Abstract
Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 engineering of the CYP3A5 *3 locus (rs776746) in human liver cell line HuH-7 (CYP3A5 *3/*3) has led to three CYP3A5 *1 cell lines by deletion of the exon 3B splice junction or point mutation. Cell lines CYP3A5 *1/*3 sd (single deletion), CYP3A5 *1/*1 dd (double deletion), or CYP3A5 *1/*3 pm (point mutation) expressed the CYP3A5 *1 mRNA and had elevated CYP3A5 mRNA (P < 0.0005 for all engineered cell lines) and protein expression compared with HuH-7. In metabolism assays, HuH-7 had less tacrolimus (all P < 0.05) or midazolam (MDZ) (all P < 0.005) disappearance than all engineered cell lines. HuH-7 had less 1-OH MDZ (all P < 0.0005) or 4-OH (all P < 0.005) production in metabolism assays than all bioengineered cell lines. We confirmed CYP3A5 metabolic activity with the CYP3A4 selective inhibitor CYP3CIDE. This is the first report of genomic CYP3A5 bioengineering in human cell lines with drug metabolism analysis.
Introduction
More than 50% of the oral drugs in the United States are enzymatically metabolized by the cytochrome P450 (P450) family of enzymes (Guengerich, 2008). The P450s and other drug-metabolizing enzymes are polymorphic, resulting in large variability in metabolic clearance of drugs. In vitro systems to study drug metabolism and genetic variation include cloned and expressed enzymes, human and animal microsomes from individual or pooled donors, and freshly isolated and cultured or cryopreserved hepatocytes; however, primary hepatocytes are not an optimal option because they require harvesting liver, they are expensive, they are not immortalized, and they are highly variable from specimen to specimen. To study genetic variants’ association with metabolism, a genotyped bank of liver microsomes (He et al., 2006), from individual donors, can be examined but cannot sustainably be engineered to study newly identified genetic variants, such as rare variants or those found in minority populations. Also, microsomes are difficult to use to study combinations of genetic variants, especially rare variants or those found in minority populations. Liver microsomes are often from Caucasians, limiting their use to understand metabolism in minority populations. Furthermore, since microsomes come from various individuals, they are genomically heterogeneous and from uncontrolled environments, whereas cell line models are, for the most part, genomically identical except for any specifically altered genetic variant. Thus, we developed genetically modified human liver cell lines that are a sustainable option to investigate the impact of genetic variants on drug metabolism.
Recent reports showed, in rats, that knockout of CYP2E1 (Wang et al., 2016) or CYP3A1/2 (Lu et al., 2017) using clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 could be used in drug metabolism studies; however, using CRISPR/Cas9 to modify human cell lines to study the association of genetic variants with drug metabolism has not been reported. We hypothesized that human liver cell lines can be engineered with CRISPR/Cas9 (Mali et al., 2013c; Ran et al., 2013) to evaluate the effect of genetic variants on drug metabolism. Single genetic variants can be engineered into cell lines that result in altered enzyme activity, gene regulation, or protein expression for drug transport or metabolism studies. Here we present evidence of this concept to study genetic variants in CYP3A5 and their effect on metabolism of two CYP3A4 and CYP3A5 enzymatic substrates: midazolam (MDZ), a sedative or anesthetic, and tacrolimus (Tac), an immune suppressant. Among the P450 enzymes, CYP3A4 and CYP3A5 are the most abundant in the liver, and their expression is highly variable. The CYP3A5 *3 (rs776746) loss of function allele is highly prevalent in people of Caucasian descent (Roy et al., 2005) (allele frequency = 0.94) and leads to low metabolism rates of Tac (de Jonge et al., 2013) compared with individuals with CYP3A5 *1 genotype; however, the CYP3A5 *1 (expresser) allele is enriched in African Americans (Bains et al., 2013) and leads to rapid metabolism of MDZ, Tac, and other drugs. Approximately, 50% of oral drugs are metabolized by CYP3A4 and CYP3A5 (Pelkonen et al., 2008; Tseng et al., 2014). Consequently, the CYP3A5 genotype is an important factor in determining the appropriate doses of drugs. People of African ancestry are often underdosed initially with Tac after organ transplantation (Jacobson et al., 2011), in part owing to the high prevalence of the CYP3A5*1 allele in the African American population (allele frequency, 0.85). Carriers of the CYP3A5 *1 allele often need higher doses of drugs that are CYP3A5 substrates to achieve therapeutic drug levels in the blood. Therefore, there is a need to develop an in vitro, cell culture–based system to understand the effects of genetic variants on drug metabolism before the clinical use of new drugs or to improve dosing of existing drugs.
The first step in development of a suitable liver cell line was to find a clinically relevant parental cell line. To date, there is no commercially available liver cell line that is diploid at chromosome 7 and expresses CYP3A5 *1. The Caco-2 cell line (Sambuy et al., 2005) is a human intestinal cell line that metabolizes drugs, but it has five copies of chromosome 7 and thus is not suitable for studying the diploid CYP3A5 seen in most patients. The HuH-7 cell line (Nakabayashi et al., 1984, 1985) was derived from a hepatic carcinoma that can convert the substrate MDZ, primarily through CYP3A4 activity, in cell culture to its metabolite products hydroxylated 1-OH MDZ and 4-OH MDZ (Choi et al., 2009; Sivertsson et al., 2010, 2013); however, HuH-7 cells are not very efficient at MDZ metabolism because they are homozygous for the slow metabolizing CYP3A5 *3 allele. Thus, there is a need to develop a liver cell line that mimics the rapid drug metabolism associated with the CYP3A5 *1 genotype in cell culture.
We hypothesized that by genetically modifying the HuH-7 cell line to the more metabolically active CYP3A5 *1/*1 or *1/*3 genotypes, the cells would have increased MDZ and Tac metabolic activity. To test the hypothesis, we used CRISPR/Cas9 bioengineering (Mali et al., 2013c; Ran et al., 2013) to develop and characterize new cell lines and then phenotypically evaluate the genotypes’ effects on MDZ and Tac metabolism. These newly engineered cells can be used as a parental cell line in future studies to assess the association of additional genetic variants with drug metabolism and metabolism of other drugs. This is the first report of genomic CYP3A5 bioengineering in human cell lines and functional analysis of associated drug metabolism phenotypes.
Materials and Methods
Selection of HuH-7 Hepatocyte Cell Line as Parental Cell Line.
We selected HuH-7, liver carcinoma cells from the Japanese Cell Research Bank (catalog no. JCRB0403) because 1) HuH-7 cells metabolized MDZ in cell culture (Sivertsson et al., 2010, 2013); 2) HuH-7 cells were diploid, at chromosome 7, where both CYP3A4 and CYP3A5 are located, and therefore are clinically relevant; 3) we sequenced the cells at the CYP3A5 *3, *6, and *7 loci, and HuH-7 cells carried *3/*3 alleles at the rs776746 locus, a defined variant to change via CRISPR/Cas9 bioengineering to the more functional *1/*1 or *1/*3 alleles; 4) these HuH-7 cells were also *1/*1 at the loss of function CYP3A4 *22 (rs35599367) allele; 5) We confirmed CYP3A4 and CYP3A5 mRNA expression by quantitative reverse transcription-polymerase chain reaction (PCR).
Parental Cell Line and Characterization.
HuH-7 (Nakabayashi et al., 1984, 1985), hepatoma cells from a 57-year-old Japanese man, were purchased from the Japanese Cell Research Bank (Osaka, Japan) and used as parental cell line for genetic modification. Cells were grown in Dulbeco’s modified Eagle’s medium (DMEM) with high glucose and pyruvate supplemented with 10% fetal bovine serum and antibiotic-antimycotic (Gibco, Carlsbad, CA), which we refer to as “media” throughout rest of this article.
Genotyping of Cell Lines.
