Abstract
Here, we describe a novel specific telomere-associated protein: TZAP (Telomeric Zinc finger-Associated Protein). TZAP binds preferentially to long telomeres that have a low concentration of shelterin complex, in virtue of competition with the telomeric repeat binding factors TRF1 and TRF2. When localized at telomeres, TZAP triggers “telomere trimming”, a process that results in the rapid deletion of telomeric repeats. Based on these results, we propose a novel model for telomere length regulation in mammalian cells. Binding of TZAP to telomeres is restricted to long telomeres and represents the switch that triggers telomere trimming, setting the upper limit of telomere length. In this model, the reduced concentration of the shelterin complex at long telomeres results in TZAP binding and initiation of telomere trimming.
Keywords: Telomere, Shelterin, Telomere Length, ZBTB48, Telomere Trimming
Main Text
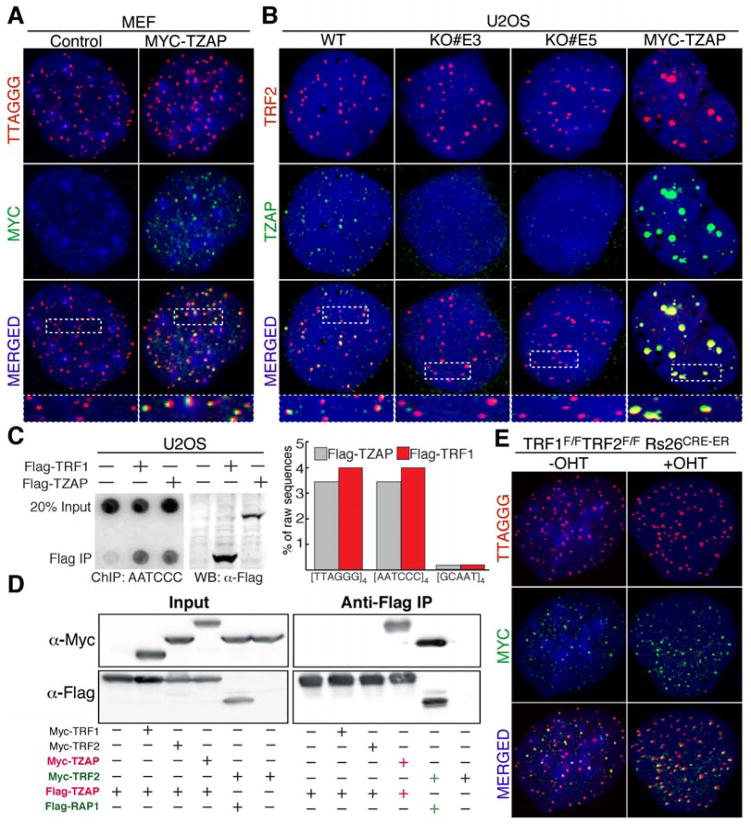
Telomere length homeostasis is essential for proper cellular function (1–3). The telomere proteome consists of ~200 proteins that have been associated with different aspects of telomere biology, including telomere protection, telomeric DNA synthesis, and telomere elongation (4–8). Here, we describe the characterization of the Kruppel-like zinc finger protein ZBTB48 as a telomere-associated factor involved in telomere length regulation. Based on the telomere-specific localization of ZBTB48 (Fig. 1A–C and Fig. S1A–C), we renamed this factor as Telomeric Zinc-finger Associated Protein (TZAP). Our data show that TZAP binds to telomeres in telomerase positive cells as well as telomerase negative cells (Fig. 1A–B and Fig. S1A–C). Using a rabbit polyclonal antibody raised against human TZAP, we confirm that endogenous TZAP resides at telomeres (Fig. 1B). The specificity of this antibody was verified using TZAP−/− U2OS clones generated by CRISPR/Cas9 genome editing (Fig. 1B and Fig. S1D–F). Using chromatin immunoprecipitation (ChIP) followed by next generation sequencing (ChIP-seq), we confirmed that TZAP associates with telomeres in U2OS cells to a level that is comparable to TRF1 (Fig. 1C). Notably, TZAP was not significantly enriched at any genetic locus beside telomeres in U2OS cells. To date, the only proteins known to uniquely and specifically associate with TTAGGG are the components of the shelterin complex. Remarkably, our data indicate that TZAP binds to telomeres independently from the shelterin complex, as shown by the fact that it does not interact with members of this complex (Fig.1D and Fig. S1G, S1H), and by its localization to telomeres in “shelterin free” MEFs (9) (Fig. 1E, Fig. S1I).
Figure 1.
TZAP, a novel telomere-associated protein. (A) FISH-IF showing localization of MYC-TZAP (green) at telomeres (Red) in MEFs. (B) IF showing endogenous TZAP at telomeres in U2OS cells. TZAP knockout clones (KO# E3 and KO# E5) and overexpressing cells are used to confirm staining specificity. (C) ChIP shows association of FLAG-TRF1 or FLAG-TZAP to telomeric DNA in U2OS cells. The bars represent the fraction of raw sequences containing 4 repeats of the indicated sequences from the total reads obtained by ChIP-seq analysis. (D) FLAG Immunoprecipitation was performed from lysates of HEK293T cells expressing the indicated Flag-tagged and Myc-tagged constructs. (E) MYC-TZAP localizes at telomeres in TRF1F/FTRF2F/F Rs26CRE-ER MEFs either untreated (-OHT) or 4 days post tamoxifen (+OHT) treatment.
