Abstract
Background
Recent climatic history has strongly impacted plant populations, but little is known about its effect on microbes. Alders, which host few and specific symbionts, have high genetic diversity in glacial refugia. Here, we tested the prediction that communities of root symbionts survived in refugia with their host populations. We expected to detect endemic symbionts and a higher species richness in refugia as compared to recolonized areas.
Methods
We sampled ectomycorrhizal (EM) root tips and the nitrogen-fixing actinomycete Frankia communities in eight sites colonized by Alnus glutinosa subsp. barbata close to the Caucasus in Georgia. Three sites were located in the Colchis, one major Eurasian climatic refugia for Arcto-Tertiary flora and alders, and five sites were located in the recolonized zone. Endemic symbionts and plant ITS variants were detected by comparing sequences to published data from Europe and another Tertiary refugium, the Hyrcanian forest. Species richness and community structure were compared between sites from refugia and recolonized areas for each symbionts.
Results
For both symbionts, most MOTUs present in Georgia had been found previously elsewhere in Europe. Three endemic Frankia strains were detected in the Colchis vs two in the recolonized zone, and the five endemic EM fungi were detected only in the recolonized zone. Frankia species richness was higher in the Colchis while the contrary was observed for EM fungi. Moreover, the genetic diversity of one alder specialist Alnicola xanthophylla was particularly high in the recolonized zone. The EM communities occurring in the Colchis and the Hyrcanian forests shared closely related endemic species.
Discussion
The Colchis did not have the highest alpha diversity and more endemic species, suggesting that our hypothesis based on alder biogeography may not apply to alder’s symbionts. Our study in the Caucasus brings new clues to understand symbioses biogeography and their survival in Tertiary and ice-age refugia, and reveals that isolated host populations could be of interest for symbiont diversity conservation.
Keywords: Frankia, Ectomycorrhiza, Colchis, Alnus, Alnicola, Glacial refugia, Tertiary refugia
Introduction
Glacial refugia are recognized sources of genetic diversity and hot spots of taxonomic diversity (Médail & Diadema, 2009). They are also key regions for studying biogeography, as recently shown for fungi (Geml et al., 2010; Ghobad-Nejhad et al., 2012; Merényi et al., 2014). In Europe, the numerous fossil records from glacial periods and the analysis of allelic diversity have contributed to identification of refugia (Taberlet & Cheddadi, 2002; Gavin et al., 2014), mainly in Italy, the Iberian Peninsula and the Balkans (Médail & Diadema, 2009). Demographic contractions and expansions, and population genetic patterns associated with ice age refugia have been deeply studied in oaks, pines, beech, and alders (Taberlet & Cheddadi, 2002; Hampe & Jump, 2011). Interestingly, all these tree species host mutualistic fungi on their roots, and are ectomycorrhizal (EM; Smith & Read, 2008). However, whether these biogeographic and genetic processes play out similarly in microbes is unknown. In obligatory host-associated microbes, the potential for joint genetic responses to extinction and recolonization is high. On the other hand, they could have also survived outside of refugia, either in spore banks, or associated with other hosts. The existing knowledge about tree biogeography offers a framework to compare their symbiont communities inside and out of refugia, and test the hypothesis that symbionts have survived with their host in glacial refugia,
The recent history of Alnus has been intensely studied in Europe (King & Ferris, 1998; Douda et al., 2014; Havrdová et al., 2015; Mandák et al., 2016a; Mandák et al., 2016b). Compared to timber trees, alders are ideal models for biogeographical studies of historical migrations because there are numerous natural populations with low human impact (Douda et al., 2014). Thanks to molecular markers, southern refugia have been detected for A. glutinosa (L.) Gaertn. in the Iberian, Apennine, Corsica, North Africa, Balkan and Anatolian Peninsulas (King & Ferris, 1998). More recently, the existence of both refugia in Belarus and western Russia has been demonstrated; they were important sources for northward post-glacial expansion of alders (Douda et al., 2014). Today, A. glutinosa populations are still marked by recent isolation in glacial refugia (King & Ferris, 1998), and several unique chloroplast haplotypes have been detected in relict populations from Morocco (Lepais et al., 2013) and Turkey (King & Ferris, 1998; Havrdová et al., 2015). Of interest, the high genetic diversity detected in Turkey concurs with the observed morphological variations of A. glutinosa in this region. Therefore, four subspecies are recognized in this region: A. glutinosa subsp. glutinosa, A. glutinosa subsp. antitaurica Yalt, A. glutinosa subsp. betuloides Ansin, and A. glutinosa subsp. barbata (CA Meyer) Yalt.
Alnus glutinosa subsp. barbata is distributed from Turkey to Iran, and is considered as a Tertiary relict plant, common in the Colchis forests of Georgia. Pollen records from the Quaternary era and distribution modeling have revealed that Alnus swamps were extended in Colchis floodplains and existed continuously over the last 10,000 years (Connor & Kvavadze, 2009). Today, stands of A. glutinosa subsp. barbata still dominate the Colchis floodplain (Nakhutsrishvili, 2012). Located in the South Caucasus, the Colchis region is one of the three major refugia of Tertiary relict taxa worldwide (Milne & Abbott, 2002; Kikvidze & Ohsawa, 2001; Denk, Frotzler & Davitashvili, 2001). Because a mountain ridge blocks rains coming from the Black Sea, the Colchis floodplain is well separated from arid lowlands and steppes from Eastern Georgia (Nakhutsrishvili, 2012). According to pollen records, alder recolonized Eastern Georgia and the central mountain ridge around 2,000 years BP, and are represented today by isolated populations (Connor & Kvavadze, 2009). Adjacent to the Colchis forests of Georgia, the other major Western Eurasia climate refugium for temperate forests is the Hyrcanian forest, located on the southern coast of the Caspian Sea in the region of Iran and southern Azerbaidjan. The two refugia lie approximately 2,000 km apart, and are now completely isolated from each other by steppe vegetation. However, they still share several Tertiary plant relict species, and are considered to be the two oldest temperate deciduous forests in western Eurasia (Maharramova et al., 2015).
As compared to other tree species, alders host few lineages of EM fungi (Molina, 1981; Moreau, Peintner & Gardes, 2006; Moreau et al., 2011; Rochet et al., 2011) and species-poor EM communities (Põlme et al., 2013; Roy et al., 2013; Kennedy, Walker & Bogar, 2015). Among frequently encountered EM taxa, Alnicola and Alpova are two genera that appear to be strictly associated with alders because they have never been found on any other tree species. Several species of Lactarius, Russula, Amanita and Cortinarius are also exclusive to alders. Alders also associate with nitrogen-fixing actinobacteria, all belonging to the genus Frankia (Weber, Nurmiaho-Lassila & Sundman, 1987) and also with strong host-specificity (Cotin-Galvan et al., 2016). At a worldwide scale, differences among Frankia communities are partly correlated with differences in EM communities (Põlme et al., 2014). From North America to Mexico, the similarities between EM communities also support a co-migration with their host (Kennedy et al., 2011). Together, these results suggest shared histories at broad scales, for the plant, Frankia and EM fungi.
If alder populations, Frankia and EM fungi communities have undergone a shared history, the bacterial and the fungal communities should have both survived in the Colchis, and alders should host more endemic symbionts in this refugium. Moreover, if South Caucasus has been a refugium and a recolonization source for Europe, European specific symbionts should be present in Georgia, together with endemic symbionts, especially in the Colchis. Therefore, the main objectives of the present study are to (1) document the diversity of the microbial symbionts communities of A. glutinosa subsp. barbata in Georgia and determine whether the Colchis floodplain hosts endemic symbionts, (2) test if EM fungi and Frankia communities are both more species rich in the Colchis, (3) examine how recent climatic history has imprinted the distribution of alder symbionts and their communities at a larger scale, by comparing our results to recent studies on the Hyrcanian forest.
