Abstract
The zebrafish has been a powerful model in forward genetic screens to identify genes essential for organogenesis and embryonic development. Conversely, using reverse genetics to investigate specific gene function requires phenotypic analysis of complete gene inactivation. Despite the availability and efficacy of morpholinos, the lack of tractable and efficient knockout technologies has impeded reverse genetic studies in the zebrafish, particularly in adult animals. The recent development of genome-editing technologies such as CRISPR/Cas9 greatly widened the scope of loss-of-function studies in the zebrafish, allowing for the rapid phenotypic assessment of gene silencing in embryos, the generation of knockout lines, and large-scale reverse genetic screens. Tissue-specific gene inactivation would be ideal for these studies given the caveats of whole-embryo gene silencing, yet spatial control of gene targeting remains a challenge. In this chapter, we focus on tissue-specific gene inactivation using the CRISPR/Cas9 technology. We first explain the rationale for this technique, including some of its potential applications to tackle important biological issues and the inability of current technologies to address these issues. We then present a method to target genes in a tissue-specific manner in the zebrafish. Finally, we discuss technical difficulties and limitations of this method as well as possible future developments.
INTRODUCTION
Zebrafish can generate large numbers of embryos that develop quickly and externally, and are therefore easy to analyze phenotypically. The zebrafish has thus been a vertebrate model of choice to perform forward genetic screens that have led to the identification of numerous genes regulating organogenesis (Driever et al., 1996; Haffter et al., 1996; Mullins, Hammerschmidt, Haffter, & Nusslein-Volhard, 1994). Reverse genetic screens as a complementary approach have been hindered by the lack of high-throughput gene-silencing technology in the zebrafish. Although morpholino antisense oligomers (morpholinos) have been used extensively to analyze loss-of-function phenotypes during early stages of embryogenesis, they present important limitations and have rarely been used for large scale screens.
The CRISPR/Cas9 technology of genome editing has opened new vistas for loss-of-function studies in zebrafish. CRISPR/Cas9 is an adaptive immune response developed by bacteria and archae to fight phage invasion through RNA-based recognition and subsequent cleavage of foreign DNA sequences (Barrangou et al., 2007; Brouns et al., 2008; Garneau et al., 2010). Deciphering this molecular mechanism rapidly led to the realization that it could be applied to genome editing (Jinek et al., 2012). Engineered short guide RNAs (gRNA) are comprised of a 20-nucleotide 5′ component (referred to as the seed) complementary to the target DNA sequence, and of a 3′ motif that forms a secondary structure capable of interacting with CRISPR-associated 9 (Cas9) endonuclease. Cas9 loads the gRNA and scans the genome in search of target sequences complementary to the seed (Sternberg, Redding, Jinek, Greene, & Doudna, 2014). When recognized sequences are followed by a proto-spacer adjacent motif (PAM), Cas9 cleaves DNA within the seed. The resulting double-strand breaks are resolved by the cell, most likely through alternative nonhomologous end joining, which leads to mutations, insertions, or deletions (indels). When occurring in coding sequences, these indels frequently disrupt the targeted gene by introducing frameshifts and early stop codons. Note however that statistically, a third of indels leave the open reading frame unaltered and may therefore not affect gene function. While the CRISPR/Cas9 technology was initially used to generate knockout cell or animal lines (Cho, Kim, Kim, & Kim, 2013; Cong et al., 2013; Hwang et al., 2013; Jiang, Bikard, Cox, Zhang, & Marraffini, 2013; Mali et al., 2013), its applications now range from the fine modulation of gene expression (Gilbert et al., 2013) to genome-wide genetic screens in vitro (Koike-Yusa, Li, Tan, Velasco-Herrera Mdel, & Yusa, 2014; Wang, Wei, Sabatini, & Lander, 2014; Zhou et al., 2014) and in vivo (Chen et al., 2015). The CRISPR technology has also joined the fight against viruses with strategies to target essential viral genes or cell surface receptors involved in viral entry, and against genetic disorders to correct disease-causing mutations.