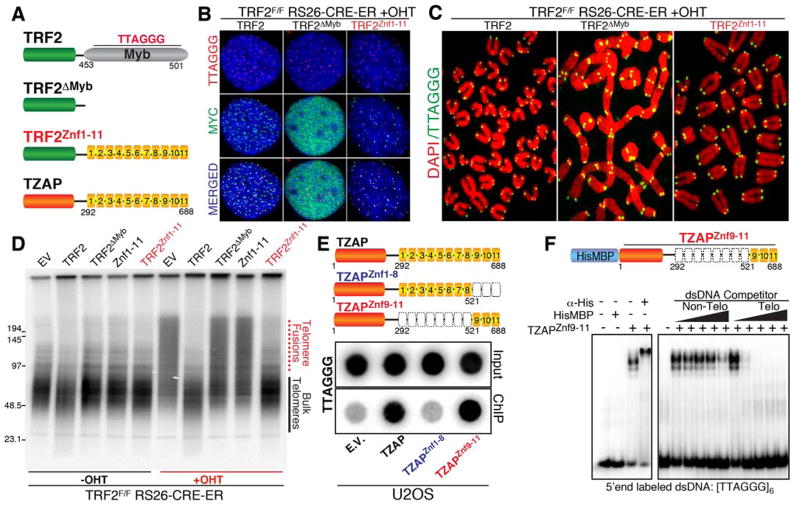
To test whether TZAP binds directly to TTAGGG repeats, we used a domain swap approach aimed at assessing whether the Zinc finger domains of TZAP could replace the DNA binding domain of the telomeric protein TRF2. We generated TRF2F/F, Rs26-CRE-ER MEFs expressing the following Myc-tagged constructs: full length TRF2 (TRF2), a TRF2 allele lacking its TTAGGG binding domain (TRF2ΔMyb), and a chimeric protein containing TRF2ΔMyb fused to the 11 zinc fingers of TZAP (TRF2Znc1–11) (Fig. 2A). As expected, in TRF2-null conditions (+OHT), full-length TRF2 localizes at telomeres while the TRF2ΔMyb allele does not (Fig. 2B). Strikingly, the TRF2Znc1–11 chimera allele localizes at telomeres (Fig. 2B) and functionally complements the loss of TRF2 in suppressing chromosome end-to-end fusions (Fig. 2C–D). Next, using an array of truncation mutants, we found that the three terminal zinc finger domains of TZAP (Znf9–11) are both required and sufficient for telomeric localization (Fig. 2E, Fig. S2E–I, Fig. S3A–D). Finally, using purified recombinant TZAP znf9–11 harboring a Hi6xs-MBP tag (Fig. 2F, Fig. S3E), we show by electrophoretic mobility shift assay (EMSA) that TZAPznf9–11 exhibits binding affinity specific to dsTTAGGG repeats in vitro (Fig. 2F, Fig. S3F). Collectively, these data show that TZAP is a novel telomere-associated protein that directly binds dsTTAGGG repeats via zinc finger domains.
Figure 2.
The Zinc Finger Domains (Znf) of TZAP directly bind telomeric repeats. (A) Schematic representation of the constructs used to show that the Zinc finger domains of TZAP are sufficient for telomeric localization. (B) IF-FISH for telomeric localization of the indicated constructs in TRF2 null MEFs (C) Metaphase spreads derived from TRF2 null MEFs expressing the indicated constructs were stained for telomeric DNA (green) and DAPI (red). (D) Genomic DNA isolated from TRF2 proficient (-OHT) or TRF2 null (+OHT) MEFs expressing the indicated constructs was digested with MboI, resolved on a pulsed field gel, and probed using radioactive telomeric probe. (E) The terminal Zinc finger domains (#9,10 and 11) of TZAP are required for telomeric localization as shown by Flag-ChIP in U2OS cells expressing the indicated Flag-tagged constructs. (F) A recombinant HisMBP-tagged TZAPZnf9–11 allele was expressed in bacteria, purified and used in gel shift assays. Left panel: HisMBP-TZAPZnf9–11 binds telomeric repeats and is supershifted by a His-tag antibody. Right panel: competition assay using increasing amounts (0x, 1x, 10x, 100x, 1000x fold molar excess) of cold non-telomeric or telomeric competitor dsDNA.