Material and Methods
Sampling sites
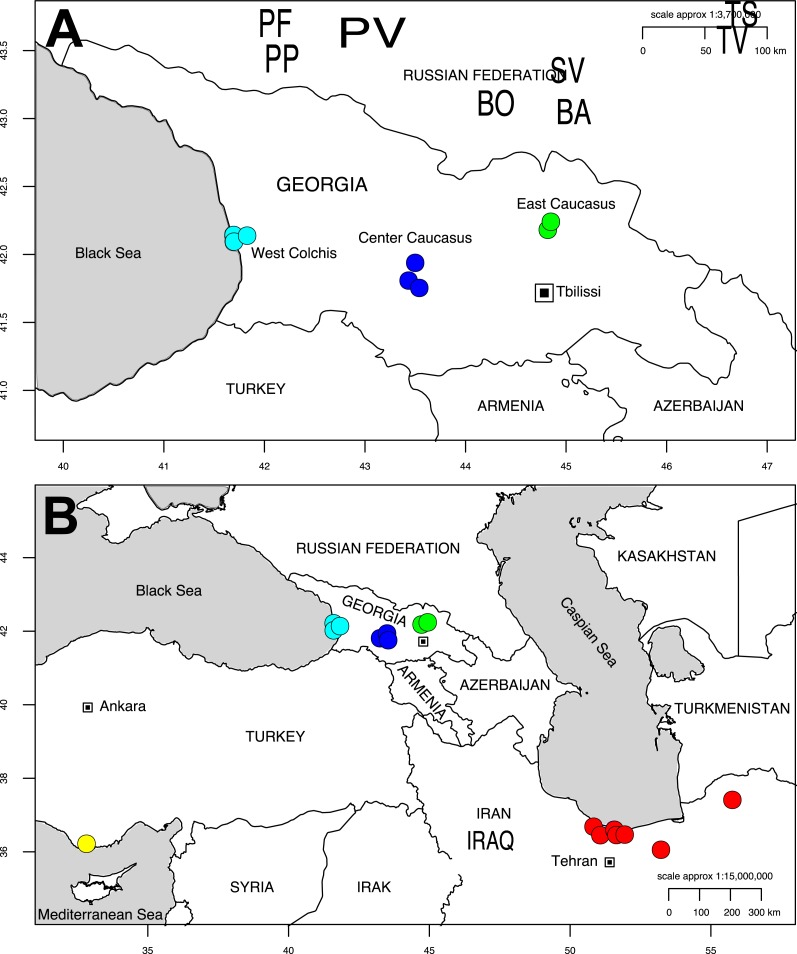
Eight sites were studied in Georgia, including sites representing both large and isolated Alnus stands located from 0 to 1,800 m elevation. Three sites were located in the Colchis, considered as a refugium (PV, PP, PF). Five sites were located in the recolonized zone, three in central Georgia near the water separation line that splits Western and Eastern Georgia (BA, BO and SV), and two in Eastern Georgia at the Caucasus footstep (TS and TR; Fig. 1A, Table S1 of Appendix S1). Climatic data (19 variables) were retrieved from the Worldclim database (http://www.worldclim.org) through R raster package (Hijmans & Van Etten, 2012).
Figure 1. Map of Georgia (A) pointing out the sampling sites for Alnus glutinosa subsp. barbata in Western (Cyan), Central (Blue) and Eastern (Green) Georgia. Only Western sites belong to the Colchis floodplain.
Map showing our study sites and sampling sites from previous studies on Alnus subcordata from the Hyrcanian forests in Iran in red, and Alnus orientalis from Turkey in Yellow (B). The two maps were produced with the R package OpenStreetMap.
Plant, ectomycorrhizal fungi and Frankia sampling
For each site, six trees were sampled, separated by at least 10 m as in Roy et al. (2013). One leaf per tree was dried in silica gel. Roots were collected after tracing roots up to one meter from the trunk and circa 50 cm of roots were collected per tree and kept in soil at 4°C until laboratory processing. In the laboratory, roots were washed under tap water over a 500 µm wide grid, and examined under a binocular microscope in distilled water. A minimum of 16 ectomycorrhizae per tree were picked, and separately kept in 2% CTAB buffer at 4°C for one week and then at −20°C. All Frankia nodules were stored in 2% CTAB buffer (100 mM Tris HCl pH8; 1.4 M NaCl; 20 mM Na2EDTA; 2% N-Acetyl-NNN-trimethyl ammonium bromide).
Alnus phylogenetic position and genetic diversity
One cm2 of leaf was ground to extract DNA using the Wizard genomic DNA purification kit (Promega, Charbonnières les Bains, France) as in Rochet et al. (2011). The Internal Transcribed Spacer (ITS) of the plant nuclear ribosomal DNA was amplified using ITS1P (5′-TTATCATTTAGAGGAAGGAG-3′)—ITS4 primers (White et al., 1990) for each tree, following conditions of Rochet et al. (2011). The chloroplast gene mat K was amplified for one tree per population using the primer pairs Matk_1R_KIM and MatK_3F_KIM (Dunning & Savolainen, 2010) and sequenced following conditions of Rochet et al. (2011). Sequences were manually corrected, and deposited in Genbank under accession numbers KX897895 –KX897935 for ITS and KX897936 –KX897944 for matK). Reference sequences of European Alnus species were downloaded from Genbank Alignments were created with MUSCLE (Edgar, 2004). Phylogenies were computed using Raxml (Stamatakis, Hoover & Rougemont, 2008), by Maximum Likelihood analysis following a GTR model of evolution, and tested through a fast-bootstrap analysis (1,000 replicates) on the CIPRES website (Miller, Pfeiffer & Schwartz, 2010). Phylogenies allowed the placing of A. glutinosa subsp. barbata among other European alders and detect the occurrence of rare ITS variant in Georgia.
Phylogenetic diversity of Frankia and comparison with European strains
The DNA was extracted from Frankia root nodules and three genes were amplified and sequenced: dnaA, ftsZ and pgk, with primers specific to Frankia as in Pozzi et al. (2015). Sequences were submitted to the EMBL (European Nucleotide Archive) under accession numbers LT616989 –LT617015 (dnaA), LT617016 –LT617048 (ftsZ), LT599862, LT599864, LT599865 and LT599870 –LT599890 (pgk). To test if Frankia sequences were phylogenetically more diverse in Georgia as compared to Europe, sequences from Pozzi et al. (2015), including strains isolated from A. glutinosa in Europe, were used as a reference. Alignments for each marker were built using MUSCLE and concatenated. A phylogeny was computed following the same methods as for the plant phylogeny.
All statistical analyses on community diversity were performed with R 2.3.4.4 (R Development Core Team, 2008). To describe the diversity of Frankia communities, pairwise Kimura’s 2-parameters distances were measured using ape (R) based on the dnaA, ftsZ and pgk alignment (concatenated). The histogram of pairwise distances measured from this alignment showed a distribution peak at 0.01 and a gap between 0.01 and 0.05 (absolute Kimura’s 2-parameters distances). Clusters of sequences more than 0.01 similar were considered as distinct molecular operational taxonomic units (MOTUs).
Phylogenetic diversity of ectomycorrhizal fungi and comparison with European sequences
DNA extraction was performed on each EM root tip, using the Wizard genomic DNA purification kit (Promega, Charbonnières les Bains, France) as described in Rochet et al. (2011). The fungal ITS region was amplified using fungal universal primers ITS-1F/ITS-4 (Gardes & Bruns, 1993; White et al., 1990). PCR conditions were the same as in Rochet et al. (2011). Amplification products were sequenced by the MilleGen company (Labège, France). Sequences were manually corrected using 4Peaks 1.7.1 (Griekspoor & Groothuis, 2017) and deposited in Genbank (accession numbers KX897613 –KX897894).
Fungal sequences were compared to Genbank and UNITE http://unite.ut.ee/ (Kõljalg et al., 2005) databases using the BLAST algorithm (Altschul et al., 1990), which allowed the identification of fungal genera. To compare our sequences with MOTUs previously detected in Europe, all sequences produced recently from Alnus roots in Europe (Põlme et al., 2013; Roy et al., 2013) were downloaded. Sequences were aligned separately for each genus with MUSCLE (http://www.drive5.com/muscle/) (Edgar, 2004), and groups of sequences more than 97% similar were delineated with MOTHUR (Schloss et al., 2009), a threshold used in previous publications on Alnus and commonly accepted to delineate fungal MOTUs (Nilsson et al., 2008). Sequences that had between 93 and 97% similarity with Genbank or Unite sequences were identified from the BLAST result at the genus level only (e.g., “genus sp.”). Sequences that had no close similarity (>97%) with Genbank or Unite sequence were considered as putatively endemic to Georgia. For the specialist genus Alnicola, reference sequences produced from European specimens were downloaded from Genbank (Moreau, Peintner & Gardes, 2006; Rochet et al., 2011) and aligned with the sequences from the present study. Based on the alignment and phylogeny, computed using methods described above, Alnicola MOTUs were identified to the species level, and rare ITS variants were detected from the alignment.
