Abstract
Blue nevi are common melanocytic tumors arising in the dermal layer of the skin. Similar to uveal melanomas, blue nevi frequently harbor GNAQ and GNA11 mutations. Recently, recurrent CYSLTR2 and PLCB4 mutations were identified in uveal melanomas not harboring GNAQ or GNA11 mutations. All four genes (GNAQ, GNA11, CYSLTR2, and PLCB4) code for proteins involved in the same signaling pathway, which is activated by mutations in these genes. Given the related functional consequences of these mutations and the known genetic similarities between uveal melanoma and blue nevi, we analyzed a cohort of blue nevi to investigate whether CYSLTR2 and PLCB4 mutations occur in tumors lacking GNAQ or GNA11 mutations (as in uveal melanoma). A targeted next-generation sequencing assay covering known activating mutations in GNAQ, GNA11, CYSLTR2, PLCB4, KIT, NRAS, and BRAF was applied to 103 blue nevi. As previously reported, most blue nevi were found to harbor activating mutations in GNAQ (59%, n = 61), followed by less frequent mutations in GNA11 (16%, n = 17). Additionally, one BRAF (1%) and three NRAS (3%) mutations were detected. In three tumors (3%) harboring none of the aforementioned gene alterations, CYSLTR2 mutations were identified. All three CYSLTR2 mutations were the same c.386T > A, L129Q mutation previously identified in uveal melanoma that has been shown to lead to increased receptor activation and signaling. In summary, our study identifies CYSLTR2 L129Q alterations as a previously unrecognized activating mutation in blue nevi, occuring in a mutually exclusive fashion with known GNAQ and GNA11 mutations. Similar to GNAQ and GNA11 mutations, CYSLTR2 mutations, when present, are likely defining pathogenetic events in blue nevi.
Blue nevi are relatively common, usually dermal, melanocytic proliferations of the skin.1 They are pigmented tumors, which appear blue when viewed clinically, due to their location beneath the skin surface, a physical phenomenon based on the Tyndall effect.2 The most frequent subtype are so-called common blue nevi, or Jadasohn-Tièche blue nevi, based on their original description.3 A range of other subtypes have been described,1,2,4 including cellular, epithelioid, sclerotic, and plaque-type blue nevi. Congenital dermal melanocytosis, which include Mongolian spots, nevi of Ota, and nevi of Ito4 are also included in the blue nevus group.
Most blue nevi are benign and rarely transform into malignant tumors. However, there are tumors designated atypical cellular blue nevi, the biological potential of which is difficult to determine from their clinicopathologic features. Additionally, so-called malignant blue nevi are clearly malignant melanomas and should be treated accordingly.5–8
Blue nevi show a distinct genetic profile from that of epidermal-derived common acquired nevi; the majority of the latter (80–90%) harbor activating BRAF V600E mutations,9 which are not commonly found in blue nevi. Blue nevi harbor highly recurrent activating GNAQ and GNA11 mutations, which are also found in uveal melanoma.10,11 The two activating mutations reported recurrently in blue nevi to date are GNAQ mutations (~55%) and GNA11 mutations (~7%).11 A number of studies have further demonstrated mutations or protein loss of the tumor suppressor BAP1 occurring in malignant blue nevi.5,12,13
In all studies to date, there remain tumors (eg, 35–40% of blue nevi) lacking GNAQ or GNA11 mutations, implying that they harbor other unrecognized activating oncogene mutations.5,10,11 All known gene alterations reported in blue nevus variants to date (GNAQ, GNA11, and BAP1) are also frequently found present in uveal melanomas.10,11,14–16 Recently, two studies demonstrated rarer mutations in PLCB417 and CYSLTR218 occurring in uveal melanomas lacking GNAQ or GNA11 mutations. These four mutations (GNA11, GNAQ, PLCB4, and CYSLTR2) affect different proteins, and although the functional consequences of the mutations will differ, they are also all involved in a common signaling pathway. CYSLTR2 codes for a seven transmembrane receptor (also termed G protein-coupled receptor), which demonstrates increased activation by the L129Q mutation.18 The CYSLTR2 receptor signals through the highly homologous heterotrimeric G proteins Gαq and Gα11, products of the GNAQ and GNA11 genes, respectively. Affecting different downstream signaling pathways, Gαq and Gα11 also activate the product of the PLCB4 gene, phospholipase-C β4 which hydrolyses PIP2 (phosphatidylinositol 4,5-bisphosphate) releasing DAG (diacylglyerol) and IP3 (inositol-1,4,5-trisphosphate) with subsequent Ca+ release.19,20 The functional similarity is highlighted by the fact that mutations in these genes were almost always found to be mutually exclusive.18 As mutations in one of the four genes (GNA11, GNAQ, PLCB4, and CYSLTR2) were identified in 96% of tumor samples (131 of 136)18 it appears likely the majority of relevant activating mutations has been identified.