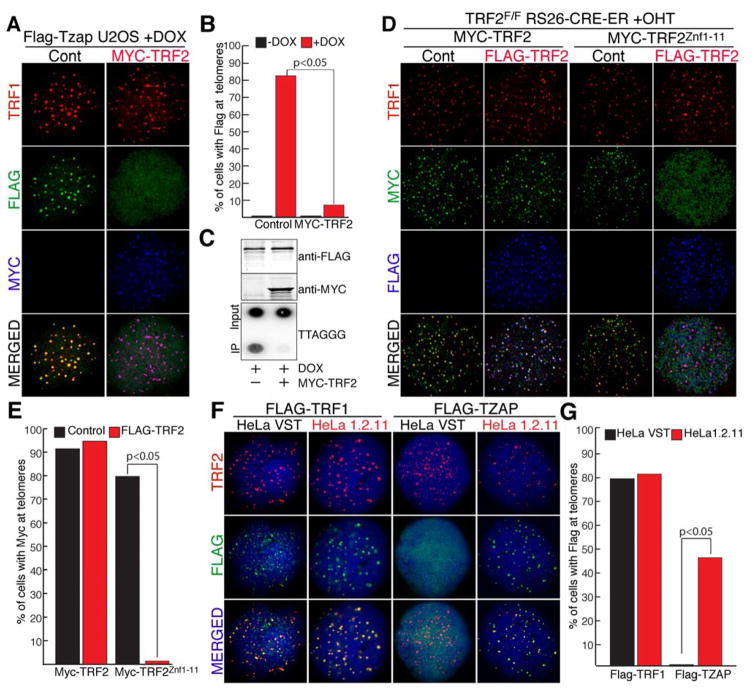
We next explored whether TZAP and TRFs compete for localization to telomeres. To test this hypothesis, we generated a U2OS cell line with doxycycline-inducible expression of Flag-TZAP (Flp-IN T-REX Flag-TZAP) (Fig. 3A). Strikingly, overexpression of TRF2 in these cells caused a drastic reduction in the localization of TZAP to telomeres, as shown by IF and ChIP (Fig. 3A–C). In contrast, induction of TZAP did not affect the localization of TRF2 to telomeres (Fig. S4A). We confirmed these data in MEFs and found that TRF2 overexpression displaces TZAP (Fig. S4D–F) as well as well as the TRF2Znc1–11 chimera (Fig. 3D–E, Fig. S4B), but not the shelterin component TRF1. Quantification of the protein levels of shelterin show that the abundance of this complex does not change in relation to telomere length (10). As a result, cells with long telomeres have a lower density of the shelterin complex compared to cells with shorter telomeres, a difference that is not detected by ChIP (11). We therefore tested whether TZAP preferentially binds long telomeres with low shelterin density. To test this hypothesis, we transduced two subclones of HeLa cells that differ in telomere length, HeLa VST (for Very Short Telomeres) with an average telomere length of approximately 5 Kb and HeLa 1.2.11 that have longer telomeres (approx. 20Kb). Strikingly, while TZAP readily localizes to telomeres in HeLa 1.2.11 cells, no distinguishable TZAP localization is detected in HeLa VST cells (Fig. 3F–G). In agreement with this, an inverse correlation between telomere length and TZAP localization at telomeres was observed in a wide array of cell lines, independently of their transformation status, mode of telomere length regulation, and specie of origin (Fig. S5A–B).
Figure 3.
TZAP binding to telomeres is inhibited by elevated TRF2 levels. (A) IF for doxycycline-inducible expression of Flag-TZAP at telomeres in FLP-IN T-Rex U2OS cells with and without Myc-TRF2 overexpression. TRF1 (red), anti-Flag IF (green), and anti-Myc IF (blue). (B) Quantification of the data shown in A, indicating the % of cells with doxycycline-inducible FLAG-TZAP localized at >90% of telomeres. (C) ChIP for Telomeric DNA associated with doxycycline-inducible expression of Flag-TZAP in FLP-IN T-Rex U2OS cells with and without MYC-TRF2 overexpression. Immunoblot of flag-tagged and myc-tagged proteins from cells used for ChIP. (D) IF for Myc-TRF2 or Myc-TRF2Znf1–11 ectopically-expressed in TRF2F/F Rs26CRE-ER MEFs at day 4 post tamoxifen treatment (+OHT) with or without overexpression of Flag-TRF2. Anti-TRF1 (red), anti-Myc (green), and anti-Flag (blue). (E) Quantification of data shown in D, indicating the % of cells with Myc localized at >90% of telomeres. (F) Localization of Flag-TRF1 or Flag-TZAP in HeLa cells with very short telomeres (HeLa VST) or with long telomeres (HeLa 1.2.11). Anti-TRF2 (red), anti-Flag (green), and DAPI (blue) (G) Quantification of the data shown in D, indicating the % of cells with Flag localized at >90% of telomeres.