Diversity patterns of symbiont communities in Georgia
For Frankia and EM fungi, the species richness (alpha diversity) was measured at the site level, and the beta diversity was measured through Bray–Curtis distance, which takes species abundance into account. Species accumulation curves at the scale of Georgia and Chao 1 diversity indices were computed to reflect gamma diversity. The spatial autocorrelation of communities was tested by a multivariate autocorrelation test (multispati r test, ade4 package, Dray & Dufour, 2007). The correlation between beta diversity and distance was tested by a Mantel test (999 permutations). Because these tests were not statistically significant, no vector of spatial weights was integrated in the following tests. The difference in community structure (measured by the Bray– Curtis distance) between the three sampling zones was tested by a permutational multivariate analysis of variance (perMANOVA). Similarities between communities were visualized by a non-metric multidimensional scaling (NMDS). The correlation of NMDS structure with bioclimatic factors, latitude, longitude, and elevation was tested by permutations tests (envfit function, vegan package in R, Oksanen et al., 2007). The same analysis was computed for Frankia and for EM fungi together and separately.
Comparison between EM fungi communities from Colchis and Hyrcan refugia
Only sequences of EM fungi were compared because our sequencing strategy for Frankia differs from Põlme et al. (2014). To compare EM communities between Colchis and the Hyrcanian forests, sequences produced by Põlme et al. (2013, sampling sites positioned on Fig. 1B) were downloaded from UNITE. Sampling strategy for EM root tips (5–6 trees sampled per site) and sequencing methods (Sanger sequencing of individual root tips, using the ITS marker) were sufficiently similar to be compared in a single analysis. Fungal sequences produced from mycorrhizae of Alnus subcordata CA Mey. and A. glutinosa in Iran, and A. orientalis Decne. in Turkey, were aligned with sequences from the present study, and MOTUs were delineated for each genus as previously described. A matrix of MOTUs x sites was built, including Turkish, Georgian and Iranian sites (see Table S2 of Appendix S1). We detected the occurrence of endemic or shared MOTU based on this matrix, We tested if Colchis communities were as species-rich as communities associated with A. subcordata in the Hyrcanian forests by analyzing alpha diversity variance (ANOVA). Finally, we tested how the geographical distance and the distinct hosts explained the differences in community structure observed in this region of Tertiary refugia (including sites in refugia and outside).
Results
Alnus phylogenetic position and genetic diversity at a regional scale
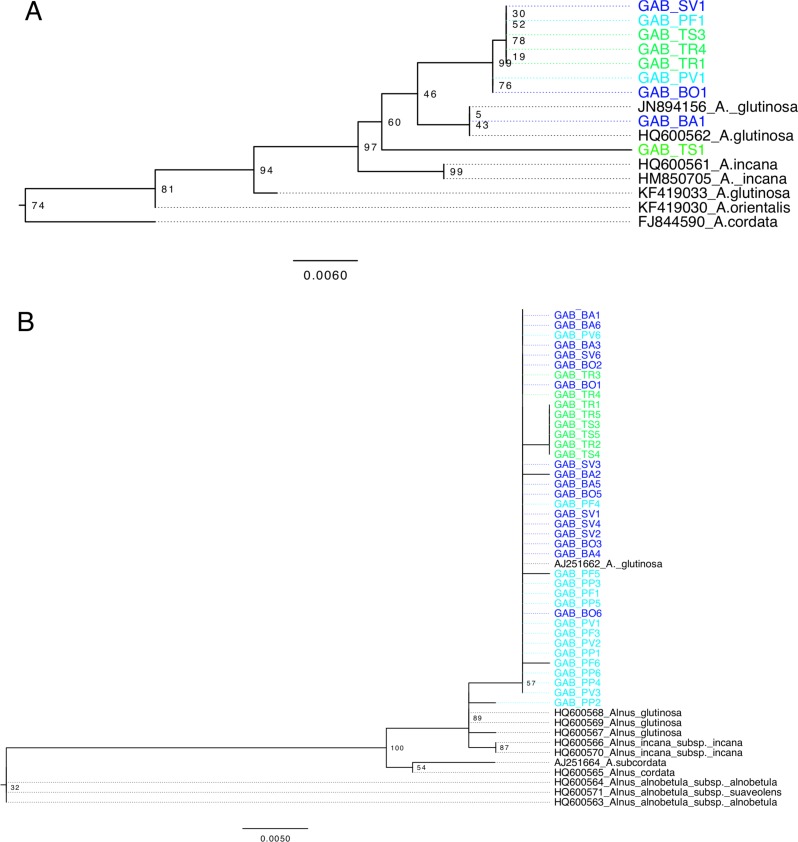
All matK sequences of A. glutinosa from Europe and A. glutinosa subsp. barbata clustered into a monophyletic clade (97% bootstrap, Fig. 2A) distinct from A. incana (L.) Moench. but monophyly of Georgian sequences was not supported. Two sequences from the Central and Eastern populations (TS1 and BA1) showed 11–27 nucleotide differences with other sequences from Georgia (Fig. 2A), and were more similar to sequences from Europe. The monophyletic clade of other Georgian sequences was characterized by eleven unique positions. Based on the ITS phylogeny, differences between A. glutinosa and A. incana were not supported, and only two nucleotides were variable between the two species (Fig. 2B). Besides a low bootstrap support, sequences from Georgia clustered in a monophyleticclade that also includes one European A. glutinosa sequence. The sequences from Eastern Georgia (all TS and part of TR) were characterized by one nucleotide difference with other sequences from Georgia.
Figure 2. Alnus phylogeny, based on matK (A) and ITS (B), computed by maximum likelihood, following a GTR + I model of evolution, and tested by 1,000 bootstrap replicates.
Sequences of Alnus isolated from Georgia are highlighted according to region as shown in Fig. 1. Sequences are named according to their sampling site.
Phylogenetic diversity of Frankia and comparison with European strains
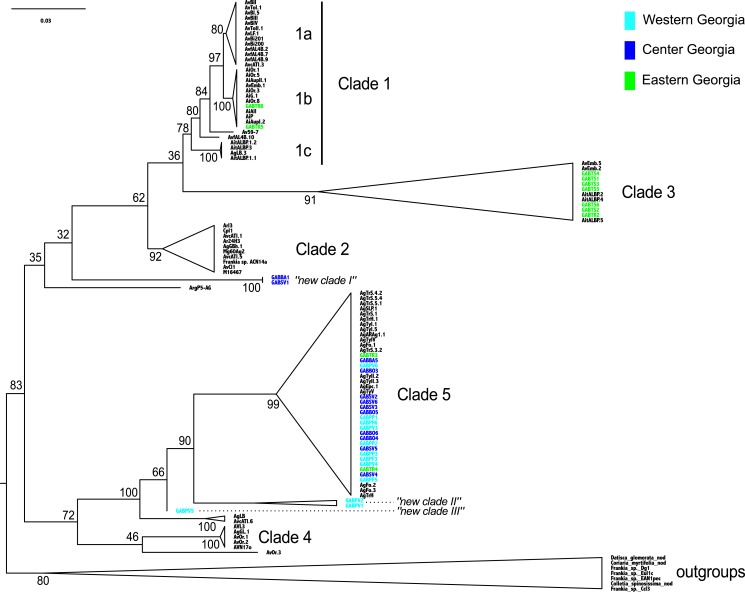
The Frankia strains detected in Georgia did not cluster into a monophyletic clade, and the twelve MOTUs were scattered throughout the phylogeny (clades 1b, 3 and 5 of Pozzi et al., 2015): clade 3 and 5 comprised three and four MOTUs respectively, and “new clade II” gathered two distinct MOTUs. All MOTUs but one belonged to clades commonly associated with A. glutinosa in Europe (Fig. 3). Two sequences from the Eastern population (TR) belonged to a monophyletic clade, frequently associated with A. alnobetula (Erhr.) K Koch (clade 1b, Fig. 3). Three sequences from Western populations were not similar to any other known sequence (new clade II and III, Fig. 3). Finally, two Central Georgia sequences clustered in an isolated lineage (new clade I, Fig. 3).
Figure 3. Frankia phylogeny, based on dnaA, ftsZ, and pgk genes, computed by maximum likelihood, following a GTR + I model of evolution, and tested by 1,000 bootstrap replicates. Sequences of Frankia isolated from Georgia are highlighted according to region as shown in Fig. 1.
Sequences are named according to their sampling sites, and reference sequences are named as in Pozzi et al. (2015).
