Abstract
Objective
Ghrelin is a stomach-derived hormone that affects food intake and regulates blood glucose. The best-characterized actions of ghrelin are mediated by its binding to and activation of the growth hormone secretagogue receptor (GHSR; ghrelin receptor). Adequate examination of the identity, function, and relevance of specific subsets of GHSR-expressing neurons has been hampered by the absence of a suitable Cre recombinase (Cre)-expressing mouse line with which to manipulate gene expression in a targeted fashion within GHSR-expressing neurons. The present study aims to characterize the functional significance and neurocircuitry of GHSR-expressing neurons in the mediobasal hypothalamus (MBH), as they relate to ghrelin-induced food intake and fasting-associated rebound hyperphagia, using a novel mouse line in which Cre expression is controlled by the Ghsr promoter.
Methods
A Ghsr-IRES-Cre mouse line that expresses Cre directed by the Ghsr promoter was generated. The line was validated by comparing Cre activity in reporter mice to the known brain distribution pattern of GHSR. Next, the requirement of MBH GHSR-expressing neuronal activity in mediating food intake in response to administered ghrelin and in response to fasting was assessed after stereotaxic delivery of inhibitory designer receptor exclusively activated by designer drugs (DREADD) virus to the MBH. In a separate cohort of Ghsr-IRES-Cre mice, stereotaxic delivery of stimulatory DREADD virus to the MBH was performed to assess the sufficiency of MBH GHSR-expressing neuronal activity on food intake. Finally, the distribution of MBH GHSR-expressing neuronal axonal projections was assessed in the DREADD virus-injected animals.
Results
The pattern of Cre activity in the Ghsr-IRES-Cre mouse line mostly faithfully reproduced the known GHSR expression pattern. DREADD-assisted inhibition of MBH GHSR neuronal activity robustly suppressed the normal orexigenic response to ghrelin and fasting-associated rebound food intake. DREADD-assisted stimulation of MBH GHSR neuronal activity was sufficient to induce food intake. Axonal projections of GHSR-expressing MBH neurons were observed in a subset of hypothalamic and extra-hypothalamic regions.
Conclusions
These results suggest that 1) activation of GHSR-expressing neurons in the MBH is required for the normal feeding responses following both peripheral administration of ghrelin and fasting, 2) activation of MBH GHSR-expressing neurons is sufficient to induce feeding, and 3) axonal projections to a subset of hypothalamic and/or extra-hypothalamic regions likely mediate these responses. The Ghsr-IRES-Cre line should serve as a valuable tool to further our understanding of the functional significance of ghrelin-responsive/GHSR-expressing neurons and the neuronal circuitry within which they act.
Keywords: GHSR, Ghrelin receptors, Ghsr-IRES-Cre, DREADD, Food intake, Mediobasal hypothalamus
Highlights
-
•
We generated a novel Ghsr-IRES-Cre knock-in mouse line.
-
•
Cre activity in the line mirrors the known GHSR expression pattern.
-
•
Chemogenetic modulation of neuronal activity reveals a required role of MBH GHSR neurons in rebound food intake after a fast.
-
•
Neuronal projections of mediobasal hypothalamic GHSR neurons are reminiscent of AgRP neuronal projections.
Abbreviations
- AgRP
Agouti-related protein
- CNO
Clozapine-N-oxide
- Cre
Cre recombinase
- DAB
3,3′-Diaminobenzidine
- DREADD
Designer receptors activated exclusively by designer drugs
- DVC
Dorsal vagal complex
- GHSR
Growth hormone secretagogue receptor; ghrelin receptor
- GOAT
Ghrelin-O-acyltransferase
- hM3Dq
AAV5-hSyn-DIO-hM3D(Gq)-mCherry virus
- hM4Di
AAV2-hSyn-DIO-hM4D(Gi)-mCherry virus
- IRES
Internal ribosome entry site
- ISHH
In situ hybridization histochemistry
- MBH
Mediobasal hypothalamus
- NPY
Neuropeptide Y
- ROSA26-YFP
B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J reporter mice
- ROSA26-ZsGreen
B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J reporter mice
- RT-PCR
Reverse transcriptase-polymerase chain reaction
- YFP
Yellow Fluorescent Protein
1. Introduction
Ghrelin is a potent orexigenic hormone and growth hormone secretagogue mainly derived from the stomach [1], [2], [3], [4], [5]. Other pleiotropic actions of the hormone include those that are glucoregulatory, food reward-enhancing, gastric motility-enhancing, and anti-depressant [6], [7], [8], [9], [10], [11], [12], [13], [14]. Most of these actions of ghrelin occur via engagement of growth hormone secretagogue receptors (GHSRs; ghrelin receptors) expressed within the brain [15]. Of the two transcripts from the Ghsr gene, GHSR type 1a (GHSR1a, commonly referred to as “GHSR”) is translated into the functional, seven transmembrane, G-protein coupled receptor for ghrelin, whereas the truncated GHSR type 1b (GHSR1b), which results from an unspliced mRNA that terminates at an intronic stop codon, does not bind to ghrelin and has no known intrinsic function other than heterodimerizing with and attenuating the cell surface expression of GHSR1a [15], [16], [17].
The pattern of GHSR expression within mouse, rat, and primate brains has been established by detection of Ghsr mRNA using in situ hybridization histochemistry (ISHH) [15], [18], [19], [20], [21], [22], [23] as well as by other techniques including receptor binding assays, Western blot analysis, reverse transcriptase-polymerase chain reaction (RT-PCR), and ribonuclease protection assay [24], [25], [26], [27], [28], [29], [30], [31], [32]. GHSR expression within the mouse brain also has been mapped using a GHSR-eGFP reporter mouse model [23], although some differences between eGFP expression in that line and GHSR expression as determined using other methods such as ISHH are apparent. Other GHSR reporter mouse models include a GHSR-knockout, in which a lacZ coding sequence replaces the GHSR coding sequence, resulting in β-galactosidase expression in the place of GHSR expression [33], and a GHSR-IRES-tauGFP line [34], although detailed expression analyses have not been published using those models. Altogether, these techniques reveal a composite pattern of brain GHSR expression that includes relatively high levels within several mediobasal hypothalamic (MBH) nuclei including the arcuate nucleus (Arc), as well as the dorsomedial hypothalamus (DMH), ventromedial hypothalamus (VMH), paraventricular hypothalamus (PVH), and ventral premamillary nucleus (PMV). In addition, GHSR expression occurs in several other hypothalamic nuclei, the midbrain [including the ventral tegmental area (VTA) and substantia nigra (SN)], the dorsal vagal complex (DVC), and the hippocampus and amygdala, to name a few. The food intake and glucoregulatory effects mediated by GHSRs in general and by specific brain GHSR subpopulations have been studied by utilizing several genetically-modified rodent models, which include those with global deletion of GHSR expression [27], [35], [36], [37], [38], [39], [40], neuron-specific GHSR deletion [41], or re-expression or deletion of GHSR expression within selective neuronal subpopulations including those expressing Agouti-related protein (AgRP), Phox2b, or tyrosine hydroxylase [8], [42], [43], [44], [45].
Of all the actions mediated through GHSRs, one of the most robust and well-characterized is the induction of an acute feeding response after peripheral or central administration of ghrelin [3], [4], [36], [42], [46], [47]. However, the orexigenic function of endogenous ghrelin under physiological conditions remains less certain. Indeed, in some studies, mice lacking ghrelin, GHSR, or the enzyme ghrelin-O-acyltransferase (GOAT), which allows ghrelin to bind to GHSR, exhibited normal food intake and/or body weight phenotypes or only subtle changes [27], [40], [48], [49], [50], [51]. However, other studies using either genetic or pharmacologic means to disrupt the endogenous ghrelin system suggest that ghrelin plays an important role in the usual feeding and/or body weight responses to certain physiological perturbations, such as following a short-term fast or chronic exposure to high-fat diet [14], [35], [52]. Efforts to resolve any uncertainties in the orexigenic actions of the endogenous ghrelin system, as well as to probe the chemical and electrophysiological properties of GHSR-expressing neurons and the neurocircuitry within which they operate, would benefit from novel mouse models that facilitate their identification and modulation of their activity.
Here, we used a designer receptors activated exclusively by designer drugs (DREADD)-based chemogenetic approach in a novel Ghsr-IRES-Cre knock-in mouse line to test whether the activity of MBH GHSR neurons is required for the normal rebound food intake following fasting. We also used the Ghsr-IRES-Cre line to map the projections of the MBH GHSR neurons and confirm the central expression pattern of GHSR. We believe that the novel Ghsr-IRES-Cre mouse line described herein will further aid in the identification of the neurocircuitry directly engaged by ghrelin and the interaction of this ghrelin-responsive neurocircuitry with other factors and inputs that regulate food intake and body weight.
2. Material and methods
2.1. Mice
All experiments were approved by the Institutional Animal Care and Use Committees of the University of Texas Southwestern (UTSW) Medical Center and Monash University. Mice were housed under a 12 h dark–light cycle in standard environmentally-controlled conditions and were provided free access to water and standard chow (either Teklad Global Diet [2016]; Envigo, Madison, WI or a 20% protein/4.8% fat chow diet; Specialty Feeds, Glen Forrest, Western Australia), unless otherwise indicated. All the studies were performed in males.
2.2. Generation of Ghsr-IRES-Cre mice
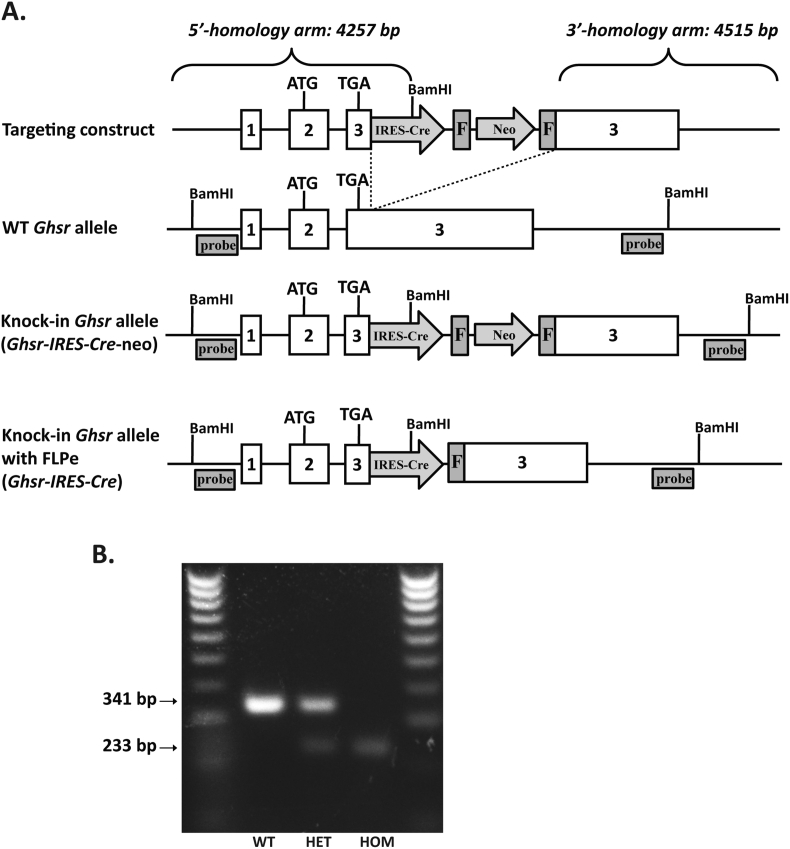
A targeting construct was generated using bacterial artificial cloning (BAC) recombineering techniques in EL250 and EL350 cells (Figure 1A) using methods described previously [53], [54]. Assembly involved insertion of an IRES-Cre-FRT-neo-FRT cassette [55] three base pairs (bp) downstream of the Ghsr STOP codon within a mouse Ghsr-containing BAC (RPCI.22 494 N7; BACPAC Resources Center at Children's Hospital Oakland Research Institute), followed by insertion of a 13,134 bp DNA fragment from the engineered BAC into a pGEM-thymidine kinase vector. The final targeting construct included 4,257 bp of Ghsr sequence upstream and 4,515 bp of Ghsr sequence downstream of the IRES-Cre-FRT-neo-FRT insertion site. The resulting construct was electroporated into 129/SvEvTac (SM-1 cells) by the UTSW Transgenic Technology Core Facility. One correctly-targeted ES cell clone was identified by Southern blot screening, and its targeted DNA sequence was further confirmed by PCR prior to blastocyst injection. The resulting pups were backcrossed once to C57BL/6N. The lines were then crossed to B6.Cg-Tg(ACTFLPe)9205Dym/J (The Jackson Laboratory) to remove the FRT-neo-FRT cassette. Mice harboring the wild-type Ghsr gene vs. the modified Ghsr-IRES-Cre gene were identified by genotyping tail snips using primers 5′-CTGGAGTTTCAATACCGGAGATCA-3′, 5′-TAGTGCTTACAGACAGCACT-3′, 5′-TCAATACATGACATCGCAGCGCAT-3′ and 5′-GGAGTTTAGTGTTACACACCAAGA-3′. The presence of the wild-type Ghsr allele results in a 341 bp band and the presence of the Ghsr-IRES-Cre allele results in a 233 bp band (Figure 1B). Mice subsequently backcrossed to C57BL/6N for 3 generations were used as breeders to generate mice used for the studies. To validate the expected pattern of Cre activity, Ghsr-IRES-Cre mice were crossed to ROSA26-YFP reporter mice [B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J] (The Jackson Laboratory; stock number 006148) and separately to ROSA26-ZsGreen reporter mice [B6.Cg-Gt(ROSA)26Sortm6(CAG-ZsGreen1)Hze/J] (The Jackson Laboratory; stock number 007906).
Figure 1.
Generation of the Ghsr-IRES-Cre allele by gene targeting. A. Schematic shows the generation of the Ghsr-IRES-Cre knock-in mouse line by homologous recombination. The IRES-Cre cassette was inserted 3 bp downstream of the GHSR stop codon (TGA) in Exon 3. Binding sites for Southern blot probes used to detect correctly-targeted genomic DNA (after restriction digest with BamHI) are shown. The mice were crossed to Flp1 recombinase mice to remove the frt-Neo-frt cassette that had been included in the targeting construct to find neomycin-resistant ES cell clones. B. PCR analysis of genomic DNA obtained by tail biopsies of representative mice carrying two copies of the wild-type Ghsr allele (“WT”), mice homozygous for the modified knock-in Ghsr-IRES-Cre allele (“HOM”), and mice with one copy of each of the alleles − heterozygotes (“HET”).