Genomic DNA was isolated from HuH-7 cells using the Roche High-Pure PCR template preparation kit. PCR and sequencing primers (Supplemental Table 1), surrounding single nucleoside polymorphisms (SNPs) CYP3A4 *22 (rs35599367, C > T), CYP3A5 *3 (rs776746, 6986A > G), *6 (rs10264272, 14690G > A), and *7 (rs41303343, nonfunctional) were designed using the NCBI primer-BLAST primer design tool. The sequences surrounding the SNPs in the genomic DNA from the HuH-7 cells were PCR-amplified using AccuPrime Pfx DNA polymerase kit (Thermo Fisher, Somerset, NJ), and then the PCR products were characterized on 1% agarose gel or purified with the Qiagen (Hilden, Germany) PCR clean-up kit and sequenced. PCR products were then Sanger-sequenced using the primers listed in Supplemental Table 1 by University of Minnesota Genomics Center.
Plasmids Guide RNA Construction and Transfection.
A plasmid that expressed a human codon-optimized Cas9 (Mali et al., 2013a–c) nuclease was purchased from Addgene (Cambridge, MA) guide RNAs (gRNAs) targeting the CYP3A5*3 locus were designed using the CRISPR design tool (http://crispr.mit.edu/). DNA gBLOCKS were designed, synthesized, and purchased from Integrated DNA Technologies (Halifax, NS) combining the gRNA from the CRISPR design tool with the gRNA synthesis protocol (Mali et al., 2013a) from Addgene. The gBLOCKs were TOPO-cloned using the Zero Blunt TOPO PCR cloning kit into pCRBlunt II-TOPO vector (Thermo Fisher Scientific). Plasmids were expanded in One Shot Stbl3 Chemically Competent Escherichia coli bacteria purchased from Thermo Fisher. Plasmids were sequence-verified. Plasmids were then prepared for transfection according to the Qiagen Plasmid Maxi kit. Plasmids were quantified and assessed for purity using a NanoDrop 2000 UV/Vis spectrophotometer. Newly designed gRNAs and hCas9 plasmid DNA were transfected into the HuH-7 cells using a Neon Transfection System (Thermo Fisher).
Surveyor Assay to Select Guide RNAs.
Genomic DNA was isolated from transfected cells, and then we performed Surveyor assay to screen gRNAs for ability to cut at CYP3A5 *3 locus using a modified protocol (Guschin et al., 2010) along with Surveyor enzyme from Surveyor Mutation Detection kit for standard gel electrophoresis from Integrated DNA Technologies. Briefly, DNA was extracted from bulk transfected cells using Roche High-Pure PCR template preparation kit. PCR was performed using AccuPrime Taq DNA polymerase, high-fidelity with CYP3A5 specific primers Cel1F*3 (5′-CAACTGCCCTTGCAGCATTT-3′) and Cel1R*3 (5′- ACCCAGGAAGCCAGACTTTG-3′) to produce a 397-base-pair (bp) product (if no deletions). Bio-Rad (Hercules, CA) thermocycler was programed as follows: 1) 94°C for 5 minutes; 2) 94°C for 15 seconds; 3) 56°C for 30 seconds; 4) 68°C for 0.5 minutes; 5) step 2, repeated 34 times; 6) 68°C for 0.5 minutes; and 7) 4°C indefinitely. PCR products were denatured and reannealed after a published protocol (Guschin et al., 2010) and visualized in a 10% Bio-Rad Criterion TBE PAGE to determine DNA heteroduplexes from heterogeneous cell cultures caused by CRISPR/Cas9 and gRNA targeting (Supplemental Fig. 3).
Transfection with Selected gRNA and hCAS9.
To create cell lines that delete the CYP3A5*3 splice junction, via nonhomologous end joining (NHEJ), two selected gRNAs (gRNA1 and gRNA2) and hCas9 plasmids were transfected into the HuH-7 cells using the Neon transfection system. The two gRNAs target each side of the CYP3A5*3 locus. The resultant cells were then single-cell–cloned to produce homogenous cell lines.
To create the point mutation (pm) cell line, gRNA2 was transfected into cells, with a homology-directed repair (HDR) template (HDR single-stranded DNA template ssODN 3A5*3_E+). The sequence of ssODN 3A5*3_E+ is 5′-gcttaacgaatgctctactgtcatttctaaccataatctctttaaagagctcttttgtctttcaaTATCTCTTCCGTGTTTGGACCACATTACCCTTCATCATATGAAGCCTTGGGTGGCTCC-3′. The underlined a indicates the *3 base that is changed from a guanine (g). The lower-case letters are the intron sequence, and the upper-case letters are the exon 3B sequence. The cells were treated with 1 µM SCR7 (Xcessbio, San Diego, CA) and 5 µM L755,507 (Xcessbio) at the time of transfection and for 7 days after transfection until single-cell cloning.
Single-Cell Cloning and Cell Line Screening via PCR and DNA Sanger Sequencing.
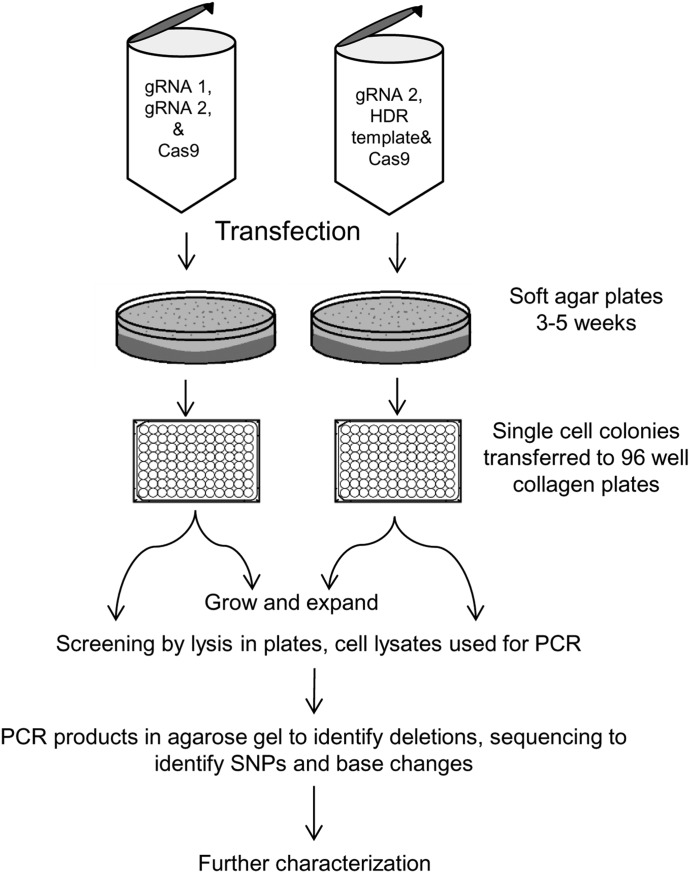
Transfected cells were plated in media/soft agar mixture as previously described (Kim et al., 2014; Dorr et al., 2015) and propagated to become homogeneous cell lines. Specifically, in 150 mm3 15 ml of a 0.6% solution of UltraPure low-melting-point agarose (Thermo Fisher Scientific, catalog no. 16520-100) in media was plated at 38.5°C and cooled until solid. Next, 15,000 transfected cells in 15 ml of 38.5°C media with 0.3% UltraPure low-melting-point agarose was layered on top and cooled. The plate was covered with 10 ml of media and incubated at 37°C with 5% CO2 for about 3–5 weeks until cell colonies were visible. Colonies were then picked with a sterile 200-µl pipette tip and transferred to individual wells of a 96-well collagen I–coated plate and cultured until confluent (approximately 3 to 4 weeks).