Phylogenetic diversity of ectomycorrhizal fungi and comparison with European sequences
DNA was extracted and PCR-amplified from a total of 732 individual root tips: 484 of them produced ITS sequences among which 328 met our quality threshold. The four Helotiales and non-EM fungi sequences were not taken into account in our analysis (one Xylariaceae, four Rhizoctonia, one Diaporthales). Twenty-nine MOTUs were recorded among which 21 belonged to Basidiomycota and 8 to Ascomycota (91.2% and 8.8% of ectomycorrhizae, respectively). Seven Alnicola species were detected, all strictly identical to species found in Europe. Of interest, two new ITS variants of Alnicola xanthophylla were detected in Central Georgia, differing by five positions from their European relatives. Among non-Alnicola EM fungi, five new MOTUs were detected in Georgia. Three were Ascomycota (Tuber sp., Tarzetta sp., Peziza sp.) and two Basidiomycota (Inocybe spp.). These five possibly endemic EM fungi were found in Central and Eastern Georgia.
Diversity patterns of symbiont communities in Georgia
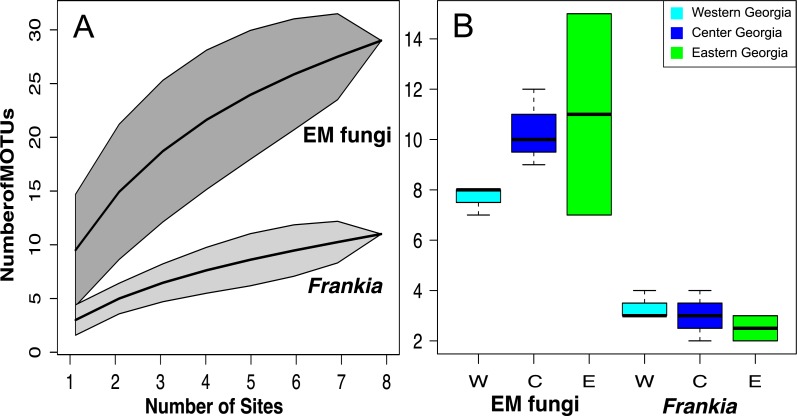
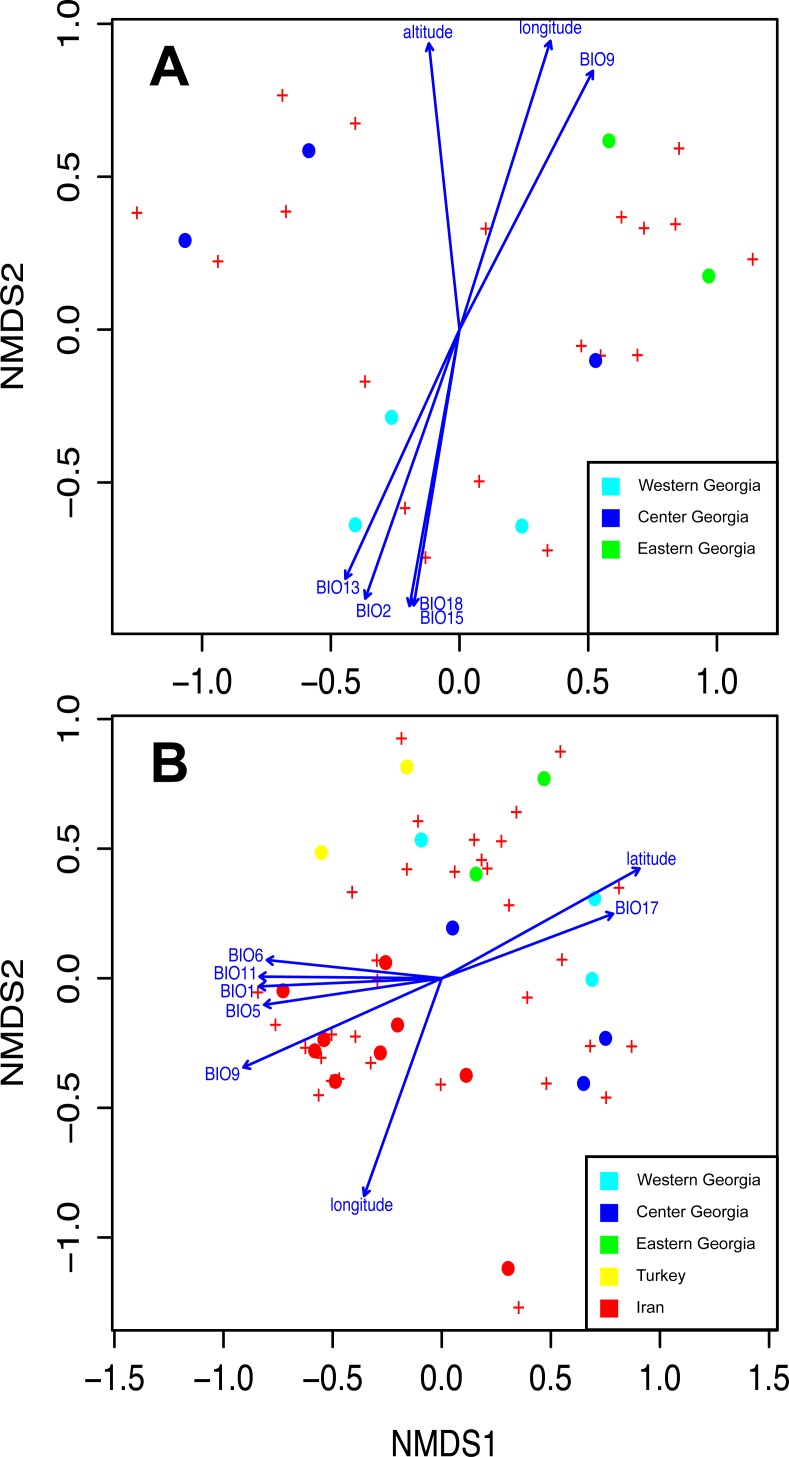
Species accumulation curves show that sampling was not saturated, especially for EM fungi. According to Chao 1 estimates, up to 26.7 Frankia and 44.7 EM MOTUs could occur in Georgia (Fig. 4A). Species richness per site varied between 7 and 15 MOTUs for fungi, and from 2 to 4 for Frankia (Fig. 4B). Compared to the recolonized zone, Frankia and EM communities were neither significantly richer (Kruskal-Wallis test, p = 0.48 and p = 0.17 respectively, Fig. 4B) nor different based on community composition (perMANOVA on Bray–Curtis distances, p = 0.40 and p = 0.19 for Frankia and EM fungi, respectively). Communities were not spatially autocorrelated (multivariate spatial autocorrelation test, p-value = 0.338 for EM fungi, p = 0.348 for Frankia), and the spatial distance did not correlate with Bray–Curtis distances at least for EM fungi (Mantel test, p = 0.16). For Frankia, the beta diversity was marginally correlated with distance (Mantel test, p = 0.0594). For the two types of symbionts, Bray–Curtis distances between sites were significantly correlated with longitude and several bioclimatic factors linked with precipitations (see Table S3 of Appendix S1, Fig. 5A).
Figure 4. MOTU accumulation curves computed for all Georgian site for Frankia and EM fungi (A) and species richness per site in West, Central and East Georgia for Frankia and EM fungi (B).
Shaded area represent 95% confidence intervals.
Figure 5. Non-Metric Multidimensional Scaling (NMDS) ordering the differences between EM and Frankia communities and showing the correlation with environmental and geographical parameters in Georgia (A) and at a wider scale between Georgia, Iran and Turkey for EM fungi only (B). Significant variables are represented (P > 0.05 according to environmental fitting tests; see Table S3 of Appendix S1).
BIO1, temperature seasonality; BIO2, maximum temperature of the warmest month; BIO5, precipitation of the warmest month; BIO6, precipitation of the driest month; BIO9, precipitation of the warmest quarter; BIO11, isothermality; BIO13, precipitation of the wettest quarter; BIO15, annual mean temperature; BIO17, mean temperature of the driest quarter and BIO18, precipitation of the coldest quarter.