The goal of our study was to investigate the frequency of activating CYSLTR2 or PLCB4 mutations in blue nevi. To do this efficiently, a custom-targeted next-generation sequencing approach was applied, assessing all potentially mutated genes in parallel with high sensitivity.
Materials and methods
Sample Selection
Blue nevus samples were obtained by searching the databases of the Department of Dermatology University Hospital Essen (n = 46), Dermatopathology Friedrichshafen (n = 27), and Dermatopathologie bei Mainz (n = 30), Germany. All cases were screened by at least two board-certified pathologists or dermatopathologists. The study was done in accordance with the guidelines set forth by the ethics committee of the University of Duisburg-Essen (IRB-number 16–6951-BO).
DNA Isolation
All DNA was isolated from formalin-fixed paraffin-embedded tissue. Ten micrometer-thick sections were cut from formalin-fixed, paraffin-embedded tumor tissues. The sections were deparaffinized and manually macrodissected according to standard procedures. Genomic DNA was isolated using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Targeted Sequencing
A custom amplicon-based sequencing panel covering 10 genes (Supplementary Table 1) was designed and prepared applying the GeneRead Library Prep Kit from QIAGEN according to the manufacturer’s instructions. Adapter ligation and barcoding of individual samples were done applying the NEBNext Ultra DNA Library Prep Mastermix Set and NEBNext Multiplex Oligos for Illumina from New England Biolabs. Up to 60 samples were sequenced in parallel on an Illumina MiSeq next-generation sequencer.
Sequencing analysis was performed applying the CLC Cancer Research Workbench from QIAGEN. In brief, the following steps were applied. The workflow in CLC included adapter trimming and read pair merging before mapping to the human reference genome (hg19). Insertions and deletions as well as single nucleotide variant detection, local realignment, and primer trimming followed. Additional information was then obtained regarding potential mutation type, known single nucleotide polymorphisms, and conservation scores by cross-referencing varying databases (COSMIC, ClinVar, dbSNP, 1000 Genomes Project, HAPMAP, and PhastCons-Conservation_scores_hg19). The resulting csv files were further analyzed manually. Mutations affecting the protein coding portion of the gene were considered if predicted to result in non-synonymous amino acid changes. Mutations were reported if the overall coverage of the mutation site was ≥ 30 reads, ≥ 5 reads reported the mutated variant and the frequency of mutated reads was ≥ considered 3%.
Associations of Oncogene Mutation Status with Clinical and Pathologic Parameters
We investigated associations of mutation status with available clinical and pathological parameters using χ2-squared tests and Fisher exact tests as appropriate. All statistical analyses were performed using IBM SPSS Statistics software (version 20.0; International Business Machines, Armonk NY, USA). A P-value of P ≤ 0.05 was considered statistically significant.
Results
Sample Cohort
The study cohort consisted of 103 blue nevus samples from 103 patients (60 females and 43 males) with an average age of 55 years (range 7–91). The tumors included 68 (66%) common, 17 (17%) cellular, 5 (5%) deep seated cellular, 10 (10%) sclerotic, and 3 (3%) amelanotic blue nevi. All samples were primary tumors. Available clinical data are listed in Table 1.
