.155 | 0.388 |
| FXA | NAT2*4/*4 | Rapid | Female | Hispanic | 0.141 | 0.303 |
| XUA | NAT2*4/*13 | Rapid | Female | Caucasian | 0.084 | 0.311 |
| VHU | NAT2*4/*5B | Intermediate | Female | African | 0.075 | 0.311 |
| UFN | NAT2*4/*5B | Intermediate | Male | Caucasian | 0.124 | 0.291 |
| ZCA | NAT2*4/*5B | Intermediate | Female | Caucasian | 0.051 | 0.256 |
| ZQM | NAT2*4/*5A | Intermediate | Female | Caucasian | 0.023 | 0.261 |
| OJE | NAT2*4/*6A | Intermediate | Female | Caucasian | 0.057 | 0.446 |
| DVR | NAT2*5B/*5B | Slow | Female | Caucasian | 0.042 | 0.329 |
| PFM | NAT2*5B/*5B | Slow | Male | Caucasian | 0.008 | 0.617 |
| ZFB | NAT2*5B/*6A | Slow | Female | Caucasian | 0.044 | 0.413 |
| MFB | NAT2*5A/*6A | Slow | Male | Hispanic | 0.005 | 0.438 |
| XMM | NAT2*6A/*6A | Slow | Male | Caucasian | 0.001 | 0.673 |
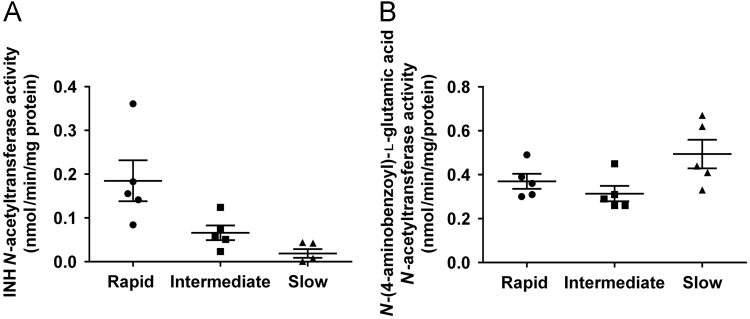
Mean ± S.E.M. for rapid, intermediate, and slow NAT2 phenotypes are illustrated in Fig. 1.
INH and ABG N-acetyltransferase activities in nmol/min/mg protein.
Table 2.
Acetyl-INH levels in human cryopreserved human hepatocytes.
| Sample ID | NAT2 genotype | Deduced NAT2 phenotype | Gender | Ethnicity | Acetyl-INH levela |
|
|---|---|---|---|---|---|---|
| 12.5 µmol/L INH | 100 µmol/L INH | |||||
| FWK | NAT2*4/*4 | Rapid | Female | Caucasian | 26.2 | 60.7 |
| ZFK | NAT2*4/*4 | Rapid | Male | Caucasian | 33.4 | 72.8 |
| YXZ | NAT2*4/*4 | Rapid | Male | Hispanic | 18.3 | 48.6 |
| JZG | NAT2*4/*4 | Rapid | Male | Hispanic | 30.1 | 69.8 |
| FAO | NAT2*4/*4 | Rapid | Female | African | 18.3 | 51.6 |
| ZFR | NAT2*4/*5B | Intermediate | Male | Caucasian | 15.3 | 41.9 |
| TUG | NAT2*4/*5B | Intermediate | Male | Caucasian | 22.5 | 60.1 |
| HUG | NAT2*4/*5B | Intermediate | Female | Caucasian | 10.2 | 33.5 |
| ASQ | NAT2*4/*5A | Intermediate | Female | Caucasian | 19.2 | 38.3 |
| NIQ | NAT2*4/*6A | Intermediate | Female | Caucasian | 12.1 | 35.6 |
| UFP | NAT2*5B/*6A | Slow | Female | Caucasian | 0.21 | 7.00 |
| DOO | NAT2*5B/*6A | Slow | Male | Caucasian | 1.70 | 13.20 |
| DNB | NAT2*5B/*6A | Slow | Male | Caucasian | 0.17 | 2.00 |
| YNS | NAT2*5A/*6A | Slow | Male | Hispanic | 0.52 | 8.05 |
| UNF | NAT2*6A/*6A | Slow | Male | African | 1.10 | 0.85 |
Hepatocyte samples were incubated 24 h with 12.5 or 100 µmol/L INH.