Comparison between EM communities from Colchis and Hyrcanian refugia
The dominant MOTUs in Georgia were rare in Turkey and Iran, and vice versa (see Table S2 of Appendix S1). Among the 68 MOTUs, three were shared between Georgia and adjacent countries (Paxillus adelphus and two MOTUs belonging to the genus Tomentella), nine between Georgia and Iran (MOTUs belonging to the genera Alnicola, Cortinarius, Inocybe, Sebacina, and Tarzetta), and none between Georgia and Turkey. Interestingly, the genus Inocybe and several Ascomycota were detected on alder roots both in Iran and in Georgia. At the community level, the Georgian EM communities were not more diverse than those on other alder species in Iran or Turkey (ANOVA, p-value = 0.14). The spatial auto-correlation test was still not significant (multivariate autocorrelation test, p = 0.072). Variation in beta-diversity was strongly correlated with geographic distances (Mantel test, r = 0.51, p < 0.0001) and was also related to host species (42% of variation explained, PERMANOVA test, p = 0.001). Finally, longitude, latitude, altitude, and bioclimatic variables related to temperature all were significantly correlated with beta-diversity (Fig. 5B, Table S3 of Appendix S1).
Discussion
Our first objective was to document the diversity of symbiont communities associated with Alnus glutinosa ssp. barbata in Georgia, especially in the Colchis floodplain, a glacial and Tertiary refugium for alders. In all, 29 EM MOTUs and 12 Frankia MOTUs were uncovered, and Chao 1 estimates showed that twice as many species may occur there. For Frankia, the common European lineages were detected, together with five new lineages, which is an exceptional pattern considering the extensive worldwide Frankia database (Pozzi et al., 2015). Georgia proved to be a region of high Frankia diversity, probably of ancient origin as suggested by the long branches supporting Caucasian lineages (Fig. 2). For EM fungi, most Georgian MOTUs (24 out of 29) were already reported from Europe, on roots of A. glutinosa, A. incana and A. cordata (Loisel.) Duby. No endemic MOTU was detected for the most specialized lineages, but two unique ITS variants of Alnicola xanthophylla were found in Central Georgia. From previous study on the genus Alnicola (Moreau, Peintner & Gardes, 2006; Rochet et al., 2011), the ITS is known to be often not variable within species, and 1.1% genetic variability is exceptional for this genus. Although a deeper sampling may reveal more fungi, these results reveal that endemism is relatively rare in the Caucasus region for EM fungi, even the more specialized ones associated with alders. Interestingly, a similar analysis on corticioid fungi in South Caucasus has also pointed out the low number of endemics, and the high similarity with European communities (Ghobad-Nejhad et al., 2012).
The presence of five endemic Frankia, but no endemic specialized EM fungi was not expected but could be explained by differences in dispersal and survival abilities between the two types of symbionts. A free-living stage (outside the host) has been reported for the Frankia strains with low host specificity (Maunuksela et al., 1999), but aside from this case, most uncultured Frankia are highly dependent of their host (Pozzi et al., 2015). As with Frankia, most EM fungi cannot survive in the soil without their host (Lindahl & Tunlid, 2015). Frankia produces spores belowground and is dispersed by water or animals (Chaia, Wall & Huss-Danell, 2010) and sometimes by wind (Dawson, 2007) leading to short-distance dispersal, and perhaps greater endemism whereas Alnicola are wind-dispersed and potentially disperse over large distances. Among fungi, two potential endemic species belong to a wind-dispersed genus (Inocybe). Although species of Inocybe are regularly detected on alder roots (Bogar & Kennedy, 2013; Roy et al., 2013; Põlme et al., 2013), current knowledge on this genus is still too limited to determine if these Inocybe sp. are specific to Alnus or if their occurrence reflects a local adaptation to a particular habitat. More generally, Ryberg et al. (2008) have shown that the ITS evolves faster in the genus Inocybe, and that environmental sequencing often reveals new species in this genus. Among the EM fungi lineages that form hypogeous (below-ground) fruitbodies, three potential endemic Ascomycota species were detected. The degree of specificity of these species towards Alnus is still unknown, and two genera, Tuber and Tarzetta, are often described as pioneer species with broad host ranges. The presence of endemics for these two genera might be due to their long-term persistence in refugia. Similarly, the persistence of the black truffle Tuber melanosporum Vittad. during glaciations has been shown in Spanish (García-Cunchillos et al., 2014) and Italian refugia (Rubini et al., 2005).
Our second objective was to assess whether the symbiont communities and their host populations follow a shared history at the scale of Georgia. We hypothesized that sites in the refugium would host more endemic and diverse communities of both Frankia and EM fungi, but we did not confirm our hypothesis. First, the host population were genetically more diverse in the recolonized zone, where several unique ITS and matK variants were detected for A. glutinosa subsp. barbata. These variants were clearly distinct from other sequences of European A. glutinosa and this result confirms that Georgia, and not only the Colchis, is a hot spot of genetic diversity for the A. glutinosa complex (pointed out by King & Ferris, 1998). Interestingly, all endemic EM MOTUs were detected in Central and Eastern sites, associated with the most genetically diverse host populations. However, the species richness of Frankia communities showed a different distribution, as three endemic strains were detected in the Colchis, and two in the Central population. These different distributions of genetic diversity and species richness suggest that all symbionts do not necessarily survive with their host in refugia. Moreover, the occurrence of unique ITS and matK variants for alders and endemic EM fungi in Central and Eastern sites show that isolated host populations could also act as refugia, especially in mountain ranges, as highlighted by recent reviews on the Mediterranean basin (Médail & Diadema, 2009; Dobrowski, 2011; Keppel et al., 2012). The differences between the Colchis and Central and Eastern populations could reflect different biogeographic histories, as shown for snails and amphibians (Tarkhnishvili, 1996; Tarkhnishvili, Thorpe & Arntzen, 2000; Pokryszko et al., 2011) but also correlate with changes in annual precipitation and summer temperatures, as pinpointed by our analyses (Fig. 5).
Our third objective was to compare the Colchis and the Hyrcanian communities, associated with two alder species, A. glutinosa subsp. barbata and A. subcordata, that are both considered to be Tertiary relics. The two refugia hosted distinct EM communities, and differences were strongly explained by both host identity and geographic distance. The lack of shared MOTUs, even abundant ones (Table S2 of Appendix S1), explains these differences, and suggests a limited spore flow between the two regions. The scarcity of alder stands between these two regions in the steppe landscapes could indeed limit dispersal, and maintain isolation between the two regions, as observed for plants (Nakhutsrishvili et al., 2015) and corticioid fungi (Ghobad-Nejhad et al., 2012). Interestingly, both A. subcordata and A. glutinosa subsp. barbata associate with several Inocybe, Ascomycota and especially Tuber. Additional Tertiary refugia should be investigated in Asia to determine if these associations are also relic from the Tertiary, or derive from local and independent adaptations in relatively dry regions for alders. Indeed, Tuber and hypogeous fungi are known to be particularly resistant to drought (Richard et al., 2011; Herzog et al., 2013 ) and abundant in dry habitats (Zambonelli et al., 2014).
For microbes, biogeographic studies are relatively recent (Martiny et al., 2006), partly because the “everything is everywhere” idiom was long said to be the rule. Now, Beringia has been pinpointed as a hotspot for high-latitude fungi (Geml et al., 2010; Bellemain et al., 2013), and could have been a glacial refugium for the fungi. In Europe, the Perigord truffle Tuber melanosporum, also an EM species, has more genetically diverse populations in southern-Italy (Rubini et al., 2005) and Spain (García-Cunchillos et al., 2014). Finally, the anther smut fungus Microbotryum sp., an obligate parasite of Caryophyllaceae, has apparently followed its host during recent glaciation (Vercken et al., 2010). Here, we focus on entire communities and show on the contrary, that alpha diversity was not higher in the refugium than in the recolonized zone. The results from analyzing communities versus populations are not necessarily coupled (Vellend & Geber, 2005), but theoretically area size and immigration have parallel effects on species richness and genetic diversity (Vellend & Geber, 2005). The Theory of Island Biogeography (MacArthur & Wilson, 1967) predicts that smaller refugia can favor local extinctions (Kadmon & Allouche, 2007), and moreover, bottleneck effects may also reduce local diversity (Bennett & Provan, 2008). On the other hand, recent experiments on alders have shown that their specificity might be controlled by pH and nitrogen availability (Huggins et al., 2014), which may reduce the influence of host biogeography on symbiont community diversity. In this framework, it is even more puzzling to observe more endemic EM fungi and rare plant ITS variant in the isolated populations from Central and Eastern Georgia. As the endemic Inocybe and Tuber are not usually associated with alders, we suspect that these taxa have only recently colonized alder roots, a host shift probably favored by the actual isolation of alders in this region.
