Abstract
Heritable loss of function mutations in the human RECQ helicase genes BLM, WRN, and RECQL4 cause Bloom, Werner, and Rothmund-Thomson syndromes, cancer predispositions with additional developmental or progeroid features. In order to better understand RECQ pathogenic and population variation, we systematically analyzed genetic variation in all five human RECQ helicase genes. A total of 3,741 unique base pair-level variants were identified, across 17,605 potential mutation sites. Direct counting of BLM, RECQL4, and WRN pathogenic variants was used to determine aggregate and disease-specific carrier frequencies. The use of biochemical and model organism data, together with computational prediction, identified over 300 potentially pathogenic population variants in RECQL and RECQL5, the two RECQ helicases that are not yet linked to a heritable deficiency syndrome. Despite the presence of these predicted pathogenic variants in the human population, we identified no individuals homozygous for any biochemically verified or predicted pathogenic RECQL or RECQL5 variant. Nor did we find any individual heterozygous for known pathogenic variants in two or more of the disease-associated RECQ helicase genes BLM, RECQL4, or WRN. Several postulated RECQ helicase deficiency syndromes—RECQL or RECQL5 loss of function, or compound haploinsufficiency for the disease-associated RECQ helicases—may remain missing, as they likely incompatible with life.
Keywords: RECQ helicase, Werner syndrome, Bloom syndrome, Rothmund-Thomson syndrome, RECQL, RECQL5, pathogenic variation, mutation functional prediction, compound haploinsufficiency, heritable cancer predisposition
Introduction
The human RECQ helicase gene family was identified in part by efforts to determine the genetic basis for three inherited human diseases, Bloom syndrome (BS; MIM# 210900), Rothmund-Thomson syndrome (RTS; MIM# 268400), and Werner syndrome (WS; MIM# 277700). All three are uncommon (≤1/50,000 live births), autosomal recessive Mendelian diseases that share an elevated risk of cancer together with more variable, syndrome-specific developmental or acquired features.
BS was identified in 1954 by David Bloom, who described three patients with congenital short stature and skin changes reminiscent of systemic lupus erythematosus [Bloom, 1954; German, 1993]. Consistent features include marked intrauterine and postnatal growth retardation; congenital short stature; a characteristic, sun-sensitive “butterfly” facial rash; cellular and humoral immune deficits; an elevated risk of diabetes mellitus; and reduced fertility. Males are typically infertile, whereas females are hypofertile but may give birth to normal offspring [German, 1979]. BS patients are predisposed to a remarkably broad range of cancers, in contrast to other inherited cancer predispositions including RTS and WS [German, 1997]. Cancer is the most common cause of death in BS patients.
RTS was described in 1868 as the familial occurrence of a sun-sensitive rash with redness, swelling, and blistering, leading to variable pigmentation with telangiectasias and focal atrophy. These skin changes were often reported in association with bilateral juvenile cataracts [Rothmund, 1868]. Characteristic skin changes typically appear first on the face at 3–6 months of age, and spread thereafter to involve the extremities while sparing the trunk [Rothmund, 1868; Wang et al., 2001; Larizza et al., 2010]. Additional features may include sparse or absent hair, eyelashes, and eyebrows; congenital short stature in conjunction with frequent bone and tooth abnormalities; cataracts; and an elevated risk of primary bone tumors—osteosarcomas—together with lymphoma [Vennos and James, 1995; Wang et al., 2001; Wang et al., 2003a; Larizza et al., 2010]. Immunologic function appears to be intact, and RTS females have given birth to normal offspring, though fertility may be reduced.
RTS is a genetically heterogenous disease. RTS Type II patients have loss of function mutations in the RECQL4 gene (MIM# 603780), and represent 60%–65% of patients. RTS Type I patients lack RECQL4 mutations, and have a higher risk of developing ectodermal dysplasia and cataracts, though they lack the elevated cancer risk seen in RTS Type II [Wang et al., 2001; Wang et al., 2003a; Larizza et al., 2010]. Two other rare syndromes resembling RTS have been reported in association with RECQL4 mutations: RA-PADILINO syndrome (MIM# 266280) and Baller-Gerold syndrome (BGS; MIM# 218600) [Siitonen et al., 2003; Van Maldergem et al., 2006; Larizza et al., 2010].
WS, alone among the RECQ helicase deficiency syndromes, has features strongly suggestive of premature aging. WS was described in 1904 by Otto Werner [Werner, 1985], who identified key clinical findings that include short stature; early graying and loss of hair; bilateral cataracts; and scleroderma-like skin changes [Epstein et al., 1966; Goto, 1997; Tollefsbol and Cohen, 1984]. WS patients are at increased risk of developing important, age-associated diseases such as atherosclerosis, myocardial infarction and stroke; osteoporosis; and diabetes mellitus. Fertility is reduced. Of note, the central nervous system is spared: WS patients are not at elevated risk of Alzheimer or other types of dementia, apart from those associated with vascular disease. In contrast to BS, WS patients are at clearly elevated risk of only five types of cancer [Goto et al., 1996b; Monnat, 2001; Lauper et al., 2013]. The leading causes of death in WS patients are cancer and premature atherosclerotic cardiovascular disease [Goto et al., 2013].
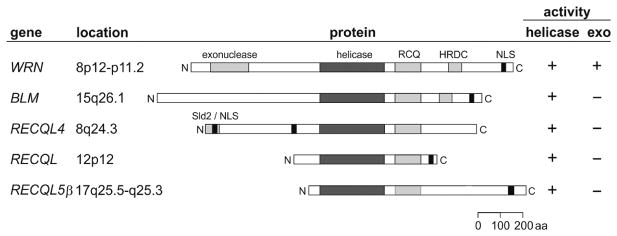
Cloning of the genes causally linked to BS, RTS, and WS identified three different members of the human RECQ helicase gene family [Ellis et al., 1995; Yu et al., 1996; Kitao et al., 1999]. The remaining two human RECQ helicase genes, RECQL (MIM# 600537) and RECQL5 (MIM# 603781), were identified, respectively, by functional cloning of the gene encoding a potent ATPase and helicase activity in human cell extracts [Puranam and Blackshear, 1994; Seki et al., 1994], or by sequence-based, homology search-driven cloning [Kitao et al., 1998]. Cloning of the disease-associated RECQ helicase genes allowed rapid identification of pathogenic mutations in BS, WS, and RTS patients. RECQL and RECQL5 have not been linked to a heritable disease state. Biochemical, cellular, and mouse model analyses of both proteins indicate that loss of function of either would lead to cellular—and potentially organismal—defects [Wang et al., 2003b; Hu et al., 2007; Sharma et al., 2007; Thangavel et al., 2009]. In similar fashion, no patient has been identified with a compound RECQ helicase deficiency syndrome. Key features of the human RECQ helicase genes and their encoded proteins are shown in Figure 1.
Figure 1.
Key features of the human RECQ helicase genes and proteins. Each of the five human RECQ helicases is presented in a row. The official HGNC gene symbol is followed by chromosomal location; a schematic representation of the protein open reading frame aligned on the RECQ helicase consensus/ATPase domain (filled center segment in each protein) and indicating the locations of additional functional domains: WRN exonuclease, RQC (RECQ consensus), HRDC (helicase and RNaseD C-terminal), and RECQL4 Sld2-like/mitochondrial-targeting domains, as well as the position of nuclear-targeting sequences. The size of each mature protein is listed after each protein model in amino acid residues, followed by a summary of biochemically verified catalytic activities for each protein.
In order to systematically analyze genetic variation in all five of the human RECQ helicase genes, we assembled a new database of clinically ascertained pathogenic variants in BLM (MIM# 604610), RECQL4 (MIM# 603780), and WRN (MIM# 604611), together with all base pair-level genetic variants reported in the > 60,000 individuals included in the Exome Sequencing Project (ESP), 1000 Genomes Project (1KGP), or Exome Aggregation Consortium (ExAC) data. We determined the frequency and spectrum (types and sites) of RECQ helicase base pair-level variation in patient and population data, together with the frequency of known pathogenic variants, to compare with previous carrier frequency estimates for BS, RTS, and WS. Finally, we extended this analysis to include the RECQL and RECQL5 genes by using experimental data and computational prediction to identify potential pathogenic variation and deficiency syndrome individuals in the human population.
Materials and Methods
Mutation Data Sources
A database of clinically ascertained mutations (hereafter referred to as RECQMutdb) was compiled for the three disease-associated genes, BLM, RECQL4, and WRN, using data from our Werner Syndrome Locus-Specific Mutation Database [Moser et al., 1999] together with additional reports of BLM, RECQL4, and WRN mutations in BS, RTS, and WS patients [Goto et al., 1996a; Oshima et al., 1996; Yu et al., 1996; Matsumoto et al., 1997; Huang et al., 2006; German et al., 2007; Zhao et al., 2008; Siitonen et al., 2009; Friedrich et al., 2010; Simon et al., 2010; Fradin et al., 2013; Agrelo et al., 2015; Yokote et al., 2016] and/or included in the ClinVar database [Landrum et al., 2016]. These germline mutation data were cross-referenced to the Catalog of Somatic Mutations in Cancer (COSMIC) [Forbes et al., 2015] to exclude somatic variants, and supplemented with additional germline mutations identified by searching Google Scholar using the following queries: “[BLM] AND [pathologic*] AND [variant*] AND [Bloom syndrome]”, “[RECQL4] AND [pathologic*] AND [variant*] AND [Rothmund*]” and “[WRN] AND [pathologic*] AND [variant*] AND [Werner syndrome]”. The final mutation capture and RECQMutdb update was completed on 19 October 2016 (Supp. Table S1).