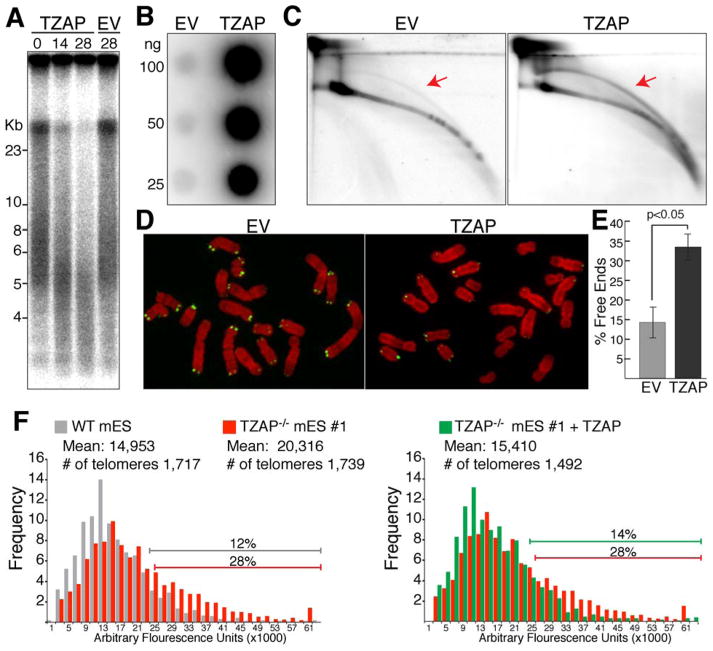
Based on its localization at long telomeres, we reasoned that TZAP might play a role in telomere length regulation. Over-expression of TZAP in U2OS cells confirmed this hypothesis showing a progressive reduction in telomere length (Fig. 4A) that culminated in the accumulation of telomere-free chromosome ends (Fig.4D–E). Since U2OS are telomerase negative these data suggest that TZAP promotes telomere rapid deletion, a mechanism of telomere length regultation identified in yeast (12) and conserved in plants (13) and mammals (14–16). Telomere trimming involves the deletion of the secondary telomeric structures known as T-loops (17), leading to telomere shortening and accumulation of Extra Chromosomal Telomeric DNA (ECT-DNA) (14–16). Strikingly, TZAP overexpression results in the accumulation of ECT-DNA as shown by single stranded C-circle assay (C-circle), telomeric circle (T-circle) assay as well as 2D-gels (18–20) (Fig. 4B–C and Fig. S6A, S6D, S6E, S6G–H). Moreover, TZAP overexpression induces ALT-associated PML-Nuclear bodies (APBs) formation, a phenotype associated with telomere trimming (14) (fig S6I–K). Conversely, reduction of the levels of TZAP resulted in a reduction of ECT-DNA in U2OS and GM847 cells (Fig. S6B–H). In conclusion, our data show that alteration in the levels of TZAP at telomeres controls the level of telomere trimming leading to significant changes in telomere length and presence of ECT-DNA.
Figure 4.
TZAP contributes to telomere length homeostasis promoting telomere trimming. (A) Telomere Restriction Fragments analysis of U2OS cells ectopically expressing the indicated constructs at the indicated time points post infection (days). (B) C-Circle analysis to detect extra chromosomal telomeric DNA (ECT-DNA) in U2OS cells expressing the indicated constructs at 6 days post infection. (C) 2-D electrophoresis to detect ECT-DNA in U2OS cells expressing the indicated constructs at 6 days post infection. Gels were denatured and probed with a telomeric probe, arrows indicate circular telomeric species. (D) Metaphase spreads of U2OS cells infected with the indicated constructs analyzed by FISH. Telomeric PNA probe (green) and DAPI (red). (E) Quantification of Chromosome ends lacking detectable telomeric signal, indicative of telomere shortening. Bars average quantification from three independent experiments. (F) Q-FISH analysis of mES cells of the indicated genotypes either with or without lentiviral infection of TZAP.
Telomere trimming has been shown to play a significant role in the control of telomere length in embryonic stem (ES) (15, 16) cells. To define whether loss of TZAP would affect telomere length homeostasis in these cells, we generated multiple clones of mouse ES (mES) cells depleted of TZAP by CRISPR/CAS9 gene editing (Fig. S7A–B). These TZAP−/− cells showed significant telomere elongation as measured by telomere restriction fragment analysis as well as q-FISH (Fig. 4). Complementation of three independent TZAP−/− mES clones with exogenous TZAP restored telomere length back to lower threshold (Fig. 4F and Fig. S7H). When measuring C-circle abundance, we found that four out of five independent TZAP−/− clonal cell lines show reduced levels of ECT-DNA (Fig. S7D–E).
In conclusion, we propose a model for the function of TZAP at telomeres in which TZAP is preferentially recruited to long telomeres containing reduced concentration of the shelterin (Fig. S8). In these settings, we find that the binding of TZAP to telomeres initiates telomere trimming, a process that prevents the accumulation of aberrantly long telomeres. Interestingly, in agreement with our data, TZAP was recently identified as a downregulated gene in mouse ES cells characterized by extremely long telomeres(21). Importantly, additional factors are likely to modulate the function of TZAP at telomeres as suggested by the fact that TZAP-mediated telomere trimming is exacerbated in ALT positive cells. As the discovery of mechanisms that control telomere elongation explains how mammalian cells bypass the end replication problem, this study provides novel insight into the mechanisms that control the upper limit of telomere length, a key determinant of lifespan and cancer susceptibility across the mammalian species (22).
Methods
Plasmids
Human cDNA for Zbtb48 was purchased from OpenBiosystems (#MHS4768-99609452), which was cloned into a pLPC retroviral vector with an N-terminal Myc or Flag tag. Mouse cDNA for Zbtb48 was derived from mouse (c57/bl6) testis and cloned into a pHAGE2 lentiviral vector with an N-terminal Myc or Flag tag. Flag-TZAP was cloned into pcDNA5/FRT/TO (Addgene) to generate doxycycline-inducible TZAP in F LP-IN T-Rex U2OS cells. Other plasmids used in this study were: pLPC-Myc-TRF1, pLPC-MYC-hTRF2, pLPC-Flag-mTin2 (kindly provided by Titia de Lange, Rockefeller University), pCDNA5-H1-sgRNA (Addgene), px330–U6-Chimeric-BB-CBh-hSpCas9n (Addgene), pOG44 (Addgene), pCDNA-FRT-TO (Addgene).