Conclusions
Wiens & Donoghue (2004) have already highlighted the chasm between historical biogeography and community ecology, and suggested that more links should be drawn between these topics. In a second step, studies on symbiont populations may help confirming the lack of isolation in refugia. The recent studies of alder refugia in Europe could be used as a framework to study more in depth alder symbiont biogeography (Douda et al., 2014; Mandák et al., 2016a; Mandák et al., 2016b). Finally, the occurrence of endemic OTUs detected for both EM fungi and Frankia outside of refugia highlight their relevance to biodiversity conservation not only in Tertiary and glacial refugia, but also in isolated host populations.
Supplemental Information
Additional Supporting Information may be found in the online version of this article: Appendix S1. Supplementary Tables. Table S1. Site characteristics, location, and occurrence of Frankia and EM MOTUs among sites. *putative endemic MOTUs Table S2. Matrix reporting occurrence of EM MOTU among sites located in Turkey, Iran and Georgia. Table S3. Correlation of climatic factors and geographic position with NMDS structure for all symbionts in Georgia, Frankia only, EM fungi only, and for EM fungi in Georgia, Turkey and Iran.
Acknowledgments
The authors are deeply grateful to Professor George Nakhutsrishvili and Dr. Angelina Jorjadze at the Tbilisi Botanical Garden and Institute of Botany, and to Joël Bastenaire and Adeline Latrille at the Institut Français de Géorgie and Lela Gogava, for their helpful support in Georgia. We thank Arndt Hampe, Corinne Vacher and Jason Hoeksema for interesting discussions and feedback. We deeply thank our anonymous referee, Dee Carter and Lee Taylor for their useful comments that contributed to improve this paper.
Funding Statement
Financial support for the sampling carried out in Georgia was provided partly by a grant from the University Toulouse 3 Paul Sabatier (AAP Relations Internationales, Aide Ponctuelle de Coopération 2011 to MG) and for the laboratory work by a grant from INSU-CNRS, (AAP programme EC2CO Initiative Structurante Ecosphère continentale et côtière 2011–2012 to MR) and the Labex TULIP. There was no additional external funding received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Melanie Roy conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Adrien C. Pozzi and Raphaëlle Gareil performed the experiments.
Melissande Nagati, Patricia Jargeat, Hervé Gryta and Pierre-Arthur Moreau analyzed the data.
Sophie Manzi, Imen Nouioui and Nino Sharikadze contributed reagents/materials/analysis tools.
Maria P. Fernandez reviewed drafts of the paper.
Monique Gardes conceived and designed the experiments, reviewed drafts of the paper.
Field Study Permissions
The following information was supplied relating to field study approvals (i.e., approving body and any reference numbers):
Studies were handled thanks to Professor George Nakhutsrishvili from Tbilisi Botany institute. No permits were required outside of parks in Georgia.
DNA Deposition
The following information was supplied regarding the deposition of DNA sequences:
Plant sequences are deposited at Genbank (KX897895 –KX897935 for ITS and KX897936 –KX897944 for matK). Frankia Sequences were submitted to the EMBL (European Nucleotide Archive) under accession numbers LT616989 –LT617015 (dnaA), LT617016 –LT617048 (ftsZ), LT599862, LT599864, LT599865 and LT599870 –LT599890 (pgk). Fungal sequences are deposited at Genbank under accession numbers KX897613 –KX897894.
Data Availability
The following information was supplied regarding data availability:
The sequences are deposited at GenBank and EMBL, and raw data is included in Appendix S1.
References
- Altschul et al. (1990).Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bellemain et al. (2013).Bellemain E, Davey ML, Kauserud H, Epp LS, Boessenkool S, Coissac E, Geml J, Edwards M, Willerslev E, Gussarova G. Fungal palaeodiversity revealed using high-throughput metabarcoding of ancient DNA from arctic permafrost. Environmental Microbiology. 2013;15:1176–1189. doi: 10.1111/1462-2920.12020. [DOI] [PubMed] [Google Scholar]
- Bennett & Provan (2008).Bennett KD, Provan J. What do we mean by “refugia”? Quaternary Science Reviews. 2008;27:2449–2455. doi: 10.1016/j.quascirev.2008.08.019. [DOI] [Google Scholar]
- Bogar & Kennedy (2013).Bogar LM, Kennedy PG. New wrinkles in an old paradigm: neighborhood effects can modify the structure and specificity of Alnus-associated ectomycorrhizal fungal communities. FEMS Microbiology Ecology. 2013;83:767–777. doi: 10.1111/1574-6941.12032. [DOI] [PubMed] [Google Scholar]
- Chaia, Wall & Huss-Danell (2010).Chaia EE, Wall LG, Huss-Danell K. Life in soil by the actinorhizal root nodule endophyte Frankia. A review. Symbiosis. 2010;51:201–226. doi: 10.1007/s13199-010-0086-y. [DOI] [Google Scholar]
- Connor & Kvavadze (2009).Connor SE, Kvavadze EV. Modelling late Quaternary changes in plant distribution, vegetation and climate using pollen data from Georgia, Caucasus. Journal of Biogeography. 2009;36:529–545. doi: 10.1111/j.1365-2699.2008.02019.x. [DOI] [Google Scholar]
- Cotin-Galvan et al. (2016).Cotin-Galvan L, Pozzi AC, Schwob G, Fournier P, Fernandez MP, Herrera-Belaroussi A. In-planta sporulation capacity enhances infectivity and rhizospheric competitiveness of Frankia Strains. Microbes and Environments. 2016;31:11–18. doi: 10.1264/jsme2.ME15090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson (2007).Dawson JO. Nitrogen-fixing actinorhizal symbioses. Springer Netherlands; Dordrecht: 2007. Ecology of actinorhizal plants; pp. 199–234. [Google Scholar]
- Denk, Frotzler & Davitashvili (2001).Denk T, Frotzler N, Davitashvili N. Vegetational patterns and distribution of relict taxa in humid temperate forests and wetlands of Georgia (Transcaucasia) Biological Journal of the Linnean Society. 2001;72:287–332. doi: 10.1111/j.1095-8312.2001.tb01318.x. [DOI] [Google Scholar]
- Dobrowski (2011).Dobrowski SZ. A climatic basis for microrefugia: the influence of terrain on climate. Global Change Biology. 2011;17:1022–1035. doi: 10.1111/j.1365-2486.2010.02263.x. [DOI] [Google Scholar]
- Douda et al. (2014).Douda J, Doudová J, Drašnarová A, Kuneš P, Hadincová V, Krak K, Zákravský P, Mandák B. Migration patterns of subgenus Alnus in Europe since the Last Glacial Maximum: a systematic review. PLOS ONE. 2014;9:e88709. doi: 10.1371/journal.pone.0088709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dray & Dufour (2007).Dray S, Dufour A-B. The ade4 package: implementing the duality diagram for ecologists. Journal of Statistical Software. 2007;22:1–20. doi: 10.18637/jss.v022.i04. [DOI] [Google Scholar]
- Dunning & Savolainen (2010).Dunning LT, Savolainen V. Broad-scale amplification of matK for DNA barcoding plants, a technical note. Botanical Journal of the Linnean Society. 2010;164:1–9. doi: 10.1111/j.1095-8339.2010.01071.x. [DOI] [Google Scholar]
- Edgar (2004).Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cunchillos et al. (2014).García-Cunchillos I, Sánchez S, Barriuso JJ, Pérez-Collazos E. Population genetics of the westernmost distribution of the glaciations-surviving black truffle Tuber melanosporum. Mycorrhiza. 2014;24:89–100. doi: 10.1007/s00572-013-0540-9. [DOI] [PubMed] [Google Scholar]
- Gardes & Bruns (1993).Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Gavin et al. (2014).Gavin DG, Fitzpatrick MC, Gugger PF, Heath KD, Rodríguez-Sánchez F, Dobrowski SZ, Hampe A, Hu FS, Ashcroft MB, Bartlein PJ. Climate refugia: joint inference from fossil records, species distribution models and phylogeography. New Phytologist. 2014;204:37–54. doi: 10.1111/nph.12929. [DOI] [PubMed] [Google Scholar]