2.3. Stereotaxic surgery
To inhibit GHSR-expressing neurons, Cre-dependent AAV2-hSyn-DIO-hM4D(Gi)-mCherry virus [56] [hereafter referred as hM4Di; UNC Gene Therapy Center Vector Core] was injected bilaterally into the MBH of 8–10 wk old male Ghsr-IRES-Cre mice while they were under ketamine/xylazine (120/16 mg/kg BW, i.p.) anesthesia and restrained in a Kopf stereotaxic apparatus. The coordinates for MBH virus injection (−1.7 mm Bregma; +/− 0.25 mm lateral; −5.6 mm ventral from surface of brain) were chosen based on a mouse brain atlas [57]. Under aseptic conditions, a small hole was drilled into the skull and the hM4Di (250 nL) was injected into each side over a period of 15 min using a glass micropipette connected to an air pressure injector system. The micropipette was then retained for an additional 10 min, after which it was slowly retracted. The incision site was closed using surgical staples. Postoperatively, the mice were singly housed, administered buprenorphine 1 mg/kg s.c. every 12 h for 24 h, and allowed free access to Carprofen wafers for 3 days to reduce post-operative pain. The mice were allowed to recover for 3 wks prior to food intake studies, at which time clozapine-N-oxide (CNO) was administered to inhibit the GHSR-expressing neurons infected with hM4Di.
Chemogenetic activation of MBH GHSR-expressing neurons was achieved by stereotaxic delivery of Cre-dependent AAV5-hSyn-DIO-hM3D(Gq)-mCherry [hereafter referred as hM3Dq; UNC Gene Therapy Center Vector Core] bilaterally to the MBH of 10–11 wk old male Ghsr-IRES-Cre mice using the same coordinates described above for delivery of hM4Di. Briefly, the mice were anesthetized using isoflurane (5% for induction, 2% for maintenance) and positioned on a Stoelting stereotaxic frame. A Neuros Syringe (Hamilton Company, Bonaduz, Switzerland) was used to manually deliver 200 nL of the virus bilaterally over 10 min. Postoperatively, the mice were administered meloxicam (Metacam) 1 mg/kg s.c. daily for 3 days to relive pain. The mice were singly housed and allowed to recover for 2 wks prior to food intake studies, at which time CNO was administered to activate the GHSR-expressing neurons infected with hM3Dq.
2.4. Food intake studies
The requirement of MBH GHSR neuronal activity for two different types of food intake was tested by stimulation of hM4Di using CNO. To assess effects on ghrelin-induced food intake, mice were first handled for 3 days to acclimate them to the handling needed at the time of injections. Food was withdrawn from the cages during the light period (at about 10 AM) for 2 h, after which CNO (Sigma–Aldrich, St. Louis, MO; 1 mg/kg BW i.p.) or vehicle was delivered. The mice were then administered ghrelin (1 mg/kg BW s.c.) 1 h after CNO or vehicle administration, after which standard chow (Teklad Global Diet [2016]) was re-introduced. Food intake was measured at 45 min, 90 min, 4 h, and 24 h post-ghrelin administration. After a 1 wk recovery period, which included another 3 days of handling, the above protocol was repeated using a crossover design to deliver CNO and vehicle.
Three wks later, using the same mice, we assessed effects on rebound food intake following a 24 h fast. Mice were again handled for 3 days prior to injections. Food was withdrawn from the cages beginning at about 10 AM, for 23 h. Mice were administered CNO (1 mg/kg BW i.p.) or vehicle. Standard chow was reintroduced 1 h after CNO or vehicle administration and food intake was measured at 45 min, 90 min, 4 h and 24 h. The protocol was repeated 1 wk later using a crossover design to deliver vehicle or CNO.
To test if chemogenetic activation of MBH GHSR neurons is sufficient to induce food intake, ad lib-fed hM3Dq-injected Ghsr-IRES-Cre mice were administered CNO (0.3 mg/kg, i.p.) after 5 consecutive days of handling. Intake of the 20% protein/4.8% fat chow diet was measured at 2 h and 5 h post-CNO administration.
2.5. Immunohistochemistry
Mice were deeply anesthetized with chloral hydrate (500 mg/kg i.p.) or isoflurane and transcardially perfused with 0.9% phosphate-buffered saline (PBS) followed by 10% neutral buffered formalin, as described previously [23]. Brains were removed immediately and stored in the same fixative for 4–6 h, at 4 °C, immersed in 20 or 30% sucrose in PBS, pH 7.0 at 4 °C overnight, and sectioned coronally into five equal series at a thickness of 25 μm (for Ghsr-IRES-Cre X ROSA26-YFP and hM4Di injected mice) or four equal series at a thickness of 30 μm (for Ghsr-IRES-Cre X ROSA26-ZsGreen and hM3Dq injected mice) on a sliding microtome. The sections were stored at −20 °C in an antifreeze solution (Simmons et al., 1989) until further processing. Sections then were mounted onto SuperFrost slides (Fisher Scientific, Pittsburgh, PA) and dried overnight.
Immunohistochemistry was performed as described previously [23]. Slides with coronal brain sections were washed three times with PBS. The sections were treated with 0.3% hydrogen peroxide in PBS, pH 7.4, for 30 min at room temperature. Antigen unmasking was performed by placing slides in a sodium citrate buffer pre-heated to 95–100 °C and then incubating them in 3% normal donkey serum (Jackson ImmunoResearch Laboratories, West Grove, PA) with 0.3% Triton X-100 in PBS (PBT) for 2 h at room temperature. To visualize Cre-mediated YFP expression, the sections were then incubated overnight with chicken anti-GFP antibody (1:5,000 dilution, Aves Labs Cat# GFP-1020, RRID:AB_10000240). After washing with PBS, sections were incubated for 2 h in biotin-conjugated donkey anti-chicken IgG (1:1,000 dilution; Jackson ImmunoResearch Labs Cat# 703-065-155, RRID:AB_2313596) secondary antibody. Thereafter, the slides were washed with PBS, immersed in a 2% ABC solution for 1 h at room temperature, and washed again in PBS. YFP expression in the sections was visualized by 3,3′-Diaminobenzidine (DAB) reaction using a metal enhanced DAB substrate kit (Thermo Scientific, Pittsburg, PA). Finally, the slides were washed in PBS, dehydrated in graded ethanol concentrations, cleared in xylenes, and cover-slipped with Permount (Thermo Scientific) mounting medium. Cre activity in Ghsr-IRES-Cre X ROSA26-ZsGreen reporter mice was examined by direct visualization of the endogenous ZsGreen fluorescence. The Cre activity expression patterns reported here were observed in at least one series from at least three different brains using both reporters.
To visualize hM4Di-infected Ghsr-IRES-Cre neurons and their projections, a similar procedure was followed. Immunohistochemical detection of mCherry was performed using an anti-DsRed, polyclonal rabbit primary antibody (1:2000, Clontech Laboratories, Inc. Cat# 632496, RRID:AB_10015246) and Biotin SP donkey anti-rabbit IgG secondary antibody (1:200 dilution, Jackson ImmunoResearch Labs Cat# 711-065-152, RRID:AB_2340593). To visualize hM3Dq-targeted Ghsr-IRES-Cre neurons, anti-DsRed polyclonal rabbit primary antibody (1:1000) and goat anti-rabbit IgG AlexaFluor 594 secondary antibody (1:400 dilution, Life Technologies; Cat# A-11037, RRID:AB_2534095) were used. For visualization of the fluorescently-labeled secondary antibody and endogenous ZsGreen, sections were coverslipped using Vectashield anti-fade mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA).
2.6. Imaging and photomicrographs
Following immunohistochemistry, the DAB-stained mounted brain sections were viewed with a Zeiss Axioskop microscope using brightfield optics. Fluorescent signals were visualized using a Zeiss Apotome microscope. Photomicrographs were captured with a Zeiss digital camera attached to the microscope using the AxioVision software. Adobe Photo-Shop CS5.1 (San Jose, CA), was used to adjust sharpness, contrast, and brightness of the photomicrographs. Images from Ghsr-IRES-Cre X ROSA26-ZsGreen reporter mice were obtained using a Zeiss Apotome.2 microscope with a motorized stage to capture tiled images. For tiled images, a 20 × 20 tiling matrix was used at 20 × magnification to capture the entire brain section. Images were captured using a MicroFiber camera using Stereoinvestigator software (MicroBrightField, Burlington, VT, USA). Photos 2.0 (Apple Inc. CA, USA) was used to adjust sharpness, contrast, and brightness of images.
2.7. Statistics
All data are expressed as mean ± SEM. Two-tailed statistical analyses and graph preparations were accomplished using GraphPad Prism 6.0 (GraphPad Software, Inc. San Diego, CA). The paired Students ‘t’ test was used to test for significant differences between test groups as indicated in figure legends. Outliers were detected by Grubb's test. P-values < 0.05 were considered statistically significant.
3. Results
3.1. Generation of Ghsr-IRES-Cre mice and assessment of their pattern of Cre activity
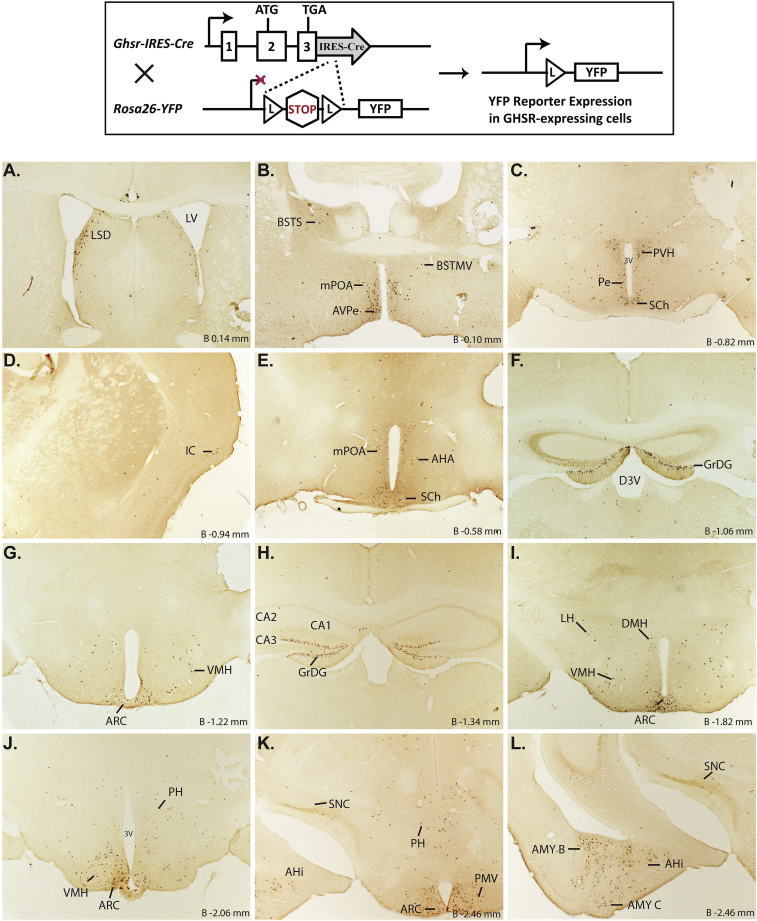
A novel Ghsr-IRES-Cre knock-in mouse line was generated by insertion of an IRES-Cre cassette into the endogenous Ghsr gene downstream of the GHSR stop codon (Figure 1). This allows for the expression of Cre-recombinase selectively in GHSR-expressing cells. To assess this selectivity, Ghsr-IRES-Cre mice were crossed to ROSA26-YFP reporter mice and separately to ROSA26-ZsGreen reporter mice, resulting in mice in which Cre-mediated removal of a loxP-flanked stop cassette in the ROSA26-YFP transgene enables YFP expression and in the ROSA26-ZsGreen transgene enables ZsGreen expression. Here, we focused on the brain, examining coronal sections from the level of the prefrontal cortex to the cervical spinal cord. Positive YFP immunolabeling within cell bodies (stained brown by DAB) or direct visualization of the endogenous ZsGreen fluorescence was used to report Cre activity. Representative photomicrographs of the pattern of Cre activity as determined using the ROSA26-YFP mouse are found in Figure 2, while those using the ROSA26-ZsGreen mouse are found in Supplemental Figure 1. Furthermore, within each brain region, Cre activity was graded subjectively based on the relative number and density of immunoreactive cell bodies in the ROSA26-YFP reporters, as summarized in Table 1. Abbreviations used to describe the brain regions within the text and figures are denoted in Table 1 and mostly follow those used in the mouse brain atlas of Paxinos and Franklin [57]. A description of this Cre activity expression pattern follows.