Cells
Cell lines were cultured in Glutamax-DMEM (Life Technologies) supplemented with 10% fetal bovine serum, at 5% CO2 and 3% O2. U2OS and IMR90 cells were obtained from ATCC. HeLa 1.2.11 and HeLa VST cells were previously described (23), TRF2F/F Rosa26 CRE-ER MEFs were previously described (24). TRF1F/F TRF2F/F Rosa26 CRE-ER MEFs were previously described (9). HCT116 cells were kindly provided by the Azzalin lab. E14.Tg2a mouse ES cells were kindly provided by the TSRI stem cell core facility. To generate doxycycline-inducible TZAP U2OS cells, pcDNA5/FRT/TO-Flag-Tzap was cotransfected with pOG44 into host FLP-IN T-Rex U2OS followed by selection in Hygromycin (200ug/ml) & Blasticidin (5ug/ml). To induce protein expression in these cells, 5ug/ml doxycycline was added to the culture medium for 24h. To generate stable cell lines with ectopic expression of myc-tagged or flag-tagged plasmids, retroviral virus were produced in pheonix cells and used to infect target cell lines. To generate TZAP overexpressing mES cells, a pHAGE lentiviral plasmid with N-terminal Myc tagged TZAP was transfected into HEK293Ts, and virus produced was used to infect target mES cells. Plasmid transfections were generally carried out using TransIT-LT1 (Mirus) transfection reagent, Lipofectamine 2000 (Invitrogen), or Polyethylenemine (Polysciences Inc).
Pulse-field gel electrophoresis (PFGE)
PFGE analysis was performed as previously described (24) using a BioRad CHEFIII system. Telomere length from the Telomere Restriction Fragment Analysis was deterimend using the open source software Telometric 1.2 (25).
CRISPR/CAS9 targeting
TZAP knockout cells were generated using CRISPR/Cas9 gene targeting of U2OS and mES cells via NHEJ and HR mediated repair, respectively. sg-RNAs were cloned into pCDNA5-H1-sgRNA (Addegene) and co-transfected with the plasmid px330–U6-Chimeric-BB-CBh-hSpCas9n (Addgene) encoding the Cas9-nuclease. Targeting of TZAP in U2OS cells was carried out with the following sgRNA: CTTCGTCCAGCACAGTGTGA (Exon 1). Targeting of mouse mES cells was carried out using the following sgRNA: TTCCGGTCGAATGTCCCACA (EXON 2). To introduce stop codons in mES cells by HR-mediated repair, a donor cassette was used with the following sequence: GTCGGATCCTTTAAACCTTAATTAAGCTGTTGTAG. Clones derived from single cell lines were genotyped to determine successful targeting.
IF and IF-FISH
IF was performed as described previously (24). Briefly: Cells grown on coverslips were fixed for 5 min in 2% paraformaldehyde, incubated in blocking solution for 30 minutes(1 mg/ml BSA, 3% goat serum, 0.1% Triton X-100, 1mM EDTA in PBS), followed by incubation with primary antibodies in blocking solution for 1 hour. Primary antibodies used in this study were as follows: Myc (9B11, Cell signaling, mouse monoclonal), Zbtb48 (H00003104-B01P, Abnova, mouse), Flag (F7425, Sigma-Aldrich, rabbit), TRF2 (NB110-57130, Novus Biologicals, rabbit), γ-H2AX (05-636, EMD Millipore, mouse), 53BP1 (Novus, NB 100–304, rabbit), PML (sc-966, Santa Cruz Biotechnology, mouse). Coverslips were then washed in PBS three times and incubated with secondary antibodies in blocking solution for 30min. Secondary antibodies used were Alexa 488, Alexa 555, and Alexa 647 (Molecular probes, Life Technologies). Cells were then washed in PBS three times and counterstained with 4.6-diamidino-2-phenylindole (DAPI). Coverslips were mounted on glass slides using ProLong Gold antifade (Sigma) and digital images were captured on Zeiss M1.
IF-FISH was performed as described above followed by the following procedures for FISH: After the last PBS wash, coverslips were dehydrated consecutively in 70%, 90%, and 100% ethanol for 5 min each. FITC-OO-[CCCTAA]3 labeled PNA probe (Applied Biosystems) was added to the coverslips in a buffer containing 70% formamide, 1 mg/ml blocking reagent (Roche), 10mM Tris-HCl pH 7.2, denatured at 80 degrees for 5 minutes, and hybridized for 2 hours at room temperature in the dark. The coverslips were washed twice with 70% formamide, 10mM Tris-HCl pH7.2 for 15 min each, and three times in PBS for 5 min each prior to mounting and image analysis.
Telomere-ChIP
Telomere-ChIP of Flag-tagged proteins was performed using Flag-M2 agarose beads (A2220, Sigma) with experimental procedures as previously described (26).
ChIP-Seq
ChIP-Seq for telomere associated factors was performed as described (27). Briefly, for each sample we used 107 cells, genomic DNA was isolated using genomic DNA isolation KIT (Qiagen) and each sample was independently processed into sequencing libraries with a ChIP-Seq sample preparation kit (Illumina) in accordance with the manufacturer’s instructions. The resulting purified DNA libraries were analyzed using an Illumina NextSeq instrument. The reads were aligned to the human reference genome using Bowtie 2, MACS was used to identified significant “peaks”. To determine the % of telomeric repeats from the raw data we used python to determine the fraction of raw reads containing telomeric sequences.