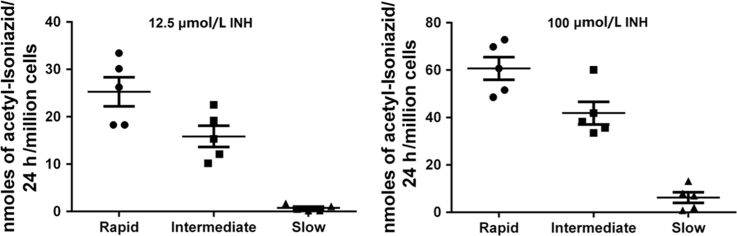
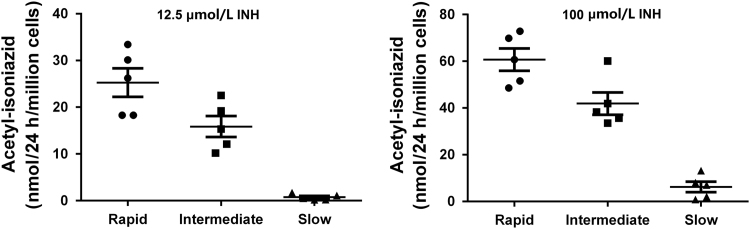
Mean ± S.E.M. for rapid, intermediate, and slow NAT2 phenotypes are illustrated in Fig. 4.
Acetyl-INH levels are nmoles acetyl-INH/24 h/million cells.
2.3. Measurement of N-acetyltransferase activity in vitro
Arylamine N-acetyltransferase activities were measured in vitro in reactions containing hepatocyte lysate (< 2 mg of protein/mL prepared from samples previously identified as rapid, intermediate, or slow NAT2 acetylator genotypes), 300 µmol/L INH or N-(4-aminobenzoyl)-L-glutamic acid (ABG) and 1 mmol/L acetyl coenzyme A (AcCoA) were incubated at 37 °C. Reactions were terminated by the addition of 1/10 volume of 1 mol/L acetic acid. The reaction tubes were centrifuged to precipitate protein. The amount of acetyl-product produced was determined following separation and quantitation by high performance liquid chromatography (HPLC). Separation of INH and acetyl-INH was accomplished using a 125 mm × 4 mm Lichrosher 100 RP-100 5 µm C18 HPLC column eluted with an isocratic gradient of 86% 25 mmol/L sodium phosphate, 10 mmol/L heptane sulfonate pH 3.0, 14% acetonitrile. Acetyl-INH was quantitated by measuring the absorbance at 266 nm. Retention times for INH and acetyl-INH were 2.63 and 2.93 min, respectively. The amount of acetyl-N-(4-aminobenzoyl)-L-glutamic acid produced was determined following separation and quantitation by HPLC. Separation of N-(4-aminobenzoyl)-L-glutamic acid and acetyl-N-(4-aminobenzoyl)-L-glutamic acid was accomplished using the same HPLC column described above eluted with a gradient of 100% 20 mmol/L sodium perchlorate pH 2.5/0% acetonitrile to 50% 20 mmol/L sodium perchlorate pH 2.5/50% acetonitrile over 5 min. N-(4-Aminobenzoyl)-L-glutamic acid and acetyl-N-(4-aminobenzoyl)-L-glutamic acid were quantitated by measuring the absorbance at 280 nm. Retention times were 8.17 and 8.83 min, respectively. The NAT2 genotypes gender and ethnicity of the samples are shown in Table 1.
2.4. Measurement of INH N-acetylation in situ
Plateable cryopreserved human hepatocyte samples (different from those used above for in vitro investigations) previously identified as rapid, intermediate, or slow NAT2 acetylator genotypes were thawed as described above and contents of the vial were transferred into a 15 mL conical tube containing 12 mL of InVitroGRO CP (Bioreclamation IVT) media pre-warmed to 37 °C. Cells (1.0 mL/well) were plated into Biocoat® collagen-coated 12-well plates (BD labware, Bedford, MA, USA) and allowed to attach overnight. The next morning media was removed (dead cells, cell debri and non-adherent live cells were washed away and removed) and attached cells washed with 1 × PBS and replaced with fresh pre-warmed InVitroGRO CP media containing 12.5–200 µmol/L INH. Hepatocytes were incubated for up to 48 h after which media was removed and protein precipitated by addition of 1/10 volume of 1 mol/L acetic acid. Media was centrifuged at 15,000 × g for 10 min and supernatant used to quantitate acetyl-INH as described above. The determination of the number of live cells per sample were determined after drug incubation was complete and were counted using Z1/Dual (Beckman, Brea, CA, USA) to get live cell numbers to determine activity in nmoles of acetylated-product over 24 h per million cells.