In the zebrafish, the CRISPR/Cas9 technology not only allows us to generate mutant lines (Hwang et al., 2013) but also to observe phenotypes rapidly in vivo by direct injection of a gRNA and Cas9 mRNA into one-cell stage embryos (Jao, Wente, & Chen, 2013). Several reports have also established knockin zebrafish lines with this technology (Auer, Duroure, De Cian, Concordet, & Del Bene, 2014; Hisano et al., 2015; Kimura, Hisano, Kawahara, & Higashijima, 2014; Li et al., 2015). Its ease of use, low cost, and the possibility of multiplexing make it a prime tool for large-scale reverse genetic screens in the zebrafish (Shah, Davey, Whitebirch, Miller, & Moens, 2015). In many cases, the investigation of gene function in vivo requires the spatiotemporal control of gene silencing. Here, we present the rationale for tissue-specific gene inactivation in zebrafish using the CRISPR/Cas9 technology and detail a method to achieve it. We also discuss some limitations and potential future developments of this technique.
1. RATIONALE
1.1 NEED FOR TISSUE-SPECIFIC TUNING OF GENE EXPRESSION
Embryonic lethality represents a major hurdle when investigating gene function in vivo. Indeed, many genes play essential roles in the proper development of an organ, and animals with mutations in these genes are not viable. In zebrafish it has been estimated that about 1400 genes are embryonic lethal (Amsterdam et al., 2004). In these cases, gene knockout in the whole organism may not be informative, requiring the development of alternative experimental approaches.
Tissue-specific gene targeting can readily address cell autonomy questions, whereas other methods are generally complex and/or indirect. For example, to prove that a gene or a pathway affects a function (eg, proliferation, self-renewal, and survival) directly in a specific cell type (eg, hematopoietic stem cell) and not via an effect on neighboring cells (eg, stromal cells), it is necessary to target the gene or pathway of interest specifically in that cell type. Transplantation between different transgenic animals or parabiosis are methods to address cell autonomy, but they present a number of technical challenges and biological caveats. Specifically inactivating a gene in different cell types allows identification of the precise cell type responsible for a phenotype observed in a knockout line or induced by systemic drug treatment. Additionally, to look for modulators of a specific trait in vivo, it may be more informative to inactivate genes specifically in the corresponding tissue.
Precise disease modeling may also require the tissue-specific inactivation of a gene. While some conditions such as muscular dystrophies are generally caused by inherited mutations and can therefore be appropriately modeled by full gene knockouts, most cancers arise from somatic mutations only present in the neoplastic tissue. The inactivation of tumor-suppressor genes can alter cell homeostasis in many different cell types. For instance, p53 mutant zebrafish predominantly develops malignant peripheral nerve sheath tumors over time (Berghmans et al., 2005). It may therefore be preferable to target tumor-suppressor genes in a tissue-restricted manner to accurately model malignant transformation in a particular organ.
1.2 LIMITATIONS OF EXISTING GENE-SILENCING METHODS
In the absence of an established knockout line for a specific gene, or if the knockout is embryonic lethal at a late stage of development, it is possible to transiently knockdown a gene to assess loss-of-function in embryos. Equivalent to the mammalian shRNA technology, morpholinos are injected into zebrafish embryos to silence a gene of interest and study its function during early development (Nasevicius & Ekker, 2000). Morpholinos are nucleotide analogs that recognize and bind short sequences (about 25 nucleotides) at the transcription start site or at splice sites of pre-mRNAs, and thus block the translation or proper splicing of the mRNA (Summerton & Weller, 1997). They can be injected into zebrafish zygotes to inhibit gene expression. Due to its ease of use and remarkable efficacy, this technology has played an instrumental role in the development of the zebrafish as a model for cell biology studies. Morpholinos have also been useful in reverse genetic screens (Huang et al., 2013). Nonspecific toxicity and off-target effects limited the utility of morpholinos and forced the zebrafish community to adopt a series of guidelines to adequately control morpholino experiments (Eisen & Smith, 2008). In addition, morpholinos function in embryos for about 3 days, which limits loss-of-function analyses to the early stages of development and precludes studies in adults. Finally, the cost of morpholino synthesis practically prevents systematic, large-scale screens.
Zinc-finger nucleases (ZFN) and transcription activator-like effector nucleases (TALEN) are enzymes engineered to recognize and cleave specific DNA sequences (Miller et al., 2011; Urnov et al., 2005). Once expressed in zebrafish embryos, they induce double-strand breaks at their target sites in the genome, which are resolved by nonhomologous end joining (Bedell et al., 2012; Doyon et al., 2008; Meng, Noyes, Zhu, Lawson, & Wolfe, 2008; Sander et al., 2011). Resulting indels often disrupt gene function. The mutations are heritable, which allows for the generation of stable knockout lines. These genome-editing technologies enable loss-of-function studies both in embryos and adults, thus complementing the transient knockdown approaches through morpholinos. Note that the phenotypes observed by acute gene down-regulation with morpholinos or permanent gene knockout with genome-editing technologies can be discordant (Kok et al., 2015), partially due to compensation mechanisms (Rossi et al., 2015), raising questions about the respective significance of either approach. Targeting a new gene with ZFN or TALEN requires the design of a new nuclease, and the mutation efficiency is generally low, which renders the intensive use of these technologies challenging.