For large-scale screening, we dissociated the cells using trypsin-EDTA (0.25%) and transferred half the culture to fresh 96-well plates with media and grown. The remaining cells in the 96-well plate were centrifuged at 350g for 5 minutes. Trypsin was removed, and the cell pellets were lysed using the QuickExtract DNA extraction solution from Epicentre Biotechnology (Madison, WI). Lysates were used as a PCR template and then PCR-amplified with an AccuPrime Pfx DNA polymerase kit in 96-well PCR plates. The primers for amplification of the CYP3A5*3 region were designed using NCBI primer design (http://www.ncbi.nlm.nih.gov/tools/primer-blast/) and are 7853F (5′-GCATTTAGTCCTTGTGAGCACTTG-3′) and 8303R (5′-CATACGTTCTGTGTGGGGACAAC-3′). Thermocycler was programed as follows: 1) 94°C for 5 minutes; 2) 94°C for 15 seconds; 3) 55°C for 30 seconds; 4) 68°C for 30 seconds; 5) go to step 2, 34 times; 6) 68°C for 7 minutes; and 7) 4°C indefinitely. PCR products were purified using the MinElute 96 UF PCR purification kit (Qiagen), characterized by electrophoresis through a 2% agarose gel and by sequencing of the PCR products. Sequencing was performed by the University of Minnesota Genomics Center using sequencing primer 7884F (5′- ACCTGCCTTCAATTTTTCACTG-3′) and 8267R (5′- CTTCACTAGCCCGATTCTGC-3′). Sequence data were analyzed using DNA Star Lasergene and Geneious software.
RNA Splicing Assay.
RNA was isolated from the confluent cells using the Qiagen RNeasy Mini Kit. RNA was quantified using the Qubit 2.0 Fluorometer and the Qubit RNA BR Assay Kit. RNA was then converted to cDNA using oligo-dT primer and ThermoScript Reverse Transcriptase kit. CYP3A5 *1/*3 or *1/*1 genotyped human liver RNA was also used as controls and cDNA made by the same method. PCR primers were developed using the NCBI primer design tool and mRNA sequence file of CYP3A5 (Genbank accession no. BC025176.1) of the 500 bp surrounding the *3 nucleotide locus. The cDNA was then used as PCR template with the following primers: CYP3A5 cDNA ex2F (5′-GTCACAATCCCTGTGACCTGAT-3′) and CYP3A5 cDNA ex5R (5′-TTGGAGACAGCAATGACCGT-3′). Thermocycler settings were as follows: 1) 94°C for 5 minutes; 2) 94°C for 15 seconds; 3) 50.5°C for 30 seconds; 4) 68°C for 30 seconds; 5) step 2, repeated 34 times; 6) 68°C for 5 minutes; 7) 4°C forever using the AccuPrime Pfx DNA polymerase kit. PCR products were purified using the QIAquick PCR purification kit, characterized by electrophoresis through a 2% agarose gel, and imaged using ethidium bromide staining and a Bio-Rad ChemiDoc Touch imaging system.
Quantitative RT-PCR to Detect CYP3A5 Transcripts.
CYP3A5 mRNA was quantified using cells from the MDZ and Tac assays. The primers were the same as for the mRNA splicing assay (Supplemental Table 1). The RNA was the same as the RNA splicing assays, and GAPDH primers were used as the reference control; 2 µg of RNA was converted to cDNA, and 5 µl of 1 in 20 diluted cDNA was used in a 20-µl reaction mix for SYBR Green assay-based quantitative RT-PCR. We used a Roche (Basel, Switzerland) light cycler and made graphs using GraphPad Prism (LaJolla, CA) software.
Immunoblot Analysis for CYP3A5 *1 and *3 Variants in Engineered Cell Lines.
CYP3A4 and CYP3A5 protein expression was determined by immunoblot analysis. Total lysates were recovered from HuH-7 cells and the new derivative cell line in RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific).
Protein was estimated by using the Bio-Rad protein assay for microsomes and Pierce BCA (Rockford, IL) protein assay kit assay for lysates with bovine serum albumin as the standard. Then 60 µg of total lysate was separated on 10% SDS-PAGE and immunoblotted with 1:10,000 dilution of monoclonal anti-CYP3A4 K03 (Schuetz et al., 1996) or 1:250 dilution of WB-3A5 (Corning, Corning, NY), followed by 1:10,000 dilution of secondary antibodies horseradish peroxidase–conjugated anti-mouse and anti-rabbit (Jackson ImmunoResearch Inc., West Grove, PA), respectively. β-Actin protein expression was determined by monoclonal anti-actin (Sigma, St. Louis, MO), followed by 1:10,000 dilution of secondary antibodies horseradish peroxidase–conjugated anti-mouse. The blot was developed with the ECL Western blotting reagents (GE Healthcare, Piscataway, NJ). Bands on film were optically scanned (Epson, Long Beach, CA).
Midazolam and Tacrolimus Metabolism Assays.
HuH-7 and engineered cells were grown to confluence in 12-well Corning BioCoat Collagen I multi-well plates for 3 to 4 weeks in media. Media were refreshed two or three times a week. Cells were then overlaid with Corning Matrigel matrix and then induced for 3 days by adding 100 µM phenytoin sodium (USP), diluted in methanol, and 10 µM rifampicin (Sigma), diluted in methanol, in cell culture. Media were changed daily with inducers rifampicin and phenytoin added. On the 4th day, 500 µl of media was added to the cells with 100 µM phenytoin and 10 µM rifampicin with either equal volumes of methanol, as a negative control, or MDZ from Cerriliant Corporation (Round Rock, TX) diluted in methanol so that MDZ’s final concentration was 5 µM (1628 ng/ml) in cell culture media. Cells were incubated overnight, and media were collected and assayed for MDZ, 1-OH MDZ, and 4-OH MDZ by high-performance liquid chromatography mass spectrometry. To determine the metabolic function of the engineered cell lines on Tac (Toronto Research Chemicals Inc., North York, ON), we used the same process except for using six-well collagen-coated plates and 1.5 ml with 13 ng/ml Tac reaction volume.
Detection Methods for Tac, MDZ, 1-OH MDZ, and 4-OH MDZ.
Detection and quantification of MDZ, 1-OH MDZ, and 4-OH MDZ in cell culture media were performed using a high-performance liquid chromatograph (Agilent 1200 Series, Santa Clara, CA) coupled with a TSQ Quantum triple-stage quadrupole mass spectrometer (Thermo-Electron, San Jose, CA). Detection and quantification of Tac were performed using chromatographic separation (Agilent 1100 HPLC, Agilent Inc.) and mass spectrometry (API 4000; Sciex Inc., Redwood City, CA). These detailed methods are in the Supplemental Material.
CYP3CIDE Experiments.
CYP3CIDE (Walsky et al., 2012; Tseng et al., 2014) (Sigma-Aldrich) was used as a selective CYP3A4 inhibitor in cell cultures and diluted in dimethylsulfoxide. To determine the concentration of CYP3CIDE to use in cell culture, we performed a dose response in HuH-7 and the CYP3A5 *1/*1 double deletion (dd) cell line between 100 nM and 1 mM using MDZ as the substrate. For further analysis, we used 50 µM CYP3CIDE in our experiments with all cell lines. Dose-response curves were assessed using GraphPad Prism software.
Statistical Analysis.
All comparisons were conducted using t test for continuous variables.
Results
Genotyping of Single-Cell Clones after CRISPR/Cas9 Bioengineering of CYP3A5.