2.5. Statistical analysis
Differences in N-acetylation rates among rapid, intermediate, and slow NAT2 acetylator genotypes were tested for significance by one way analysis of variance across rapid, intermediate, and slow NAT2 genotypes followed when significant (P < 0.05) by Tukey-Kramer multiple comparisons tests between rapid, intermediate, and slow NAT2 genotypes.
3. Results
3.1. N-Acetyltransferase activity in vitro
N-acetylation rates of ABG in vitro, a substrate highly selective for human N-acetyltransferase 1 versus NAT223 did not differ significantly (P > 0.05) between rapid, intermediate, and slow NAT2 acetylator cryopreserved human hepatocytes (Fig. 1). In contrast, N-acetylation rates towards INH in the identical samples of cryopreserved human hepatocytes from rapid, intermediate, and slow NAT2 acetylators exhibited a robust and highly significant (P < 0.005) NAT2 genotype-dependent metabolism (Fig. 1).
Figure 1.
In vitro N-acetyltransferase catalytic activities towards INH (A) and N-(4-aminobenzoyl)-L-glutamic acid (B) in cryopreserved human hepatocytes. Individual data points represent different human hepatocyte samples. Error bars illustrate mean ± S.E.M. in rapid, intermediate, and slow acetylators (n = 5). INH N-acetyltransferase activities differed significantly (P < 0.005) between rapid, intermediate, and slow acetylators (rapid vs. intermediate P < 0.05; rapid vs. slow P < 0.01) whereas N-(4-aminobenzoyl)-L-glutamic acid N-acetyltransferase activities did not (P > 0.05).
3.2. N-acetylation capacity in situ
N-acetylation of INH in situ was shown to be both INH concentration- and time-dependent up to concentrations of 100 µmol/L INH (Fig. 2) and 48 h incubation (Fig. 3) in the cryopreserved human hepatocytes. As shown in Fig. 4, robust and significant NAT2 genotype-dependent patterns (rapid > intermediate > slow) were exhibited for the N-acetylation of INH in situ following 24 h incubations with either 12.5 (P = 0.0023) or 100 (P = 0.0002) µmol/L INH. Cytotoxicity of isoniazid over 24 h was tested by Alamar Blue and IC50 was determined to be greater than 1 mmol/L for both rapid and slow NAT2 acetylator hepatocytes which is much higher than the concentration used for the N-acetylation assays.
Figure 2.
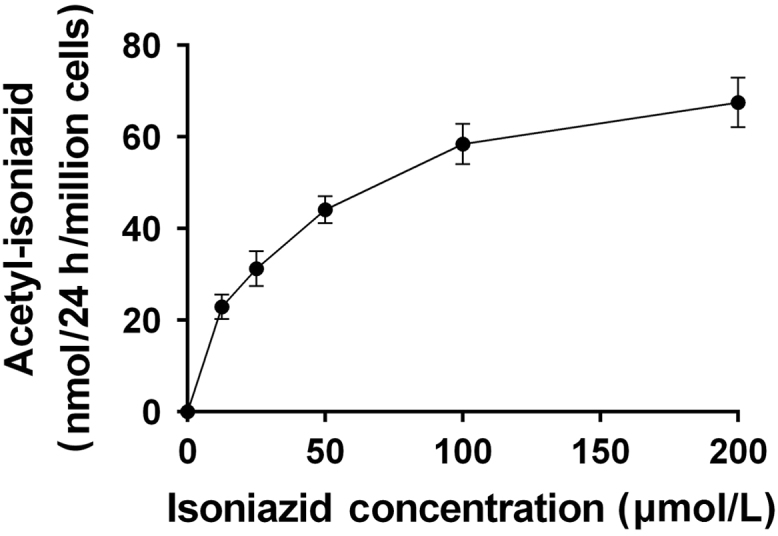
Concentration-dependent N-acetylation of INH in cryoplateable human hepatocytes. Cryoplateable human hepatocytes with a rapid NAT2 genotype were plated on collagen coated 12-well plate and allowed to attach. After 24 h plating media was removed and replaced with media containing (0–200 µmol/L) INH for 24 h. The amount of acetyl-INH produced was measured in the media. Each data point is mean ± S.E.M. for three independent measurements. Data shows concentration-dependent increase in the production of INH.