In contrast, the CRISPR/Cas9 technology uses a single endonuclease, Cas9, to target specific genomic sequences in an RNA-dependent manner (Jinek et al., 2012). The co-injection of Cas9 mRNA or protein and a short guide RNA (gRNA) recognizing a 20-bp genomic sequence into one-cell stage zebrafish embryos yields frequent indels at the target locus (Hwang et al., 2013). The relative ease of use and efficiency of this technology have made it a prime tool to assess loss-of-function phenotypes in injected embryos and to establish mutant lines quickly. It does not allow, however, for the spatiotemporal control of gene inactivation.
Of note, alternatives to direct gene editing have been developed to perform tissue-specific loss-of-function studies. The conditional Cre/lox technology has been adapted to the zebrafish for lineage tracing experiments (Mosimann et al., 2011) and could serve the same purpose as in mice (ie, spatiotemporal control of gene knockout) but the generation of floxed alleles is complex and time consuming. Interestingly, tissue-specific overexpression of dominant-negative mutants has also been used to circumvent the lack of spatial control of gene inactivation. However, gene silencing remains the method of choice to study gene function because it most faithfully recapitulates physiologic expression levels.
2. METHODS
Here we detail a method based on the CRISPR/Cas9 technology to target genes in a tissue-specific fashion in the zebrafish (Ablain, Durand, Yang, Zhou, & Zon, 2015). Developments and alternatives to this method are discussed in the next section.
2.1 IDENTIFICATION OF CRISPR TARGET SITES
The first step consists of identifying gRNAs that efficiently mutate the gene of interest. This can be done following published protocols.
Prediction algorithms such as CHOPCHOP (https://chopchop.rc.fas.harvard.edu/) are based on rules inferred from in vivo measurement of gRNA efficiency and can be used to design gRNAs (Montague, Cruz, Gagnon, Church, & Valen, 2014; Moreno-Mateos et al., 2015). The presence of a G as the last base of the seed sequence and a high GC content seem to be the two most important rules. The number of predicted off-target sites is also taken into account in the ranking of potential target sequences. The choice of the first two bases will determine the type of in vitro transcription enzyme needed in the next step. We recommend picking target sequences that start with a G since the U6 promoter used in the tissue-specific vector (see Section 2.2) requires a G as a transcription start site. If the proposed target sequence does not start with GG or GA, it is possible to replace the second base by a G or an A since the presence of mismatches at the 5′ end of the gRNA is generally well tolerated by Cas9 for the recognition of the cleavage site (Fu et al., 2013). Depending on the structure of the gene of interest, it may or may not be suitable to target early exons preferentially. In general, if a gene contains known functional domains (eg, DNA-binding or catalytic or ligand-binding domains), we recommend targeting these in priority. In our experience, about a third of gRNAs designed following these guidelines show a good targeting efficiency, although we and others have observed significant gene-dependent variability. Designing at least three gRNAs per gene may therefore be warranted to find an efficient seed sequence.
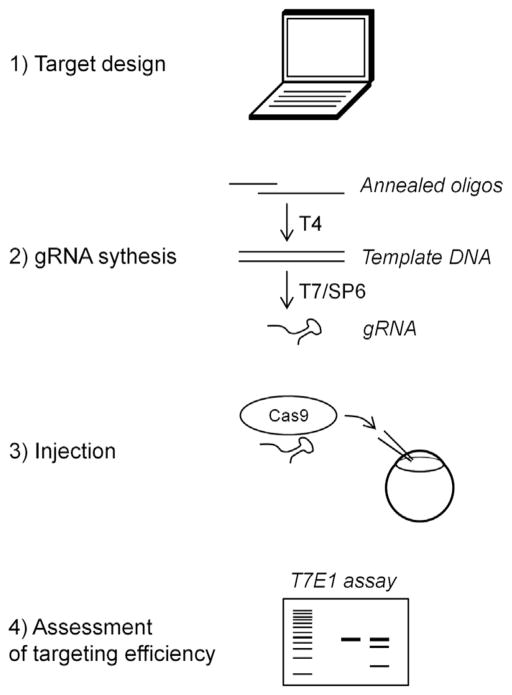
Once potential CRISPR target sequences have been chosen, a DNA template for in vitro transcription can be prepared either by cloning the seed sequence into a vector that contains both a T7 or SP6 promoter and the constant 3′ part of the gRNA (Hwang et al., 2013), or by T4 DNA polymerase (or PCR) synthesis following annealing of two long oligonucleotides (Gagnon et al., 2014) (Fig. 1). The purified DNA template can then be transcribed in vitro into a gRNA using either T7 (for target sequences starting with GG) or SP6 (for sequences starting with GA) RNA polymerases.