Human genetic variation data for the five RECQ genes were obtained from the ESP and 1KGP data archives, supplemented with non-TCGA data from ExAC. ESP data were generated as part of the NHLBI-funded GO (Grand Opportunity) Exome Sequencing Project using samples drawn from several well-phenotyped populations (i.e., 4,298 European Americans and 2,217 African Americans) who were sequenced with the goal of discovering new genes and mechanisms contributing to heart, lung, and blood disorders. All study participants provided written informed consent for the use of their DNA in studies aimed at identifying genetic risk variants for disease, and for broad data sharing [Fu et al., 2013]. 1KGP Phase III data includes 2,504 individuals sampled from 26 populations drawn from sub-Saharan Africa, Europe, East Asia, South Asia, and the Americas. The 1KGP Project aimed to identify genetic variation data broadly representative of the vast majority of individuals within a continent, though no phenotype or clinical information is publicly available [Consortium, 2010; Consortium, 2015]. Neither source of genetic variation data would have systematically excluded individuals with a RECQ helicase deficiency syndrome, though it is unlikely such individuals would have been included unless they were asymptomatic (e.g., a younger WS patient). In order to achieve a tradeoff between variant identification sensitivity and genotyping accuracy, only variants with the call rate of ≥ 80% were used in the following analysis.
Aggregated data on the five human RECQ helicase genes was also downloaded from the ExAC database. This includes exome sequencing data from 60,706 unrelated individuals who were analyzed as part of different disease-specific or population-based genetic studies [Lek et al., 2016]. We excluded overlap samples and data from TCGA to preclude analyzing somatic mutations identified in tumor exomes.
We took the following quality control steps prior to performing analyses. First, where there was a choice we used the highest quality data from respective projects. For example, from 1000 Genomes we used the high quality targeted exome data that had a mean depth of coverage of 65.7× (The 1000 Genomes Project Consortium, 2015), unlike the lower-coverage of the 1000 Genomes whole-genome sequencing data. Second, we ensured that we used data from all three sources where the false discovery rates (FDRs) of base pair-level genetic variants was controlled under 0.05, as estimated and reported in their original publications (The 1000 Genomes Project Consortium (2015) for 1KGP; Fu et al. (2014) for ESP; and Lek et al. (2016) for ExAC). Third, in order to focus on germline variation, we used accessible sample information from 1KGP, ESP, and ExAC to ensure that we had excluded overlap samples and data from TCGA to avoid including somatic mutations, and any individual annotated to have a RECQ helicase deficiency syndrome. Fourth, for second level of analysis where we needed to use individual genotype data (e.g., the estimation of carrier frequency and carrier patterns) we again applied stringent coverage and quality metrics, using only individuals with deep exome sequencing data from 1KGP (with the mean depth of 65.7×), or corresponding data ESP (with the median depth more than 100×) and equivalent FDRs. As a final quality check, we determined whether the use of data from specific studies might be introducing subtle biases in our combined datasets analyses. This was done by comparing the concordance of results across analyses first performed separately for each data source and ancestry. This analysis made use of allele frequencies from 1KGP and ESP data.
Molecular Spectrum of RECQ Mutations
In order to characterize base pair-level variation in RECQ genes, we focused first on the disease-associated RECQ genes BLM, RECQL4, and WRN, then performed, whenever possible, equivalent analyses on all five of the human RECQ helicase genes (Fig. 1). These gene sets are referred to below as the disease-associated “three-gene set” and the inclusive “five-gene set.” Base pair-level genetic variation was categorized by molecular type: single base substitutions; single base insertions or deletions (indels); and variants of > 1 bp that were either “simple” (e.g., insertion or deletion of multiple base pairs, with no other changes) or “complex” (e.g., combinations of base pair changes, insertions, deletions, or other rearrangements). The largest variant class as defined at the base pair level was indel variants of ≤ 10 bp. The reference DNA sequence, open reading frame, and inferred protein sequence for each RECQ gene were taken from the most recent RefSeq gene, mRNA, and protein accessions for each gene (accession date: 8 March 2016; see Supp. Table S1 footnote for more detail). The positions of amino acid residues were annotated according to the largest isoform for each gene, and genome sequences from human genome assembly GRCh37.p13. Variants were plotted on the open reading frame and intron/exon junction sequences of each RECQ gene in order to determine the spectrum–the distribution of molecular types and sites–of variation within each gene. We did not systematically analyze RECQ gene copy number variants due to the absence of data and/or a precise molecular characterization of putative pathogenic mutations of >10 bp. Our final dataset included 3,741 unique base pair-level variants, across 17,605 potential mutation sites, in the five human RECQ helicase genes.
Mutation Hotspot Analysis
We used a probabilistic, gene-specific model to determine whether mutations were randomly distributed, and whether there were mutation “hotspots” in the human RECQ genes. This model was developed using population data in ESP, 1KGP, and ExAC, and used to predict the relative probability of observing mutations in a specific gene region. The mutation model incorporates an overall rate of mutation, relative locus-specific rates based on fixed differences between humans and chimpanzees for each gene coding and splice junction sequence, and also takes into account codon structure and transition/transversion ratios (for additional detail see [O’Roak et al., 2012]). This probabilistic model was also used to determine whether there is enrichment or depletion of variants by gene, functional consequences or location (e.g., in the helicase domain) of a given RECQ protein.
We applied this model by taking a total of NT RECQ variants from our population data (i.e., the number of RECQ variants identified in ESP, 1KGP, and ExAC data), corresponding to NT mutation events assuming an infinite alleles model, and randomly distributed these mutation events, according to their relative probabilities, to obtain the expected mutation distribution in the absence of hotspots. Then, we compared the observed number of genetic mutations identified in ESP, 1KGP, and ExAC data with the expected number by a chi-square test. Mutational hotspots were defined by use of a small sliding window (of 10 or 20 bp) to determine if significantly more genetic variants were observed in a window than expected by chance alone in population data (the chi-square test, P < 0.05).
We assessed the relationship between the distribution of pathogenic and population variants and mutational hotspots using a Mann–Whitney U test. In order to determine whether there might be an underlying sequence-dependent basis for mutational hotspots, we downloaded the coordinates of simple tandem repeats identified by Tandem Repeats Finder [Benson, 1999] from the UCSC Genome Browser [Kuhn et al., 2013]. We also determined the coordinates of CpG sites and their methylation status using ENCODE project data. Reduced representation bisulfite sequencing data were used to quantitatively profile DNA methylation at an average of 1.2 million CpGs in each of 82 cell lines and tissues [Consortium, 2012]. We defined a CpG site as “methylated” when the site was methylated in >80% of molecules sequenced. Fisher’s exact test was then used to compare the distribution of simple tandem repeats, CpG and methylated CpG sites across mutational hotspots and non-hotspots. Previous studies have observed an increase in base miscalls and other errors in GC-rich regions sequenced using next-generation sequencing (NGS) technologies [Allhoff et al., 2013]. We thus excluded from further analysis hotspots that contained one or more NGS error-inducing motif.
Imputation of Potentially Pathogenic Human RECQ Gene Variants
Accurately predicting the functional consequences of genetic variation plays a central role in the identification of causal variants for human diseases or traits. In order to identify potential pathogenic variants in the human RECQ helicase genes, we evaluated the ability of the existing annotation methods to correctly identify known pathogenic variants in RECQMutdb and discriminate these from benign variants in WRN, RECQL4, and BLM. A total of 72 benign variants were used for this analysis including (1) “benign” or “likely benign” variants from ClinVar [Landrum et al., 2016]; (2) likely benign common variants with derived allele frequencies (DAF) > 0.05 in at least one population from ESP, 1KGP, and ExAC data; and (3) fixed substitutions whose allele in human reference sequences is different from the ancestral allele inferred from the six primate EPO alignments [Consortium, 2010].
Nine functional annotation metrics were evaluated: CADD v1.3 [Kircher et al., 2014]; http://cadd.gs.washington.edu); DANN [Quang et al., 2015]; https://cbcl.ics.uci.edu/public_data/DANN/); Eigen v1.1[Ionita-Laza et al., 2016]; http://www.columbia.edu/~ii2135/eigen.html); FATHMM[Shihab et al., 2015]; http://fathmm.biocompute.org.uk/inherited.html); PhyloPNH[Fu et al., 2014]; http://akeylab.gs.washington.edu/wqfu_files/phyloP_NH.tar.gz); GERP++[Davydov et al., 2010]; http://mendel.stanford.edu/SidowLab/downloads/gerp/hg19.GERP_scores.tar.gz); LRT[Chun and Fay, 2009]; http://www.genetics.wustl.edu/jflab/data5.html); SIFT[Kumar et al., 2009]; http://sift.jcvi.org/www/SIFT_chr_coords_submit.html); and PolyPhen2[Adzhubei et al., 2013]; http://genetics.bwh.harvard.edu/pph2/). Comparative evaluation of the performance of these metrics identified CADD as having the best ability to distinguish known pathogenic variants from benign variants across different molecular types of variation. In this analysis, a CADD score of ≥ 24 lead to a FDR of <0.1.