Co-IP
HEK293T cells were plated at 20% confluency in 10cm dishes 20–24h prior to transfection. Cells were transfected using PEI (Polyethylenimine, Linear, #9002-98-6, Polysciences Inc) with the indicated Flag and/or Myc-tagged plasmids at a 3:1 PEI to DNA ratio. 48 hours after transfection, cells were harvested by trypsinization and lysed in a buffer containing 0.1% SDS, 150mM NaCl, 1mM EDTA, 1% Triton-X, complete protease inhibitor mix (#11873580001, Roche), 1mM DTT, and 1mM PMSF. Lysates were centrifuged at maximum speed, the soluble fraction was incubated with Flag-M2 beads (A2220, Sigma) for 90 min at 4 degree, IP samples were washed in lysis buffer three times prior to elution in 2xLaemli sample. Samples were analyzed by western blot using Flag and Myc antibodies and detected by the Li-Cor imaging system (Odyssey).
Recombinant protein purification
TZAPznf9–11 was cloned into the expression vector pSV272 encoding an N-terminal 6xHis-MBP tag (kind gift from MacRae Lab). Recombinant HisMBP as well as HisMBP-TZAPZnf9–11 proteins were expressed in E. coli BL21 (DE3) competent cells. Cells were grown at 37°C in LB supplemented with 150μM zinc sulfate to an OD600 of 0.7, at which protein expression was induced with 1mM IPTG, followed by overnight growth at 18°C for 16hrs. Cells were harvested and lysed by sonication in buffer containing 20mM Tris pH 8.0 and 1M NaCl. Soluble lysate was incubated with Roche cOmplete His-Tag Purification Resin #05893682001 for 10min rotating at 4°C. Resin was washed with buffer containing 20mM Tris pH 8.0, 1M NaCl, and 5mM imidazole. Protein was eluted with buffer containing 20mM Tris pH 8.0, 0.20M NaCl, and 200mM imidazole. Elutions were analyzed for purify by SDS/PAGE.
Electrophoretic mobility shift assays
DNA binding activity of TZAP was analyzed as follows: unless indicated otherwise 40 fmol of end [γ-32P] labeled Telo-6 dsDNA template was incubated with 30 ng of recombinant HisMBP or HisMBP-TZAP in 20 μl reactions containing 20 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 100 μg/ml BSA, 5 mM DTT, 150 mM NaCl, 10% Glycerol. Reactions were incubated for 15 minutes at 23 °C before separation on a 6% polyacrylamide native gel in 0.5 xTBE for 180 min at 130 V. For the supershift experiment 1 μl of His-Tag (27E8) mouse mAb #2366 (Cell Signaling Technology) was included in the mixture.
DNA probes and competitors
The PAGE-purified Telo-6 was end labeled (γ-P32) using T4 polynucleotide kinase (NEB) and subsequently annealed with its complementary strand in a 1:5 molar ratio. Annealing was confirmed by PAGE. Cold competitor dsDNA templates (see below) were generated by annealing synthetic oligonucleotides with their corresponding reverse complementary strands. List of DNA templates used are as follows:
Doule Stranded DNAs
Telo6
GGTCGACTCTAGAGATCGAAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTAGGGTTACGATCCCGGGTACCGAGCTCGA
Non-Telo:
TCGAAGCATGGGTCGAAGCATGGGTCGAAGTCGAAGCATGGGTCGAAGCATGGGTCGAAG TCGAAGCATGGGTCGAAG
Telo-0:
GGTCGACTCTAGAGATCGAAACGTCGACTCTACCGGGTACCCCCCTCTAGAGTCGACGATC
Telo-1:
GGTCGACTCTAGAGATCGAAACGTCGACTCTAGGGTTAGGGCCCCTCTAGAGTCGACGATC
Telo-3:
GGTCGACTCTAGAGATCGAAGGGTTAGGGTTAGGGTTAGGGCCCCTCTAGAGTCGACGATC
Single Stranded oligos
ssTeloG: TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG
ssTeloC: CCCAATCCCAATCCCAATCCCAATCCCAAT
Plasmid template
270 telomeric repeats-containing plasmid used as competitor corresponds to the modified pSXneo-1.6-T2AG3 used in (28).
Metaphase-FISH
At the indicated time points, ~80% confluent cells were incubated for 30 minutes with 0.2ug/ml colcemid (Sigma). Cells were harvested by trypsinization, resuspended in 0.075M KCl at 37°C for 15 minutes, and fixed in methanol/acetic acid (3:1) O/N. Cells were dropped onto glass slides, incubated at 80°C for 5 min, and glass slides were left to dry O/N. The next day, slides were rehydrated with 1xPBS for 10 min and dehydrated consecutively with 75%, 95%, and 100% ethanol. Slides were left to air dry before applying hybridization solution (70% formamide, 1mg/ml blocking reagent (Roche), 10mM Tris-HCl pH7.2) containing FITC-OO-[CCCTAA]3 PNA probes (Applied Biosystems). Slides were denatured by heating at 80°C for 3 minutes before hybridizing for 2 hrs at room temperature. Following hybridization, slides were washed twice for 15 min each in 70% formamide/10mM Tris-HCl, followed by three 5 min washs in 0.1M Tris-HCl, pH7.0/0.15 M NaCl/0.08% Tween-20. DNA was counterstained with 4, 6-diamidino-2-phenylindole (DAPI) applied in the second wash. Slides were mounted in antifade reagent (ProLong Gold, Invitrogen) and digital images were captured on a Zeis microscope.