FIGURE 1.
CRISPR sequences against any gene of interest can be identified in silico through published algorithms. gRNAs can then be prepared by in vitro transcription (using T7 or SP6 RNA polymerases) from DNA templates comprising an RNA polymerase promoter, the gene-specific seed sequence, and the sequence encoding the constant 3′ part of the gRNA. Purified gRNAs can be injected into single-cell zebrafish embryos along with Cas9 mRNA or protein. Finally, genomic DNA can be extracted from injected embryos and analyzed for CRISPR-induced mutations, for example, by direct sequencing, HRM, or T7E1 mutagenesis assay.
After the gRNAs have been purified (on column or by ammonium acetate precipitation), their quality can be controlled by gel electrophoresis or using a fragment analyzer. The presence of shorter transcripts is expected due to impurity of the oligonucleotides used to synthesize the DNA template and to incomplete RNA elongation, but the major band on the gel has to correspond to the full-size transcript. gRNAs can then be co-injected into one-cell stage zebrafish embryos with Cas9 mRNA or protein. The reported gRNA concentration in the injection mix greatly varies in the literature (from 12.5 ng/μL to almost 1 μg/μL), but it seems that even very high doses show limited toxicity to the embryos while the impact of concentration on targeting efficiency is still debated. Cas9 mRNA can be easily synthesized in vitro from plasmid DNA (Jao et al., 2013) and injected at 300 ng/μL, while Cas9 protein can be produced (Gagnon et al., 2014) or purchased and used at 500 ng/μL.
Genomic DNA can be extracted from embryos 24–48 h post-injection, and several methods can be used to assess mutagenesis efficiency including: enzymatic methods (restriction, T7E1, or Surveyor assays (Kim, Lee, Kim, Cho, & Kim, 2009; Qiu et al., 2004)), PCR-based methods (High Resolution Melt (HRM) (Parant, George, Pryor, Wittwer, & Yost, 2009), direct PCR in the case of expected deletions between two CRISPR target sites), or sequencing of the CRISPR target site. Only the latter is truly quantitative and gives access to the full range of mutations. Enzymatic methods are arguably cheaper and easier to perform, but also more susceptible to technical variability. In our experience, about half of gRNAs display poor targeting efficiency (less than 10% of mutated alleles, which is generally under the detection threshold of enzymatic methods), and a third of gRNAs target 20% or more of alleles. Only the latter category should be further considered for loss-of-function studies.