Table 1.
Associations of activating gene mutation status with clinicopathologic features
| Factor |
Total N |
WT | GNAQ | GNA11 | NRAS | CYSLTR2 | BRAF | P-value | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Median (range) | Median (range) | Median (range) | Median (range) | Median (range) | Median (range) | ||||
|
| |||||||||
| Age | 103 | 41 (17–76) | 53 (7–91) | 67 (22–85) | 70 (31–83) | 78 (73–81) | a | 0.054 | |
| N (%) | N (%) | N (%) | N (%) | N (%) | N (%) | ||||
| Sex | |||||||||
| Female | 60 | 14 (23) | 33 (55) | 9 (15) | 3 (5) | 0 | 1 (2) | 0.06 | |
| Male | 43 | 4 (9) | 28 (65) | 8 (19) | 0 | 3 (7) | 0 | ||
| Anatomic site | |||||||||
| Head/neck | 26 | 6 (23) | 13 (50) | 5 (19) | 0 | 2 (8) | 0 | 0.08 | |
| Upper limbs | 27 | 3 (11) | 19 (70) | 3 (11) | 0 | 1 (4) | 1 (4) | ||
| Trunk | 28 | 7 (25) | 19 (68) | 1 (4) | 1 (4) | 0 | 0 | ||
| Lower limbs | 17 | 1 (6) | 8 (47) | 7 (41) | 1 (6) | 0 | 0 | ||
| Missing data | 5 | 1 | 2 | 1 | 0 | 1 | 0 | ||
| Subtype | |||||||||
| Common BN | 81 | 13 (16) | 48 (59) | 15 (19) | 2 (3) | 3 (4) | 0 | 0.29 | |
| Cellular BNb | 22 | 5 (23) | 13 (59) | 2(9) | 1 (5) | 0 | 1 (5) | ||
| Depth | |||||||||
| Upper dermis | 11 | 3 (27) | 4 (36) | 3 (27) | 0 | 1 (9) | 0 | 0.28 | |
| Lower dermis | 64 | 15 (23) | 37 (58) | 8 (13) | 2 (3) | 1 (2) | 1 (2) | ||
| Subcutis | 20 | 0 | 16 (80) | 3 (15) | 0 | 1 (5) | 0 | ||
| Missing data | 8 | 0 | 5 | 3 | 0 | 0 | 0 | ||
| Cell type | |||||||||
| Spindle | 77 | 13 (17) | 48 (62) | 12 (16) | 1 (1) | 3 (4) | 0 | 0.02 | |
| Epithelioidc | 7 | 1 (14) | 5 (71) | 0 | 0 | 0 | 1 (14) | ||
| Mixed | 10 | 4 (40) | 3 (30) | 2 (20) | 1 (10) | 0 | 0 | ||
| Missing data | 9 | 0 | 5 | 3 | 1 | 0 | 0 | ||
| Pigmentation | |||||||||
| Absent | 9 | 2 (22) | 6 (67) | 0 | 0 | 1 (11) | 0 | 0.06 | |
| Mild-moderate | 70 | 15 (21) | 42 (60) | 10 (14) | 0 | 2 (3) | 1 (1) | ||
| Marked | 15 | 1 (7) | 8 (53) | 4 (27) | 2 (13) | 0 | 0 | ||
| Missing data | 9 | 0 | 5 | 3 | 1 | 0 | 0 | ||
1 patient, age 46 years.
All tumors demonstrating a cellular component
Epithelioid signifies cell morphology with oval to rounded (rather than elongated/spindle) nuclei. Mixed refers to nevi having both a spindle cell and epithelioid component.
WT, wild-type for all tested genes; epith., epithelioid.