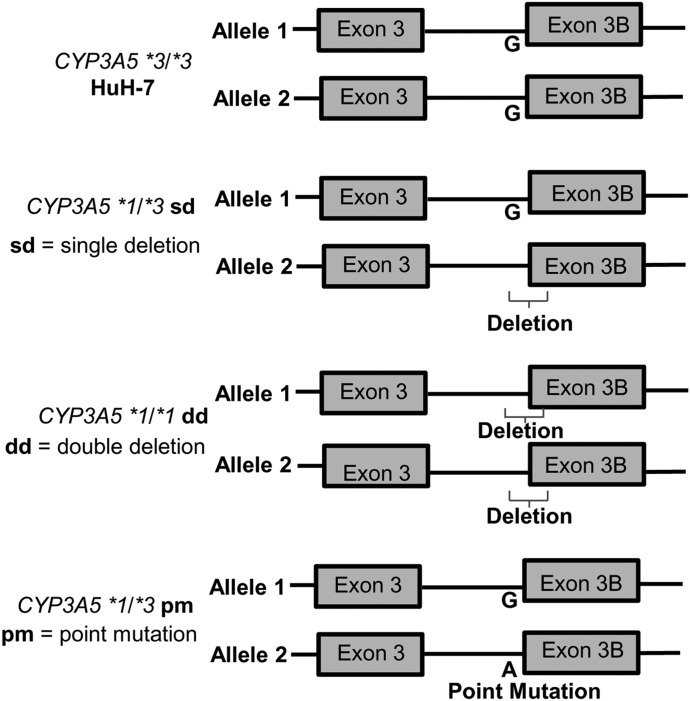
To create CYP3A5*1 cells from the HuH-7 cells that were CYP3A*3, we altered CYP3A5 based on CRISPR proto-spacer adjacent motifs (PAM) near the exon 3B splice junction (Fig. 1). To identify the cell line that had the exon 3B 5′ splice junction deleted, we screened single-cell clones by PCR amplifying, then forward and reverse Sanger sequencing of a 451-bp region flanking the splice junction (Fig. 1). The splice junction contains the *3 locus (rs776746), and we deleted this junction to express CYP3A5 *1 mRNA via alternative splicing. These cells were transfected with gRNA1, gRNA2, and hCas9 plasmids (Fig. 2). We screened 235 single-cell clones, and 74 (32%) were mutated. Mutations in modified cell lines were as follows: 22 (9.4%) had a heterozygous frameshift near one of the two gRNA cut sites, 23 (9.8%) were heterozygous for the 77-bp deletion between gRNA sites, 15 (6.4%) cell lines were homozygous for the 77-bp deletion, and 14 (6.0%) cell lines were classified as “other.” The “other” mutations in cell lines include cell lines that had multiple frameshifts or were heterozygous for a deletion and frameshift or had other mutations. The heterozygous deletion cell line was designated CYP3A5 *1/*3 sd (sd = single deletion), and the homozygous deletion cell line was designated CYP3A5 *1/*1 dd (Fig. 3).
Fig. 1.
CYP3A5 *3 allele (rs776746) and guide RNA targeting strategy. Guide RNAs (gRNAs) were targeted to PAM sequences on each side of the CYP3A5 *3 allele (gRNA1 or gRNA2 locus). Exon 3B sequence is in capital letters, and the upstream intron sequence is in lower-case letters. There are 77 bp between the gRNA guided Cas9 cut sites.
Fig. 2.
Workflow for development of CYP3A5 genetically modified cell lines using CRISPR/Cas9 and clonal selection. The CYP3A5 *3 splice junction was deleted with gRNA1, gRNA2, and Cas9, or the CYP3A5 *3 SNP was point mutated using gRNA2 and an HDR template to convert the *3 guanine to a *1 adenine. After transfection, the cells were single-cell cloned by plating in soft agar. The single-cell clones were transferred to collagen I–coated plates until confluent. Cells were then expanded or screened by PCR of the CYP3A5 *3 locus and sequencing of the PCR products.
Fig. 3.
CYP3A5 maps of cell lines used in this study: HuH-7 (CYP3A5 *3/*3); CYP3A5 *1/*3 sd, which has a deletion of *3 splice junction at one allele; CYP3A5 *1/*1 dd, which has a deletion of *3 splice junction at two alleles; and the CYP3A5 *1/*3 pm, which has a guanine to adenine point mutation converting one allele from *3 to *1.
To identify the cell line CYP3A5 *1/*3 pm (pm = point mutation) (Fig. 3), we screened 212 single-cell clones using DNA sequencing that were transfected with hCAS9, gRNA2 and HDR template (Fig. 2); 33 (16%) were mutated. Mutations in cell lines were as follows: 23 (11%) had a heterozygous frameshift near the gRNA2 cut site, 2 (0.9%) had homozygous frameshift, 1 (0. 5%) had heterozygous point mutation at *3 locus, 0 (0.0%) had homozygous point mutations at *3 locus, and 8 (4.0%) were classified as “other.” These other mutants include cell lines that had multiple frameshifts, were heterozygous for a deletion and frameshift, or had other mutations.
PCR Characterization of Genomic DNA in New Cell Lines.
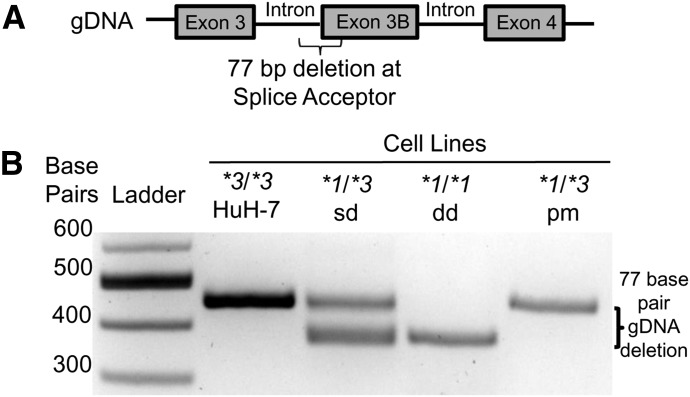
Of the cell lines that were sequenced, select cell lines that were CYP3A5 *1/*3 or *1/*1 by Sanger sequencing screening were validating by PCR characterization of genomic DNA at the CYP3A5 *3 locus (Fig. 4, A and B). The 77-bp deletions were characterized by PCR amplification of genomic DNA and visualized by 2% agarose gel electrophoresis (Fig. 4B). Both heterozygous (CYP3A5 *1/*3 sd) and homozygous (CYP3A5 *1/*1 dd) cell lines were developed with the 77 bp CYP3A5 exon 3B 5′ splice junction deleted. The point mutation heterozygous cell line, CYP3A5*1/*3 pm, did not have a deletion at the splice junction (Fig. 4B). These cell lines were then further validated with Sanger sequencing (Supplemental Fig. 1).
Fig. 4.
Characterization of genomic DNA at CYP3A5 *3 loci in genetically modified cell lines. (A) Map of genomic DNA spanning CYP3A5 exons 3, 3B, and 4. (B) Electrophoresis through 2% agarose gel of PCR products spanning the CYP3A5 *3 locus (rs776746) of genomic DNA from the cell lines. Data show HuH-7 genomic DNA as the reference. CYP3A5 *1/*3 sd had two different alleles, one allele being the same as the HuH-7 reference and one allele being 77 bp shorter than the reference. The 77-bp deletion indicated deletion of the exon 3B splice junction. CYP3A5 *1/*1 1dd showed no reference allele and thus deletion of the exon 3B in both alleles. CYP3A5 *1/*3 pm has one PCR product the same size as the reference, which is expected in a point mutant.
CYP3A5 mRNA Splicing Assay and Sequencing of Engineered Cells.
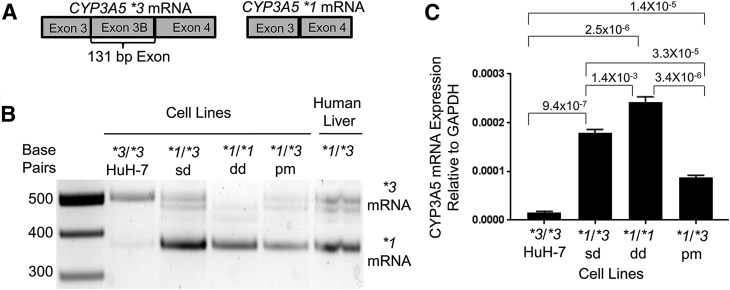
CYP3A5 mRNA splice variants were evaluated by gel electrophoresis at the *3 locus to ensure that the deletion, or point mutation, of the CYP3A5 exon 3B 5′ splice junction changed the cells to express the *1 mRNA instead of the *3 mRNA (Fig. 5, A and B). Keuhl et al. (2001) have previously shown that the 131-bp exon 3B was present in the CYP3A5 *3 mRNA and absent in the *1 mRNA, which was confirmed by Busi and Cresteil (2005a, b). Total mRNA from the cell lines was converted to cDNA and then PCR-amplified with primers that flanked the CYP3A5 exon 3B to determine whether the exon 3B was absent in the engineered cell lines. RNA from human liver cDNA genotyped as *1/*3 was used as the control (Fig. 5B). This mRNA splicing assay confirmed the absence of the *3 mRNA in the newly developed cell line, CYP3A5 *1/*1 dd. Additionally, the *1/*3 heterozygote cell lines, CYP3A5 *1/*3 sd and CYP3A5 *1/*3 pm, expressed both the *1 and the *3 mRNA splice variants as compared with the human liver cDNA controls (Fig. 5B). To further validate the identity of the CYP3A5 splice variants in the cell lines, we sequenced the CYP3A5 mRNA via Sanger sequencing of the CYP3A5 RT-PCR products (Supplemental Fig. 2). The sequences confirmed that the exon 3B was absent in the CYP3A5 *1/*1 dd cell line when aligned to a *3 sequence. The CYP3A5 *1/*3 cell lines CYP3A5 *1/*3 sd and CYP3A5 *1/*3 pm sequences became jumbled at exon 3B, as expected when sequencing a heterozygote (Supplemental Fig. 2); however, sequencing in forward and reverse directions confirmed that the CYP3A5 *1/*3 heterozygote-engineered cells expressed both the CYP3A5 *3 and *1 mRNAs.