Figure 3.
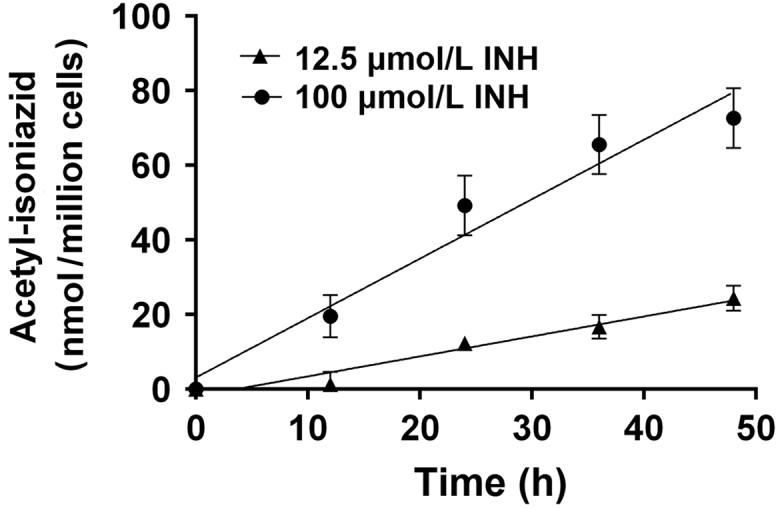
Time-dependent N-acetylation of INH in cryoplateable human hepatocytes. Cryoplateable human hepatocytes with a rapid NAT2 genotype were plated on collagen coated 12-well plate and allowed to attach. After 24 h plating media was removed and replaced with media containing 12.5 or 100 µmol/L INH. Cells were incubated with media containing INH for 0–48 h. The amount of acetyl-INH produced was measured in the media. Each data point is mean ± S.E.M. for three independent measurements. Data shows R-squared for goodness of fit for the time-dependent increase in the production of acetylated-INH was 0.9612 and 0.9599 for 12.5 and 100 µmol/L, respectively.
Figure 4.
N-Acetylation of INH by cryoplateable human hepatocytes with rapid, intermediate or slow NAT2 acetylator genotypes. Individual data points represent different human hepatocyte samples. Error bars illustrate mean ± S.E.M. acetyl-INH levels in rapid, intermediate and slow acetylators (n = 5), following 24 h cell culture with 12.5 or 100 µmol/L INH. The levels of acetyl-INH differed significantly between the acetylator genotypes following incubation with 12.5 µmol/L (P =0.0023; rapid vs. intermediate P < 0.05; intermediate vs. slow P < 0.01) or 100 µmol/L (P = 0.0002; rapid vs intermediate P < 0.05; intermediate vs. slow P < 0.001) INH.
4. Discussion
Robust NAT2 genotype-dependent N-acetylation of INH was noted in human cryopreserved hepatocytes both in vitro and in situ following incubations at different concentrations of INH. The INH NAT2 enzyme activities in the rapid, intermediate, and slow NAT2 genotype hepatocytes in vitro was quite consistent with NAT2 enzyme activities in cryopreserved human hepatocytes reported previously towards arylamine drugs including sulfamethazine and solithromycin and carcinogens including 4-aminobiphenyl18, 19.
Although recent meta-analyses on the role of NAT2 acetylator polymorphism in INH hepatoxicity have reported increased risk among slow NAT2 acetylators, they have not as yet reported differences in risk between rapid and intermediate acetylators. Although the results of our study show significant differences in the N-acetylation of INH between rapid and intermediate acetylator hepatocytes both in vitro and in situ, the magnitude of the differences between rapid and intermediate acetylators were smaller than between intermediate and slow acetylators. Previous human in vivo studies have reported tri-modal distributions of INH N-acetylation reflective of rapid, intermediate, and slow NAT2 acetylator phenotypes24, 25, 26, 27, 28, 29.