Mutation Analysis for Activating Oncogene Driver Mutations
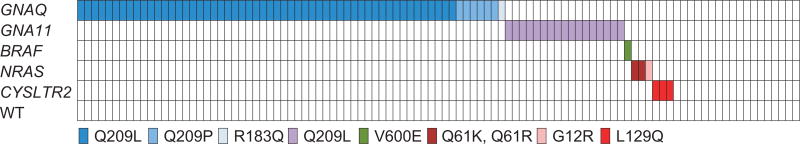
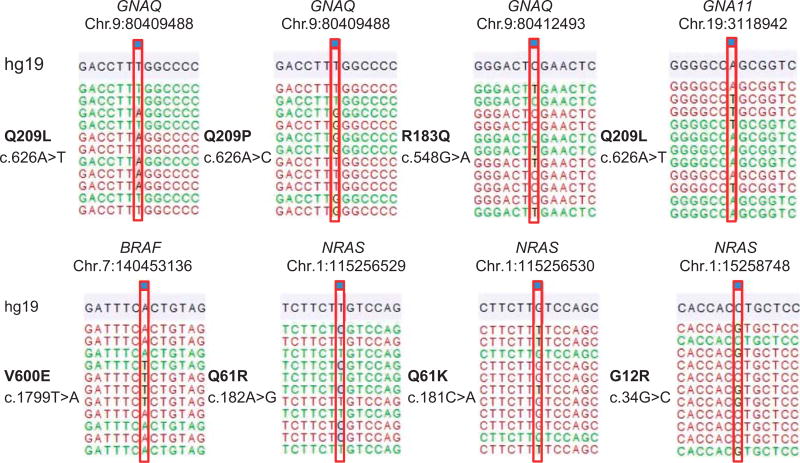
Targeted amplicon sequencing of all genes as described in the material and methods section was performed for the entire tumor cohort. In line with previous results, the most frequent activating mutations identified were in GNAQ, 59% (54c.626A > T Q209L, 6c.626A > C Q209P, and 1c.548G > A R183Q) and GNA11, 16% (17c.626A > T Q209L) (Figures 1 and 2). One (1%) BRAF (c.1799A > T, V600E) and 3 (3%) NRAS mutations (1c.182A > G Q61R, 1c.181C > A Q61K, and 1c.34G > C G12R) were detected. No PLCB4 mutations were identified. Three tumors (3%) harbored activating CYSLTR2 c.386T > A L129Q mutations (Figure 3, Supplementary Figures 1 and 2). All mutations were found to be mutually exclusive (Figure 1).
Figure 1.
Distribution of activating mutations identified in blue nevi. Distribution of activating mutations identified in different oncogenes in the blue nevus cohort. The resulting amino acid changes are color-coded according to the scheme underneath the illustration.
Figure 2.
Examples of activating mutations identified in the tumor cohort. Demonstrated are representative examples of the various mutations in GNAQ, GNA11, BRAF, and NRAS identified in the analyzed cohort of blue nevi. The notation is according to human genome assembly 19 (hg19). (The depicted NRAS mutations show the complimentary strand.)
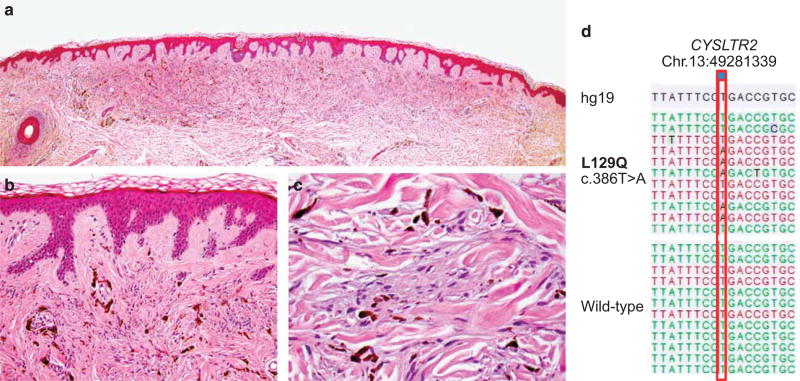
Figure 3.