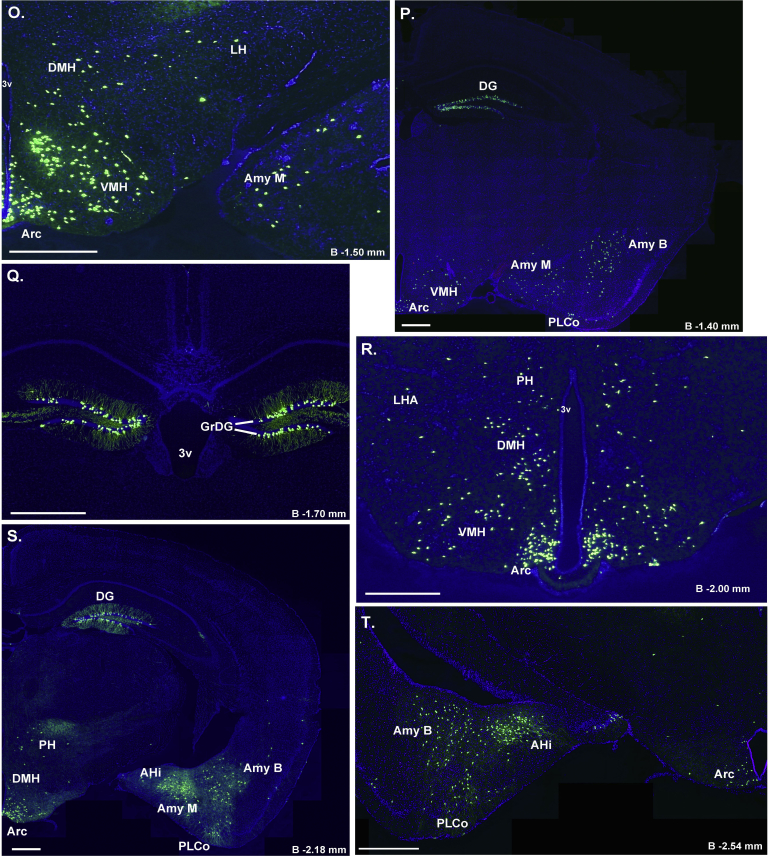
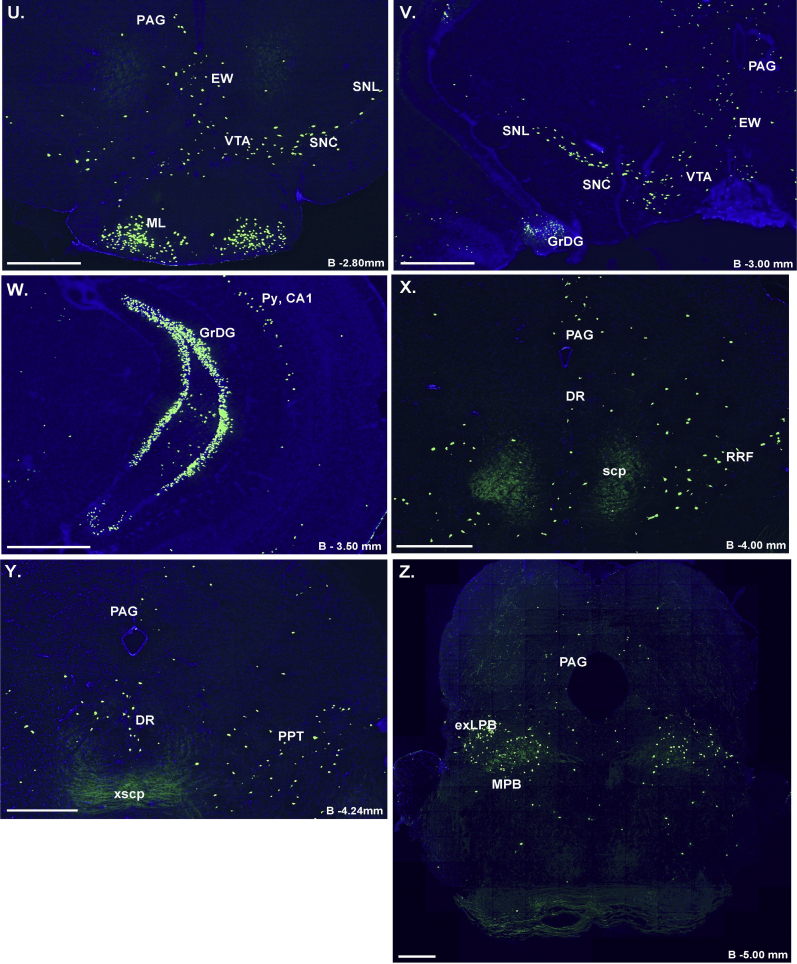
Figure 2.
Assessment of Cre-activity in Ghsr-IRES-Cre mice. Inset shows the schematic of YFP reporter expression in GHSR-expressing cells in mice derived from the cross of Ghsr-IRES-Cre mice with Rosa26-YFP reporter mice. Cre-mediated excision of the loxP-flanked STOP cassette allows for YFP reporter expression in cells with Cre activity. A–Z. Series of low-power bright-field representative photomicrographs summarizing YFP-immunoreactive cell bodies in coronal brain sections of Ghsr-IRES-Cre X ROSA26-YFP reporter mice. Immunoreactivity is demonstrated by the dark brown DAB stain. Scale bar in panel Z = 500 μm (applies to A–Z). Approximate distance of the coronal section from bregma (B) is indicated for panels A–Y. 3V – third ventricle, 4V – fourth ventricle, Aq – Aqueduct of Sylvius, BSTS – supracapsular part and BSTMV – medial division, ventral part of the BNST, D3V – Dorsal 3rd ventricle, GM – gray matter of the spinal cord, GrDG – Granular layer of Dentate Gyrus, LV – lateral ventricle, Py – Pyramidal cells. See Table 1 for explanation of abbreviations of the brain regions.
Table 1.
Histologic assessment of YFP expression in the Ghsr-IRES-Cre X Rosa26-YFP mice.
| Region of the brain | YFP-IR |
|---|---|
| Cerebral cortex | |
| Prelimbic cortex – PrL | – |
| Cingulate cortex – Cg | – |
| Insular cortex – IC | ± |
| Ectorhinal cortex – Ect | ± |
| Piriform cortex – Pir | ++ |
| Hippocampus | |
| Ammon's horn, CA1 | ± |
| Ammon's horn, CA2 | – |
| Ammon's horn, CA3 | ++ |
| Dentate gyrus | ++++ |
| Basal Ganglia and Septum | |
| Amygdaloid hippocampal area – AHi | ++ |
| Bed nucleus of the stria terminalis – BNST | ++ |
| Basolateral amygdaloid nucleus – Amy B | +++ |
| Cortical amygdaloid nucleus – Amy C | + |
| Medial amygdaloid nucleus – Amy M | ++ |
| Lateral septal nucleus, Dorsal – LSD | ++ |
| Medial globus pallidus – MGP | – |
| Hypothalamus | |
| Anterior Hypothalamic Area – AHA | + |
| Anteromedial nucleus – Ant | – |
| Anteroventral periventricular nucleus – AVPe | ++ |
| Arcuate Nucelus – Arc | ++++ |
| Dorsomedial nucleus – DMH | + |
| Lateral hypothalamus – LH | + |
| Magnocellular nucleus of the posterior commissure – MCPC | ++ |
| Medial mamillary nucleus, Lateral – ML | +++ |
| Medial Preoptic Area – mPOA | ++ |
| Posterior hypothalamus – PH | + |
| Paraventricular nucleus – PVH | ++ |
| Periventricular hypothalamic nucleus – Pe | + |
| Premamillary nucleus, Ventral – PMV | ++ |
| Parvicellular red nucleus – RPC | + |
| Suprachiasmatic nucleus, Dorsomedial – Sch | ++ |
| Ventromedial nucleus – VMH | ++ |
| Midbrain, Pons and Medulla Oblongata | |
| Area postrema – AP | + |
| Central gray, alpha – CGa | + |
| Dorsal motor nucleus of the vagus – DMNV | + |
| Dorsal raphe nucleus – DR | + |
| Dorsal tegmental nucleus – DTg | + |
| Laterodorsal tegmental nucleus – LDTg | + |
| Edinger Westphal nucleus – EW | ++ |
| Facial Motor Nucleus – 7 | + |
| Lateral parabrachial nucleus, external – exLPB | ++++ |
| Medial parabrachial nucleus – MPB | ++ |
| Medullary reticular nucleus, Dorsal – MdD | + |
| Nucleus ambiguous – Amb | + |
| Nucleus of the solitary tract – NTS | + |
| Pedunculopontine tegmental nucleus – PPT | ++ |
| Periaqueductal gray – PAG | ++ |
| Raphe magnus nucleus – RMg | + |
| Retrorubral field – RRF | ++ |
| Reuniens thalamic nucleus – Re | – |
| Substantia nigra, pars compacta – SNC | ++ |
| Substantia nigra, pars lateralis – SNL | ++ |
| Substantia nigra, pars reticulata – SNR | + |
| Superior salivatory nucleus – SuS | – |
| Ventral cochlear nucleus, Anterior – VCA | + |
| Ventral tegmental area – VTA | ++ |
| Cerebellum | |
| Cerebellar nucleus, interposed – ICb | +++ |
| Cerebellar nucleus, Medial – MCb | ++ |
| Vestibulocerebellar nucleus – VeCb | ++ |
Expression of YFP was estimated qualitatively based on the number and density of labeled cells: ++++, highest expression; +++, high expression; ++, moderate expression; +, low expression; –, background; ±, inconsistent expression. The list includes brain regions that showed YFP expression as well as regions previously demonstrated to have either significant GHSR mRNA expression or GHSR-eGFP transgene expression for comparison purposes (Zigman et al., 2006; Mani et al., 2014).
Starting with the cortical regions, expression was scattered throughout the piriform cortex, particularly in its more caudal extent. Within the insular cortex and ectorhinal cortex, YFP-expressing cells were observed sparsely, and only in occasional cases. Although not examined using the ROSA26-YFP reporter mice, Cre activity as evaluated by crossing Ghsr-IRES-Cre mice to the ROSA26-ZsGreen reporter mouse model, also was observed within the granular, glomerular and mitral layers of the main olfactory bulb, as well as the accessory olfactory bulb (Supplemental Figure 1). Within the hippocampus, the CA3 field of Ammon's horn contained many YFP-expressing cells, whereas little to no expression was observed in the CA1 and CA2 fields, except for the CA1 pyramidal cell layer. Also, the granular cell layer of both the dorsal and ventral dentate gyrus (DG) demonstrated very strong expression of YFP. YFP expression also was observed in the amygdala, where its expression was highest in the basolateral amygdaloid nucleus (Amy B), followed by less intense expression in the medial (Amy M) and cortical amygdaloid nuclei (Amy C). Furthermore, the bed nucleus of stria terminalis (BNST) demonstrated YFP expression, most prominently within the medial and lateral subdivisions, as did both the dorsal aspect of the lateral septal nucleus (LSD) and the ventral aspect (LSV; observed only in the ROSA26-ZsGreen reporter line; Supplemental Figure 1).
The distribution of YFP-expressing cells was most extensive within the hypothalamus, particularly the MBH. The highest expression was observed within the Arc followed by the VMH, DMH, lateral hypothalamus area (LH), PVH, PMV, and magnocellular nucleus of the posterior commissure (MCPC). More rostrally, YFP expression was observed in the suprachiasmatic nucleus (Sch), anterior hypothalamic area (AHA), anteroventral periventricular nucleus (AVPe), and medial preoptic area (mPOA).
YFP expression also was observed within the midbrain, medulla, and pons. This included the VTA and substantia nigra [pars compacta (SNC), pars reticularis (SNR), and pars lateralis (SNL)]. Other notable regions with YFP-immunoreactive cells included the external lateral parabrachial nucleus (exLPB), Edinger-Westphal Nucleus (EW), facial motor nucleus (7), pedunculopontine tegmental nucleus (PPT), retrorubral field (RRF), and the DVC [comprising the area postrema (AP), nucleus of the solitary tract (NTS), and dorsal motor nucleus of the vagus (DMNV)]. YFP immunoreactivity also was observed in the anterior interposed nucleus of the cerebellum (IntA) and the vestibulocerebellar nucleus (VecB). Finally, YFP-immunoreactive cells were sparsely distributed within the gray matter of the cervical spinal cord.
It also should be noted that while the above pattern of Ghsr-IRES-Cre mice was observed in at least three different cases in both the YFP and ZsGreen reporter lines, occasional cases exhibited a somewhat asymmetric pattern (more expression on one side of the brain) or a less extensive bilateral pattern of Cre-activity (See Supplemental Figure 2 for an example). The reasons for these alternate patterns of Cre-activity in a subset of the reporter mice are as yet unclear. Also, there were some slight differences in numbers of observed cells containing Cre-activity within certain brain regions, that seemed dependent on the reporter line used (e.g. the VMH, Figure 2J vs. Supplemental Figure 1O).