Fig. 5.
Characterization of CYP3A5 RNA expression in genetically modified cell lines. (A) mRNA map of CYP3A5 exons 3, 3B, and 4. (B) mRNA splicing assay showed expression of CYP3A5 *1 mRNA in modified cell lines. Genotyped human liver from a CYP3A5 *1/*3 genotyped patient was used as control. (C) Quantitative RT-PCR showed expression levels of CYP3A5 in cell lines relative to GAPDH. P value comparing CYP3A5 mRNA expression levels between cell lines are from a paired two-sample t test.
Quantitative RT-PCR Resulted in Elevated CYP3A5 Transcripts in *1 Expressing Cell Lines.
Quantitative RT-PCR showed significantly elevated CYP3A5 mRNA expression in CYP3A5 *1/*1 dd compared with HuH-7 CYP3A5 *3/*3 cells (P = 2.5 × 10−6). All engineered CYP3A5 *1 cell lines had elevated CYP3A5 mRNA compared with HuH-7 (P ≤ 0.0001) (Fig. 5C). The CYP3A5 *3 mRNA, expressed in cell lines, was targeted for nonsense-mediated decay30 as the likely cause of reduced CYP3A5 mRNA in *3 cell lines. The CYP3A5 *1/*3 cell lines had intermediate CYP3A5 mRNA expression compared with HuH-7 CYP3A5 *3/*3 and CYP3A5 *1/*1 dd (Fig. 5C).
Immunoblot Confirms for CYP3A5 Expression in Engineered Cell Lines.
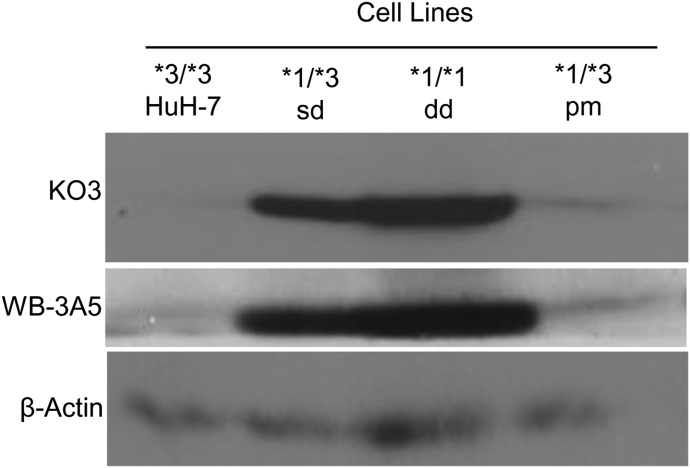
We immunoblotted for the CYP3A5 protein expression in the cell lines using two separate primary antibodies: KO3 (Schuetz et al., 1996), which detects CYP3A family (including CYP3A4 and CYP3A5), or the CYP3A5-specific WB-3A5 (Schmidt et al., 2004) (Fig. 6). The CYP3A5*1/*3 sd cell line visually expressed less CYP3A5 protein expression than CYP3A5 *1/*1 dd but higher expression than the CYP3A5 *3/*3 HuH-7 cells. The CYP3A5*1/*3 pm cell line had poor CYP3A5 protein expression compared with the CYP3A5 *1/*3 sd cell line (Fig. 6). These results were consistent with both the KO3 and WB-3A5 antibodies. It is likely that the K03 antibody did not detect much CYP3A4 protein owing to low CYP3A4 expression in the HuH-7 cell line. Thus, the bioengineered cell lines express CYP3A5 protein.
Fig. 6.
Characterization of CYP3A5 protein expression in genetically modified cell lines. Immunoblot with primary (1°) antibodies K03 antibody (Schuetz et al., 1996) that recognizes P450 family proteins, including CYP3A4 and CY3A5, and WB-3A5 (Schmidt et al., 2004), which is specific for CYP3A5 or β-actin as a reference.
Metabolism Assays Show CYP3A5 *1 Expressing Cells Have Increased MDZ and Tac Metabolism Compared with CYP3A5 *3/*3 HuH-7 Cells.
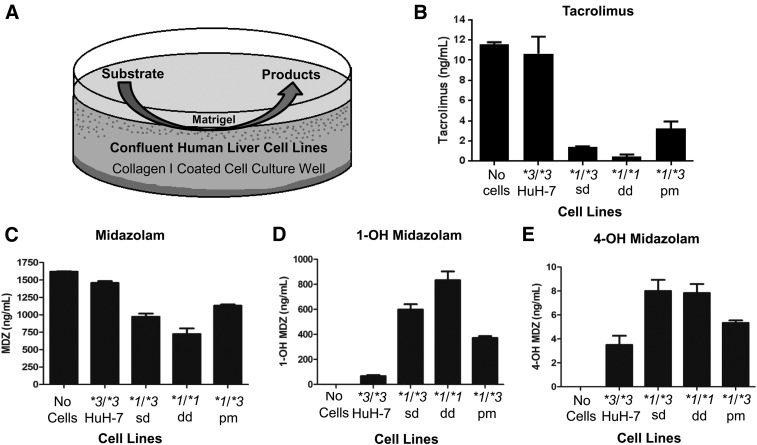
As shown in Fig. 7A, we performed an MDZ metabolism assay to determine whether the new cell lines metabolized MDZ to the products 1-OH MDZ and 4-OH MDZ. We also quantitated Tac disappearance using the metabolism assay. As expected, the CYP3A5 *3/*3 (HuH-7) cells had higher levels of Tac (Fig. 7B) and MDZ (Fig. 7C) in cell culture after overnight incubations than CYP3A5 *1/*3 sd, CYP3A5 *1/*3 pm, or CYP3A5 *1/*1 dd cell lines because of the decreased metabolism by the CYP3A5 *3/*3 cells. Furthermore, increased production of 1-OH MDZ (Fig. 7D) and 4-OH MDZ (Fig. 7E) was observed by the engineered cell lines compared with the parental HuH-7 cell line. Significant metabolic differences between each of the cell lines were found by comparing substrate disappearance or product formation between the engineered cell lines and the HuH-7 parental cell line (all P < 0.05) (Table 1). Thus, the engineered CYP3A5 *1-expressing cell lines were more efficient at converting MDZ to its hydroxylated metabolites compared with the CYP3A5 *3/*3 parental HuH-7 cell line. The engineered CYP3A5 expressing cells are also more active at metabolizing Tac than the HuH-7 cells, which coincides with previous studies with cloned, expressed CYP3A4 and CYP3A5 that demonstrated that the intrinsic clearance of Tac is higher for CYP3A5 than CYP3A4 (Dai et al., 2006).
Fig. 7.