Activating CYSLTR2 mutation in a superficial common blue nevus. (a–c) Histological pictures of a common blue nevus harboring an activating CYSLTR2 mutation, showing the characteristic proliferation of bland pigmented spindle cells in the dermis with no epidermal involvement ((a) ×20, (b) ×200, and (c) ×400 magnification). The tumor was removed from the upper right arm of a 78-yea-rold male. (d) The CYSLTR2 c.386T > A mutation identified in the tumor with a wild-type sequence for comparison shown underneath. Annotation according to human genome assembly 19 (hg19).
Associations of Clinical and Pathological Parameters with Oncogene Mutation Status
An analysis with available clinicopathological data was performed. The only statistically significant association found was that of oncogene mutation status with cell type (P = 0.02). Complete details are presented in Table 1.
Discussion
We analyzed a cohort of blue nevi and found that in addition to previously recognized recurrent activating mutations in GNAQ and GNA11, mutually exclusive activating hotspot mutations in the CYSLTR2 gene occur. The mutation type and frequency are essentially identical to the initial report of CYSLTR2 mutations in uveal melanoma, once again underlying the genetic similarities between these two melanocytic tumor entities, affecting different organ systems.
Similar to previous reports, we found GNAQ mutations to be by far the most common (59%). Frequent Q209L or Q209P mutations with only a rare occurrence of one R183Q mutation is in line with previous data.10,11 The frequency of GNA11 mutations with 16% is somewhat higher than the 7.5% frequency (exon 4 and 5 mutations) initially reported in blue nevi.11 Almost all mutations in both studies were Q209L. The ratio of GNAQ to GNA11 mutations in our cohort is 3.7, which is lower than the initial report of 8.4; however, the clear predominance of GNAQ mutations remains well documented.
Our study identified one BRAF (V600E) and three NRAS (one Q61R, one Q61K, and one G12V) mutations. Neither mutation is frequently found in blue nevi. BRAF mutations are frequent in acquired nevi,9 and NRAS mutations in congenital nevi.21 We excluded cases in our study which were clearly combined nevi, harboring a common nevus cell clone, or cases showing a clear epidermal melanocytic component. The BRAF- and NRAS-mutant cases in our study were all diagnosed as blue nevi initially and on histological re-review. They showed no apparent epidermal or acquired nevus component. However, the possibility that such regions were present but not represented in the assessed histological slides cannot be entirely excluded. Potentially, future studies will be able to further clarify if presence of BRAF or NRAS mutations is clearly associated with an epidermal-derived (acquired or combined type) nevus component, leaving GNAQ, GNA11, and CYSLTR2 mutations as genetic markers of truly dermal-derived blue nevi.
The frequency of CYSLTR2 mutations (3%) in our study is identical to that reported in uveal melanoma.18 Additionally, all mutations identified were c.386T > A alterations resulting in a L129Q amino acid change, which has been shown to lead to increased receptor activation and signaling output.18 In conjunction with the genetic evidence of CYSLTR2 mutations occurring in a mutually exclusive fashion to other known activating mutations (GNAQ and GNA11) activating the same pathway, this makes a strong case for CYSLTR2 L129Q mutations being bona-fide driver mutations in the pathogenesis of blue nevi as well as uveal melanoma.
We identified one PLCB4 D630 mutation using our assay in a cutaneous metastasis of a uveal melanoma (data not shown), but failed to find a PLCB4 D630 mutation in our cohort of blue nevi. Considering in uveal melanoma, PLCB4 mutations are rare with a frequency of ~ 4%,17,18 selection bias may be a factor. Additional studies analyzing considerably larger numbers of tumors will be required before a potential role of activating PLCB4 D630 mutations in blue nevi can be excluded.