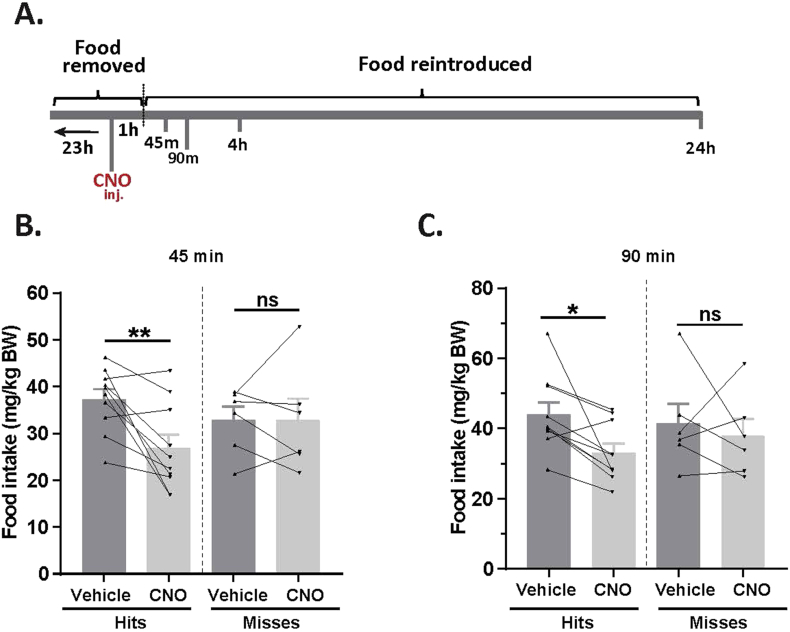
3.2. Inhibition of MBH GHSR neurons attenuates ghrelin-induced food intake
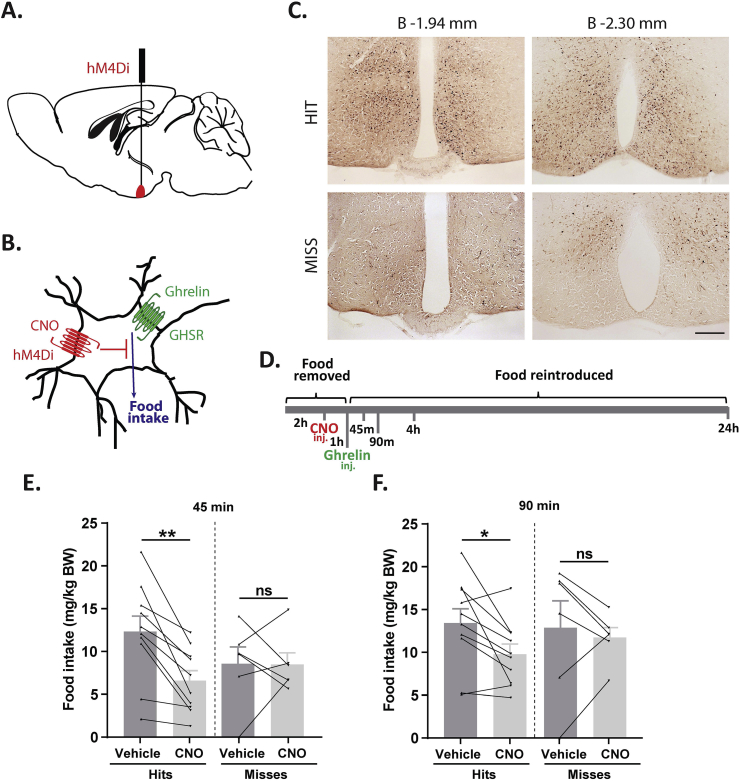
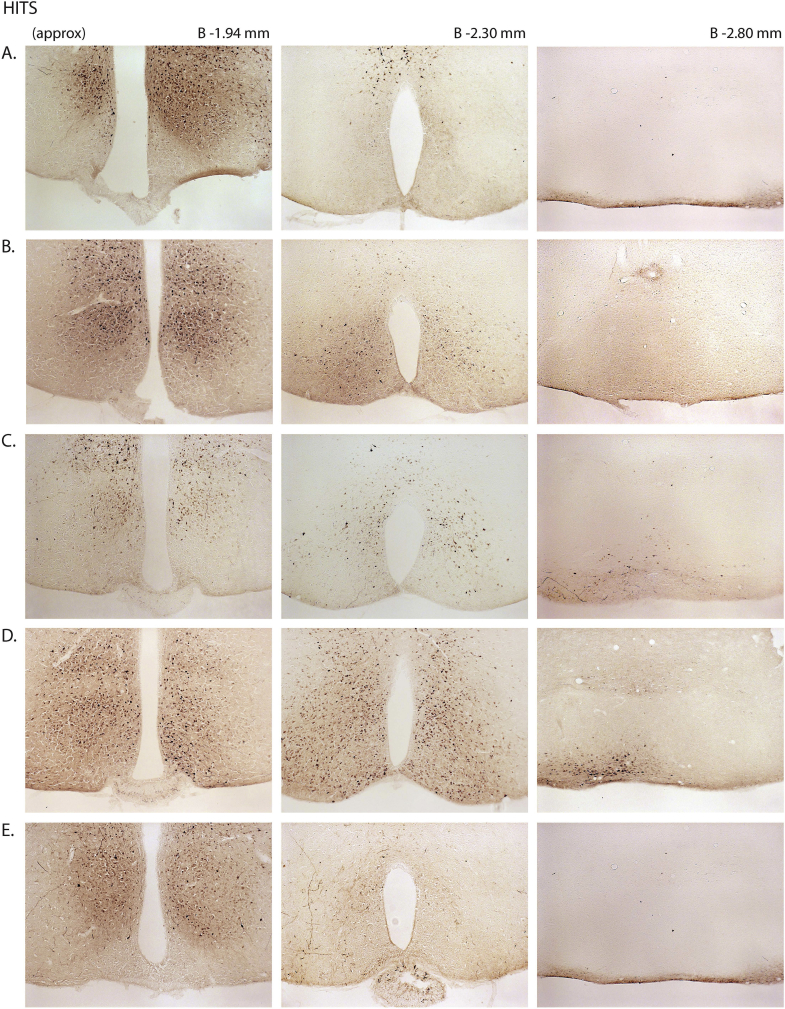
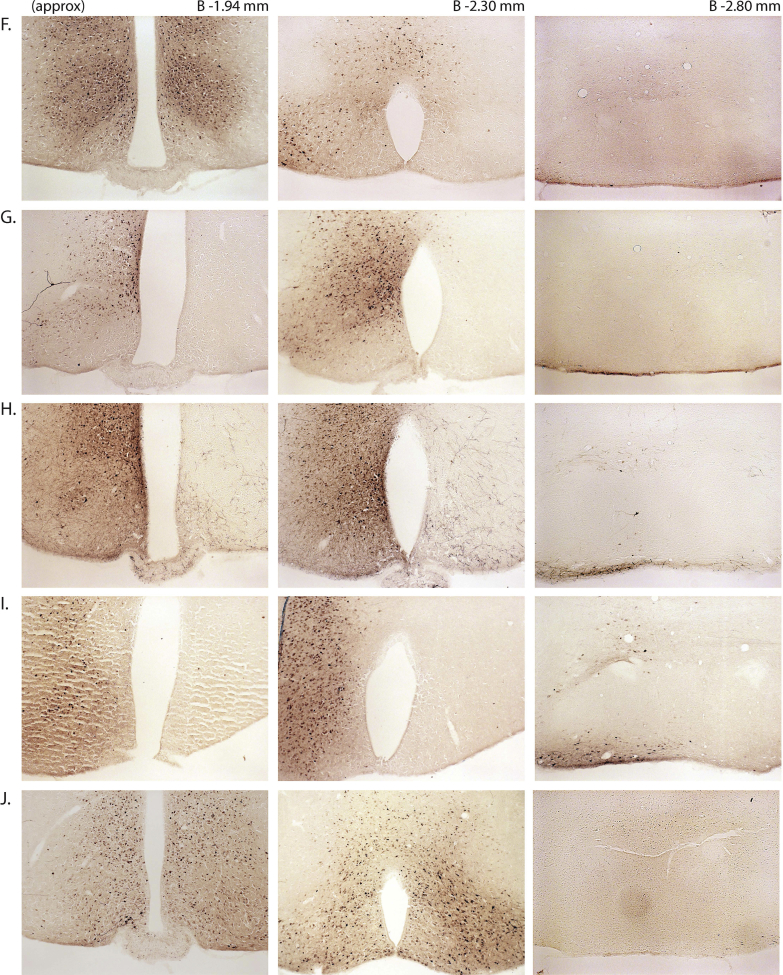
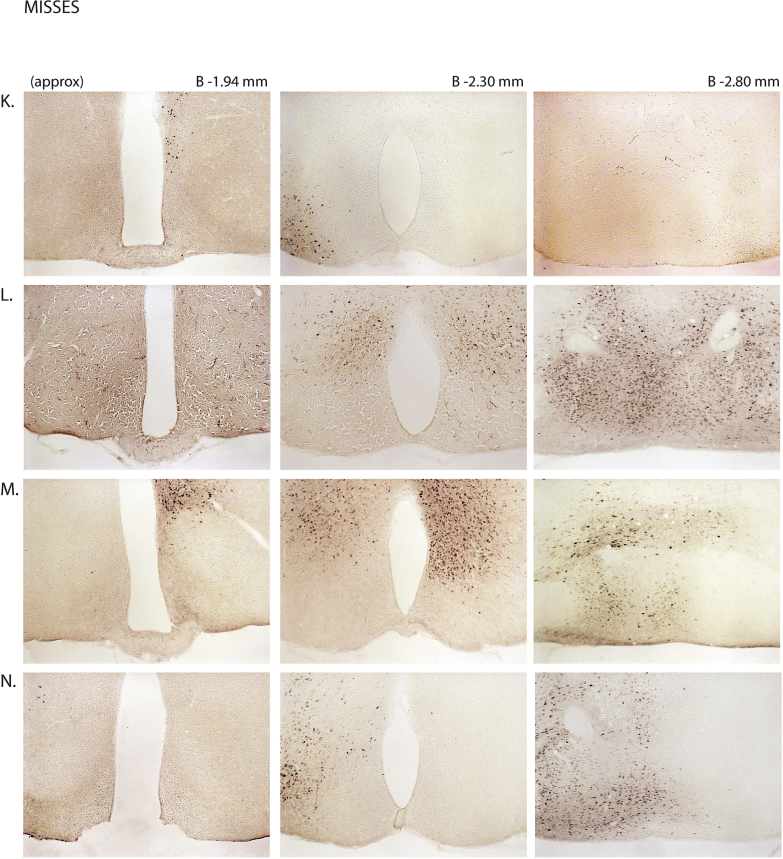
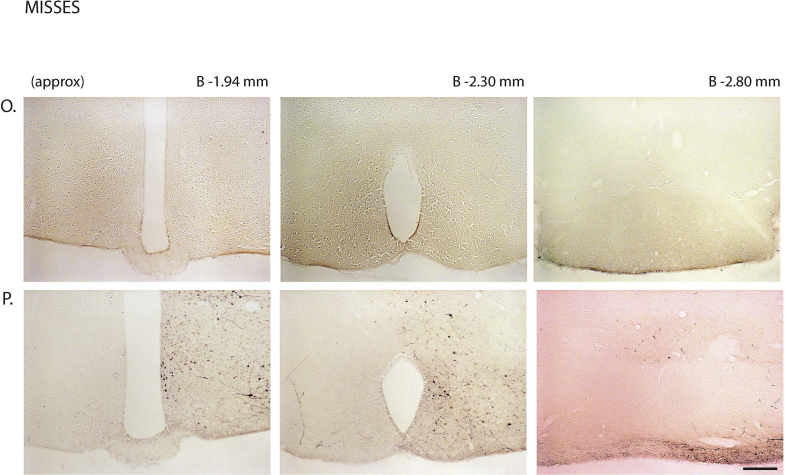
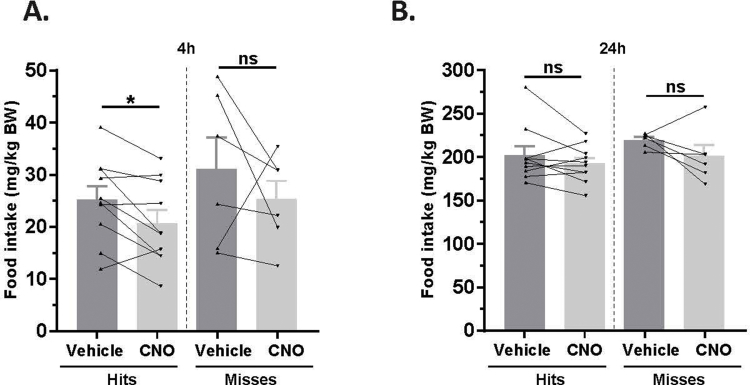
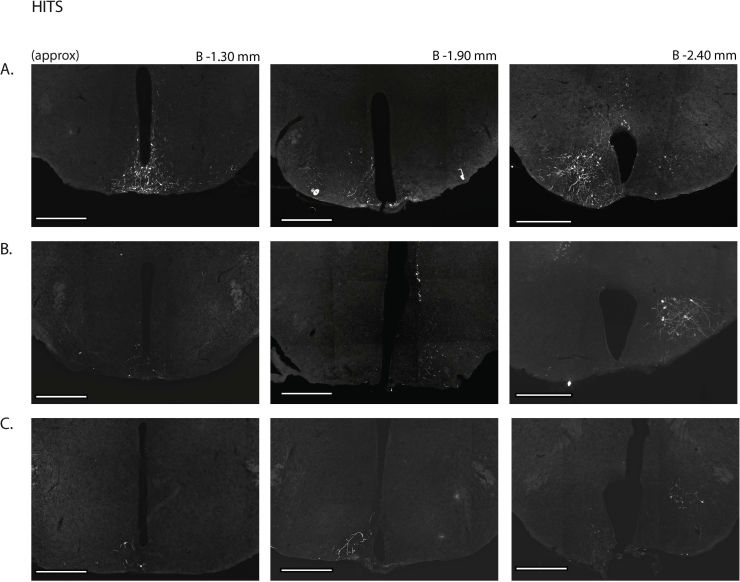
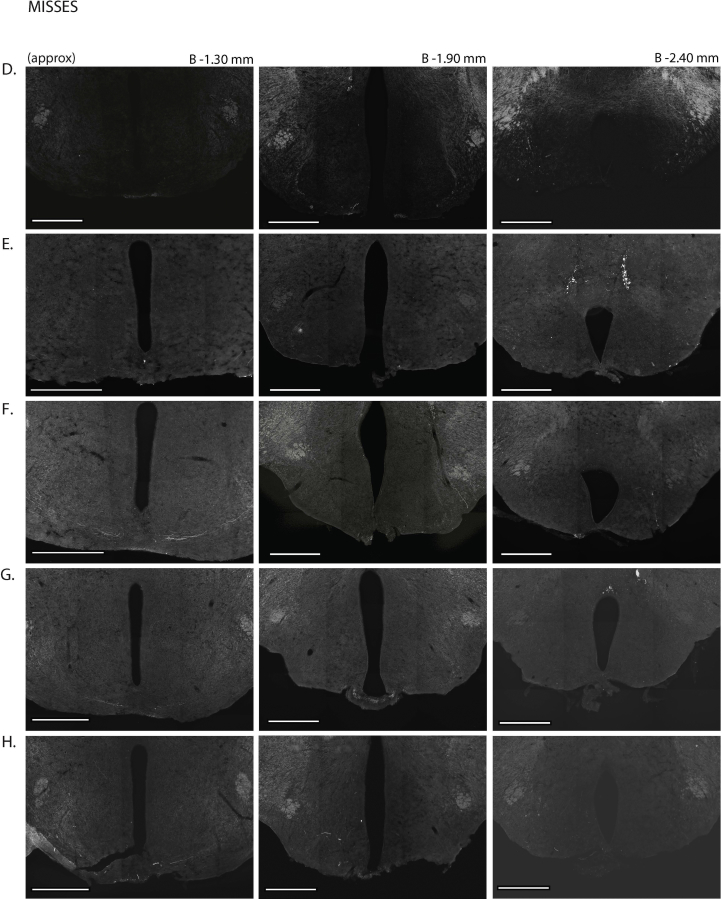
Next, we assessed the functional significance of GHSR-expressing MBH neurons by determining if inhibiting their activity, as achieved using a Cre-dependent chemogenetic system, would attenuate the well-characterized acute orexigenic response to peripherally-administered ghrelin. Stereotaxic surgery was used to deliver an inhibitory DREADD virus (hM4Di) to the MBH of Ghsr-IRES-Cre, using coordinates that were chosen to target the Arc and adjacent nuclei bilaterally (Figure 3A). Those Cre-expressing neurons infected with hM4Di express a designer receptor that engages downstream Gi-coupled signaling cascades, which, in turn, inhibit the activity of those neurons upon engagement by CNO [56], [58]. In this case, engagement of hM4Di with CNO would be expected to inhibit the activity of GHSR-expressing neurons in the MBH (Figure 3B). Also, those Cre-expressing neurons at the site of viral microinjection infected with hM4Di express a mCherry reporter, enabling their identification and characterization of their axonal projections. mCherry expression was determined at the end of the study to classify those cases with appropriately targeted virus injections as “hits” (defined here as cases in which mCherry expression was present either bilaterally or unilaterally within the MBH centered at the level of bregma −1.94 mm) or as “misses” (defined here as cases in which mCherry expression was either not present, present only in a few MBH cells, or not targeted to the MBH). Notably, for the majority of the “misses”, the injections appeared to have been mis-targeted to a region slightly caudal to the MBH (Figure 3C; Supplemental Figure 3). Ghrelin (1 mg/kg BW s.c.) was administered 1 h following CNO (1 mg/kg BW i.p.) or vehicle administration (Figure 3D). Ghrelin-induced food intake was significantly attenuated with CNO treatment when compared to vehicle treatment in the “hits”, when measured 45 min, 90 min, and 4 h post-ghrelin administration (46.4%, 27.1%, and 17.9% reductions at 45 min, 90 min, and 4 h, respectively; Figure 3E,F, Supplemental Figure 4A). Cumulative food intake was not significantly different between the 2 treatment groups, when measured 24 h after treatment (Supplemental Figure 4B). CNO treatment did not affect ghrelin-induced food intake in the “misses” group (Figure 3E,F, Supplemental Figure 4).