MDZ and Tac metabolism assays confirm that CYP3A5 *1–expressing cells have increased metabolic activity compared with HuH-7 CYP3A5 *3/*3–expressing cells. (A) Metabolism assay used in this study. Cells were plated on collagen I–coated plates and grown at confluence for 2 to 3 weeks and then layered with Matrigel. Cells were then induced with rifampicin and phenytoin for 3 days; then a substrate of Tac or MDZ was added over night. Cell culture media were collected and assayed for Tac, MDZ, or the MDZ products 1-OH MDZ or 4-OH MDZ by liquid chromatography-mass spectrometry. (B) Tac was used as the substrate and was assayed to assess its disappearance. Each column represents five biologic replicates of a representative experiment and shows the disappearance of the Tac caused by the cells’ metabolism. (C) MDZ was used as the substrate to assess its metabolism. Each column represents six biologic replicates of a representative experiment and shows the disappearance of the MDZ caused by the cells’ metabolism. (D) The corresponding 1-OH MDZ products from the MDZ experiments and the (E) corresponding 4-OH MDZ products.
TABLE 1 .
P values comparing Tac or MDZ metabolic activity between cell lines
P values were calculated based on the paired two-sample t test comparing between each cell line. Substrates and products were quantified by liquid chromatography-mass spectrometry of the media from the cell cultures.
| Cell Lines Compared |
P Values |
|||
|---|---|---|---|---|
| Substrates |
Products |
|||
| Tac | MDZ | 1-OH MDZ | 4-OH MDZ | |
| HuH-7 a *3/*3 vs. CYP3A5 *1/*3 sdb | 5.4 × 10−3 | 2.4 × 10−5 | 2.3 × 10−6 | 7.3 × 10−4 |
| HuH-7 *3/*3 vs. CYP3A5 *1/*1 ddc | 2.7 × 10−3 | 5.3 × 10−4 | 8.6 × 10−5 | 2.7 × 10−4 |
| HuH-7 *3/*3 vs. CYP3A5 *1/*3 pmd | 2.7 × 10−2 | 5.0 × 10−7 | 3.8 × 10−8 | 1.3 × 10−3 |
| CYP3A5 *1/*3 sd vs. CYP3A5 *1/*1 dd | 1.0 × 10−2 | 3.6 × 10−1 | 3.5 × 10−2 | 1.6 × 10−2 |
| CYP3A5 *1/*3 sd vs. CYP3A5 *1/*3 pm | 5.3 × 10−2 | 5.3 × 10−1 | 1.8 × 10−3 | 9.4 × 10−1 |
| CYP3A5 *1/*1 dd vs. CYP3A5 *1/*3 pm | 3.1 × 10−2 | 4.1 × 10−1 | 2.0 × 10−3 | 2.4 × 10−2 |
HuH-7 was the parental cell line with CYP3A5 *3/*3 alleles.
CYP3A5 *1/*3 sd was a bioengineered cell line with the CYP3A5 *1 allele made by deletion of a splice acceptor on one of the alleles.
CYP3A5 *1/*1 dd was a bioengineered cell line with both CYP3A5 *1 alleles made by deletion of a splice acceptor on two of the alleles.
CYP3A5 *1/*3 pm was a bioengineered cell line with the CYP3A5 *1 allele made by a point mutation of a splice acceptor on one of the alleles.
CYP3CIDE as a Selective CYP3A4 Inhibitor in MDZ Assays.
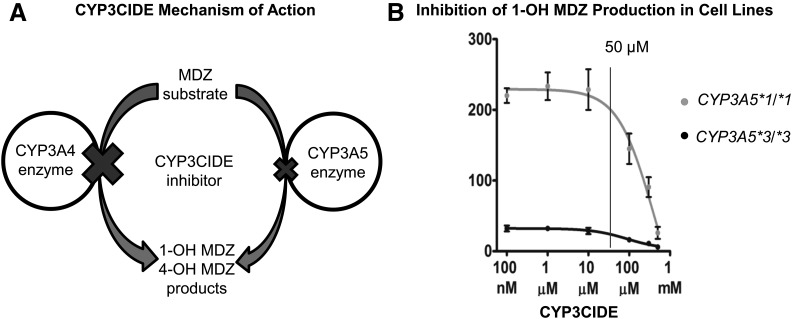
CYP3CIDE (Walsky et al., 2012; Tseng et al., 2014) is a selective inhibitor of CYP3A4 that can also inhibit CYP3A5 at higher concentrations. The concentration-dependent effects of CYP3CIDE are shown in Fig. 8A. To determine the concentration of CYP3CIDE to use in cell culture, we did a dose-response study with the HuH-7 (CYP3A5 *3/*3) and CYP3A5 *1/*1 dd cell lines (Fig. 8B). After the dose-response study, the cell lines were incubated with MDZ with or without 50 µM CYP3CIDE to assess CYP3A5 activity in modified cell lines.
Fig. 8.
CYP3CIDE-selective inhibition of CYP3A4 and CYP3A5 and dose response in cell lines. (A) CYP3CIDE inhibition of CYP3A4 and CYP3A5 enzymatic activity. CYP3CIDE has a higher affinity on CYP3A4 inhibition than on CYP3A5 inhibition. The substrate in this experiment was MDZ, and the products of the reactions were 1-OH MDZ and 4-OH MDZ. (B) CYP3CIDE dose-response curve in HuH-7 (CYP3A5 *3/*3) and CYP3A5 *1/*1 dd cell lines with the 1-OH MDZ product as metabolite. The concentration of 50 µM CYP3CIDE was chosen for further study in other cell lines.
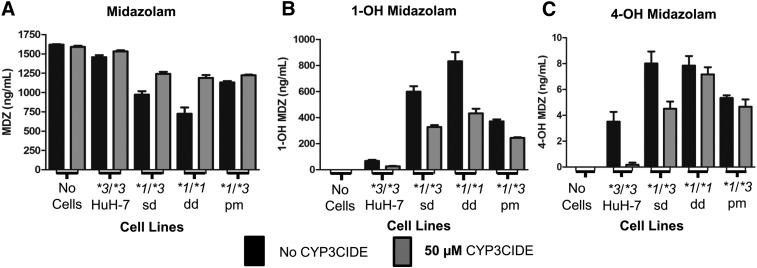
When CYP3CIDE was present, there was slight difference in the MDZ reduction by the HuH-7 cell line (P = 0.044). The MDZ reduction was more pronounced in all the CYP3A5*1-expressing cell lines comparing those with and those without CYP3CIDE (P ≤ 0.005) (Fig. 9A). The 1-OH MDZ production by HuH-7 cells was lower with CYP3CIDE (P < 0.05), whereas all three CYP3A5 *1-expressing cell lines had even more significant reduction of 1-OH MDZ production (all P ≤ 0.005) (Fig. 9B). Further analysis of MDZ metabolism by the cell lines with 4-OH MDZ as the minor metabolic product (Fig. 9C) showed that CYP3CIDE almost completely halted 4-OH MDZ production by HuH-7 cells (P = 0.007). Figure 9C also showed that HuH-7 and CYP3A5 *1/*3 sd cell lines had less 4-OH MDZ production with CYP3CIDE (P ≤ 0.01 and P ≤ 0.05, respectively). Neither CYP3A5 *1/*1 dd nor CYP3A5 *1/*3 pm had significant 4-OH MDZ production differences with CYP3CIDE (P > 0.05). These CYP3CIDE experiments showed that these cell lines had differential activity when a selective CYP3A4 inhibitor was present, indicating phenotypically active CYP3A5, which was not present in the parental HuH-7 (CYP3A5 *3/*3) cell line.
Fig. 9.