We found no activating oncogene mutation in 17% of blue nevi. The frequency of wild-type tumors (harboring neither a mutation in GNAQ, GNA11, PLCB4, or CYSLTR2) in uveal melanoma is ~ 5%.18 There are different potential explanations for the still relatively high number of wild-type tumors in our study. In contrast to uveal melanomas, in which there is generally a considerable amount of available tumor material, blue nevi can be smaller, less cellular proliferations, frequently interspersed with surrounding connective tissue. This means that the percentage of tumor DNA in the isolated DNA can be low. We applied a next-generation sequencing approach with low frequency mutation cutoffs (3%) to make certain that we detected even low mutation frequencies, in an attempt to compensate for potentially low tumor purity; nevertheless, it is possible that in some of the tumors analyzed and reported as wild-type, the mutation present was not detected due to little or no tumor material obtained in the DNA isolated, or to inadequate sensitivity of our sequencing assay. Another possibility is the presence of additional activating mutations in genes not represented in our assay.
The three CYSLTR2 mutations we identified in our study occurred in morphologically benign common blue nevi (Figure 3, Supplementary Figures 1 and 2). In such unequivocally benign cases, genetically analyzing tumors to determine oncogene mutation status is not clinically relevant. However, although rare, there are atypical and malignant blue nevi (also termed ‘blue nevus-like melanoma’) that have the potential to metastasize and result in patient death.5–7 Although the cases are not presented here (manuscript in preparation), we were able to identify and analyze eight tumors of the rare entity malignant blue nevus. No CYSLTR2 mutations were identified. Considering the low mutation frequency determined in blue nevi and uveal melanoma, higher numbers of these tumors will need to be analyzed to definitively ascertain the presence of CYLSTR2 mutations in these tumors. In the meantime, it would appear prudent to evaluate CYLSTR2 mutation status in these tumors, in particular if they have been found to lack mutations in GNAQ and GNA11. Should CYSLTR2 mutations prove to be of therapeutic value, identifying such a mutation could potentially be of immediate benefit to affected patients.
Our analysis of oncogene status with clinical and pathological parameters showed a few trends with respect to cell type, age, sex, anatomic site, and pigmentation, although the majority of these associations did not reach statistical significance. Considering five different oncogene mutations were identified, some of them being rare, a considerably larger tumor cohort will be necessary to allow meaningful statistical analyses.
In summary, our findings identify blue nevi as a second tumor type harboring activating mutations in the newly identified CYLSTR2 oncogene. The mutation frequency of 3% is identical to that in uveal melanoma. CYSLTR2 mutations being mutually exclusive of oncogenic driver GNAQ and GNA11 mutations in blue nevi is additional supportive evidence of this mutation being a bona-fide driver mutation with clear oncogenic potential. The genetic evidence suggests that similar to activating GNAQ and GNA11 mutations, CYSLTR2 L129Q mutations may be the only driver mutation present in these tumors and fully sufficient to induce a blue nevus.
Acknowledgments
We thank Nadine Stadler and Marion Schwamborn for their excellent technical support. The research was supported by a grant from the Deutsche Forschungsgemeinschaft (GR 3671/3-1) and the Deutsche Stiftung für Dermatologie (Stipendium zur Förderung der Dermatohistologie). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DS is on the advisory board or has received honararia from Roche, Genetech, Novartis, Amgen, GSK, BMS, Boehringer Ingelheim, and Merck. BS received honoraria from Roche, Bristol-Myers Squibb, and MSD Sharp & Dohme, research funding from Bristol-Myers Squibb and MSD Sharp & Dohme, and travel support from Roche, Bristol-Myers Squibb, and AMGEN.
Footnotes
Disclosure/conflict of interest
All other authors have nothing to declare.