Figure 3.
Inhibition of MBH GHSR neurons attenuates ghrelin-induced food intake: A. Schematic of sagittal section of the brain (modified from mouse brain atlas of Paxinos and Franklin) demonstrates the site of injection of the hM4Di virus within the MBH of the Ghsr-IRES-Cre mice. B. Graphical representation shows the principle of the experiment: activation of hM4Di with CNO inhibits the activity GHSR-expressing neurons in the MBH, thereby attenuating food intake, which would otherwise normally be induced by stimulation of GHSR by ghrelin. C. Representative coronal sections of the brains from virus-injected Ghsr-IRES-Cre mice demonstrating Cre-dependent expression of mCherry (or lack thereof) in the MBH. The top row shows brain sections from an animal with a correctly-targeted MBH (classified as a “hit”) and the bottom row represents a “miss”. The approximate distances from bregma are indicated. Scale bar = 200 μm. D. Schematic shows the experimental procedure. See detailed description in the methods section. E&F. Effect of CNO administration on ghrelin-induced food intake at 45 min (E) and 90 min (F) following food reintroduction in the “hits” and “misses”. *P < 0.05, **P < 0.01, statistically significant reduction in ghrelin-induced food intake with CNO administration; n.s., no significant difference. Data were analyzed by paired Students “t” test and represented as mean ± SE.
3.3. Inhibition of MBH GHSR neurons attenuates rebound food intake following fasting
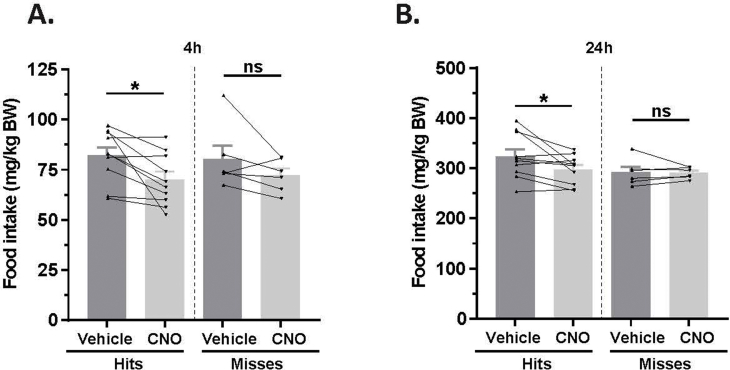
Next, we tested if activity of MBH GHSR neurons is required for rebound food intake stimulated physiologically following a 24 h fast. The hM4Di-injected Ghsr-IRES-Cre mice were fasted for 23 h, after which CNO (1 mg/kg BW i.p.) or vehicle was administered, and then food re-introduced 1 h later (Figure 4A). Rebound food intake was attenuated with CNO treatment at all the time points measured in the “hits” (28.1%, 25.0%, 14.8% and 8.1% reductions at 45 min, 90 min, 4 h, and 24 h, respectively; Figure 4B,C; Supplemental Figure 5). CNO treatment did not affect rebound food intake in the “misses” (Figure 4B,C; Supplemental Figure 5).
Figure 4.
Inhibition of MBH GHSR neurons attenuates fasting-induced rebound food intake. A. Schematic shows the experimental procedure. B&C. Effect of CNO administration on 24 h fasting-induced food intake at 45 min (B) and 90 min (C) following food reintroduction in the hM4Di-injected Ghsr-IRES-Cre “hits” and “misses”. *P < 0.05, **P < 0.01, statistically significant reduction in fasting-induced food intake with CNO administration; n.s., no significant difference. Data were analyzed by paired Students “t” test and represented as mean ± SE.
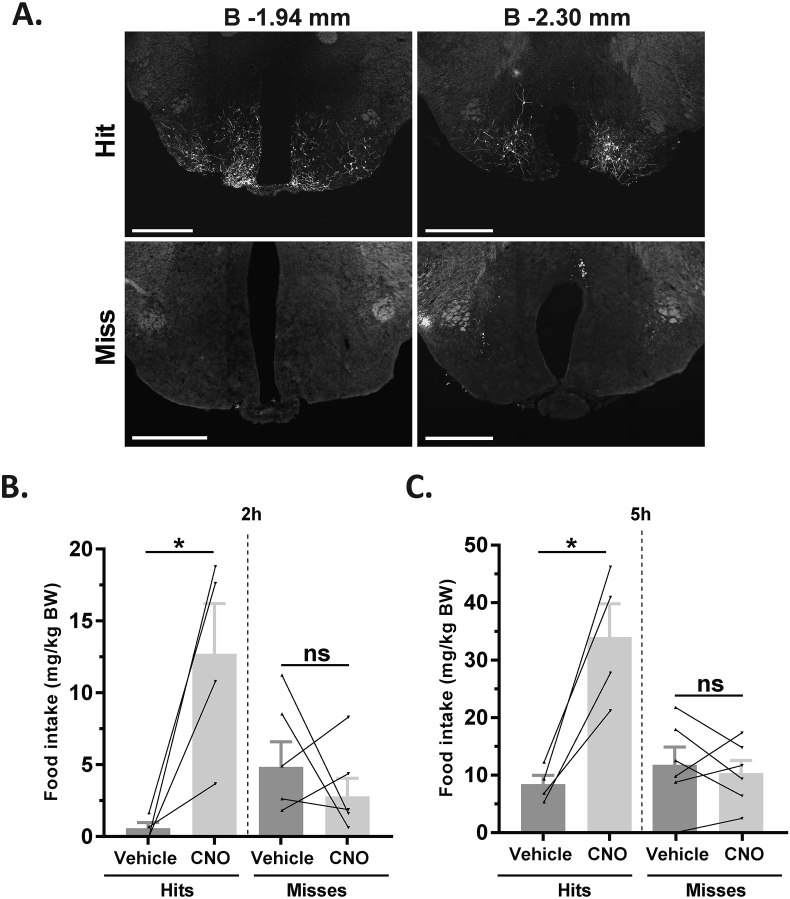
3.4. Stimulation of MBH GHSR neurons is sufficient to stimulate food intake
To determine if stimulating MBH GHSR neuron activity is sufficient to increase food intake, Ghsr-IRES-Cre mice were bilaterally injected into the MBH with a stimulatory DREADD virus (hM3Dq). In contrast to hM4Di-infected neurons, those Cre-expressing neurons infected with hM3Dq express a designer receptor that engages downstream Gq-coupled signaling cascades, which, in turn, stimulate the activity of those neurons upon engagement by CNO [56], [58]. Once again, injection sites were confirmed at the end of the study by mCherry immunofluorescence, allowing the cases to be grouped as “hits” or “misses” (Figure 5A; Supplemental Figure 6). Ad lib-fed mice were administered CNO (0.3 mg/kg BW i.p.) or vehicle and subsequent food intake was measured. CNO induced a significant increase in food intake in the “hits” when measured 2 h and 5 h post-CNO injection (21-fold and 3-fold increases at 2 h and 5 h, respectively; Figure 5B,C). No significant increase in food intake was observed in the “misses” (Figure 5B,C).
Figure 5.
Activation of MBH GHSR neurons is sufficient to induce food intake. A. Representative coronal sections of brains from hM3Dq-mCherry virus-injected Ghsr-IRES-Cre mice demonstrating Cre-dependent expression of mCherry (or lack thereof) in the MBH. The top row of brain sections represents a “hit” and the bottom row represents a “miss”. The approximate distances from bregma are indicated. Scale bar = 500 μm. B&C. Effect of CNO-induced food intake at 2 h (B) and 5 h (C) following food reintroduction in the “hits” and “misses”. *P < 0.05, statistically significant reduction in ghrelin-induced food intake with CNO administration; n.s., no significant difference. Data were analyzed by paired Students “t” test and represented as mean ± SE.
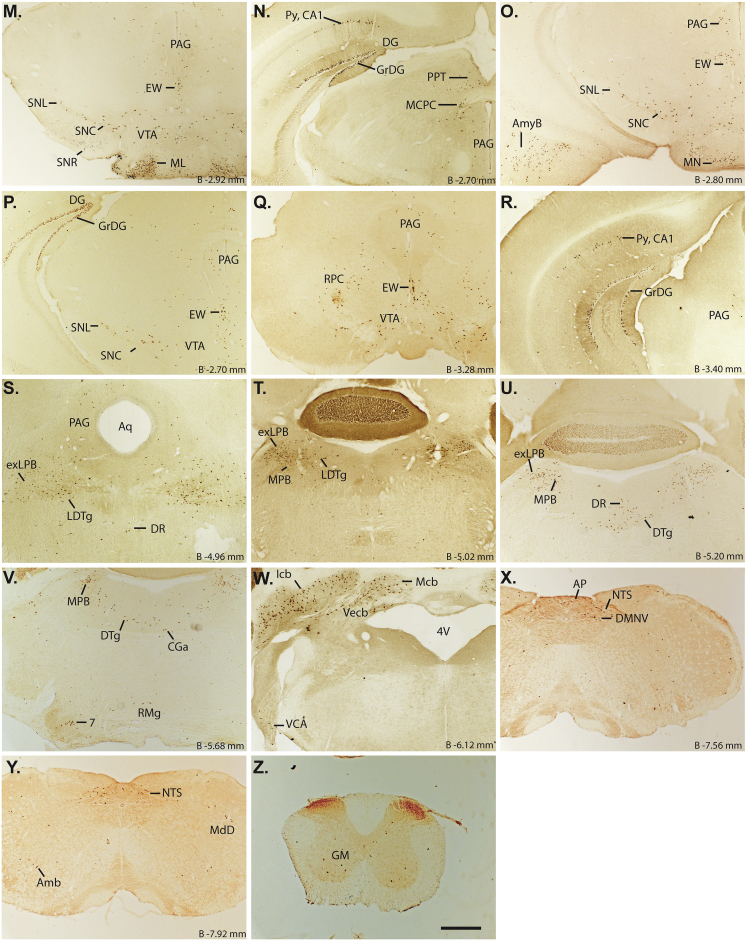
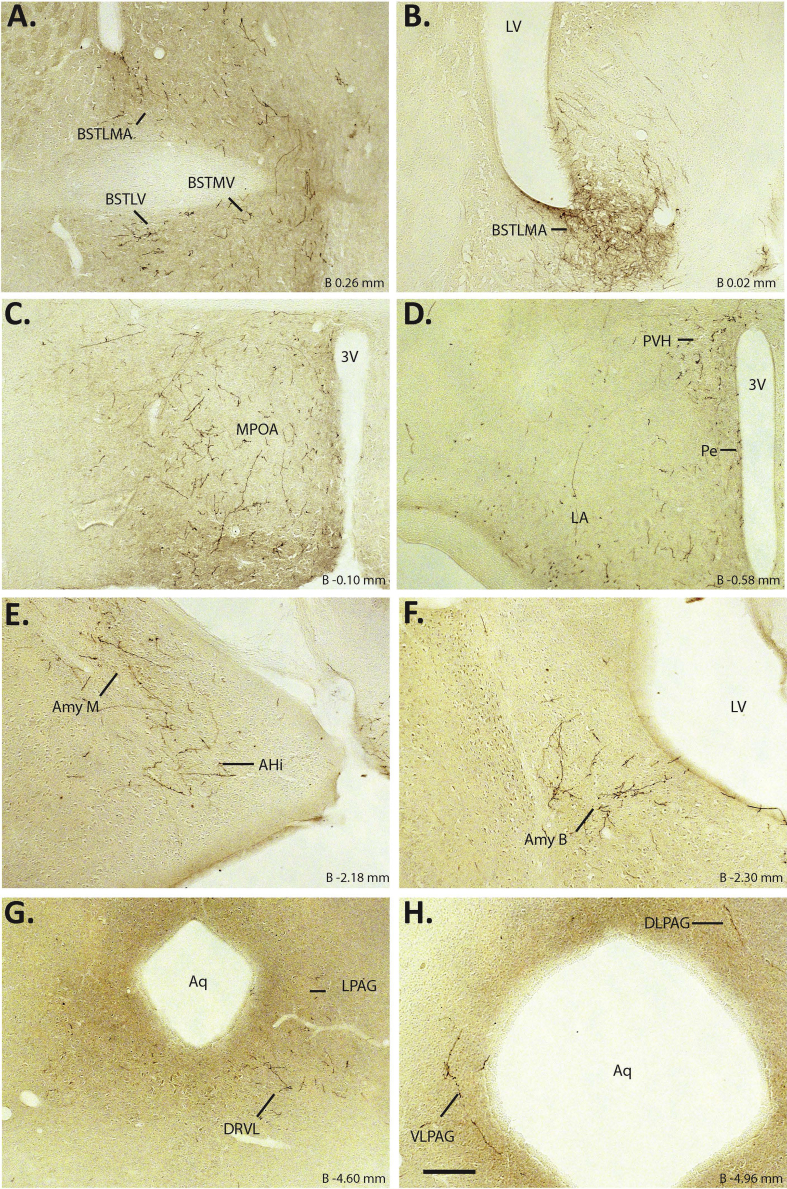
3.5. Hypothalamic and extrahypothalamic projects of MBH GHSR neurons
In order to begin to understand those second order neurons participating in neuronal circuits downstream of MBH GHSR neurons, we determined the expression pattern of neuronal projections emanating from MBH GHSR neurons. For these studies, the neuronal projection patterns were determined using those hM4Di-injected Ghsr-IRES-Cre cases classified as “hits”. Among the “hits”, axonal projections were observed consistently in the medial and lateral divisions of the BNST, mPOA, periventricular hypothalamic nucleus (Pe), PVH, LH, and the lateral, dorsolateral (DLPAG) and ventrolateral (VLPAG) divisions of periaqueductal gray (PAG) (Figure 6). Projections to the basolateral and medial amygdala were observed in 4 out of 10 cases. Projections to the ventrolateral dorsal raphe (DRVL) were observed in 5 out of 10 cases. There were no obvious distinguishing features about the extent of the MBH targeted in those cases that projected to the amygdala and DRVL.