Effect of CYP3CIDE on MDZ metabolism in CYP3A5 genetically modified cells. Cells were treated with (gray bars) or without 50 µM CYP3CIDE (black bars) and assayed for MDZ metabolism. Each column in (C–E) represents six biologic replicates of a representative experiment. (C) MDZ disappearance by cell lines after incubation of MDZ in the presence of CYP3CIDE. (D) Production of 1-OH MDZ by cell lines in the presence of CYP3CIDE. (E) Production of 4-OH MDZ by cell lines in the presence of CYP3CIDE. P values in (C–E) were calculated based on the paired two-sample t test comparing with or without CYP3CIDE are ns, not significant; P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Discussion
This study showed the successful CRISPR/Cas9 bioengineering of a human liver cell line, HuH-7, to create new cell lines that express the common CYP3A5 *1 variant that is known to be highly relevant toward drug metabolism. Unlike recent reports using CRISPR/Cas9 to knock out CYP2E1 (Wang et al., 2016) or CYP3A1/2 (Lu et al., 2017) function in rats, we used CRISPR/Cas9 to activate CYP3A5 expression in human cells lines by conversion of *3 to *1 genotype. This is the first report of engineered cell lines for both heterozygous and homozygous CYP3A5 *1 expression in human liver cell culture and phenotypic analysis.
This study showed that it is possible to use two methods of CRISPR/Cas9 biotechnology to modify the HuH-7 cells to express CYP3A5 *1 by splice junction deletion using two gRNAs or with one gRNA and a homology-directed repair template. Without the need for fluorescent- activated cell sorting or less precise limiting dilution techniques, a soft agar clonal selection with expansion on collagen I–coated plates technique was used to isolate unique human hepatocyte cell lines. This technique is important in isolating hepatocyte cell lines because the cells do not grow well as single cells on standard plastic cell culture dishes. Also, growing the cell lines at confluence for 2 to 3 weeks, layering with Matrigel (Corning), and inducing the cells with rifampicin and phenytoin increased the hepatocytes’ metabolic activity. We determined the impact of induction while developing the MDZ metabolism assay. Since induction increased MDZ metabolism in the HuH-7 cells, we did not change the protocol for the genetically modified cell lines or when using Tac as the substrate. In a previous study, 1-OH MDZ production by HuH-7 cells was low, ∼2.5 pmol/mg of protein/minute with rifampicin induction and lower at ∼1 pmol/mg or protein/minute without induction (Sivertsson et al., 2010). In comparison, our induction method in HuH-7 parental cell line produced ∼67 ng/ml 1-OH MDZ in media. Although these units are not the same, our method does have higher MDZ metabolism. In our genetically modified cell lines, such as CYP3A5 *1/*1 dd, 833 ng/ml 1-OH MDZ was formed. Relative to the unmodified HuH-7 (CYP3A5 *3/*3) cells, these new cell lines express elevated CYP3A5 mRNA and led to significantly higher metabolism of Tac and MDZ, which are well known substrates for CYP3A5. The genetically modified cells also produce the expected MDZ metabolites (1-OH MDZ and 4-OH MDZ). Thus, these cells show promise as cell lines for drug metabolism assessment. Since these cells are diploid at chromosome 7 by karyotype (Ding et al., 2013) and by copy number of the CYP3A5 locus (Forbes et al., 2015) and these new cells have increased drug metabolism, the cells would be ideal to use as parental cell lines to functionally study other genetic variants and metabolism of other drugs. Newly discovered variants can also be genetically engineered into the cells to study combinations of genetic variants.
Differential response to drug therapy is common. These differences are due in part to the presence of genetic variants in P450 enzymes that alter drug metabolism and pharmacokinetics. Ethnic minority populations may carry alternative variants or the same variant as Caucasians but with a different allele frequency (Yasuda et al., 2008). Because ethnic minority populations are generally underrepresented in clinical trials in the United States, differences in pharmacokinetics and response rates are often not detected until the drug has reached the market (Hamel et al., 2016). Therefore, efficient preclinical methods to study how metabolism may be altered in the presence of alternative alleles would be extremely useful. The cell lines that we developed show the potential of the CRISPR/Cas9 biotechnology in the drug metabolism field. We chose CYP3A5 *3 and *1 as model alleles to genetically engineer since the effect of these alleles on metabolism is well established in vitro both by traditional methods and clinically. Specifically, this technology will allow for the creation of cell lines with varying combinations of common alleles, a platform to study rare alleles and those occurring in populations not represented in clinical trials.
A limitation of this study is that we did not investigate the metabolic activity of the newly developed cell lines in the absence of induction. We know induction increases MDZ metabolism in HuH-7 cells (Sivertsson et al., 2010); thus, the induction process is necessary to study robust metabolism in the genetically modified cells. Another limitation of this study is that there are possibly off-target effects of the CRISPR/Cas9 engineering that are common in this type of genetic engineering (Cho et al., 2014). We addressed this limitation by selecting gRNAs with the least amount of potential off targets, using the Massachusetts Institute of Technology CRISPR algorithm, and confirmed with sequencing that the DNA was cut at the desired site surrounding the CYP3A5*3 locus. Additionally, this study showed that we can produce the CYP3A5 *1 mRNA in human liver cell lines, as seen in human samples from previous studies by Kuehl and colleagues (2001) and by Busi and Cresteil (2005b). We showed that these cells definitively express CYP3A5 *1 mRNA and active CYP3A5 enzyme as either heterozygous or homozygous compared with control HuH-7 cells; however, the two CYP3A5 *1/*3 heterozygote cell lines have either a 77-bp splice junction deletion or a point mutation, and these cell lines have CYP3A5 expression and activity that are different from those of HuH-7. The CYP3A5 *1/*1 dd was vastly more metabolically active than the other cell lines and would be the most useful for comparative studies with the CYP3A5 *1 genotype.
Future work using these methods and developed cell lines includes a number of directions. We envision panels of cell lines developed using genetic modification that can be used to study genetic variants associated with drug metabolism. These panels could include variants of particular metabolism genes, gene families, common variants, rare variants or population specific variants. Common variants or combinations of common variants could be engineered into cell lines and used in preclinical drug metabolism screens to predict pharmacokinetics. Rare variants could also be engineered into the cell lines. If rare alleles were found to alter metabolism, such results may predict subjects at risk for drug failure or toxicity and may also allow for early testing of alternative doses for trial subjects carrying combination of variants or rare alleles. There are substantial challenges in studying rare alleles in human clinical trials owing to inadequate sample size; therefore, engineered cells could bring extreme value to drug development. In addition, drugs already on the market could be rapidly screened. Since the new cells in this study express CYP3A5, the cells are especially useful to studying genetic variants that effect CYP3A5 expression. Finally, it may also be possible that multiple variants could be engineered into a single cell line that would more closely emulate the specific human populations. This technology can potentially lead to faster preclinical development that can save time and money. As the use of this technology expands, we will be able to more accurately predict substrate metabolism, pharmacokinetics, toxicities, and efficacy, especially in minority patients with rare genetic variants.
Acknowledgments
The authors thank the University of Minnesota Genomics Center for numerous molecular biology services. Ajay Israni and Casey Dorr, along with Minneapolis Medical Research Foundation, have filed a provisional patent with the US Patent and Trademark Office titled: “Genetically Modified Cells for Metabolic Studies.” The patent application number is 62/459,749. The cell line CYP3A5 *1/*3 sd is deposited at American Type Culture Collection (ATCC) patent depository under the name “Human Liver Cell Line, 97 CYP3A5 *1/*3” with ATCC patent deposit designation PTA-123710.
Abbreviations
- bp
base pair
- CRISPR
clustered regularly interspaced short palindromic repeats
- dd
double deletion
- gRNA
guide RNA
- HDR
homology directed repair
- MDZ
midazolam
- NHEJ
nonhomologous end joining
- 1-OH MDZ
1-hydroxy midazolam
- 4-OH MDZ
4- hydroxyl midazolam
- pm
point mutation, P450, cytochrome P450
- PAM
proto-spacer adjacent motif, PCR, polymerase chain reaction
- sd
single deletion
- SNP
single nucleotide polymorphism
- Tac
tacrolimus
Authorship Contributions
Participated in research design: Dorr, Remmel, Muthusamy, Moriarity, Wu, Guan, Oetting, Jacobson, Israni.
Conducted experiments: Dorr, Muthusamy, Wu, Fisher, Schuetz, Kazuto.
Contributed new reagents or analytic tools: Dorr, Remmel, Muthusamy, Wu, Fisher, Moriarity, Israni.