We further imputed all possible single base changes for all five human RECQ helicase gene coding regions, together with their functional consequences (i.e., synonymous, missense, nonsense, and splice-altering). Deleterious predictions were made for all variants identified in ESP, 1KGP, and ExAC exome-sequencing data (n = 3,581), and all possible single base substitutions (n = 32,815). We then examined the distribution of potentially pathogenic variants in ESP, 1KGP, and ExAC over mutational hotspots, and investigated the carrier frequencies and patterns of these predicted deleterious variants in ESP/1KGP data.
Human RECQ Helicase Variant Carrier Frequencies
In order to determine the distribution and frequency of pathogenic RECQ variants in population sequencing data, we focused on 4,298 European Americans and 2,217 African Americans included in ESP data [Fu et al., 2013], and on 2,504 unrelated individuals with ancestry from five geographic regions (sub-Saharan Africa, Europe, East Asia, South Asia, and the Americas) included in 1KGP data [Consortium, 2015]. In order to avoid the uncertainty of phasing, especially for rare variants, we used only unphased genotyping data. The observed frequencies of pathogenic variants were then used to determine how direct counting of pathogenic alleles compared with previous carrier frequency estimates from BS, RTS, and WS disease data.
We also asked whether individuals with multiple pathogenic variants, in either the same or in different RECQ genes, were observed at the expected frequency in the ESP/1KGP cohorts. Permutation testing was used to evaluate the significance of whether multiple pathogenic variants were more or less frequent than expected. Since only genotype information was considered here, we assumed the independence of loci during the permutation process. Briefly, we randomly resampled individual genotypes for each variant, and summarized the number of pathogenic variants found in each simulated individual. After 10,000 replicates, we compared the observed distribution of pathogenic variants for each individual with the simulated distribution from permutation.
Functional Rescue Potential of Pathogenic RECQ Variants
We examined all pathogenic variants in RECQMutdb in order to determine what subset might be candidates for functional rescue by exon skipping, premature termination codon (PTC or “stop” codon) read-through or small molecule functional rescue. Exon skipping therapies aim to exclude mutant exons from mature mRNAs, and thus have the potential to recover partial function by restoring the downstream protein open reading frame. The potential of exon skipping to restore function was determined by mapping of disease-associated variants onto exons for the three disease-associated RECQ helicases, and then determining whether the skipping of the variant-containing exons would restore the downstream open reading frame. For simplicity and with an eye to potential clinical applicability, we investigated only instances in which the skipping of single exons both removed a pathogenic variant and restored the downstream open reading frame. The potential of PTC read-through function was assessed by determining the fraction of pathogenic premature termination codons that were within 50 nucleotides of the last exon–intron junction, and thus had the highest likelihood of being rescued by PTC read-through to escape nonsense-mediated decay [Huang and Wilkinson, 2012; Keeling et al., 2014].
Results
Molecular Spectrum of RECQ Mutations
We first assembled a dataset consisting of known pathogenic RECQ variants in BLM, RECQL4, and WRN, together with variants in all five genes included in ESP, 1KGP, and ExAC project data. This approach identified 211 pathogenic variants that we captured in a newly constructed database of pathogenic RECQ variants (RECQMutdb; Supp. Table S1), and a non-redundant set of 3,581 additional genetic variants from ESP (n = 674), 1KGP (n = 611), and ExAC (n = 3,479) data. Among these population variants, 2,606 were in the three disease-associated RECQ helicase genes BLM, RECQL4, and WRN. The vast majority of variants in ESP, 1KGP, and ExAC data—95.6%—were single base substitutions. In contrast, the distribution of pathogenic variants in BLM, RECQL4, and WRN in RECQMutdb data displayed a broader distribution of molecular variant types with a substantially larger fraction of variants, 40.2% indels or more complex variants, with a higher likelihood of leading to loss of function (Table 1).
Table 1.
RECQ Genetic Variants by Type in RECQMutdB and ESP, 1KGP, and ExAC Data
| Gene | RECQMutdB (n = 211) | ESP, 1KGP, and ExAC (n = 3,581) | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| 1 bp Subs | ±1 bp Indel | >1 bp Simple Indel | >1 bp Complex Indel | 1 bp Subs | ±1 bp Indel | >1 bp Simple Indel | >1 bp Complex Indel | |
| WRN | 59 | 12 | 18 | 2 | 723 | 18 | 18 | 0 |
| RECQL4 | 29 | 12 | 15 | 0 | 1,133 | 11 | 19 | 0 |
| BLM | 38 | 17 | 8 | 1 | 642 | 19 | 23 | 0 |
| RECQL | 326 | 9 | 13 | 0 | ||||
| RECQL5 | 599 | 11 | 17 | 0 | ||||
| Total (%) | 126 (59.8%) | 41 (19.4%) | 41 (19.4%) | 3 (1.4%) | 3,423 (95.6%) | 68 (1.9%) | 90 (2.5%) | 0 |
In order to determine whether we might be systematically under sampling specific gene regions by virtue of capture or sequencing methods [Clark et al., 2011; Sulonen et al., 2011; Sims et al., 2014], we compared variant densities based on a 10-bp window analysis between the first or last exon with middle exons of each RECQ gene. Except for a significantly low density in the first exon of RECQL4, no significant difference was observed for any other human RECQ helicase gene region (Supp. Fig. S2). A second analysis we performed showed that the distribution of base pair positions where we had “no calls”(no identified variant at that bp position) was uniformly distributed across the RECQ genes (additional results not shown). These results indicate that we were not systematically or significantly under-sampling RECQ gene regions for variation.
As a final quality check we compared the allele frequencies for variants identified in both 1KGP and ESP data, and found they were highly concordant, with a Pearson coefficients of 0.998 between individuals with African ancestry in 1KGP and African American ESP samples, and 0.999 between individuals with European ancestry in 1KGP and European American ESP data (P < 10−15). These results, shown in Supp. Figure S1, indicate that the use of data from different studies was unlikely to be introducing subtle or systemic biases in our combined datasets analyses.
Functional Consequences of RECQ Variants
Among 211 RECQMutdb variants, all, except two, were predicted to affect protein structure and/or function by creating a frameshift (79, 37.4%); introducing a nonsense codon (53, 25.1%) or missense substitution (43, 20.4%); by disrupting splicing (30, 14.2%); or by inserting or deleting codons while preserving the open reading frame (4, 1.9%). In contrast, BLM, RECQL4 and WRN variants in ESP, 1KGP, and ExAC data were enriched for missense-generating variants (Chi-square test, P = 3.33 × 10−27) and depleted for synonymous and nonsense variants (Chi-square test, P = 6.42 × 10−33 and 0.002, respectively). Indels were enriched among population variants, with 71.3% of these predicted to lead to a frameshift (Chi-square test, P = 5.08 × 10−8). Most of these indels were only reported in ExAC data.
The same distribution of predicted variant consequences was observed when all five RECQ helicases were examined using ESP, 1KGP, and ExAC data (Fig. 2A): a high proportion of missense-generating variants (2,280, 63.7%), followed by synonymous (1,006, 28.1%), nonsense (76, 2.1%), splice-altering (61, 1.7%) and indel-inducing variants (158, 4.4%). A majority of indel variants (73.4%) were predicted to lead to frame shift or splice site disruptions, while 26.6% resulted in the addition or removal of a codon while preserving the open reading frame (Fig. 2A).
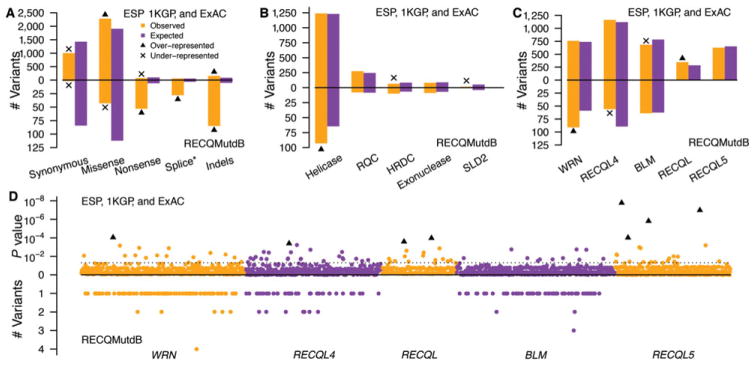
Figure 2.