2.2 CONSTRUCTION OF VECTORS
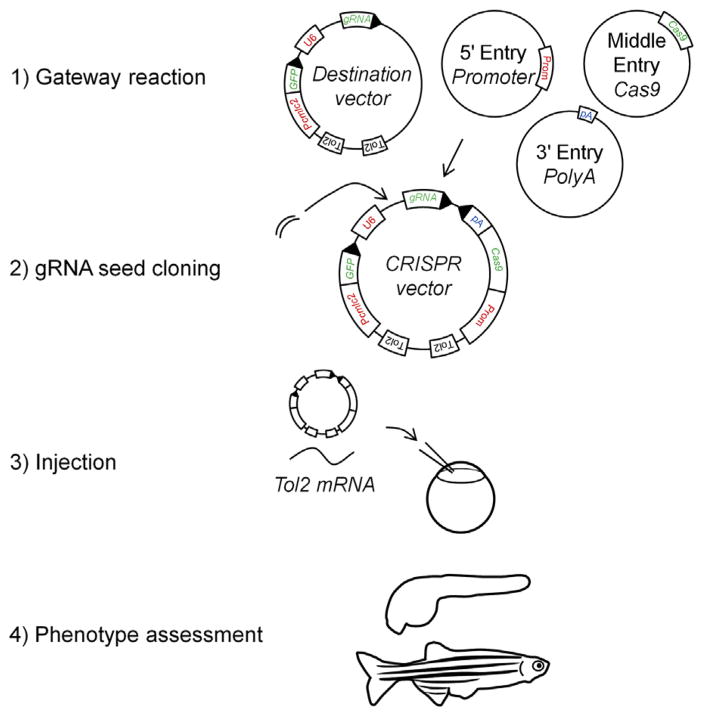
Here we describe vectors based on the Tol2 transposase technology (Kawakami et al., 2004) that can be injected into zebrafish embryos along with Tol2 mRNA for integration into the genome. Other vectors (eg, viral vectors) can be constructed on the same model. Gateway technology (Hartley, Temple, & Brasch, 2000) or regular cloning methods can easily insert a Cas9 sequence codon-optimized for zebrafish (Jao et al., 2013) under the control of a tissue-specific promoter (Fig. 2). It is possible to adapt a vector already containing a transgenesis marker (eg, cmlc2:GFP that yields green fluorescent hearts) or to add GFP linked to Cas9 via a 2A peptide in order to visualize the cells that express the vector in vivo. Once efficient CRISPR seed sequences have been identified, they can be cloned under the control of a zebrafish U6 promoter at the 5′ end of a gRNA scaffold (Ablain et al., 2015). A protocol for gRNA and Cas9 cloning into Tol2 vectors is detailed in (Ablain et al., 2015).
FIGURE 2.
A Gateway reaction allows to assemble a Cas9 sequence under the control of a tissue-specific promoter and followed by polyA into a backbone containing a U6:gRNA cassette and possibly a transgenesis marker. One can then clone any gene-specific seed sequence into the U6:gRNA cassette of the resulting vector. This tissue-specific CRISPR vector can be injected with Tol2 mRNA into one cell–stage embryos that will later be submitted to phenotype evaluation. (See color plate)
Note: In some cases, it may be possible to skip the identification of efficient gRNAs (Section 2.1) and either identify them after injection of the vectors (this requires mutation detection in the targeted tissue) or pool several vectors directed against the same gene to statistically increase targeting.
2.3 INJECTION, EVALUATION OF MOSAICISM, AND PHENOTYPE ASSESSMENT
The generated CRISPR vectors are then integrated into the zebrafish genome by direct co-injection with Tol2 mRNA (20 ng/μL) into one-cell stage embryos. The DNA concentration in the injection mix varies according to the size of the plasmid and is typically between 10 and 25 ng/μL. Depending on the transgenesis marker used, mosaicism can be approximated by the ratio of fluorescent cells to the total number of cells in the tissue of interest. For example, in the case of the vector expressing GFP under the control of the heart-specific promoter cmlc2, the proportion of the heart that displays green fluorescence at 48 hpf provides an estimate of mosaicism. When using a Cas9-T2A-GFP construct, injection into a transgenic line expressing a different fluorescent protein in the tissue of interest, or comparison of injected embryos with embryos of such a transgenic line, may allow a better assessment of mosaicism.
We showed that the zebrafish U6-3 promoter drives early, continuous, and ubiquitous expression of gRNAs (Ablain et al., 2015). Thus, the regulation and specificity of gene targeting stem from Cas9 expression. Theoretically, the consequences of gene inactivation can be observed shortly after the tissue-specific promoter has turned on. Indeed, Cas9 transcription and translation; subsequent gene targeting; and the decrease in mRNA and protein levels of the targeted gene presumably all occur within a few hours. Some phenotypes may be noticed at the level of the whole embryo (eg, survival and defects in organ development). Phenotypes at the cellular (eg, proliferation and migration) or molecular levels (eg, gene expression and pathway activation) may be more difficult to detect because of the mosaicism of vector expression in F0-injected embryos. To overcome this issue, it may be possible to image or analyze the cells that express the Cas9-T2A-GFP construct in situ, or sort them by fluorescence-activated cell sorting (FACS) and proceed with molecular assays.
3. DISCUSSION
3.1 TECHNICAL CAVEATS AND TROUBLESHOOTING
The method for tissue-specific gene targeting described earlier presents several caveats that may result in poor gene targeting.
One of the main potential limitations resides in the tissue-specific promoter used to drive Cas9 expression. Weak promoters may not produce enough Cas9 in order to recognize and cleave the CRISPR target. Measures of Cas9 mRNA levels by whole mount in situ hybridization or qPCR in the tissue of interest may provide insights into promoter activity.