- Geml et al. (2010).Geml J, Tulloss RE, Laursen GA, Sazanova NA, Taylor DL. Relict Species. Springer; Berlin, Heidelberg: 2010. Phylogeographic analyses of a boreal-temperate ectomycorrhizal basidiomycete, Amanita muscaria, suggest forest refugia in Alaska during the last glacial maximum; pp. 173–186. [Google Scholar]
- Ghobad-Nejhad et al. (2012).Ghobad-Nejhad M, Hallenberg N, Hyvönen J, Yurchenko E. The Caucasian corticioid fungi: level of endemism, similarity, and possible contribution to European fungal diversity. Fungal Diversity. 2012;52:35–48. doi: 10.1007/s13225-011-0122-0. [DOI] [Google Scholar]
- Griekspoor & Groothuis (2017).Griekspoor A, Groothuis T. 4Peaks. Version 1.7.1http://nucleobytes.com/index.php/4peaks 2017
- Hampe & Jump (2011).Hampe A, Jump AS. Climate relicts: past, present, future. Annual Review of Ecology, Evolution, and Systematics. 2011;42:313–333. doi: 10.1146/annurev-ecolsys-102710-145015. [DOI] [Google Scholar]
- Havrdová et al. (2015).Havrdová A, Douda J, Krak K, Vít P, Hadincová V, Zákravský P, Mandák B. Higher genetic diversity in recolonized areas than in refugia of Alnus glutinosa triggered by continent-wide lineage admixture. Molecular Ecology. 2015;24:4759–4777. doi: 10.1111/mec.13348. [DOI] [PubMed] [Google Scholar]
- Herzog et al. (2013).Herzog C, Peter M, Pritsch K, Günthardt-Goerg MS, Egli S. Drought and air warming affects abundance and exoenzyme profiles of Cenococcum geophilum associated with Quercus robur, Q. petraea and Q. pubescens. Plant Biology. 2013;15:230–237. doi: 10.1111/j.1438-8677.2012.00614.x. [DOI] [PubMed] [Google Scholar]
- Hijmans & Van Etten (2012).Hijmans RJ, Van Etten J. 2012 Raster: geographic analysis and modeling with raster data. R package version 2.0–12. [Google Scholar]
- Huggins et al. (2014).Huggins JA, Talbot J, Gardes M, Kennedy PG. Unlocking environmental keys to host specificity: differential tolerance of acidity and nitrate by Alnus-associated ectomycorrhizal fungi. Fungal Ecology. 2014;12:52–61. [Google Scholar]
- Kadmon & Allouche (2007).Kadmon R, Allouche O. Integrating the effects of area, isolation, and habitat heterogeneity on species diversity: a unification of island biogeography and niche theory. The American Naturalist. 2007;170:443–454. doi: 10.1086/519853. [DOI] [PubMed] [Google Scholar]
- Kennedy et al. (2011).Kennedy PG, Garibay-Orijel R, Higgins LM, Angeles-Arguiz R. Ectomycorrhizal fungi in Mexican Alnus forests support the host co-migration hypothesis and continental-scale patterns in phylogeography. Mycorrhiza. 2011;21:559–568. doi: 10.1007/s00572-011-0366-2. [DOI] [PubMed] [Google Scholar]
- Kennedy, Walker & Bogar (2015).Kennedy PG, Walker JK, Bogar LM. Mycorrhizal Networks. Springer Netherlands; Dordrecht: 2015. Interspecific mycorrhizal networks and non-networking hosts: exploring the ecology of the host genus Alnus; pp. 227–254. [Google Scholar]
- Keppel et al. (2012).Keppel G, Van Niel KP, Wardell-Johnson GW, Yates CJ, Byrne M, Mucina L, Schut AG, Hopper SD, Franklin SE. Refugia: identifying and understanding safe havens for biodiversity under climate change. Global Ecology and Biogeography. 2012;21:393–404. doi: 10.1111/j.1466-8238.2011.00686.x. [DOI] [Google Scholar]
- Kikvidze & Ohsawa (2001).Kikvidze Z, Ohsawa M. Richness of Colchic vegetation: comparison between refugia of south-western and East Asia. BMC Ecology. 2001;1:1. doi: 10.1186/1472-6785-1-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King & Ferris (1998).King RA, Ferris C. Chloroplast DNA phylogeography of Alnus glutinosa (L.) Gaertn. Molecular Ecology. 1998;7:1151–1161. doi: 10.1046/j.1365-294x.1998.00432.x. [DOI] [Google Scholar]
- Kõljalg et al. (2005).Kõljalg U, Larsson K-H, Abarenkov K, Nilsson RH, Alexander IJ, Eberhardt U, Erland S, Høiland K, Kjøller R, Larsson E. UNITE: a database providing web-based methods for the molecular identification of ectomycorrhizal fungi. New Phytologist. 2005;166:1063–1068. doi: 10.1111/j.1469-8137.2005.01376.x. [DOI] [PubMed] [Google Scholar]
- Lepais et al. (2013).Lepais O, Muller SD, Saad-Limam SB, Benslama M, Rhazi L, Belouahem-Abed D, Daoud-Bouattour A, Gammar AM, Ghrabi-Gammar Z, Bacles CFE. High genetic diversity and distinctiveness of rear-edge climate relicts maintained by ancient tetraploidisation for Alnus glutinosa. PLOS ONE. 2013;8:e75029. doi: 10.1371/journal.pone.0075029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl & Tunlid (2015).Lindahl BD, Tunlid A. Ectomycorrhizal fungi–potential organic matter decomposers, yet not saprotrophs. New Phytologist. 2015;205:1443–1447. doi: 10.1111/nph.13201. [DOI] [PubMed] [Google Scholar]
- MacArthur & Wilson (1967).MacArthur R, Wilson E. The theory of biogeography. Princeton University Press; New Jersey: 1967. pp. 19–67. [Google Scholar]
- Maharramova et al. (2015).Maharramova EH, Safarov HM, Kozlowski G, Borsch T, Muller LA. Analysis of nuclear microsatellites reveals limited differentiation between Colchic and Hyrcanian populations of the wind-pollinated relict tree Zelkova carpinifolia (Ulmaceae) American Journal of Botany. 2015;102:119–128. doi: 10.3732/ajb.1400370. [DOI] [PubMed] [Google Scholar]
- Mandák et al. (2016a).Mandák B, Havrdová A, Krak K, Hadincová V, Vít P, Zákravský P, Douda J. Recent similarity in distribution ranges does not mean a similar postglacial history: a phylogeographical study of the boreal tree species Alnus incana based on microsatellite and chloroplast DNA variation. New Phytologist. 2016a;210:1395–1407. doi: 10.1111/nph.13848. [DOI] [PubMed] [Google Scholar]
- Mandák et al. (2016b).Mandák B, Vít P, Krak K, Trávníček P, Havrdová A, Hadincová V, Zákravský P, Jarolímová V, Bacles CFE, Douda J. Flow cytometry, microsatellites and niche models reveal the origins and geographical structure of Alnus glutinosa populations in Europe. Annals of Botany. 2016b;117:107–120. doi: 10.1093/aob/mcv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny et al. (2006).Martiny JBH, Bohannan BJ, Brown JH, Colwell RK, Fuhrman JA, Green JL, Horner-Devine MC, Kane M, Krumins JA, Kuske CR. Microbial biogeography: putting microorganisms on the map. Nature Reviews Microbiology. 2006;4:102–112. doi: 10.1038/nrmicro1341. [DOI] [PubMed] [Google Scholar]
- Maunuksela et al. (1999).Maunuksela L, Zepp K, Koivula T, Zeyer J, Haahtela K, Hahn D. Analysis of Frankia populations in three soils devoid of actinorhizal plants. FEMS Microbiology Ecology. 1999;28:11–21. doi: 10.1111/j.1574-6941.1999.tb00556.x. [DOI] [Google Scholar]
- Médail & Diadema (2009).Médail F, Diadema K. Glacial refugia influence plant diversity patterns in the Mediterranean Basin. Journal of Biogeography. 2009;36:1333–1345. doi: 10.1111/j.1365-2699.2008.02051.x. [DOI] [Google Scholar]
- Merényi et al. (2014).Merényi Z, Varga T, Geml J, Orczán ÁK, Chevalier G, Bratek Z. Phylogeny and phylogeography of the Tuber brumale aggr. Mycorrhiza. 2014;24:101–113. doi: 10.1007/s00572-014-0566-7. [DOI] [PubMed] [Google Scholar]
- Miller, Pfeiffer & Schwartz (2010).Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE), 2010; 2010. pp. 1–8. [Google Scholar]
- Milne & Abbott (2002).Milne RI, Abbott RJ. The origin and evolution of Tertiary relict floras. Advances in Botanical Research. 2002;38:281–314. doi: 10.1016/S0065-2296(02)38033-9. [DOI] [Google Scholar]
- Molina (1981).Molina R. Ectomycorrhizal specificity in the genus Alnus. Canadian Journal of Botany. 1981;59:325–334. doi: 10.1139/b81-045. [DOI] [Google Scholar]