Functional consequences and domain mapping of RECQ variants and the identification of RECQ mutation hotspots. A: The number of observed variants in RECQMutdb, ESP, 1KGP, and ExAC data, respectively, was compared with expectation as a function of predicted functional consequence. Splice-altering variants were counted based on SNVs only. Indels were counted based on the generation of splice alterations, frameshifts, or the addition/loss of a codon while preserving reading frame. B: The distribution of RECQ variants by variant source across RECQ protein (helicase, RQC, HRDC, exonuclease, and SLD2-line/mitochondrial targeting) domains. C: The mutability of human RECQ helicase genes in population data (i.e., ESP, 1KGP, and ExAC) and in RECQMutdb, respectively. The number of observed variants in RECQMutdb or in ESP, 1KGP, and ExAC, respectively, was compared with expectation in RECQ genes (i.e., WRN, RECQL4, BLM, RECQL, and RECQL5) by chi-square tests between the observed and expected numbers of variants. Significant results (P < 0.05) were marked with over-represented variants by triangles and under-represented variants by “x” for each gene. D: Mutation hotspots identified using population variation data. A total of 87 hotspots were identified by a 10-bp window analysis in the five RECQ genes, listed left to right along a horizontal axis in their chromosomal location order. The location of each hotspot (identified by a chi-square test P < 0.05 indicated by the dotted line) is indicated by gene-specific, color-coded dots, with hotspot significance in population data indicated by the –log10 (P) value above the horizontal line. The top eight mutation hotspots indicated by their –log10 (P) values are highlighted by black triangles. RECQ pathogenic variants are plotted by location below the horizontal line, with the number of different variants per window indicated.
We also mapped variants onto major RECQ helicase functional domains: the RECQ helicase consensus (RQC or RECQ consensus), HRDC (helicase and RNaseD C-terminal) WRN exonuclease, and RECQL4 Sld2-like/mito-targeting domains (Fig. 1). RECQMutdb variants were enriched in the helicase domains of BLM, RECQL4, and WRN (Chi-square test, P = 2.26 × 10−5). In contrast, no variant class was overrepresented in functional domains in ESP, 1KGP, or ExAC data, whereas population variants were depleted from HRDC and Sld2 domains (Chi-square test, P = 0.03 and 3.90 × 10−7, respectively) (Fig. 2B).
Mutability and Mutation Hotspot Analysis
The distribution of sequence variants was comparatively uniform by visual inspection across the open reading frames of all five RECQ helicases (results not shown). We used population variation data to determine whether specific RECQ helicase genes or gene regions were disproportionately mutable, and to identify mutation hotspots in all five RECQ helicase genes. We also determined whether specific DNA sequence features might be promoting specific molecular classes of mutations. WRN was the most mutable (Chi-square test, P = 8.90 × 10−7) and RECQL4 the least mutable (Chi-square test, P = 3.10 × 10−6) among the three disease-associated RECQ helicase genes (Fig. 2C). In ESP, 1KGP, and ExAC data, WRN was not enriched or depleted in variants (Chi-square test, P = 0.18), whereas RECQL4 was slightly enriched (Chi-square test, P = 0.022), and BLM significantly depleted (Chi-square test, P = 1.59 × 10−4). A five gene analysis confirmed the significant depletion of BLM variants in ESP, 1KGP, and ExAC data (Chi-square test, P = 5.59 × 10−5), and identified RECQL as the most mutable of the five RECQ helicase genes (Chi-square test, P = 9.63 × 10−5; Fig. 2C).
Mutational hotspots were next identified using a previously developed, gene-specific probabilistic mutation model [O’Roak et al., 2012]. Hotspots of both 10 and 20-bp were identified based on significant enrichment for variants compared with expectation (Chi-square test, P < 0.05). The 10 and 20-bp window sizes were chosen to avoid very short windows that might exclude hotspots consisting of nucleotide runs or repeats, as well as longer windows (≥50-bp) that might reduce the sensitivity of hotspot detection by averaging out signal over a large number of nucleotides. Hotspots, once identified, were examined for an over-representation of known mutable sequences including tandem repeats; CpG and methylated CpG dinucleotides within or bridging hotspot boundaries; and sequence motifs associated with elevated NGS error frequencies.
In a three-gene analysis using a 10-bp window, 52 mutational hotspots were identified in ESP, 1KGP, and ExAC data. The corresponding number identified using a 20-bp window was 43 hotspots. A five-gene analysis using a 10-bp window identified 87 mutational hotspots in ESP, 1KGP, and ExAC data that included the 52 mutational hotspots identified in BLM, RECQL4, and WRN. The corresponding number identified using a 20-bp window was 67 (Fig. 2D). Pathogenic variants were significantly enriched in both 10-bp (Mann–Whitney test, P = 0.016) and 20-bp (Mann–Whitney test, P = 7.98 × 10−4) hotspot windows defined using ESP, 1KGP, and ExAC data. When using a 10-bp window, 13.9% of all RECQ variants from ESP, 1KGP, and ExAC were located in hotspots despite their representing only 4.7% of the total mutable sequence, a 3.25-fold enrichment. Among pathogenic variants contained in RECQMutdb, 14 (6.6%) were located in 14 mutational hotspots defined by ESP, 1KGP, and ExAC data, a 1.66-fold enrichment.
Neither hotspot length class was significantly enriched for repeat sequences (Fisher’s exact test, P = 0.68 and 0.17 for the 10 and 20-bp window, respectively), though both were significantly enriched in CpG dinucleotides (Fisher’s exact test, P = 0.0025 and 0.046 for 10 and 20-bp windows, respectively) and methylated CpG sites (Fisher’s exact test, P = 0.0015 and 0.019 for 10 and 20-bp windows, respectively).
Previous studies have observed an increase in base miscalls and other errors in GC-rich regions analyzed by NGS [Dohm et al., 2008]. Thus, we determined whether 91 NGS error-prone sequence motifs [Allhoff et al., 2013] were present or enriched in our 10 bp hotspots. Four of 87 mutational hotspots contained an error-inducing sequence motif, with modest enrichment of error-inducing sequence motifs across all mutation hotspots compared with non-hotspots (Fisher’s exact test P = 0.03, OR = 3.91 (1.31~11.64)). Mutation hotspots of 10 bp that lacked NGS error sequence motifs were still enriched for CpG and methylated CpG sites (Fisher’s exact test, P = 0.0037 for CpG sites, and 0.004 for methylated CpG sites, respectively).
All of the RECQ helicase genes, except BLM, harbored at least one of the “hottest” 10% of hotspots as determined by variant enrichment. These eight hotspots are GC-rich (66% GC), and contained 56 population and 1 RECQMutB variants with 5 to 10 variants mapping in each hotspot window. Hottest hotspot variants were nearly all (98.3%) base substitutions, with nearly equal numbers of transitions (26) and transversions (29). This high proportion of transversions is unusual, and was significantly different from the transition: transversion ratio in other mutation hotspots (Fisher’s exact test, P = 0.002, OR = 0.393 (0.222–0.694) or in RECQ gene sequence mapping outside hotspots (P = 0.0001, OR = 0.347 (0.203–0.592)).
Human RECQ Helicase Carrier Frequencies and Patterns
A quarter (51, 24.2%) of the pathogenic variants contained in RECQMutdb was also present in ESP, 1KGP and ExAC population data (Supp. Table S1). There were 20 nucleotide positions at which we observed both pathogenic and population variants that led to different predicted protein changes, and four nucleotide positions at which different variants observed in RECQMutdb and in population data were predicted to lead to the same protein change.
We determined the distribution of pathogenic RECQ variants in 9,019 individuals included in ESP and 1KGP data in order to identify individuals who carried zero, one, or more than one pathogenic variant in one or more RECQ gene. Among the 51 variants present in RECQMutdb and population data, 25 were found in ESP and 1KGP data. Among 9,019 ESP/1KGP individuals, 2.28% (206/9,019) carried at least one pathogenic derived allele. Carrier frequencies ranged from 2.12% (191/9,019) for WRN, through 0.11% (10/9,019) for RECQL4 to 0.06% (5/9,019) for BLM. This variability in carrier frequency is largely driven by population-specific high frequency alleles in WRN. For example, when we evaluated allele frequencies in different populations in populations with African ancestry, we observed a high DAF of > 0.01 that was driven largely by rs11574395:G>A only in African populations, whereas the frequency of this allele was < 0.01 in other populations. A similar situation pertained for rs3087425:C>T, which predominated only in American samples, where the DAF was again >0.01.
Eight putative RECQ deficiency syndrome individuals (8/9,019, 0.089%) were identified, including four individuals homozygous for pathogenic mutations in both the WRN or RECQL4 genes (Table 2). Among 206 carriers of pathogenic variants, two carried more than one pathogenic variant in WRN, and two carried more than one pathogenic variant in RECQL4. No individual carried multiple pathogenic variants in more than one RECQ gene. These observed carrier frequencies and patterns did not differ from expectation (10,000 permutations, P > 0.05) (Table 2).
Table 2.