A study conducted in Brazil found that the incidence of INH-induced hepatotoxicity differed among rapid (2.9%), intermediate (9.8%), and slow (22%) NAT2 acetylators30. Other studies also have shown an effect of NAT2 genotype on INH efficacy31, 32 and pharmacogenetic-based therapy for tuberculosis has been proposed33, 34, 35, 36, 37. Isoniazid concentrations of 3–6 μg/mL are the usual target for tuberculosis therapy36, 37. Our findings in cryopreserved human hepatocytes following incubations with 12.5 (1.71 µg/mL) and 100 (13.7 µg/mL) µmol/L INH reflect this therapeutic range of INH concentrations supporting use of NAT2 genotype in isoniazid dosing strategies (i.e., rapid > intermediate> slow acetylators). An NAT2 genotype-guided regime (INH dose 7.5 mg/kg for rapid acetylators; 5.0 mg/kg for intermediate acetylators; 2.5 mg/kg for slow acetylators) reduced INH-induced liver injury and -early treatment failure in a randomized controlled trial38. Although it would be an over-extension to conclude that quantitative INH dosing strategies should be determined by INH N-acetylation rates in cryopreserved human hepatocytes, the results of the present study provide experimental metabolic support towards the use of NAT2 genotype to guide INH dosing strategies in the treatment and prevention of tuberculosis.
Acknowledgments
This research project was supported by National Institutes of Health grants R25-CA134283 and P20-GM113226 (USA). We thank Timothy Moeller and Bioreclamation IVT (Baltimore, MD, USA) for valuable contributions towards this study. The authors declare no conflict of interest in this work.
Footnotes
Peer review under responsibility of Institute of Materia Medica, Chinese Academy of Medical Sciences and Chinese Pharmaceutical Association.
References
- 1.Weber W.W., Hein D.W. N-Acetylation pharmacogenetics. Pharmacol Rev. 1985;37:25–79. [PubMed] [Google Scholar]
- 2.McDonagh E.M., Boukouvala S., Aklillu E., Hein D.W., Altman R.B., Klein T.E. PharmGKB summary: very important pharmacogene information for N-acetyltransferase 2. Pharmacogenet Genom. 2014;24:409–425. doi: 10.1097/FPC.0000000000000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weber W.W., Hein D.W. Clinical pharmacokinetics of isoniazid. Clin Pharmacokinet. 1979;4:401–422. doi: 10.2165/00003088-197904060-00001. [DOI] [PubMed] [Google Scholar]
- 4.Klein D.J., Boukouvala S., McDonagh E.M., Shuldiner S.R., Laurieri N., Thorn C.F. PharmGKB summary: isoniazid pathway, pharmacokinetics. Pharmacogenet Genom. 2016;26:436–444. doi: 10.1097/FPC.0000000000000232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization Global Tuberculosis Report; 2016. Available from: 〈http://www.who.int/tb/publications/global_report/en/〉 [Accessed 21 January 2017].
- 6.Hayashi P.H., Fontana R.J., Chalasani N.P., Stolz A.A., Talwalkar J.A., Navarro V.J. Under-reporting and poor adherence to monitoring guidelines for severe cases of isoniazid hepatotoxicity. Clin Gastroenterol Hepatol. 2015;13:1676–1682. doi: 10.1016/j.cgh.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du H., Chen X., Fang Y., Yan O., Xu H., Li L. Slow N-acetyltransferase 2 genotype contributes to anti-tuberculosis drug-induced hepatotoxicity: a meta-analysis. Mol Biol Rep. 2013;40:3591–3596. doi: 10.1007/s11033-012-2433-y. [DOI] [PubMed] [Google Scholar]
- 8.Shi J., Xie M., Wang J., Xu Y., Liu X. Susceptibility of N-acetyltransferase 2 slow acetylators to antituberculosis drug-induced liver injury: a meta-analysis. Pharmacogenomics. 2015;16:2083–2097. doi: 10.2217/pgs.15.144. [DOI] [PubMed] [Google Scholar]