Supplementary Information accompanies the paper on Modern Pathology website (http://www.nature.com/modpathol)
References
- 1.Zembowicz A, Phadke PA. Blue nevi and variants: an update. Arch Pathol Lab Med. 2011;135:327–336. doi: 10.5858/2009-0733-RA.1. [DOI] [PubMed] [Google Scholar]
- 2.Zembowicz A, Mihm MC. Dermal dendritic melanocytic proliferations: an update. Histopathology. 2004;45:433–451. doi: 10.1111/j.1365-2559.2004.01975.x. [DOI] [PubMed] [Google Scholar]
- 3.Tièche M. Über benigne Melanome (‘Chromatophorome’) der Haut—‘blaue Neavi’. Virch Arch für path Anat. 1906;1186:212–229. [Google Scholar]
- 4.Barnhill RL, Cerroni L, Cook M, et al. State of the art, nomenclature, and points of consensus and controversy concerning benign melanocytic lesions: outcome of an international workshop. Adv Anat Pathol. 2010;17:73–90. doi: 10.1097/PAP.0b013e3181cfe758. [DOI] [PubMed] [Google Scholar]
- 5.Costa S, Byrne M, Pissaloux D, et al. Melanomas associated with blue nevi or mimicking cellular blue nevi: clinical, pathologic, and molecular study of 11 cases displaying a high frequency of gna11 mutations, bap1 expression loss, and a predilection for the scalp. Am J Surg Pathol. 2016;40:368–377. doi: 10.1097/PAS.0000000000000568. [DOI] [PubMed] [Google Scholar]
- 6.Loghavi S, Curry JL, Torres-Cabala CA, et al. Melanoma arising in association with blue nevus: a clinical and pathologic study of 24 cases and comprehensive review of the literature. Mod Pathol. 2014;27:1468–1478. doi: 10.1038/modpathol.2014.62. [DOI] [PubMed] [Google Scholar]
- 7.Martin RC, Murali R, Scolyer RA, et al. So-called ‘malignant blue nevus’: a clinicopathologic study of 23 patients. Cancer. 2009;115:2949–2955. doi: 10.1002/cncr.24319. [DOI] [PubMed] [Google Scholar]
- 8.Murali R, McCarthy SW, Scolyer RA. Blue nevi and related lesions: a review highlighting atypical and newly described variants, distinguishing features and diagnostic pitfalls. Adv Anat Pathol. 2009;16:365–382. doi: 10.1097/PAP.0b013e3181bb6b53. [DOI] [PubMed] [Google Scholar]
- 9.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 10.Van Raamsdonk CD, Bezrookove V, Green G, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Raamsdonk CD, Griewank KG, Crosby MB, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dai J, Tetzlaff MT, Schuchter LM, et al. Histopathologic and mutational analysis of a case of blue nevus-like melanoma. J Cutan Pathol. 2016;43:776–780. doi: 10.1111/cup.12731. [DOI] [PubMed] [Google Scholar]
- 13.Perez-Alea M, Vivancos A, Caratu G, et al. Genetic profile of GNAQ-mutated blue melanocytic neoplasms reveals mutations in genes linked to genomic instability and the PI3K pathway. Oncotarget. 2016;7:28086–28095. doi: 10.18632/oncotarget.8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels AB, Lee JE, MacConaill LE, et al. High throughput mass spectrometry-based mutation profiling of primary uveal melanoma. Invest Ophthalmol Vis Sci. 2012;53:6991–6996. doi: 10.1167/iovs.12-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin M, Masshofer L, Temming P, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harbour JW, Onken MD, Roberson ED, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansson P, Aoude LG, Wadt K, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore AR, Ceraudo E, Sher JJ, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016;48:675–680. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suh PG, Park JI, Manzoli L, et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008;41:415–434. doi: 10.5483/bmbrep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- 20.Waldo GL, Ricks TK, Hicks SN, et al. Kinetic scaffolding mediated by a phospholipase C-beta and Gq signaling complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bauer J, Curtin JA, Pinkel D, et al. Congenital melanocytic nevi frequently harbor NRAS mutations but no BRAF mutations. J Invest Dermatol. 2007;127:179–182. doi: 10.1038/sj.jid.5700490. [DOI] [PubMed] [Google Scholar]