Q-FISH
Metaphase-FISH was performed as described above. HeLa 1.2.11 metaphases were spiked into mES metaphases as internal control in all experiments. Quantitative-FISH analysis was performed using TFL-Telo V2 image analysis software as described by Poon et al.. Arbitrary fluorescence intensity values of mES metaphases were normalized to HeLa 1.2.11 values for each slide.
Two-dimensional gel electrophoresis
C-circle assay
The C-circle assay protocol was slightly modified from that performed in Henson et al. (19). Genomic DNA was purified and digested with AluI and MboI and cleaned by phenol-chloroform extraction and precipitation. DNA was mixed with 10 μl 0.2 mg/ml BSA (NEB), 0.1 % Tween, 1mM each dATP, dGTP, dTTP, 1x phi29 Buffer (NEB) and 7.5 U phi29 DNA polymerase (NEB). Reactions were incubated for 8 h at 30°C followed by 20 min at 65°C, diluted to 60 μl with 2XSSC and dot-blotted onto a nylon membrane (Amersham Hybond- N+, GE Healthcare) previously soaked in 2XSSC. After UV-crosslinking the membrane was hybridized with a P32 end-labeled (CCCTAA)4 oligonucleotide probe at 55°C for 16 h. The membrane was then washed (3 times with 4XSSC for 30 minutes at 55°C, 1 time with 4XSSC + 0.1% SDS, 30 min 55°C), exposed and scanned using a Typhoon FLA 9500 Phosphoimager (GE Healthcare) and analyzed using ImageQuant software.
T-circle assay
The method for T-circle amplification was performed as described in (20) with some modifications. Briefly, genomic DNA was denatured at 96°C and annealed to 50 pmol of a Thio-TelC primer (AATCCC)5 for 60 min at RT in 1X annealing buffer containing 200 mM Tris-HCl pH 8.0, 200 mM KCl and 1 mM EDTA in a total volume of 20 μl. The annealed sample was divided in two and half (10 μl) was incubated at 30°C for 12 h in the presence of dNTPs (200 μM each), 0.2 mg/ml BSA (NEB), 1X phi29 Buffer (NEB) and 7.5 U phi29 DNA polymerase (NEB) in a total volume of 20 μl. The remaining half was incubated under the same conditions but excluding phi29 DNA polymerase from the reaction. Reactions were heat inactivated (65°C) for 20 min and separated on 0.6% agarose gels at 100 V for 1 h followed by 40 V 16 h in 0.5X TBE in the presence of ethidium bromide. T-circle products were visualized by in-gel hybridization using a P32 end-labeled (TTAGGG)4 oligonucleotide probe.
siRNA transfection
We used On-Target plus human ZBTB48 SMARTpool siRNA (Dharmacon, GE Healthcare; cat. L-008953-00-0005) for knockdown experiments. ON-TARGET plus Non-targeting pool (cat. D-001810-10-20) was used as siRNA control. Human TZAP (ZBTB48) targeting sequences: GCAAUGAACCGCUCGGAACA; GAUCUUUGGCCUCUUGUUG; GCUACAAGUUUACCCGACA; CAGGAGACAUUCCGCCGAA.
Two consecutive siRNA transfections were carried out as follows: ~700,000 cells were seeded in 10 cm dishes containing 9 ml growth medium in the absence of antibiotics 3hrs prior to transfection. For one transfection, 10μl DharmaFECT-1 transfection reagent (cat. #T-2001-03) and siRNAs (50nM final transfection concentration) were diluted in 1 ml Opti-MEM (Life Technologies) and added dropwise onto cells. 24 hrs later, transfection medium was replaced with complete media or cells were split as needed. 72 hours post transfection cells were harvested for downstream experiments.
Knockdown efficiency was deterimed by qPCR using the following primers:
Forward(494–513): 5′-AGCCCAGAGGCAGTCATAGT and Reverse (630–610): 5′-GCAGTCCTCGGGTTTCTTGTC.
Supplementary Material
Acknowledgments
We thank Agnel Sfeir for performing experiment involving shelterin-free cells and for comments on the manuscript. We thank Peter Wright and members of his lab for critical support in the purification of TZAP. This work was supported by a grant from the American Cancer Society (RSG-14-186-01) (E.L.D.), the Swedish Research Council International postdoc grant (D0730801) (J.M.F) and the NIH (R01GM087476, R01CA174942) (J.K.).