- Moreau, Peintner & Gardes (2006).Moreau P-A, Peintner U, Gardes M. Phylogeny of the ectomycorrhizal mushroom genus Alnicola (Basidiomycota, Cortinariaceae) based on rDNA sequences with special emphasis on host specificity and morphological characters. Molecular Phylogenetics and Evolution. 2006;38:794–807. doi: 10.1016/j.ympev.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Moreau et al. (2011).Moreau P-A, Rochet J, Richard F, Chassagne F, Manzi S, Gardes M. Taxonomy of Alnus-associated hypogeous species of Alpova and Melanogaster (Basidiomycota, Paxillaceae) in Europe. Cryptogamie, Mycologie. 2011;32:33–62. doi: 10.7872/crym.v32.iss1.2012.033. [DOI] [Google Scholar]
- Nakhutsrishvili (2012).Nakhutsrishvili G. The vegetation of Georgia (South Caucasus) Springer Science & Business Media; 2012. [Google Scholar]
- Nakhutsrishvili et al. (2015).Nakhutsrishvili G, Zazanashvili N, Batsatsashvili K, Montalvo CS. Colchic and Hyrcanian forests of the Caucasus: similarities, differences and conservation status. Flora Mediterranea. 2015;25:185–192. doi: 10.7320/flmedit25si.185. [DOI] [Google Scholar]
- Nilsson et al. (2008).Nilsson RH, Kristiansson E, Ryberg M, Hallenberg N, Larsson K-H. Intraspecific ITS variability in the kingdom Fungi as expressed in the international sequence databases and its implications for molecular species identification. Evolutionary Bioinformatics Online. 2008;4:193. doi: 10.4137/ebo.s653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen et al. (2007).Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, Suggests M. The vegan package. Community Ecology Package. 2007;10:631–637. [Google Scholar]
- Pokryszko et al. (2011).Pokryszko BM, Cameron RA, Mumladze L, Tarkhnishvili D. Forest snail faunas from Georgian Transcaucasia: patterns of diversity in a Pleistocene refugium. Biological Journal of the Linnean Society. 2011;102:239–250. doi: 10.1111/j.1095-8312.2010.01575.x. [DOI] [Google Scholar]
- Põlme et al. (2014).Põlme S, Bahram M, Kõljalg U, Tedersoo L. Global biogeography of Alnus-associated Frankia actinobacteria. New Phytologist. 2014;204:979–988. doi: 10.1111/nph.12962. [DOI] [PubMed] [Google Scholar]
- Põlme et al. (2013).Põlme S, Bahram M, Yamanaka T, Nara K, Dai YC, Grebenc T, Kraigher H, Toivonen M, Wang P-H, Matsuda Y. Biogeography of ectomycorrhizal fungi associated with alders (Alnus spp.) in relation to biotic and abiotic variables at the global scale. New Phytologist. 2013;198:1239–1249. doi: 10.1111/nph.12170. [DOI] [PubMed] [Google Scholar]
- Pozzi et al. (2015).Pozzi AC, Bautista-Guerrero HH, Nouioui I, Cotin-Galvan L, Pepin R, Fournier P, Menu F, Fernandez MP, Herrera-Belaroussi A. In-planta sporulation phenotype: a major life history trait to understand the evolution of Alnus-infective Frankia strains. Environmental Microbiology. 2015;17:3125–3138. doi: 10.1111/1462-2920.12644. [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2008).R Development Core Team . Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Richard et al. (2011).Richard F, Roy M, Shahin O, Sthultz C, Duchemin M, Joffre R, Selosse M-A. Ectomycorrhizal communities in a Mediterranean forest ecosystem dominated by Quercus ilex: seasonal dynamics and response to drought in the surface organic horizon. Annals of Forest Science. 2011;68:57–68. [Google Scholar]
- Rochet et al. (2011).Rochet J, Moreau PA, Manzi S, Gardes M. Comparative phylogenies and host specialization in three mushroom-forming genera of Alnus-associated ectomycorrhizal fungi: Alnicola, Alpova and Lactarius (Basidiomycota) BMC Evolutionary Biology. 2011;11:40. doi: 10.1186/1471-2148-11-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy et al. (2013).Roy M, Rochet J, Manzi S, Jargeat P, Gryta H, Moreau P-A, Gardes M. What determines Alnus-associated ectomycorrhizal community diversity and specificity? A comparison of host and habitat effects at a regional scale. New Phytologist. 2013;198:1228–1238. doi: 10.1111/nph.12212. [DOI] [PubMed] [Google Scholar]
- Rubini et al. (2005).Rubini A, Paolocci F, Riccioni C, Vendramin GG, Arcioni S. Genetic and phylogeographic structures of the symbiotic fungus Tuber magnatum. Applied and Environmental Microbiology. 2005;71:6584–6589. doi: 10.1128/AEM.71.11.6584-6589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryberg et al. (2008).Ryberg M, Nilsson RH, Kristiansson E, Töpel M, Jacobsson S, Larsson E. Mining metadata from unidentified ITS sequences in GenBank: a case study in Inocybe (Basidiomycota) BMC Evolutionary Biology. 2008;8:50. doi: 10.1186/1471-2148-8-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss et al. (2009).Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Applied and Environmental Microbiology. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith & Read (2008).Smith SE, Read DJ. Mycorrhizal symbiosis. edn. Academic Press, Elsevier; Cambridge: 2008. [Google Scholar]
- Stamatakis, Hoover & Rougemont (2008).Stamatakis A, Hoover P, Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Taberlet & Cheddadi (2002).Taberlet P, Cheddadi R. Quaternary refugia and persistence of biodiversity. Science. 2002;297:2009–2010. doi: 10.1126/science.297.5589.2009. [DOI] [PubMed] [Google Scholar]
- Tarkhnishvili (1996).Tarkhnishvili D. The distribution and ecology of the amphibians of Georgia and the Caucasus: a biogeographical analysis. Zeitschrift für Feldherpetologie. 1996;3:167–196. [Google Scholar]
- Tarkhnishvili, Thorpe & Arntzen (2000).Tarkhnishvili DN, Thorpe RS, Arntzen JW. Pre-Pleistocene refugia and differentiation between populations of the Caucasian salamander (Mertensiella caucasica) Molecular Phylogenetics and Evolution. 2000;14:414–422. doi: 10.1006/mpev.1999.0718. [DOI] [PubMed] [Google Scholar]
- Vellend & Geber (2005).Vellend M, Geber MA. Connections between species diversity and genetic diversity. Ecology Letters. 2005;8:767–781. doi: 10.1111/j.1461-0248.2005.00775.x. [DOI] [Google Scholar]
- Vercken et al. (2010).Vercken E, Fontaine MC, Gladieux P, Hood ME, Jonot O, Giraud T. Glacial refugia in pathogens: European genetic structure of anther smut pathogens on Silene latifolia and Silene dioica. PLOS Pathog. 2010;6:e1001229. doi: 10.1371/journal.ppat.1001229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, Nurmiaho-Lassila & Sundman (1987).Weber A, Nurmiaho-Lassila E-L, Sundman V. Features of the intrageneric Alnus-Frankia specificity. Physiologia Plantarum. 1987;70:289–296. doi: 10.1111/j.1399-3054.1987.tb06146.x. [DOI] [Google Scholar]
- White et al. (1990).White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a Guide to Methods and Applications. 1990;18:315–322. doi: 10.1016/B978-0-12-372180-8.50042-1. [DOI] [Google Scholar]
- Wiens & Donoghue (2004).Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends in Ecology & Evolution. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Zambonelli et al. (2014).Zambonelli A, Donnini D, Rana GL, Fascetti S, Benucci GMN, Iotti M, Morte A, Khabar L, Bawadekji A, Piattoni F. Hypogeous fungi in Mediterranean maquis, arid and semi-arid forests. Plant Biosystems. 2014;148:329–401. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found in the online version of this article: Appendix S1. Supplementary Tables. Table S1. Site characteristics, location, and occurrence of Frankia and EM MOTUs among sites. *putative endemic MOTUs Table S2. Matrix reporting occurrence of EM MOTU among sites located in Turkey, Iran and Georgia. Table S3. Correlation of climatic factors and geographic position with NMDS structure for all symbionts in Georgia, Frankia only, EM fungi only, and for EM fungi in Georgia, Turkey and Iran.
Data Availability Statement
The following information was supplied regarding data availability:
The sequences are deposited at GenBank and EMBL, and raw data is included in Appendix S1.