Distribution of Pathogenic Variants across the Three RECQ Disease Genes in ESP/1KGP Individuals
| Population | Individuals Who Carried At Least One Derived Allele | Individuals Who Carried Homozygous Derived Allele | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| 0,0 | S,S | M,S | M,M | 0,0 | S,S | M,S | M,M | ||
| ESP_AA | Obs. | 2,121 | 94 | 2 | 0 | 2,214 | 3 | 0 | 0 |
| Exp. | 2,120.6 | 95.3 | 1.1 | 0 | 2,214.8 | 2.2 | 0 | 0 | |
| P | 0.96 | 0.89 | 0.40 | NA | 0.60 | 0.60 | NA | NA | |
| ESP_EA | Obs. | 4,247 | 50 | 1 | 0 | 4,295 | 3 | 0 | 0 |
| Exp. | 4,251.6 | 46.0 | 0.43 | 0 | 4,293.3 | 4.7 | 0 | 0 | |
| P | 0.50 | 0.55 | 0.38 | NA | 0.43 | 0.43 | NA | NA | |
| 1KGP_AFR | Obs. | 628 | 32 | 1 | 0 | 659 | 2 | 0 | 0 |
| Exp. | 627.1 | 33.3 | 0.6 | 0 | 659.1 | 1.8 | 0 | 0 | |
| P | 0.87 | 0.82 | 0.60 | NA | 0.91 | 0.91 | NA | NA | |
| 1KGP_EUR | Obs. | 500 | 3 | 0 | 0 | 503 | 0 | 0 | 0 |
| Exp. | 499.1 | 3.9 | 0 | 0 | 503 | 0 | 0 | 0 | |
| P | 0.64 | 0.64 | NA | NA | NA | NA | NA | NA | |
| 1KGP_EAS | Obs. | 497 | 7 | 0 | 0 | 504 | 0 | 0 | 0 |
| Exp. | 496.6 | 7.3 | 0.05 | 0 | 504 | 0 | 0 | 0 | |
| P | 0.89 | 0.91 | 0.82 | NA | NA | NA | NA | NA | |
| 1KGP_SAS | Obs. | 485 | 4 | 0 | 0 | 489 | 0 | 0 | 0 |
| Exp. | 485.2 | 3.8 | 0 | 0 | 489 | 0 | 0 | 0 | |
| P | 0.90 | 0.90 | NA | NA | NA | NA | NA | NA | |
| 1KGP_AME | Obs. | 335 | 12 | 0 | 0 | 347 | 0 | 0 | 0 |
| Exp. | 335.3 | 11.6 | 0.07 | 0 | 347.0 | 0 | 0 | 0 | |
| P | 0.93 | 0.91 | 0.79 | NA | NA | NA | NA | NA | |
This analysis was performed based on 2,217 African Americans (ESP_AA), 4,298 European Americans (ESP_EA), 661 Africans (1KGP_AFR), 503 Europeans (1KGP_EUR), 504 East Asians (1KGP_EAS), 489 South Asians (1KGP_SAS), and 347 Americans (1KGP_AME). The observed (Obs.) and expected (Exp.) number of individuals who carried none, one, or more than one pathogenic variant were categorized into four groups: (0,0), (S, S), (M,S), and (M,M) individuals carried, respectively, no variants in any RECQ gene, 1 variant in one gene, >1 variant in one gene, or >1 variant in multiple genes, respectively. The observed distribution was compared with the expected one from 10,000 permutations.
Identification of Potential Pathogenic Variants in RECQL or RECQL5
Known pathogenic variants of RECQL or RECQL5, defined by their occurrence in a heritable Mendelian disease phenotype mapping to the RECQL or RECQL5 genes, have not yet been identified. In order to determine whether these two genes might harbor cryptic human disease-promoting variation, we used computational prediction to identify potential pathogenic variants, then searched for these variants in our three population sample datasets.
There is abundant evidence that the loss of function of RECQL or RECQL5 can confer deleterious organismal and/or cellular phenotypes. The RECQL gene has homologues in several model organisms including Xenopus, mouse and chicken. Among these organisms, only RECQL knockout mice have been generated thus far. Though viable, embryonic fibroblast of these mice display aneuploidy, spontaneous chromosomal breakage and frequent translocations [Sharma et al., 2007]. Depletion of RECQL protein also sensitizes human cells to ionizing radiation and camptothecin, and increases both spontaneous (-H2AX foci and sister chromatin exchanges [Sharma et al., 2007]. Recent breast cancer sequencing analyses have revealed several deleterious RECQL variants that may be strong breast cancer predisposition alleles. These include five missense mutations that have been verified to disrupt the helicase activity of RECQL protein (p.Ala195Ser, p.Arg215Gln, p.Arg455Cys, p.Met458Kys, and p.Thr562Ile), and several additional variants with a high likelihood of disrupting RECQL structure and/or function: c.634C>T, p.Arg215*; and c.1667 1667+3delAGTA, p.Lys555delinsMetTyrLysLeuIleHisTyrSerPheArg [Cybulski et al., 2015], together with nonsense mutations (p.Leu128*, p.Trp172*, and p.Gln266*) and a single variant predicted to affect mRNA splicing (c.395-2A>G)[Sun et al., 2015].
The human RECQL5 gene has homologues in many model organisms, including mouse, C. elegans, Drosophila melanogaster, and Xenopus. Among these, only knockout mice have been generated. Embryo fibroblasts from Recql5 knockout mice display chromosomal instability and high levels of sister chromatin exchange though a normal LOH frequency, elevated levels of Rad51 and (-H2AX foci and a predisposition to cancer [Hu et al., 2007]. D. melanogaster lacking RecQ5 displays mitotic defects, chromosomal aberrations, and nuclear dysmorphology during early development [Nakayama et al., 2009]. As was observed for RECQL, the depletion of RECQL5 from human cells leads to transcriptional stress and associated genomic instability [Saponaro et al., 2014]. One RECQL5 substitution of p.Lys58Arg in the helicase consensus domain has also been shown to abolish RECQL5 ATPase and helicase activity [Garcia et al., 2004].
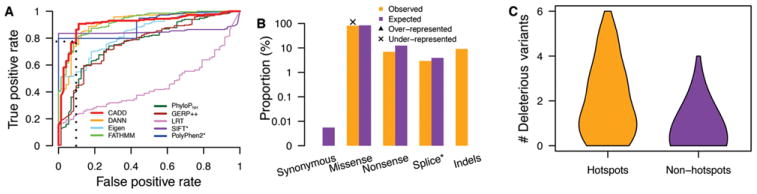
In order to identify additional, potentially pathogenic human RECQL and RECQL5 variants, we first determined the performance of several widely employed algorithms to correctly identify known pathogenic variants contained in RECQMutdb. Our rationale was that the best algorithm(s), as assessed by their ability to correctly identify known pathogenic human RECQ variants, would have a higher likelihood of predicting additional, potentially deleterious variants in RECQL and RECQL5. For this assessment we used area under the curve (AUC) values to discriminate between pathogenic variants in RECQMutdb (n = 211) and known benign variants (n = 72). Nine annotation metrics were tested individually: CADD, DANN, Eigen, FATHMM, PhyloPNH, GERP++, LRT, SIFT, and PolyPhen2. With the exception of CADD, all of these metrics can only annotate single base variants or, as in the case of SIFT and PolyPhen2, only missense variants. From Receiver Operating Curve (ROC) plots, four of these methods displayed the highest discrimination power: FATHMM with an AUC of 0.92, CADD with an AUC of 0.91, DANN with an AUC of 0.91, and PolyPhen2 with an AUC of 0.90 (Fig. 3A). We chose to use CADD for subsequent analyses as it performed nearly as well as the best alternative approaches, and was more versatile in being able to annotate variants of different molecular and functional types. Predicted pathogenic variants were defined by a CADD score of ≥24, corresponding to the FDR of 0.1 (Fig. 3A).
Figure 3.
Prediction of potentially pathogenic mutations in the five human RECQ helicase genes. A: Sensitivity analysis of ability of nine prediction metrics to distinguish pathogenic variants in RECQMutdb from benign variants as indicated by ROC analysis. The values of AUC for the nine metrics are 0.91 (CADD), 0.91 (DANN), 0.84 (Eigen), 0.92 (FATHMM), 0.77 (PhyloPNH), 0.76 (GERP++), 0.50 (LRT), 0.85 (SIFT), and 0.90 (PolyPhen2), respectively. All metrics, except CADD, annotate only point mutations/SNVs, with SIFT and PolyPhen2 further restricted to missense variants. B: The proportion of potentially pathogenic variants in ESP, 1KGP, and ExAC data (CADD ≥ 24) as a function of functional consequences compared with expectation by imputing all possible single base changes by Fisher’s exact test. Only missense-generating variants were significantly under-represented (P < 0.05) compared with expectation (marked by “x”). C: The distribution of the number of potentially pathogenic variants across all five human RECQ helicase genes residing in either 10-bp mutational hotspots or in non-hotspots.
This approach identified 1,053 potentially pathogenic RECQ variants in ESP, 1KGP, and ExAC: 263 in RECQL4, 255 in WRN, 211 in BLM, 180 in RECQL5, and 144 in RECQL. The type distribution of variants by predicted consequence included 853 (81.0%) missense, 73 (6.9%) nonsense, 32 (3.0%) splicing-altering, 92 (8.7%) frameshift, and 3 (0.28%) codon insertion/deletion variants (Fig. 3B). When compared with all imputed single-base variants in the five human RECQ helicase genes, predicted pathogenic missense variants were significantly underrepresented in population data (Fisher’s exact test, P = 0.016, OR = 0.899 (0.823–0.98)) (Fig. 3B). These predicted pathogenic variants were also significantly enriched in mutational hotspots (Mann–Whitney test, P < 10−16 for a 10-bp hotspot window, and P = 7.52 × 10−9 for a 20-bp hotspot window): 16.6% of the potentially pathogenic variants were located in 10-bp hotspots, corresponding to a 4.02-fold enrichment (Fig. 3C).