References
- 1.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005 Sep 15;19:2100. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 3.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006 Dec;115:413. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 4.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic Loci. Cell. 2009 Jan 9;136:175. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grolimund L, et al. A quantitative telomeric chromatin isolation protocol identifies different telomeric states. Nat Commun. 2013;4:2848. doi: 10.1038/ncomms3848. [DOI] [PubMed] [Google Scholar]
- 6.Bartocci C, et al. Isolation of chromatin from dysfunctional telomeres reveals an important role for Ring1b in NHEJ-mediated chromosome fusions. Cell Rep. 2014 May 22;7:1320. doi: 10.1016/j.celrep.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee OH, et al. Genome-wide YFP fluorescence complementation screen identifies new regulators for telomere signaling in human cells. Mol Cell Proteomics. 2011 Feb;10:M110 001628. doi: 10.1074/mcp.M110.001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xin H, Liu D, Songyang Z. The telosome/shelterin complex and its functions. Genome Biol. 2008;9:232. doi: 10.1186/gb-2008-9-9-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sfeir A, de Lange T. Removal of shelterin reveals the telomere end-protection problem. Science. 2012 May 4;336:593. doi: 10.1126/science.1218498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takai KK, Hooper S, Blackwood S, Gandhi R, de Lange T. In vivo stoichiometry of shelterin components. J Biol Chem. 2010 Jan 8;285:1457. doi: 10.1074/jbc.M109.038026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cristofari G, Lingner J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006 Feb 8;25:565. doi: 10.1038/sj.emboj.7600952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li B, Lustig AJ. A novel mechanism for telomere size control in Saccharomyces cerevisiae. Genes Dev. 1996 Jun 1;10:1310. doi: 10.1101/gad.10.11.1310. [DOI] [PubMed] [Google Scholar]
- 13.Watson JM, Shippen DE. Telomere rapid deletion regulates telomere length in Arabidopsis thaliana. Molecular and cellular biology. 2007 Mar;27:1706. doi: 10.1128/MCB.02059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickett HA, Cesare AJ, Johnston RL, Neumann AA, Reddel RR. Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 2009 Apr 8;28:799. doi: 10.1038/emboj.2009.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickett HA, Henson JD, Au AY, Neumann AA, Reddel RR. Normal mammalian cells negatively regulate telomere length by telomere trimming. Hum Mol Genet. 2011 Dec 1;20:4684. doi: 10.1093/hmg/ddr402. [DOI] [PubMed] [Google Scholar]
- 16.Pickett HA, Reddel RR. The role of telomere trimming in normal telomere length dynamics. Cell Cycle. 2012 Apr 1;11:1309. doi: 10.4161/cc.19632. [DOI] [PubMed] [Google Scholar]
- 17.Griffith JD, et al. Mammalian telomeres end in a large duplex loop. Cell. 1999 May 14;97:503. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 18.Cesare AJ, Griffith JD. Telomeric DNA in ALT cells is characterized by free telomeric circles and heterogeneous t-loops. Molecular and cellular biology. 2004 Nov;24:9948. doi: 10.1128/MCB.24.22.9948-9957.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henson JD, et al. DNA C-circles are specific and quantifiable markers of alternative-lengthening-of-telomeres activity. Nat Biotechnol. 2009 Dec;27:1181. doi: 10.1038/nbt.1587. [DOI] [PubMed] [Google Scholar]
- 20.Zellinger B, Akimcheva S, Puizina J, Schirato M, Riha K. Ku suppresses formation of telomeric circles and alternative telomere lengthening in Arabidopsis. Mol Cell. 2007 Jul 6;27:163. doi: 10.1016/j.molcel.2007.05.025. [DOI] [PubMed] [Google Scholar]
- 21.Varela E, Munoz-Lorente MA, Tejera AM, Ortega S, Blasco MA. Generation of mice with longer and better preserved telomeres in the absence of genetic manipulations. Nat Commun. 2016;7:11739. doi: 10.1038/ncomms11739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes NM, et al. Comparative biology of mammalian telomeres: hypotheses on ancestral states and the roles of telomeres in longevity determination. Aging Cell. 2011 Oct;10:761. doi: 10.1111/j.1474-9726.2011.00718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Sullivan RJ, et al. Rapid induction of alternative lengthening of telomeres by depletion of the histone chaperone ASF1. Nature structural & molecular biology. 2014 Feb;21:167. doi: 10.1038/nsmb.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okamoto K, et al. A two-step mechanism for TRF2-mediated chromosome-end protection. Nature. 2013 Feb 28;494:502. doi: 10.1038/nature11873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant JD, et al. Telometric: a tool providing simplified, reproducible measurements of telomeric DNA from constant field agarose gels. BioTechniques. 2001 Dec;31:1314. doi: 10.2144/01316bc02. [DOI] [PubMed] [Google Scholar]
- 26.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003 Jun 26;423:1013. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 27.Garrobo I, Marion RM, Dominguez O, Pisano DG, Blasco MA. Genome-wide analysis of in vivo TRF1 binding to chromatin restricts its location exclusively to telomeric repeats. Cell Cycle. 2014;13:3742. doi: 10.4161/15384101.2014.965044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunham MA, Neumann AA, Fasching CL, Reddel RR. Telomere maintenance by recombination in human cells. Nat Genet. 2000 Dec;26:447. doi: 10.1038/82586. [DOI] [PubMed] [Google Scholar]
- 29.Raices M, et al. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008 Mar 7;132:745. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 30.Nabetani A, Ishikawa F. Unusual telomeric DNAs in human telomerase-negative immortalized cells. Molecular and cellular biology. 2009 Feb;29:703. doi: 10.1128/MCB.00603-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.