Figure 6.
Hypothalamic and extrahypothalamic axonal projects of MBH GHSR neurons: A–H. Representative coronal sections of the brain show mCherry immunoreactivity, a marker of the axonal projections from the MBH GHSR neurons in the Ghsr-IRES-Cre mice injected with hM4Di virus. BSTMA – Bed nucleus of stria terminalis, medial division, anterior; BSTLV – Bed nucleus of stria terminalis, lateral division, ventral, BSTMV – Bed nucleus of stria terminalis, medial division, ventral; LPAG – lateral periaqueductal gray; DLPAG – Dorsolateral periaqueductal gray; VLPAG – Ventrolateral periaqueductal gray. Scale bar in panel H = 200 μm (applies to all panels). Approximate distance of the coronal section from bregma (B) is indicated in each photomicrograph.
4. Discussion
In this study, we describe a novel Ghsr-IRES-Cre knock-in mouse line that has allowed the molecular manipulation of cells characterized by their expression of GHSR. Here, we focused on GHSR-expressing neurons, first investigating their central distribution. In particular, neuroanatomical characterization of Cre-dependent reporter expression in Ghsr-IRES-Cre mice crossed to either ROSA26-YFP or ROSA26-ZsGreen mice revealed a pattern of Cre activity that was largely consistent with the GHSR expression pattern observed previously using a combination of ISHH [19], [20], [23] plus a GHSR-eGFP transgenic reporter mouse line [23] and confirmed using other methods. Although those previous methods have been meritorious, the patterns of GHSR expression they reveal underrepresent the full extent of GHSR expression in the brain. For instance, GHSR mRNA expression in the dorsomedial and central aspects of the VMH as assessed by ISHH is noticeably absent in the mouse using ISHH and is sparse in the GHSR-eGFP reporter, whereas it had been expected to be marked based on ISHH findings in the rat [20], [23]. Similarly, while Arc expression of GHSR is marked by ISHH in the mouse, it is sparse in the GHSR-eGFP reporter [20], [23]. More broadly, ISHH seems to under represent GHSR expression in the cortex, hippocampus, VMH, and amygdala, whereas the GHSR-eGFP reporter underrepresents GHSR expression within the MBH and the midbrain [20], [23]. As described here, the pattern of Cre activity in the Ghsr-IRES-Cre line encompasses the regions missed or underrepresented by either ISHH or the GHSR-eGFP model alone. The Ghsr-IRES-Cre line also demonstrates Cre activity within the BNST and LSD, neither of which had been reported with ISHH or the GHSR-eGFP model [20], [23]. The BNST and LSD are important neural centers that receive and integrate inputs from the limbic system and are involved in the regulation of metabolism, mood, motivation, and behavioral stress responses [59], [60], [61], [62]. Many of these responses are also influenced by GHSR action [6], [63], [64], and future studies should assess whether GHSRs expressed within the BNST and/or LSD are involved in mediating those responses. Thus, the novel Ghsr-IRES-Cre line sensitively reports the central expression pattern of GHSR and as such should serve as an effective tool to investigate the chemical and electrophysiological properties and neurocircuitry of ghrelin-responsive, GHSR-expressing neurons, as we have begun here.
A possible caveat in using the Ghsr-IRES-Cre driver line to manipulate GHSR-expressing cells derives from the potential for temporal variance in GHSR expression, and thus Cre activity, during development vs. adult life and for differential stability of GHSR and Cre. Thus, a scenario could exist in which strong, transient GHSR expression in the fetal period within a particular region could lead to strong Cre expression-induced changes in that region that persist into adulthood. Such temporal variance could influence the fidelity of the Ghsr-IRES-Cre driver line, potentially contributing to some of the observed differences with the GHSR expression pattern as revealed by ISHH. That said, the overall similarity of the GHSR expression pattern in the Ghsr-IRES-Cre line to the expression pattern as determined, for instance using ISHH, suggests that any such influence is likely not widespread.
The novel Ghsr-IRES-Cre knock-in mouse line allowed us to deconstruct the involvement of MBH GHSR-expressing neurons in mediating food intake responses using chemogenetic modulation of their neuronal activity. In our studies, hM4Di DREADD virus injections to inhibit GHSR neurons targeted several nuclei within the MBH area including the Arc, VMH, DMH, and the PMV, all of which are involved in the regulation of food intake [20], [23], [32], [46], [65], [66], [67]. Within these regions, we predict that the inhibition of GHSR-expressing Arc AgRP/Neuropeptide Y (NPY) neurons is likely key to the reduced food intake response following fasting. This prediction is based on the known function of Arc AgRP/NPY neurons in homeostatic and fasting-induced rebound feeding and the previously-described high responsiveness of Arc ARP/NPY neurons to ghrelin, as discussed below. Arc AgRP/NPY neurons are activated during fasted conditions [65], [68], [69] and are essential for feeding, as ablation of these neurons in adult animals leads to cessation of eating [70], [71]. Numerous other studies have indicated key roles for Arc AgRP/NPY neurons in food intake, including those in which optogenetic and chemogenetic methodologies were used to stimulate them [56], [72]. In contrast, chemogenetic inhibition of Arc AgRP neurons reduces food intake [56] and diphtheria toxin-induced ablation of AgRP neurons from neonates significantly blunts fasting-induced rebound feeding of standard chow [73].
GHSR expression and c-fos induction – a marker of neuronal activation – by ghrelin or ghrelin mimetics are very high in these orexigenic Arc AgRP/NPY neurons [46], [47], [74], [75], [76]. As would be expected based on this high level of GHSR expression within and ghrelin responsiveness of Arc AgRP/NPY neurons, several studies also have implicated Arc AgRP/NPY neurons as direct mediators of ghrelin's orexigenic actions [3], [32], [42], [45], [46], [55], [73], [77], [78], [79], [80], [81]. For example, chemical ablation of the Arc and selective ablation of Arc AgRP neurons both abolish ghrelin-induced feeding [73], [79]. Also, selective expression of GHSRs in Arc AgRP neurons reestablishes the acute orexigenic response to peripherally-administered ghrelin in animals that are otherwise completing lacking GHSR [42], while selective deletion of GHSR expression from Arc AgRP neurons inhibits the orexigenic response to ghrelin [45]. Importantly, many parallels in the food intake responses exist between our current studies targeting MBH GHSR neurons and the above-described previous studies targeting Arc AgRP/NPY neurons. For example, our studies suggest that chemogenetic activation of the MBH GHSR neurons is sufficient to increase food intake, as observed with optogenetic and chemogenetic stimulation of Arc AgRP neurons [56], [72]. Also, chemogenetic inhibition of the MBH GHSR neurons reduced food intake, as shown for Arc AgRP neurons [56]. Moreover, chemogenetic inhibition of MBH GHSR neurons blunted fasting-induced rebound feeding, as described for mice with AgRP neuronal ablation [73]. Hypothalamic and extrahypothalamic axonal projection sites of the Cre expressing MBH GHSR neurons revealed here, which include the PVN, PAG, BNST, mPOA, and LH, mirror the axonal projection sites observed for Arc AgRP neurons [82], [83], [84]. Collectively, the above similarities point to the likely involvement of Arc AgRP/NPY feeding circuits in mediating the food intake responses that we observe with modification of the MBH GHSR neuronal activity. Further studies are needed to dissect out the roles of the other targeted GHSR-expressing MBH neuronal populations.
As introduced above, an area of disagreement in the literature is the significance of the endogenous ghrelin system as it relates to food intake [13], [35], [49], [50], [85], [86], [87]. There is no doubt that ghrelin can dose-dependently increase food intake acutely when administered peripherally or centrally; however, some studies argue that this occurs only when doses resulting in supraphysiological plasma ghrelin levels are used [51]. However, other studies suggest that food intake does occur when administered ghrelin leads to similar plasma ghrelin levels as achieved physiologically upon caloric restriction or psychosocial stress [88]. Furthermore, several models with genetic deletion of ghrelin, GOAT, or GHSR exhibit no or only minimal reductions in body weight and/or food intake [27], [40], [48], [49], [50], [51]. These stand in contrast to other studies in which GHSR deletion lowered both body weight and food intake [35], [89]. Even in those studies in which there was a decrease in body weight, the effect on food intake was minimal [52]. It could be argued that the lack of a food intake phenotype in some loss-of-function models could be a result of developmental or physiological adaptations to the lack of GHSR and/or ghrelin. That said, even in a mouse model in which ghrelin cells were ablated from adult mice − presumably after feeding circuits had developed normally − acute effects on food intake and body weight were not observed [51].
Here, we used chemogenetic modulation of MBH GHSR neurons to address the role of the endogenous ghrelin system in the normal, hyperphagic response to a short-term fast. Studies demonstrating the requirement of an intact ghrelin-GHSR system for the rebound food intake following fasting have been inconsistent. For instance, fasting-induced rebound food intake was unaltered in mice lacking ghrelin [49] or GHSR [14], [36], but became apparent in GHSR-knockout mice following repetitive fasting [36]. While one approach to immunoneutralize ghrelin did not affect rebound food intake following a fast [90], another reduced food intake [91], as did pharmacological antagonism of GHSRs [14]. While our results here do not prove that an intact, endogenous ghrelin system is required for the usual rebound food intake response to a 24 h fast, they do demonstrate that the activity of GHSR-expressing MBH neurons is required and suggest that any existing parallel feeding circuits that might play a role in the rebound hyperphagic response to fasting are unable to fully compensate for the deficient activity of the MBH GHSR neurons. It also remains to be seen if activity of these neurons is required for normal food intake when food availability is plentiful, a question that could be addressed by chronic DREADD-assisted inhibition of these neurons.
In summary, we present a novel Ghsr-IRES-Cre knock-in mouse line. The GHSR expression pattern revealed here by crossing the Ghsr-IRES-Cre to reporter mice appears to be more complete than that apparent from other individual methods. In addition, the reported physiological data achieved by chemogenetic manipulations of the Ghsr-IRES-Cre line suggest that the activity of MBH GHSR neurons is required for the full acute orexigenic response to administered ghrelin, is required for the usual rebound food intake response following a 24 h fast, and is sufficient to induce spontaneous food intake. The Ghsr-IRES-Cre line should serve as a valuable tool in future studies aimed at investigating the functional significance of other populations of ghrelin-responsive/GHSR-expressing neurons and the neuronal circuitries within which they act.
Acknowledgments
This work was supported through an International Research Alliance with the Novo Nordisk Foundation Center for Basic Metabolic Research at the University of Copenhagen, the Diana and Richard C. Strauss Professorship in Biomedical Research, the Mr. and Mrs. Bruce G. Brookshire Professorship in Medicine, the Kent and Jodi Foster Distinguished Chair in Endocrinology, in Honor of Daniel Foster, M.D., and institutional funds from the University of Texas Southwestern Medical Center to J.M.Z., the Hilda and Preston Davis Foundation Postdoctoral Fellowship Program in Eating Disorders Research to B.K.M., and a National Health and Medical Research Council Fellowship and grants to Z.B.A (APP1084344, APP1030037, APP1125690). We thank Prasanna Vijayaraghavan, Nathan Metzger, and Connor Lawrence for technical assistance with animal breeding.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.06.011.
Contributor Information
Zane B. Andrews, Email: zane.andrews@monash.edu.
Jeffrey M. Zigman, Email: jeffrey.zigman@utsouthwestern.edu.
Conflicts of interest
No conflict of interest exists.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
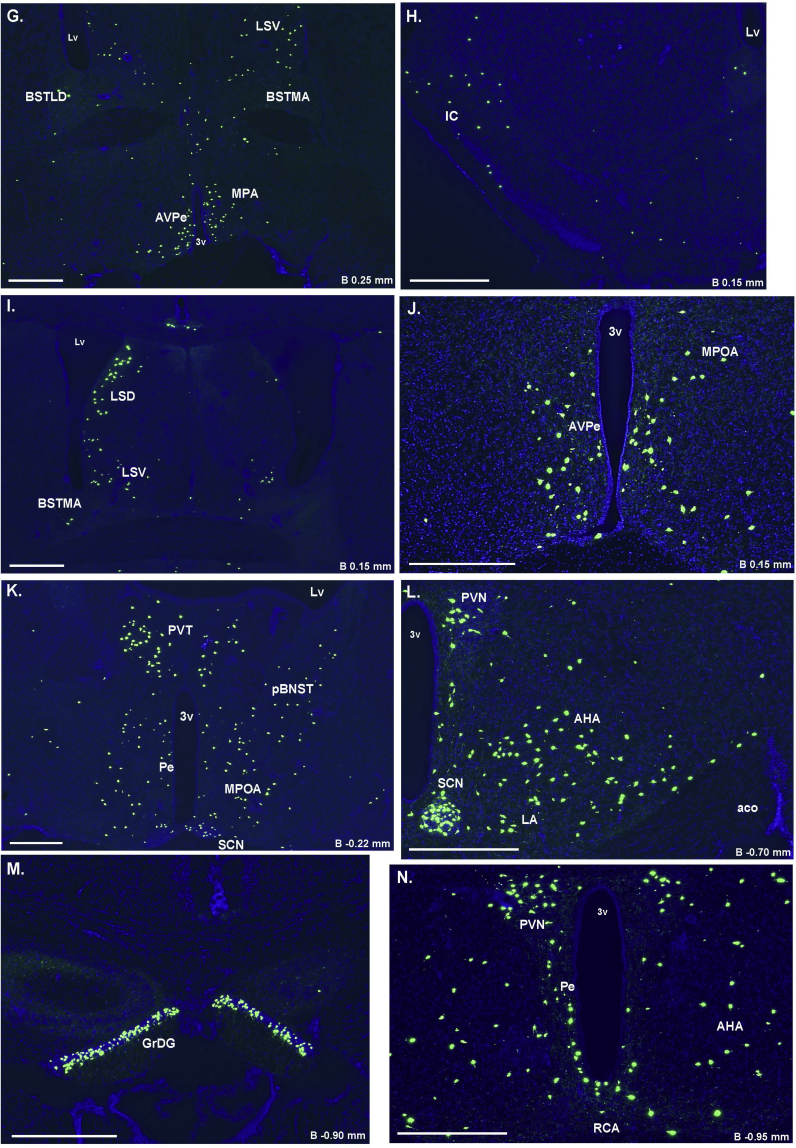
Supplemental Figure 1: Assessment of Cre-activity in Ghsr-IRES-Cre X ROSA26-ZsGreen mice. A–AC. Series of representative photomicrographs summarizing endogenous ZsGreen fluorescent cell bodies in the coronal brain sections of Ghsr-IRES-Cre X ROSA26-ZsGreen reporter mice. Scale bars in each panel = 500 μm. Approximate distance of the coronal section from bregma (B) is indicated in each photomicrograph. 3V – third ventricle, 4V – fourth ventricle, Aq – Aqueduct of Sylvius, AOB – accessory olfactory bulb, AON – anterior olfactory nucleus, granular (gr), mitral (ml) and glomerular (gl) layers of the main olfactory bulb, BSTMA-anteromedial division of the BNST, pBNST-posterior division of the BNST, DTT2-layer 2 of dorsal tenia tecta, GrDG-granular layer of dentate gyrus, LSV – lateral septal nucleus (ventral), LV – lateral ventricle, PCLO – posterolateral cortical amygdaloid nucleus, Py – Pyramidal cells of CA1, PVT-paraventricular thalamic nucleus, scp-superior cerebellar peduncle, xscp-decussation of superior cerebellar peduncle.