A total of 294 potentially pathogenic variants were found in ESP (n = 185) and 1KGP (n = 151) data. The number carried by individuals displayed a wide range: 1.304 and 0.639 for African American and European American samples in ESP, respectively; and 1.633, 0.728, 0.28, 0.914, and 1.15 for African, European, East Asian, South Asian, and samples from the Americas in 1KGP, respectively. Nearly one third (26.5%, 2,394/9,019) of individuals from ESP/1KGP carried at least one predicted pathogenic derived allele, and 63 individuals (0.69%, 63/9,019) carried a predicted pathogenic variant in homozygous form: 46 in BLM, 15 in RECQL4 and 2 in WRN. Among the predicted deleterious variants in RECQL and RECQL5 was a single substitution in RECQL5 that had been experimentally verified to suppress WRN helicase activity (p.Lys58Arg) [Popuri et al., 2013] and multiple missense substitutions known to interfere with RECQL5-RNA polymerase II binding: p.Glu584Asp, p.Glu584Ala, p.Tyr597Ala, p.Cys553Ala, p.Leu556Asp, p.Leu602Asp, p.Lys939Ala, and p.Arg943Ala. Also identified as potentially deleterious was an p.Arg550Ala substitution that appears to destabilize RECQL5 protein by interfering with folding [Islam et al., 2010]. Of note, we identified no individuals (0/9,019) homozygous for biochemically verified or predicted pathogenic variants in RECQL or RECQL5 (Table 3 and additional results not shown).
Table 3.
Distribution of Potentially Pathogenic Variants across the Five RECQ Disease Genes in ESP/1KGP Individuals
| Population | Individuals who Carried at Least One Derived Allele | Individuals Who Carried Homozygous Derived Allele | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||
| 0,0 | S,S | M,S | M,M | 0,0 | S,S | M,S | M,M | ||
| ESP_AA | Obs. | 1,398 | 677 | 26 | 116 | 2,203 | 14 | 0 | 0 |
| Exp. | 1,403.6 | 677.7 | 38.1 | 97.6 | 2,205.5 | 11.5 | 0 | 0 | |
| P | 0.81 | 0.97 | 0.05 | 0.06 | 0.47 | 0.47 | NA | NA | |
| ESP_EA | Obs. | 3,433 | 798 | 9 | 58 | 4,274 | 24 | 0 | 0 |
| Exp. | 3,431.1 | 809.7 | 6.4 | 50.7 | 4,270.5 | 27.5 | 0 | 0 | |
| P | 0.94 | 0.65 | 0.31 | 0.30 | 0.50 | 0.50 | NA | NA | |
| 1KGP_AFR | Obs. | 363 | 233 | 12 | 53 | 649 | 12 | 0 | 0 |
| Exp. | 364.8 | 227.4 | 13.8 | 55.0 | 648.5 | 12.4 | 0 | 0.1 | |
| P | 0.89 | 0.65 | 0.62 | 0.78 | 0.89 | 0.90 | NA | 0.80 | |
| 1KGP_EUR | Obs. | 387 | 106 | 0 | 10 | 497 | 6 | 0 | 0 |
| Exp. | 388.2 | 105.9 | 0.4 | 8.5 | 497.4 | 5.6 | 0 | 0 | |
| P | 0.90 | 0.99 | 0.55 | 0.62 | 0.88 | 0.88 | NA | NA | |
| 1KGP_EAS | Obs. | 461 | 40 | 0 | 3 | 504 | 0 | 0 | 0 |
| Exp. | 460.0 | 41.9 | 0.3 | 1.9 | 504 | 0 | 0 | 0 | |
| P | 0.87 | 0.76 | 0.58 | 0.40 | NA | NA | NA | NA | |
| 1KGP_SAS | Obs. | 356 | 117 | 3 | 13 | 489 | 0 | 0 | 0 |
| Exp. | 359.6 | 113.0 | 2.6 | 13.8 | 489 | 0 | 0 | 0 | |
| P | 0.72 | 0.67 | 0.80 | 0.83 | NA | NA | NA | NA | |
| 1KGP_AME | Obs. | 227 | 101 | 2 | 17 | 340 | 7 | 0 | 0 |
| Exp. | 227.8 | 100.5 | 2.8 | 15.9 | 340.0 | 7.0 | 0 | 0 | |
| P | 0.93 | 0.96 | 0.63 | 0.77 | 0.99 | 0.99 | NA | NA | |
This analysis was performed based on 2,217 African Americans (ESP_AA), 4,298 European Americans (ESP_EA), 661 Africans (1KGP_AFR), 503 Europeans (1KGP_EUR), 504 East Asians (1KGP_EAS), 489 South Asians (1KGP_SAS), and 347 Americans (1KGP_AME). The observed (Obs.) and expected (Exp.) number of individuals who carried none, one, or more than one potentially pathogenic variant were categorized into four groups: (0,0), (S, S), (M,S), and (M,M) individuals carried, respectively, no variants in any RECQ gene, 1 variant in one gene, >1 variant in one gene, or >1 variant in multiple genes, respectively. The observed distribution was compared with the expected one from 10,000 permutations.
Potential for Mutation-Specific Functional Rescue
In order to determine whether pathogenic RECQ variants might be good candidates for mutation type-specific therapies, we determined which of the variants in BLM, RECQL4, and WRN might be amenable to therapeutic exon skipping or stop codon read-through [Huang and Wilkinson, 2012; Keeling et al., 2014; Touznik et al., 2014; Veltrop and Aartsma-Rus, 2014]. BLM, RECQL4 and WRN collectively consist of 78 exons, of which 20 (or 25.6%) can be “skipped” or excluded from a mature, spliced mRNA without disrupting the downstream protein open reading frame. While these “skippable” exons collectively contained 19% (40/211) of RECQ disease-associated mutations, the skipping of only four of these exons would restore a protein open reading frame without deleting a known, functionally important RECQ protein domain (Supp. Table S1).
Among the 3,506 base substitutions identified for all five RECQ helicases in RECQMutdb, ESP/1KGP, and ExAC data, there were 113 (3.2%) that created a new nonsense codon (stop-gain variant). Of these, 51 were in RECQMutB, where they represented a quarter (24.2%) of all variants (Supp. Table S1). Despite their prevalence, none of these stop-gain variants in a disease-associated RECQ helicase was located within 50 nucleotides of the terminal intron–exon junction where they would present the best target for premature termination codon read-through. There has been one report of aminoglycoside and ataluren/PTC-124 treatment leading to stop codon read-through of a novel homozygous c.3767C>G, p.Ser1256* WRN mutation, with apparent partial restoration of full length WRN protein and the recovery of WRN-related functions [Agrelo et al., 2015]. In light of this report, we extended our analysis to identify eight additional nonsense codons in the last 600 bp/200 residues of the disease-associated RECQ helicases that might be suitable targets for stop codon read through: 1 in BLM, 3 in RECQL4, and 4 in WRN (Supp. Table S1). Further biochemical characterization of these alleles may identify a subset that can be rescued by stop codon read-through. There have been no reports thus far of targeted RECQ exon skipping, or of other small molecule rescue pharmacologic or chemical chaperone treatment in the context of a RECQ helicase deficiency syndrome [Leidenheimer and Ryder, 2014].
Discussion
The human RECQ helicase proteins play a number of important roles in DNA metabolism. Heritable loss of function mutations in three family members lead to the recessive disorders BS, WS, and RTS and related disorders (RAPADILINO and BGS)[Larsen and Hickson, 2013; Croteau et al., 2014]. Our aim here was to determine the frequency, spectrum (types and sites), and predicted functional consequences of base pair-level genetic variation for all five of the human RECQ helicase genes.
Base pair-level variants of three molecular types—base substitutions or single nucleotide variants (SNVs), and insertion-deletion (indel) mutations of ≥ 1 bp—were present in all five RECQ helicase genes, with an ninefold higher frequency of variants predicted to affect function among variants ascertained in RECQ syndrome patients than in population data (40.2% of patient-ascertained variants vs. 4.4% in population samples). One molecular class of variant—complex mutations involving the insertion and/or deletion of > 1 bp—was observed only in WS and BS patients (Table 1). All of the human RECQ helicase genes contained mutation hotspots identified by a probabilistic gene-specific mutation model, though only a minority of these hotspots contained additional clues (e.g., methylated CpG dinucleotides) that might explain enhanced mutability.
RECQ pathogenic genetic variation was also readily identified in population data: over 2% of ESP/1KGP individuals (2.28%, or 206/9,019) carried at least one pathogenic derived allele. WRN had the highest carrier frequency for pathogenic variants (2.12%; 191/9,019), followed by RECQL4 (0.11%; 10/9,019) and BLM (0.06%; 5/9,019). The carrier frequency for WRN was approximately double the highest previous estimate of the frequency of pathogenic alleles, whereas the estimates for BLM and RECQL4 more closely approximated prior estimates for these diseases derived from case-based counting and/or consanguinity-based estimation methods [Epstein et al., 1966; Goto et al., 1981; Cerimele et al., 1982; Oddoux et al., 1999; Shahrabani-Gargir et al., 1998; Fares et al., 2008; Olry, 2015]. The higher WRN carrier frequency was being driven largely by population-specific alleles that varied in frequency, as noted above. We identified potential WS and RTS patients among 9,019 ESP/1KGP individuals, though failed to identify any individual who carried one or more known pathogenic variant in different RECQ genes (Table 2).