- 9.Wang P., Pradhan K., Zhong X.B., Ma X. Isoniazid metabolism and hepatotoxicity. Acta Pharm Sin B. 2016;6:384–392. doi: 10.1016/j.apsb.2016.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell J.R., Thorgeirsson U.P., Black M., Timbrell J.A., Snodgrass W.R., Potter W.Z. Increased incidence of isoniazid hepatitis in rapid acetylators: possible relation to hydranize metabolites. Clin Pharmacol Ther. 1975;18:70–79. doi: 10.1002/cpt197518170. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell J.R., Zimmerman H.J., Ishak K.G., Thorgeirsson U.P., Timbrell J.A., Snodgrass W.R. Isoniazid liver injury: clinical spectrum, pathology, and probable pathogenesis. Ann Int Med. 1976;84:181–192. doi: 10.7326/0003-4819-84-2-181. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto T., Suou T., Hirayama C. Elevated serum aminotransferase induced by isoniazid in relation to isoniazid acetylator phenotype. Hepatology. 1986;6:295–298. doi: 10.1002/hep.1840060223. [DOI] [PubMed] [Google Scholar]
- 13.Huang Y.S. Recent progress in genetic variation and risk of antituberculosis drug-induced liver injury. J Chin Med Assoc. 2014;77:169–173. doi: 10.1016/j.jcma.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Gomez-Lechon M.J., Donato M.T., Castell J.V., Jover R. Human hepatocytes as a tool for studying toxicity and drug metabolism. Curr Drug Metab. 2003;4:292–312. doi: 10.2174/1389200033489424. [DOI] [PubMed] [Google Scholar]
- 15.Hengstler J.G., Ringel M., Biefang K., Hammel S., Milbert U., Gerl M. Cultures with cryopreserved hepatocytes: applicability for studies of enzyme induction. Chem Biol Interact. 2000;125:51–73. doi: 10.1016/s0009-2797(99)00141-6. [DOI] [PubMed] [Google Scholar]
- 16.Hengstler J.G., Utesch D., Steinberg P., Platt K.L., Diener B., Ringel M. Cryopreserved primary hepatocytes as a constantly available in vitro model for the evaluation of human and animal drug metabolism and enzyme induction. Drug Metab Rev. 2000;32:81–118. doi: 10.1081/dmr-100100564. [DOI] [PubMed] [Google Scholar]
- 17.Alexandre E., Viollon-Abadie C., David P., Gandillet A., Coassolo P., Heyd B. Cryopreservation of adult human hepatocytes obtained from resected liver biopsies. Cryobiology. 2002;44:103–113. doi: 10.1016/s0011-2240(02)00011-1. [DOI] [PubMed] [Google Scholar]
- 18.Doll M.A., Zang Y., Moeller T., Hein D.W. Codominant expression of N-acetylation and O-acetylation activities catalyzed by N-acetyltransferase 2 in human hepatocytes. J Pharmacol Exp Ther. 2010;334:540–544. doi: 10.1124/jpet.110.168567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hein D.W., Doll M.A. Role of N-acetylation polymorphism in solithromycin metabolism. Pharmacogenomics. 2017 doi: 10.2217/pgs-2017-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hein D.W., Doll M.A. Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics. 2012;13:31–41. doi: 10.2217/pgs.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hein D.W. N-Acetyltransferase SNPs: emerging concepts serve as a paradigm for understanding complexities of personalized medicine. Expert Opin Drug Metab Toxicol. 2009;5:353–366. doi: 10.1517/17425250902877698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doll M.A., Hein D.W. Comprehensive human NAT2 genotype method using single nucleotide polymorphism-specific polymerase chain reaction primers and fluorogenic probes. Anal Biochem. 2001;288:106–108. doi: 10.1006/abio.2000.4892. [DOI] [PubMed] [Google Scholar]
- 23.Minchin R.F. Acetylation of p-aminobenzoylglutamate, a folic acid catabolite, by recombinant human arylamine N-acetyltransferase and U937 cells. Biochem J. 1995;307:1–3. doi: 10.1042/bj3070001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parkin D.P., Vandenplas S., Botha F.J., Vandenplas M.L., Seifart H.I., van Helden P.D. Trimodality of isoniazid elimination: phenotype and genotype in patients with tuberculosis. Am J Respir Crit Care Med. 1997;155:1717–1722. doi: 10.1164/ajrccm.155.5.9154882. [DOI] [PubMed] [Google Scholar]