Supplemental Figure 2: Example of asymmetrical Cre-activity in brain sections from Ghsr-IRES-Cre mice. Coronal brain section from a Ghsr-IRES-Cre X ROSA26-YFP mouse (A) and a Ghsr-IRES-Cre X ROSA26-ZsGreen mouse (B) show relatively modest Cre-activity in one side of the dentate gyrus compared to the other side. The sections are approximately −1.46 mm from bregma.
Supplemental Figure 3: mCherry expression in the MBH of hM4Di virus-injected Ghsr-IRES-Cre mice: Coronal sections of the brains from all hM4Di virus-injected Ghsr-IRES-Cre mice demonstrating Cre-dependent expression (or lack thereof) of mCherry in the MBH. The brain sections in panels A–J were classified as “hits” and those in K–P were classified as “misses”. The approximate distances from bregma are indicated. Scale bar in last panel = 200 μm (applies to all the panels).
Supplemental Figure 4: Inhibition of MBH GHSR neurons attenuates ghrelin-induced food intake: Effect of CNO administration on ghrelin-induced food intake at 4 h (A) and 24 h (B) following food reintroduction in the “hits” and “misses”. *P < 0.05, statistically significant reduction in ghrelin-induced food intake with CNO administration; n.s., no significant difference. Data was analyzed by paired Students “t” test and represented as mean ± SE.
Supplemental Figure 5: Inhibition of GHSR neurons within the MBH attenuates fasting-induced rebound food intake. Effect of CNO administration on fasting-induced food intake at 4 h (A) and 24 h (B) following food reintroduction in the “hits” and “misses”. *P < 0.05, statistically significant reduction in fasting-induced food intake with CNO administration; n.s., no significant difference. Data was analyzed by paired Students “t” test and represented as mean ± SE.
Supplemental Figure 6: mCherry expression in the MBH of hM3Dq virus-injected Ghsr-IRES-Cre mice: Coronal sections of the brains from all hM3Dq virus-injected Ghsr-IRES-Cre mice demonstrating Cre-dependent expression of mCherry (or lack thereof) in the MBH. The brain sections in panels A–C were classified as “hits” and those in D–H were classified as “misses”. The approximate distances from bregma are indicated. Scale bar in each panel = 500 μm.
References
- 1.Kojima M., Hosoda H., Date Y., Nakazato M., Matsuo H., Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402(6762):656–660. doi: 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 2.Asakawa A., Inui A., Kaga T., Yuzuriha H., Nagata T., Ueno N. Ghrelin is an appetite-stimulatory signal from stomach with structural resemblance to motilin. Gastroenterology. 2001;120(2):337–345. doi: 10.1053/gast.2001.22158. [DOI] [PubMed] [Google Scholar]
- 3.Wren A.M., Small C.J., Abbott C.R., Dhillo W.S., Seal L.J., Cohen M.A. Ghrelin causes hyperphagia and obesity in rats. Diabetes. 2001;50(11):2540–2547. doi: 10.2337/diabetes.50.11.2540. [DOI] [PubMed] [Google Scholar]
- 4.Tschop M., Smiley D.L., Heiman M.L. Ghrelin induces adiposity in rodents. Nature. 2000;407(6806):908–913. doi: 10.1038/35038090. [DOI] [PubMed] [Google Scholar]
- 5.Kamegai J., Tamura H., Shimizu T., Ishii S., Sugihara H., Wakabayashi I. Central effect of ghrelin, an endogenous growth hormone secretagogue, on hypothalamic peptide gene expression. Endocrinology. 2000;141(12):4797–4800. doi: 10.1210/endo.141.12.7920. [DOI] [PubMed] [Google Scholar]
- 6.Muller T.D., Nogueiras R., Andermann M.L., Andrews Z.B., Anker S.D., Argente J. Ghrelin. Molecular Metabolism. 2015;4(6):437–460. doi: 10.1016/j.molmet.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castaneda T.R., Tong J., Datta R., Culler M., Tschop M.H. Ghrelin in the regulation of body weight and metabolism. Frontiers in Neuroendocrinology. 2010;31(1):44–60. doi: 10.1016/j.yfrne.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Chuang J.C., Perello M., Sakata I., Osborne-Lawrence S., Savitt J.M., Lutter M. Ghrelin mediates stress-induced food-reward behavior in mice. The Journal of Clinical Investigation. 2011;121(7):2684–2692. doi: 10.1172/JCI57660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dickson S.L., Egecioglu E., Landgren S., Skibicka K.P., Engel J.A., Jerlhag E. The role of the central ghrelin system in reward from food and chemical drugs. Molecular and Cellular Endocrinology. 2011;340(1):80–87. doi: 10.1016/j.mce.2011.02.017. [DOI] [PubMed] [Google Scholar]
- 10.Delhanty P.J., van der Lely A.J. Ghrelin and glucose homeostasis. Peptides. 2011;32(11):2309–2318. doi: 10.1016/j.peptides.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Masuda Y., Tanaka T., Inomata N., Ohnuma N., Tanaka S., Itoh Z. Ghrelin stimulates gastric acid secretion and motility in rats. Biochemical and Biophysical Research Communications. 2000;276(3):905–908. doi: 10.1006/bbrc.2000.3568. [DOI] [PubMed] [Google Scholar]
- 12.Ueno H., Yamaguchi H., Kangawa K., Nakazato M. Ghrelin: a gastric peptide that regulates food intake and energy homeostasis. Regulatory Peptides. 2005;126(1–2):11–19. doi: 10.1016/j.regpep.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 13.Zhao T.J., Liang G., Li R.L., Xie X., Sleeman M.W., Murphy A.J. Ghrelin O-acyltransferase (GOAT) is essential for growth hormone-mediated survival of calorie-restricted mice. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(16):7467–7472. doi: 10.1073/pnas.1002271107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perello M., Sakata I., Birnbaum S., Chuang J.C., Osborne-Lawrence S., Rovinsky S.A. Ghrelin increases the rewarding value of high-fat diet in an orexin-dependent manner. Biological Psychiatry. 2010;67(9):880–886. doi: 10.1016/j.biopsych.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howard A.D., Feighner S.D., Cully D.F., Arena J.P., Liberator P.A., Rosenblum C.I. A receptor in pituitary and hypothalamus that functions in growth hormone release. Science. 1996;273(5277):974–977. doi: 10.1126/science.273.5277.974. [DOI] [PubMed] [Google Scholar]
- 16.McKee K.K., Palyha O.C., Feighner S.D., Hreniuk D.L., Tan C.P., Phillips M.S. Molecular analysis of rat pituitary and hypothalamic growth hormone secretagogue receptors. Molecular Endocrinology. 1997;11(4):415–423. doi: 10.1210/mend.11.4.9908. [DOI] [PubMed] [Google Scholar]
- 17.Chow K.B.S., Sun J., Man Chu K., Tai Cheung W., Cheng C.H.K., Wise H. The truncated ghrelin receptor polypeptide (GHS-R1b) is localized in the endoplasmic reticulum where it forms heterodimers with ghrelin receptors (GHS-R1a) to attenuate their cell surface expression. Molecular and Cellular Endocrinology. 2012;348(1):247–254. doi: 10.1016/j.mce.2011.08.034. [DOI] [PubMed] [Google Scholar]
- 18.Bennett P.A., Thomas G.B., Howard A.D., Feighner S.D., van der Ploeg L.H., Smith R.G. Hypothalamic growth hormone secretagogue-receptor (GHS-R) expression is regulated by growth hormone in the rat. Endocrinology. 1997;138(11):4552–4557. doi: 10.1210/endo.138.11.5476. [DOI] [PubMed] [Google Scholar]
- 19.Guan X.M., Yu H., Palyha O.C., McKee K.K., Feighner S.D., Sirinathsinghji D.J. Distribution of mRNA encoding the growth hormone secretagogue receptor in brain and peripheral tissues. Brain Research Molecular Brain Research. 1997;48(1):23–29. doi: 10.1016/s0169-328x(97)00071-5. [DOI] [PubMed] [Google Scholar]
- 20.Zigman J.M., Jones J.E., Lee C.E., Saper C.B., Elmquist J.K. Expression of ghrelin receptor mRNA in the rat and the mouse brain. The Journal of Comparative Neurology. 2006;494(3):528–548. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bron R., Yin L., Russo D., Furness J.B. Expression of the ghrelin receptor gene in neurons of the medulla oblongata of the rat. The Journal of Comparative Neurology. 2013;521(12):2680–2702. doi: 10.1002/cne.23309. [DOI] [PubMed] [Google Scholar]
- 22.Tannenbaum G.S., Lapointe M., Beaudet A., Howard A.D. Expression of growth hormone secretagogue-receptors by growth hormone-releasing hormone neurons in the mediobasal hypothalamus. Endocrinology. 1998;139(10):4420–4423. doi: 10.1210/endo.139.10.6330. [DOI] [PubMed] [Google Scholar]
- 23.Mani B.K., Walker A.K., LopezSoto E.J., Raingo J., Lee C.E., Perello M. Neuroanatomical characterization of a growth hormone secretagogue receptor-green fluorescent protein reporter mouse. The Journal of Comparative Neurology. 2014;522(16):3644–3666. doi: 10.1002/cne.23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamegai J., Wakabayashi I., Kineman R.D., Frohman L.A. Growth hormone-releasing hormone receptor (GHRH-R) and growth hormone secretagogue receptor (GHS-R) mRNA levels during postnatal development in male and female rats. Journal of Neuroendocrinology. 1999;11(4):299–306. doi: 10.1046/j.1365-2826.1999.00330.x. [DOI] [PubMed] [Google Scholar]
- 25.Gnanapavan S., Kola B., Bustin S.A., Morris D.G., McGee P., Fairclough P. The tissue distribution of the mRNA of ghrelin and subtypes of its receptor, GHS-R, in humans. The Journal of Clinical Endocrinology and Metabolism. 2002;87(6):2988. doi: 10.1210/jcem.87.6.8739. [DOI] [PubMed] [Google Scholar]
- 26.Sun Y., Garcia J.M., Smith R.G. Ghrelin and growth hormone secretagogue receptor expression in mice during aging. Endocrinology. 2007;148(3):1323–1329. doi: 10.1210/en.2006-0782. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y., Wang P., Zheng H., Smith R.G. Ghrelin stimulation of growth hormone release and appetite is mediated through the growth hormone secretagogue receptor. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(13):4679–4684. doi: 10.1073/pnas.0305930101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cabral A., Suescun O., Zigman J.M., Perello M. Ghrelin indirectly activates hypophysiotropic CRF neurons in rodents. PloS One. 2012;7(2):e31462. doi: 10.1371/journal.pone.0031462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tong J., Mannea E., Aime P., Pfluger P.T., Yi C.X., Castaneda T.R. Ghrelin enhances olfactory sensitivity and exploratory sniffing in rodents and humans. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2011;31(15):5841–5846. doi: 10.1523/JNEUROSCI.5680-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabral A., Fernandez G., Perello M. Analysis of brain nuclei accessible to ghrelin present in the cerebrospinal fluid. Neuroscience. 2013;253:406–415. doi: 10.1016/j.neuroscience.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaeffer M., Langlet F., Lafont C., Molino F., Hodson D.J., Roux T. Rapid sensing of circulating ghrelin by hypothalamic appetite-modifying neurons. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(4):1512–1517. doi: 10.1073/pnas.1212137110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowley M.A., Smith R.G., Diano S., Tschop M., Pronchuk N., Grove K.L. The distribution and mechanism of action of ghrelin in the CNS demonstrates a novel hypothalamic circuit regulating energy homeostasis. Neuron. 2003;37(4):649–661. doi: 10.1016/s0896-6273(03)00063-1. [DOI] [PubMed] [Google Scholar]
- 33.Diano S., Farr S.A., Benoit S.C., McNay E.C., da Silva I., Horvath B. Ghrelin controls hippocampal spine synapse density and memory performance. Nature Neuroscience. 2006;9(3):381–388. doi: 10.1038/nn1656. [DOI] [PubMed] [Google Scholar]
- 34.Jiang H., Betancourt L., Smith R.G. Ghrelin amplifies dopamine signaling by cross talk involving formation of growth hormone secretagogue receptor/dopamine receptor subtype 1 heterodimers. Molecular Endocrinology. 2006;20(8):1772–1785. doi: 10.1210/me.2005-0084. [DOI] [PubMed] [Google Scholar]