We extended this analysis of population-level variation to all five of the human RECQ helicase genes by systematically predicting putative deleterious variants across all five human RECQ helicase genes. This approach revealed that a quarter (26.5%, 2,394/9,019) of ESP/1KGP individuals carried at least one predicted deleterious derived RECQ allele, with 63 individuals (0.69%, 63/9,019) homozygous for predicted pathogenic variants in BLM (n = 46), RECQL4 (n = 15), or WRN (n = 2). In contrast, we identified no individuals homozygous for verified or predicted pathogenic variants in RECQL or RECQL5, or who carried ≥ 1 known or predicted pathogenic variant in more than one RECQ helicase gene.
Our results suggest that RECQL or RECQL5 loss of function or compound haploinsufficiency for two or more of the disease-associated RECQ helicase genes may remain among plausible but “missing” diseases by virtue of embryonal lethality. We identified few known pathogenic variants in BLM, RECQL4, or WRN that were good candidates for either exon-skipping or stop codon read-through therapies, though note that this question has not been systematically examined as yet for any RECQ helicase gene.
In this study, we used newly available human genetic variation data from high-quality exome sequencing of more than 60,000 individuals. Despite this large sample size, we note that our population sample captured only a quarter of clinically ascertained pathogenic RECQ gene variants, and that current exome capture and variant calling technologies are less than perfect. Additional analyses of the questions raised here, using improved capture and variant calling methods and substantially larger sample sizes, would be welcome to help confirm and extend our results.
Supplementary Material
Acknowledgments
Contract Grant Sponsors: NIH Pathway to Independence Award K99HG008122 to WF and NCI award P01CA077852 to RJMJr.
We thank Drs. Junko Oshima, Fuki Hisama, and George M. Martin of the University of Washington for contributing information from the Werner Syndrome International Registry to help construct the RECQMutdb. KJR’s participation in this project was supported by a UW-HHMI Integrative Research Internship, a UW Mary Gates Undergraduate Research Fellowship and a Herschel and Caryl Roman Undergraduate Science Scholarship award through the Department of Genome Sciences, University of Washington.
Footnotes
Disclosure statement The authors declare no conflict of interest.
Additional Supporting Information may be found in the online version of this article.
References
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;07(Unit7.20) doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrelo R, Sutz MA, Setien F, Aldunate F, Esteller M, Da Costa V, Achenbach R. A novel Werner Syndrome mutation: Pharmacological treatment by read-through of nonsense mutations and epigenetic therapies. Epigenetics. 2015;10:329–341. doi: 10.1080/15592294.2015.1027853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allhoff M, Schonhuth A, Martin M, Costa I, Rahmann S, Marschall T. Discovering motifs that induce sequencing errors. BMC Bioinformatics. 2013;14(Suppl 5):S1. doi: 10.1186/1471-2105-14-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom D. Congenital telangiectatic erythema resembling lupus erythematosus in dwarfs. Am J Dis Child. 1954;88:754–758. [PubMed] [Google Scholar]
- Cerimele D, Cottoni F, Scappaticci S, Rabbiosi G, Borroni G, Sanna E, Zei G, Fraccaro M. High prevalence of Werner’s syndrome in Sardinia: Description of six patients and estimate of the gene frequency. Hum Genet. 1982;62:25–30. doi: 10.1007/BF00295600. [DOI] [PubMed] [Google Scholar]
- Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–1561. doi: 10.1101/gr.092619.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MJ, Chen R, Lam HYK, Karczewski KJ, Chen R, Euskirchen G, Butte AJ, Snyder M. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TGP. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TTGP. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu Rev Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cybulski C, Carrot-Zhang J, Kluzniak W, Rivera B, Kashyap A, Wokolorczyk D, Giroux S, Nadaf J, Hamel N, Zhang S, Huzarski T, Gronwald J. Germline RECQL mutations are associated with breast cancer susceptibility. Nat Genet. 2015;47:643–646. doi: 10.1038/ng.3284. [DOI] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner’s syndrome: A review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine. 1966;45:177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- Fares F, Badarneh K, Abosaleh M, Harari-Shaham A, Diukman R, David M. Carrier frequency of autosomal-recessive disorders in the Ashkenazi Jewish population: Should the rationale for mutation choice for screening be reevaluated? Prenat Diagn. 2008;28:236–241. doi: 10.1002/pd.1943. [DOI] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, Kok CY, Jia M. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(D1):D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin M, Merklen-Djafri C, Perrigouard C, Aral B, Muller J, Stoetzel C, Frouin E, Flori E, Doray B, Dollfus H, Lipsker D. Long-term follow-up and molecular characterization of a patient with a RECQL4 mutation spectrum disorder. Dermatology. 2013;226:353–357. doi: 10.1159/000351311. [DOI] [PubMed] [Google Scholar]
- Friedrich K, Lee L, Leistritz D, Nürnberg G, Saha B, Hisama F, Eyman D, Lessel D, Nürnberg P, Li C, Garcia-F-Villalta MJ, Kets CM. WRN mutations in Werner syndrome patients: Genomic rearrangements, unusual intronic mutations and ethnic-specific alterations. Hum Genet. 2010;128:103–111. doi: 10.1007/s00439-010-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Gittelman RM, Bamshad MJ, Akey JM. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am J Hum Genet. 2014;95:421–436. doi: 10.1016/j.ajhg.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Altshuler D, Shendure J, Nickerson DA, Bamshad MJ, Akey JM NHLBI Exome Sequencing Project. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5β, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German J. Bloom’s syndrome. VIII. Review of clinical and genetic aspects. In: Goodman RM, Motulsky AG, editors. Genetic diseases among Askenazi Jews. New York: Raven Press; 1979. pp. 121–139. [Google Scholar]
- German J. Bloom syndrome: A Mendelian prototype of somatic mutational disease. Medicine. 1993;72:393–406. [PubMed] [Google Scholar]
- German J. Bloom’s syndrome: XX. The first 100 cancers. Cytogenet Cell Genet. 1997;93:100–106. doi: 10.1016/s0165-4608(96)00336-6. [DOI] [PubMed] [Google Scholar]
- German J, Sanz MM, Ciocci S, Ye TZ, Ellis NA. Syndrome-causing mutations of the BLM gene in persons in the Bloom syndrome registry. Hum Mutat. 2007;28:743–753. doi: 10.1002/humu.20501. [DOI] [PubMed] [Google Scholar]
- Goto M. Hierarchical deterioration of body systems in Werner’s syndrome: Implications for normal ageing. Mech Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- Goto M, Imamura O, Kuromitsu J, Matsumoto T, Yamabe Y, Tokutake Y, Suzuki N, Mason B, Drayna D, Sugawara M, Sugimoto M, Furuichi Y. Analysis of helicase gene mutations in Japanese Werner’s syndrome patients. Hum Genet. 1996a;99:191–193. doi: 10.1007/s004390050336. [DOI] [PubMed] [Google Scholar]
- Goto M, Ishikawa Y, Sugimoto M, Furuichi Y. Werner syndrome: A changing pattern of clinical manifestations in Japan (1917–2008) BioSci Trends. 2013;7:13–22. [PubMed] [Google Scholar]
- Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996b;5:239–246. [PubMed] [Google Scholar]
- Goto M, Tanimoto K, Horiuchi Y, Sasazuki T. Family analysis of Werner’s syndrome: A survey of 42 Japanese families with a review of the literature. Clin Genet. 1981;19:8–15. doi: 10.1111/j.1399-0004.1981.tb00660.x. [DOI] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wilkinson MF. Regulation of nonsense-mediated mRNA decay. Wiley Interdiscip Rev RNA. 2012;3:807–828. doi: 10.1002/wrna.1137. [DOI] [PubMed] [Google Scholar]
- Huang S, Lee L, Hanson NB, Lenaerts C, Hoehn H, Poot M, Rubin CD, Chen D-F, Yang C-C, Juch H, Dorn T, Spiegel R. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27:558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionita-Laza I, McCallum K, Xu B, Buxbaum JD. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Fox D, Guo R, Enomoto T, Wang W. RecQL5 promotes genome stabilization through two parallel mechanisms—interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol. 2010;30:2460–2472. doi: 10.1128/MCB.01583-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon read through. Annu Rev Genomics Hum Genet. 2014;15:371–394. doi: 10.1146/annurev-genom-091212-153527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao S, Ohsugi I, Ichikawa K, Goto M, Furuichi Y, Shimamoto A. Cloning of two new human helicase genes of the RecQ family: Biological significance of multiple species in higher eukaryotes. Genomics. 1998;54:443–452. doi: 10.1006/geno.1998.5595. [DOI] [PubMed] [Google Scholar]
- Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQ4L cause a subset of cases of Rothmund-Thomson syndrome. Nat Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- Kuhn RM, Haussler D, Kent WJ. The UCSC genome browser and associated tools. Brief Bioinform. 2013;14:144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larizza L, Roversi G, Volpi L. Rothmund-Thomson syndrome. Orphanet J Rare Dis. 2010;5:2. doi: 10.1186/1750-1172-5-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Hickson ID. RecQ helicases: Conserved guardians of genomic integrity. Adv Exp Med Biol. 2013;767:161–184. doi: 10.1007/978-1-4614-5037-5_8. [DOI] [PubMed] [Google Scholar]
- Lauper JM, Krause A, Vaughan TL, Monnat RJ., Jr Spectrum and risk of neoplasia in Werner syndrome: A systematic review. PLoS ONE. 2013;8:e59709. doi: 10.1371/journal.pone.0059709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidenheimer NJ, Ryder KG. Pharmacological chaperoning: A primer on mechanism and pharmacology. Pharmacol Res. 2014;83:10–19. doi: 10.1016/j.phrs.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Imamura O, Yamabe Y, Kuromitsu J, Tokutake Y, Shimamoto A, Suzuki N, Satoh M, Kitao S, Ichikawa K, Kataoka H, Sugawara K. Mutation and haplotype analyses of the Werner’s syndrome gene based on its genomic structure: Genetic epidemiology in the Japanese population. Hum Genet. 1997;100:123–130. doi: 10.1007/s004390050477. [DOI] [PubMed] [Google Scholar]
- Monnat RJ., Jr . Cancer pathogenesis in the human RecQ helicase deficiency syndromes. In: Goto M, Miller RW, editors. From Premature Gray Hair to Helicase Werner Syndrome: Implications for Aging and Cancer. Tokyo: Japan Scientific Societies Press; 2001. pp. 83–94. [Google Scholar]
- Moser MJ, Oshima J, Monnat RJ., Jr WRN mutations in Werner syndrome. Hum Mutat. 1999;13:271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Yamaguchi S-I, Sagisu Y, Sakurai H, Ito F, Kawasaki K. Loss of RecQ5 leads to spontaneous mitotic defects and chromosomal aberrations in Drosophila melanogaster. DNA Repair. 2009;8:232–241. doi: 10.1016/j.dnarep.2008.10.007. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, Munson J, Hiatt JB. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oddoux C, Clayton CM, Nelson HR, Ostrer H. Prevalence of Bloom syndrome heterozygotes among Ashkenazi Jews. Am J Hum Genet. 1999;64:1241–1243. doi: 10.1086/302312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olry AE. Prevalence and incidence of rare diseases: bibliographic data. Number 1: Diseases listed in alphabetical order. Orphanet Report Series Rare Diseases collection. 2015:1–55. http://www.orpha.net/orphacom/cahiers/docs/GB/Prevalence of rare diseases by alphabetical list.pdf.