Performed data analysis: Dorr, Muthusamy, Fisher, Kazuto, Wu, Oetting.
Wrote or contributed to the writing of the manuscript: Dorr, Remmel, Muthusamy, Fisher, Moriarity, Wu, Guan, Oetting, Jacobson, Israni.
Footnotes
This study was funded by a Minneapolis Medical Research Foundation Postdoctoral Career Development Award (to C.D.) and grant support from United States National institutes of Health National Institute for Allergy and Infectious Diseases [Grant AI U19 AI070119] (to A.I.).
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Bains RK, Kovacevic M, Plaster CA, Tarekegn A, Bekele E, Bradman NN, Thomas MG. (2013) Molecular diversity and population structure at the cytochrome P450 3A5 gene in Africa. BMC Genet 14:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi F, Cresteil T. (2005a) CYP3A5 mRNA degradation by nonsense-mediated mRNA decay. Mol Pharmacol 68:808–815. [DOI] [PubMed] [Google Scholar]
- Busi F, Cresteil T. (2005b) Phenotyping-genotyping of alternatively spliced genes in one step: study of CYP3A5*3 polymorphism. Pharmacogenet Genomics 15:433–439. [DOI] [PubMed] [Google Scholar]
- Caco2 [Caco2] (ATCC® HTB37™).
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, Kim JS. (2014) Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24:132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Sainz B, Jr, Corcoran P, Uprichard S, Jeong H. (2009) Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica 39:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COSMIC : Catalogue of somatic mutations in cancer.
- Dai Y, Hebert MF, Isoherranen N, Davis CL, Marsh C, Shen DD, Thummel KE. (2006) Effect of CYP3A5 polymorphism on tacrolimus metabolic clearance in vitro. Drug Metab Dispos 34:836–847. [DOI] [PubMed] [Google Scholar]
- de Jonge H, de Loor H, Verbeke K, Vanrenterghem Y, Kuypers DR. (2013) Impact of CYP3A5 genotype on tacrolimus versus midazolam clearance in renal transplant recipients: new insights in CYP3A5-mediated drug metabolism. Pharmacogenomics 14:1467–1480. [DOI] [PubMed] [Google Scholar]
- Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, Kuperwasser N, Motola DL, Meissner TB, Hendriks WT, et al. (2013) A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell 12:238–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorr C, Janik C, Weg M, Been RA, Bader J, Kang R, Ng B, Foran L, Landman SR, O’Sullivan MG, et al. (2015) Transposon mutagenesis screen identifies potential lung cancer drivers and CUL3 as a tumor suppressor. Mol Cancer Res 13:1238–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Beare D, Gunasekaran P, Leung K, Bindal N, Boutselakis H, Ding M, Bamford S, Cole C, Ward S, et al. (2015) COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res 43:D805–D811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich FP. (2008) Cytochrome p450 and chemical toxicology. Chem Res Toxicol 21:70–83. [DOI] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. (2010) A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol 649:247–256. [DOI] [PubMed] [Google Scholar]
- Hamel LM, Penner LA, Albrecht TL, Heath E, Gwede CK, Eggly S. (2016) Barriers to clinical trial enrollment in racial and ethnic minority patients with cancer. Cancer Contr 23:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Xie HG, Collins X, Edeki T, Yan Z. (2006) Effects of individual ginsenosides, ginkgolides and flavonoids on CYP2C19 and CYP2D6 activity in human liver microsomes. Clin Exp Pharmacol Physiol 33:813–815. [DOI] [PubMed] [Google Scholar]
- Jacobson PA, Oetting WS, Brearley AM, Leduc R, Guan W, Schladt D, Matas AJ, Lamba V, Julian BA, Mannon RB, et al. DeKAF Investigators (2011) Novel polymorphisms associated with tacrolimus trough concentrations: results from a multicenter kidney transplant consortium. Transplantation 91:300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Shin HJ, Ha HL, Park YH, Kwon TH, Jung MR, Moon HB, Cho ES, Son HY, Yu DY. (2014) Methylsulfonylmethane suppresses hepatic tumor development through activation of apoptosis. World J Hepatol 6:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391. [DOI] [PubMed] [Google Scholar]
- Lu J, Shao Y, Qin X, Liu D, Chen A, Li D, Liu M, Wang X. (2017) CRISPR knockout rat cytochrome P450 3A1/2 model for advancing drug metabolism and pharmacokinetics research. Sci Rep 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. (2013a) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 31:833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Esvelt KM, Church GM. (2013b) Cas9 as a versatile tool for engineering biology. Nat Methods 10:957–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. (2013c) RNA-guided human genome engineering via Cas9. Science 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Yamane T, Miyazaki M, Miyano K, Sato J. (1984) Phenotypical stability of a human hepatoma cell line, HuH-7, in long-term culture with chemically defined medium. Gan 75:151–158. [PubMed] [Google Scholar]
- Nakabayashi H, Taketa K, Yamane T, Oda M, Sato J. (1985) Hormonal control of alpha-fetoprotein secretion in human hepatoma cell lines proliferating in chemically defined medium. Cancer Res 45:6379–6383. [PubMed] [Google Scholar]
- Pelkonen O, Turpeinen M, Hakkola J, Honkakoski P, Hukkanen J, Raunio H. (2008) Inhibition and induction of human cytochrome P450 enzymes: current status. Arch Toxicol 82:667–715. [DOI] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy JN, Lajoie J, Zijenah LS, Barama A, Poirier C, Ward BJ, Roger M. (2005) CYP3A5 genetic polymorphisms in different ethnic populations. Drug Metab Dispos 33:884–887. [DOI] [PubMed] [Google Scholar]
- Sambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F. (2005) The Caco-2 cell line as a model of the intestinal barrier: influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol Toxicol 21:1–26. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Baumann F, Knüpfer H, Brauckhoff M, Horn LC, Schönfelder M, Köhler U, Preiss R. (2004) CYP3A4, CYP2C9 and CYP2B6 expression and ifosfamide turnover in breast cancer tissue microsomes. Br J Cancer 90:911–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz EG, Schinkel AH, Relling MV, Schuetz JD. (1996) P-glycoprotein: a major determinant of rifampicin-inducible expression of cytochrome P4503A in mice and humans. Proc Natl Acad Sci USA 93:4001–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivertsson L, Edebert I, Palmertz MP, Ingelman-Sundberg M, Neve EP. (2013) Induced CYP3A4 expression in confluent Huh7 hepatoma cells as a result of decreased cell proliferation and subsequent pregnane X receptor activation. Mol Pharmacol 83:659–670. [DOI] [PubMed] [Google Scholar]
- Sivertsson L, Ek M, Darnell M, Edebert I, Ingelman-Sundberg M, Neve EP. (2010) CYP3A4 catalytic activity is induced in confluent Huh7 hepatoma cells. Drug Metab Dispos 38:995–1002. [DOI] [PubMed] [Google Scholar]
- Tseng E, Walsky RL, Luzietti RA, Jr, Harris JJ, Kosa RE, Goosen TC, Zientek MA, Obach RS. (2014) Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). Drug Metab Dispos 42:1163–1173. [DOI] [PubMed] [Google Scholar]
- Walsky RL, Obach RS, Hyland R, Kang P, Zhou S, West M, Geoghegan KF, Helal CJ, Walker GS, Goosen TC, et al. (2012) Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug Metab Dispos 40:1686–1697. [DOI] [PubMed] [Google Scholar]
- Wang X, Tang Y, Lu J, Shao Y, Qin X, Li Y, Wang L, Li D, Liu M. (2016) Characterization of novel cytochrome P450 2E1 knockout rat model generated by CRISPR/Cas9. Biochem Pharmacol 105:80–90. [DOI] [PubMed] [Google Scholar]
- Yasuda SU, Zhang L, Huang SM. (2008) The role of ethnicity in variability in response to drugs: focus on clinical pharmacology studies. Clin Pharmacol Ther 84:417–423. [DOI] [PubMed] [Google Scholar]