- 35.Zigman J.M., Nakano Y., Coppari R., Balthasar N., Marcus J.N., Lee C.E. Mice lacking ghrelin receptors resist the development of diet-induced obesity. The Journal of Clinical Investigation. 2005;115(12):3564–3572. doi: 10.1172/JCI26002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abizaid A., Liu Z.W., Andrews Z.B., Shanabrough M., Borok E., Elsworth J.D. Ghrelin modulates the activity and synaptic input organization of midbrain dopamine neurons while promoting appetite. The Journal of Clinical Investigation. 2006;116(12):3229–3239. doi: 10.1172/JCI29867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blum I.D., Patterson Z., Khazall R., Lamont E.W., Sleeman M.W., Horvath T.L. Reduced anticipatory locomotor responses to scheduled meals in ghrelin receptor deficient mice. Neuroscience. 2009;164(2):351–359. doi: 10.1016/j.neuroscience.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma X., Lin L., Qin G., Lu X., Fiorotto M., Dixit V.D. Ablations of ghrelin and ghrelin receptor exhibit differential metabolic phenotypes and thermogenic capacity during aging. PLoS One. 2011;6(1):e16391. doi: 10.1371/journal.pone.0016391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin L., Saha P.K., Ma X., Henshaw I.O., Shao L., Chang B.H. Ablation of ghrelin receptor reduces adiposity and improves insulin sensitivity during aging by regulating fat metabolism in white and brown adipose tissues. Aging Cell. 2011;10(6):996–1010. doi: 10.1111/j.1474-9726.2011.00740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun Y., Butte N.F., Garcia J.M., Smith R.G. Characterization of adult ghrelin and ghrelin receptor knockout mice under positive and negative energy balance. Endocrinology. 2008;149(2):843–850. doi: 10.1210/en.2007-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee J.H., Lin L., Xu P., Saito K., Wei Q., Meadows A.G. Neuronal deletion of ghrelin receptor almost completely prevents diet-induced obesity. Diabetes. 2016;65(8):2169–2178. doi: 10.2337/db15-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q., Liu C., Uchida A., Chuang J.C., Walker A., Liu T. Arcuate AgRP neurons mediate orexigenic and glucoregulatory actions of ghrelin. Molecular Metabolism. 2014;3(1):64–72. doi: 10.1016/j.molmet.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott M.M., Perello M., Chuang J.C., Sakata I., Gautron L., Lee C.E. Hindbrain ghrelin receptor signaling is sufficient to maintain fasting glucose. PloS One. 2012;7(8):e44089. doi: 10.1371/journal.pone.0044089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuto Y., Shibasaki T., Otagiri A., Kuriyama H., Ohata H., Tamura H. Hypothalamic growth hormone secretagogue receptor regulates growth hormone secretion, feeding, and adiposity. The Journal of Clinical Investigation. 2002;109(11):1429–1436. doi: 10.1172/JCI13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu C.S., Bongmba O.Y.N., Yue J., Lee J.H., Lin L., Saito K. Suppression of GHS-R in AgRP neurons mitigates diet-induced obesity by activating thermogenesis. International Journal of Molecular Sciences. 2017;18(4):E832. doi: 10.3390/ijms18040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakazato M., Murakami N., Date Y., Kojima M., Matsuo H., Kangawa K. A role for ghrelin in the central regulation of feeding. Nature. 2001;409(6817):194–198. doi: 10.1038/35051587. [DOI] [PubMed] [Google Scholar]
- 47.Willesen M.G., Kristensen P., Romer J. Co-localization of growth hormone secretagogue receptor and NPY mRNA in the arcuate nucleus of the rat. Neuroendocrinology. 1999;70(5):306–316. doi: 10.1159/000054491. [DOI] [PubMed] [Google Scholar]
- 48.Kirchner H., Gutierrez J.A., Solenberg P.J., Pfluger P.T., Czyzyk T.A., Willency J.A. GOAT links dietary lipids with the endocrine control of energy balance. Nature Medicine. 2009;15(7):741–745. doi: 10.1038/nm.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wortley K.E., Anderson K.D., Garcia K., Murray J.D., Malinova L., Liu R. Genetic deletion of ghrelin does not decrease food intake but influences metabolic fuel preference. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(21):8227–8232. doi: 10.1073/pnas.0402763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun Y., Ahmed S., Smith R.G. Deletion of ghrelin impairs neither growth nor appetite. Molecular and Cellular Biology. 2003;23(22):7973–7981. doi: 10.1128/MCB.23.22.7973-7981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McFarlane M.R., Brown M.S., Goldstein J.L., Zhao T.J. Induced ablation of ghrelin cells in adult mice does not decrease food intake, body weight, or response to high-fat diet. Cell Metabolism. 2014;20(1):54–60. doi: 10.1016/j.cmet.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wortley K.E., del Rincon J.P., Murray J.D., Garcia K., Iida K., Thorner M.O. Absence of ghrelin protects against early-onset obesity. The Journal of Clinical Investigation. 2005;115(12):3573–3578. doi: 10.1172/JCI26003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muyrers J.P., Zhang Y., Stewart A.F. Techniques: recombinogenic engineering–new options for cloning and manipulating DNA. Trends in Biochemical Sciences. 2001;26(5):325–331. doi: 10.1016/s0968-0004(00)01757-6. [DOI] [PubMed] [Google Scholar]
- 54.Lee E.C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D.A., Court D.L. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73(1):56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 55.Tong Q., Ye C.P., Jones J.E., Elmquist J.K., Lowell B.B. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nature Neuroscience. 2008;11(9):998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krashes M.J., Koda S., Ye C., Rogan S.C., Adams A.C., Cusher D.S. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. The Journal of Clinical Investigation. 2011;121(4):1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paxinos G., Franklin K.B.J. Elsevier Academin Press; San Diego, CA: 2004. The mouse brain in stereotaxic coordinates. [Google Scholar]
- 58.Atasoy D., Aponte Y., Su H.H., Sternson S.M. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2008;28(28):7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sheehan T.P., Chambers R.A., Russell D.S. Regulation of affect by the lateral septum: implications for neuropsychiatry. Brain Research Reviews. 2004;46(1):71–117. doi: 10.1016/j.brainresrev.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 60.Lebow M.A., Chen A. Overshadowed by the amygdala: the bed nucleus of the stria terminalis emerges as key to psychiatric disorders. Molecular Psychiatry. 2016;21(4):450–463. doi: 10.1038/mp.2016.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Steculorum S.M., Ruud J., Karakasilioti I., Backes H., Engstrom Ruud L., Timper K. AgRP neurons control systemic insulin sensitivity via myostatin expression in brown adipose tissue. Cell. 2016;165(1):125–138. doi: 10.1016/j.cell.2016.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jennings J.H., Rizzi G., Stamatakis A.M., Ung R.L., Stuber G.D. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science. 2013;341(6153):1517–1521. doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chuang J.C., Zigman J.M. Ghrelin's roles in stress, mood, and anxiety regulation. International Journal of Peptides. 2010 doi: 10.1155/2010/460549. 2010:pii:460549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spencer S.J., Emmerzaal T.L., Kozicz T., Andrews Z.B. Ghrelin's role in the hypothalamic-pituitary-adrenal axis stress response: implications for mood disorders. Biological Psychiatry. 2015;78(1):19–27. doi: 10.1016/j.biopsych.2014.10.021. [DOI] [PubMed] [Google Scholar]
- 65.Sternson S.M., Atasoy D. Agouti-related protein neuron circuits that regulate appetite. Neuroendocrinology. 2014;100(2–3):95–102. doi: 10.1159/000369072. [DOI] [PubMed] [Google Scholar]
- 66.Toshinai K., Date Y., Murakami N., Shimada M., Mondal M.S., Shimbara T. Ghrelin-induced food intake is mediated via the orexin pathway. Endocrinology. 2003;144(4):1506–1512. doi: 10.1210/en.2002-220788. [DOI] [PubMed] [Google Scholar]
- 67.Kwon Jeong J., Dae Kim J., Diano S. Ghrelin regulates hypothalamic prolyl carboxypeptidase expression in mice. Molecular Metabolism. 2013;2(1):23–30. doi: 10.1016/j.molmet.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takahashi K.A., Cone R.D. Fasting induces a large, leptin-dependent increase in the intrinsic action potential frequency of orexigenic arcuate nucleus neuropeptide Y/Agouti-related protein neurons. Endocrinology. 2005;146(3):1043–1047. doi: 10.1210/en.2004-1397. [DOI] [PubMed] [Google Scholar]
- 69.Liu T., Kong D., Shah B.P., Ye C., Koda S., Saunders A. Fasting activation of AgRP neurons requires NMDA receptors and involves spinogenesis and increased excitatory tone. Neuron. 2012;73(3):511–522. doi: 10.1016/j.neuron.2011.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luquet S., Perez F.A., Hnasko T.S., Palmiter R.D. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310(5748):683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 71.Gropp E., Shanabrough M., Borok E., Xu A.W., Janoschek R., Buch T. Agouti-related peptide-expressing neurons are mandatory for feeding. Nature Neuroscience. 2005;8(10):1289–1291. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- 72.Aponte Y., Atasoy D., Sternson S.M. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nature Neuroscience. 2011;14(3):351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luquet S., Phillips C.T., Palmiter R.D. NPY/AgRP neurons are not essential for feeding responses to glucoprivation. Peptides. 2007;28(2):214–225. doi: 10.1016/j.peptides.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 74.Dickson S.L., Luckman S.M. Induction of c-fos messenger ribonucleic acid in neuropeptide Y and growth hormone (GH)-releasing factor neurons in the rat arcuate nucleus following systemic injection of the GH secretagogue, GH-releasing peptide-6. Endocrinology. 1997;138(2):771–777. doi: 10.1210/endo.138.2.4907. [DOI] [PubMed] [Google Scholar]
- 75.Wang L., Saint-Pierre D.H., Tache Y. Peripheral ghrelin selectively increases Fos expression in neuropeptide Y – synthesizing neurons in mouse hypothalamic arcuate nucleus. Neuroscience Letters. 2002;325(1):47–51. doi: 10.1016/s0304-3940(02)00241-0. [DOI] [PubMed] [Google Scholar]
- 76.Hewson A.K., Dickson S.L. Systemic administration of ghrelin induces Fos and Egr-1 proteins in the hypothalamic arcuate nucleus of fasted and fed rats. Journal of Neuroendocrinology. 2000;12(11):1047–1049. doi: 10.1046/j.1365-2826.2000.00584.x. [DOI] [PubMed] [Google Scholar]
- 77.Shintani M., Ogawa Y., Ebihara K., Aizawa-Abe M., Miyanaga F., Takaya K. Ghrelin, an endogenous growth hormone secretagogue, is a novel orexigenic peptide that antagonizes leptin action through the activation of hypothalamic neuropeptide Y/Y1 receptor pathway. Diabetes. 2001;50(2):227–232. doi: 10.2337/diabetes.50.2.227. [DOI] [PubMed] [Google Scholar]
- 78.Kamegai J., Tamura H., Shimizu T., Ishii S., Sugihara H., Wakabayashi I. Chronic central infusion of ghrelin increases hypothalamic neuropeptide Y and Agouti-related protein mRNA levels and body weight in rats. Diabetes. 2001;50(11):2438–2443. doi: 10.2337/diabetes.50.11.2438. [DOI] [PubMed] [Google Scholar]
- 79.Tamura H., Kamegai J., Shimizu T., Ishii S., Sugihara H., Oikawa S. Ghrelin stimulates GH but not food intake in arcuate nucleus ablated rats. Endocrinology. 2002;143(9):3268–3275. doi: 10.1210/en.2002-220268. [DOI] [PubMed] [Google Scholar]
- 80.Chen H.Y., Trumbauer M.E., Chen A.S., Weingarth D.T., Adams J.R., Frazier E.G. Orexigenic action of peripheral ghrelin is mediated by neuropeptide Y and agouti-related protein. Endocrinology. 2004;145(6):2607–2612. doi: 10.1210/en.2003-1596. [DOI] [PubMed] [Google Scholar]
- 81.Chen Y., Lin Y.C., Kuo T.W., Knight Z.A. Sensory detection of food rapidly modulates arcuate feeding circuits. Cell. 2015;160(5):829–841. doi: 10.1016/j.cell.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Betley J.N., Cao Z.F., Ritola K.D., Sternson S.M. Parallel, redundant circuit organization for homeostatic control of feeding behavior. Cell. 2013;155(6):1337–1350. doi: 10.1016/j.cell.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang D., He X., Zhao Z., Feng Q., Lin R., Sun Y. Whole-brain mapping of the direct inputs and axonal projections of POMC and AgRP neurons. Frontiers in Neuroanatomy. 2015;9:40. doi: 10.3389/fnana.2015.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dietrich M.O., Zimmer M.R., Bober J., Horvath T.L. Hypothalamic Agrp neurons drive stereotypic behaviors beyond feeding. Cell. 2015;160(6):1222–1232. doi: 10.1016/j.cell.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Smet B., Depoortere I., Moechars D., Swennen Q., Moreaux B., Cryns K. Energy homeostasis and gastric emptying in ghrelin knockout mice. The Journal of Pharmacology and Experimental Therapeutics. 2006;316(1):431–439. doi: 10.1124/jpet.105.091504. [DOI] [PubMed] [Google Scholar]
- 86.Dezaki K., Sone H., Koizumi M., Nakata M., Kakei M., Nagai H. Blockade of pancreatic islet-derived ghrelin enhances insulin secretion to prevent high-fat diet-induced glucose intolerance. Diabetes. 2006;55(12):3486–3493. doi: 10.2337/db06-0878. [DOI] [PubMed] [Google Scholar]
- 87.Pfluger P.T., Kirchner H., Gunnel S., Schrott B., Perez-Tilve D., Fu S. Simultaneous deletion of ghrelin and its receptor increases motor activity and energy expenditure. American Journal of Physiology Gastrointestinal and Liver Physiology. 2008;294(3):G610–G618. doi: 10.1152/ajpgi.00321.2007. [DOI] [PubMed] [Google Scholar]
- 88.Chuang J.C., Sakata I., Kohno D., Perello M., Osborne-Lawrence S., Repa J.J. Ghrelin directly stimulates glucagon secretion from pancreatic alpha-cells. Molecular Endocrinology. 2011;25(9):1600–1611. doi: 10.1210/me.2011-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Longo K.A., Charoenthongtrakul S., Giuliana D.J., Govek E.K., McDonagh T., Qi Y. Improved insulin sensitivity and metabolic flexibility in ghrelin receptor knockout mice. Regulatory Peptides. 2008;150(1–3):55–61. doi: 10.1016/j.regpep.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 90.Zakhari J.S., Zorrilla E.P., Zhou B., Mayorov A.V., Janda K.D. Oligoclonal antibody targeting ghrelin increases energy expenditure and reduces food intake in fasted mice. Molecular Pharmaceutics. 2012;9(2):281–289. doi: 10.1021/mp200376c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu S.C., Xu J., Chinookoswong N., Liu S., Steavenson S., Gegg C. An acyl-ghrelin-specific neutralizing antibody inhibits the acute ghrelin-mediated orexigenic effects in mice. Molecular Pharmacology. 2009;75(4):901–907. doi: 10.1124/mol.108.052852. [DOI] [PubMed] [Google Scholar]