- Oshima J, Yu CE, Piussan C, Klein G, Jabkowski J, Balci S, Miki T, Nakura J, Ogihara T, Ells J, Smith M, Melaragno MI. Homozygous and compound heterozygous mutations at the Werner syndrome locus. Hum Mol Genet. 1996;5:1909–1913. doi: 10.1093/hmg/5.12.1909. [DOI] [PubMed] [Google Scholar]
- Popuri V, Huang J, Ramamoorthy M, Tadokoro T, Croteau DL, Bohr VA. RECQL5 plays co-operative and complementary roles with WRN syndrome helicase. Nucleic Acids Res. 2013;41:881–899. doi: 10.1093/nar/gks1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam KL, Blackshear PJ. Cloning and characterization of RecQL, a potential human homologue of the Escherichia coli DNA helicase RecQ. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- Quang D, Chen Y, Xie X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothmund A. Ueber cataracten in verbindung mit einer eigent mlichen hautde-generation. Arch Klin Exp Ophtal. 1868;14:159–182. [Google Scholar]
- Saponaro M, Kantidakis T, Mitter R, Kelly GP, Heron M, Williams H, Soding J, Stewart A, Svejstrup JQ. RECQL5 controls transcript elongation and suppresses genome instability associated with transcription stress. Cell. 2014;157:1037–1049. doi: 10.1016/j.cell.2014.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino Si, Ozawa K, Eki T, Nogami M, Okumura K, Taguchi H, Hanaoka F, Enomoto T. Molecular cloning of cDNA encoding human DNA helicase Q1 which has homology to Escherichia coli RecQ helicase and localization of the gene at chromosome 12p12. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrabani-Gargir L, Shomrat R, Yaron Y, Orr-Urtreger AVI, Groden J, Legum C. High frequency of a common Bloom syndrome Ashkenazi mutation among Jews of Polish origin. Genet Test. 1998;2:293–296. doi: 10.1089/gte.1998.2.293. [DOI] [PubMed] [Google Scholar]
- Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr, Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihab HA, Rogers MF, Gough J, Mort M, Cooper DN, Day INM, Gaunt TR, Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum Mol Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- Siitonen HA, Sotkasiira J, Biervliet M, Benmansour A, Capri Y, Cormier-Daire V, Crandall B, Hannula-Jouppi K, Hennekam R, Herzog D, Keymolen K, Lipsanen-Nyman M. The mutation spectrum in RECQL4 diseases. Eur J Hum Genet. 2009;17:151–158. doi: 10.1038/ejhg.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon T, Kohlhase J, Wilhelm C, Kochanek M, De Carolis B, Berthold F. Multiple malignant diseases in a patient with Rothmund–Thomson syndrome with RECQL4 mutations: Case report and literature review. Am J Med Genet Part A. 2010;152A:1575–1579. doi: 10.1002/ajmg.a.33427. [DOI] [PubMed] [Google Scholar]
- Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. Sequencing depth and coverage: Key considerations in genomic analyses. Nat Rev Genet. 2014;15:121–132. doi: 10.1038/nrg3642. [DOI] [PubMed] [Google Scholar]
- Sulonen A-M, Ellonen P, Almusa H, Lepistö M, Eldfors S, Hannula S, Miettinen T, Tyynismaa H, Salo P, Heckman C, Joensuu H, Raivio T. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Wang Y, Xia Y, Xu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, Lou H, Xie Y. Mutations in RECQL gene are associated with predisposition to breast cancer. PLoS Genet. 2015;11:e1005228. doi: 10.1371/journal.pgen.1005228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr, Falaschi A, Vindigni A. The human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol. 2009;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsbol TO, Cohen HJ. Werner’s sydrome: An underdiagnosed disorder resembling premature aging. Age. 1984;7:75–88. [Google Scholar]
- Touznik A, Lee JJA, Yokota T. New developments in exon skipping and splice modulation therapies for neuromuscular diseases. Expert Opin Biol Ther. 2014;14:809–819. doi: 10.1517/14712598.2014.896335. [DOI] [PubMed] [Google Scholar]
- Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, Delague V, Muenke M, Jabs EW, Cai J, Wang LL, Plon SE, Fourneau C. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J Med Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltrop M, Aartsma-Rus A. Antisense-mediated exon skipping: Taking advantage of a trick from Mother Nature to treat rare genetic diseases. Exp Cell Res. 2014;325:50–55. doi: 10.1016/j.yexcr.2014.01.026. [DOI] [PubMed] [Google Scholar]
- Vennos EM, James WD. Rothmund-Thomson syndrome. Dermatol Clin. 1995;13:143–150. [PubMed] [Google Scholar]
- Wang LL, Gannavarapu A, Kozinetz CA, Levy ML, Lewis RA, Chintagumpala MM, Ruiz-Maldanado R, Contreras-Ruiz J, Cunniff C, Erickson RP, Lev D, Rogers M. Association between osteosarcoma and deleterious mutations in the RECQL4 gene in Rothmund-Thomson syndrome. J Natl Cancer Inst. 2003a;95:669–674. doi: 10.1093/jnci/95.9.669. [DOI] [PubMed] [Google Scholar]
- Wang LL, Levy ML, Lewis RA, Chintagumpala MM, Lev D, Rogers M, Plon SE. Clinical manifestations in a cohort of 41 Rothmund-Thomson syndrome patients. Am J Hum Genet. 2001;102:11–17. doi: 10.1002/1096-8628(20010722)102:1<11::aid-ajmg1413>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Wang W, Seki M, Narita Y, Nakagawa T, Yoshimura A, Otsuki M, Kawabe T, Tada S, Yagi Y, Ishii Y, Enomoto T. Functional relation among RecQ family helicases RecQL1, RecQL5, and BLM in cell growth and sister chromatid exchange formation. Mol Cell Biol. 2003b;23:3527–3535. doi: 10.1128/MCB.23.10.3527-3535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner O. On cataract in conjunction with scleroderma. In: Hoehn H, translator; Salk D, Fujiwara Y, Martin GM, editors. Werner’s Syndrome and Human Aging. New York: Plenum Press; 1985. pp. 1–14. [Google Scholar]
- Yokote K, Chanprasert S, Lee L, Eirich K, Takemoto M, Watanabe A, Koizumi N, Lessel D, Mori T, Hisama FM, Ladd PD, Angle B. WRN mutation update: Mutation spectrum, patient registries, and translational prospects. Human Mutation. 2016 Sep 26; doi: 10.1002/humu.23128. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Ouais S, Nakura J, Miki T, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- Zhao N, Hao F, Qu T, Zuo YG, Wang BX. A novel mutation of the WRN gene in a Chinese patient with Werner syndrome. Clin Exp Dermatol. 2008;33:278–281. doi: 10.1111/j.1365-2230.2007.02641.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.