- 25.Smith C.A., Wadelius M., Gough A.C., Harrison D.J., Wolf C.R., Rane A. A simplified assay for the arylamine N-acetyltransferase 2 polymorphism validated by phenotyping with isoniazid. J Med Genet. 1997;34:758–760. doi: 10.1136/jmg.34.9.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B., Zhang W.X., Cai W.M. The influence of various genotypes on the metabolic activity of NAT2 in a Chinese population. Eur J Clin Pharmacol. 2006;62:355–359. doi: 10.1007/s00228-006-0110-6. [DOI] [PubMed] [Google Scholar]
- 27.Singh N., Dubey S., Chinnaraj S., Golani A., Maitra A. Study of NAT2 gene polymorphisms in an Indian population: association with plasma isoniazid concentration in a cohort of tuberculosis patients. Mol Diag Ther. 2009;13:49–58. doi: 10.1007/BF03256314. [DOI] [PubMed] [Google Scholar]
- 28.Bing C., Xiaomeia C., Jinhenga L. Gene dose effect of NAT2 variants on the pharmacokinetics of isoniazid and acetylisoniazid in healthy Chinese subjects. Drug Metab Drug Interact. 2011;26:113–118. doi: 10.1515/DMDI.2011.016. [DOI] [PubMed] [Google Scholar]
- 29.Kiser J.J., Zhu R., D׳Argenio D.Z., Cotton M.F., Bobat R., McSherry G.D. Isoniazid pharmacokinetics, pharmacodynamics, and dosing in South African infants. Ther Drug Monit. 2012;34:446–451. doi: 10.1097/FTD.0b013e31825c4bc3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teixeira R.L., Morato R.G., Cabello P.H., Muniz L.M., Moreira Ada S., Kritski A.L. Genetic polymorphisms of NAT2, CYP2E1 and GST enzymes and the occurrence of antituberculosis drug-induced hepatitis in Brazilian TB patients. Memorias Inst Oswaldo Cruz. 2011;106:716–724. doi: 10.1590/s0074-02762011000600011. [DOI] [PubMed] [Google Scholar]
- 31.Donald P.R., Sirgel F.A., Venter A., Parkin D.P., Seifart H.I., van de Wal B.W. The influence of human N-acetyltransferase genotype on the early bactericidal activity of isoniazid. Clin Infect Dis. 2004;39:1425–1430. doi: 10.1086/424999. [DOI] [PubMed] [Google Scholar]
- 32.Pasipanodya J.G., Srivastava S., Gumbo T. Meta-analysis of clinical studies supports the pharmacokinetic variability hypothesis for acquired drug resistance and failure of antituberculosis therapy. Clin Infect Dis. 2012;55:169–177. doi: 10.1093/cid/cis353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinzig-Schippers M., Tomalik-Scharte D., Jetter A., Scheidel B., Jakob V., Rodamer M. Should we use N-acetyltransferase type 2 genotyping to personalize isoniazid doses? Antimicrob Agents Chemother. 2005;49:1733–1738. doi: 10.1128/AAC.49.5.1733-1738.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zabost A., Brzezińska S., Kozińska M., Błachnio M., Jagodziński J., Zwolska Z. Correlation of N-acetyltransferase 2 genotype with isoniazid acetylation in Polish tuberculosis patients. Biomed Res Inter. 2013;2013:853602. doi: 10.1155/2013/853602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto T., Ohno M., Azuma J. Future of pharmacogenetics-based therapy for tuberculosis. Pharmacogenomics. 2014;15:601–607. doi: 10.2217/pgs.14.38. [DOI] [PubMed] [Google Scholar]
- 36.Jung J.A., Kim T.E., Lee H., Jeong B.H., Park H.Y., Jeon K. A proposal for an individualized pharmacogenetic-guided isoniazid dosage regimen for patients with tuberculosis. Drug Des Dev Ther. 2015;9:5433–5438. doi: 10.2147/DDDT.S87131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi R., Jeong B.H., Koh W.J., Lee S.Y. Recommendations for optimizing tuberculosis treatment: therapeutic drug monitoring, pharmacogenetics, and nutritional status considerations. Ann Lab Med. 2017;37:97–107. doi: 10.3343/alm.2017.37.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azuma J., Ohno M., Kubota R., Yokota S., Nagai T., Tsuyuguchi K. NAT2 genotype guided regimen reduces isoniazid-induced liver injury and early treatment failure in the 6-month four-drug standard treatment of tuberculosis: a randomized controlled trial for pharmacogenetics-based therapy. Eur J Clin Pharmacol. 2013;69:1091–1101. doi: 10.1007/s00228-012-1429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]