It remains unclear whether the intrinsic quality of gRNAs (ie, their efficiency at inducing mutations at the CRISPR site after co-injection with Cas9 mRNA or protein into one-cell embryos) impacts targeting efficiency by the vector. Yet, it is conceivable that the continuous production of Cas9 mRNA and gRNA by the vector will force targeting over time, normalizing the efficiency of different gRNAs. Along the same line, the vector may favor homozygous over heterozygous targeting in each cell, thus circumventing one of the issues associated with transient Cas9 injection. Single-cell sequencing of the CRISPR site would be required to evaluate whether one or both alleles have been hit.
If the CRISPR vector fails to induce mutations at the target locus, injection of the vector with either the gRNA itself or Cas9 mRNA, and subsequent assessment of targeting may help determine whether the problem stems from the gRNA or from the promoter driving Cas9 expression. Injection of a vector producing the same gRNA and expressing Cas9 under a ubiquitous promoter such as ubi (Mosimann et al., 2011) may also prove informative in that case.
A consequence of continual vector targeting is the higher likelihood of off-target effects. While the issue of off-target effects by the CRISPR technology is still debated (Fu et al., 2013; Hsu et al., 2013; Kuscu, Arslan, Singh, Thorpe, & Adli, 2014; Wang et al., 2015), a clear assessment of the number and frequency of off-targets may be warranted in the case of the tissue-specific CRISPR vectors.
3.2 PHENOTYPES IN F0 ANIMALS VERSUS STABLE LINES
The mosaicism associated with Tol2-mediated insertion of the vectors may also be an issue. Indeed, some phenotypes may require a high frequency of gene inactivation to be detected. For example, deleterious phenotypes that lead to the death or proliferation arrest of the targeted cells may be masked by the growth of cells in the same tissue that do not express the vector. This problem is exacerbated by the semirandom nature of DNA double-strand break resolution by alternative nonhomologous end-joining mechanisms, which induce insertions and deletions at the CRISPR target site that can either result in frameshift or retain the reading frame of the gene. In some cells, CRISPR targeting may thus lead to “silent” mutations that leave a functional copy of the gene (eg, point mutations or short inframe indels). These cells, although expressing the vector, may escape the deleterious consequences of gene inactivation and obscure the phenotype. One way to minimize this risk is to design gRNAs against domains essential for the activity of the corresponding protein, so that even in-frame mutations may disrupt gene function. Additionally, it is possible to generate large deletions of entire exons by combining several gRNAs directed against the same gene.
To circumvent the impact of mosaicism, it may be beneficial to grow injected F0 embryos up to adulthood and cross them to establish a line of zebrafish stably expressing the vector. This would allow virtually every cell in the tissue of interest to target the gene. In addition, this would eliminate the ectopic expression occasionally observed with Tol2 vectors in F0. The main drawbacks of this method are the time required to generate stable lines, which makes it unsuitable for a screen (particularly if the phenotype is to be observed in adults), and the possible silencing of Tol2 vectors in the F1 generation, which complicates the selection of founders. In addition, due to the relatively large size of the CRISPR vectors, germline transmission rates may be low. Taken together, these considerations point to the importance of adapting the method according to the expected phenotype.
3.3 DEVELOPMENTS AND ALTERNATIVES
The CRISPR vector system described earlier provides a modular platform for a range of developments and improvements. It can easily be adapted for use in species other than the zebrafish by modifying the codon-optimized Cas9 and U6 promoter sequences. Note that systems for tissue-specific gene inactivation have also been developed in fly and worm (Shen et al., 2014; Xue et al., 2014). We have also added a second U6:gRNA cassette in the vector backbone to allow for multiplexing. This is particularly useful to perform reverse genetic studies and analyze the loss-of-function phenotypes of human genes in the zebrafish since many human genes have more than one zebrafish orthologue (due to the whole-genome duplication that occurred in the ray-finned fish lineage after its separation from the tetrapods (Taylor, Braasch, Frickey, Meyer, & Van de Peer, 2003)).
The power of the Gal4-UAS system (Brand & Perrimon, 1993) may also be harnessed for tissue-specific gene targeting. Indeed, a vector containing a U6:gRNA cassette and Cas9 under the control of a UAS element can be injected into a stable zebrafish line expressing Gal4 in a specific tissue. Alternatively, a stable line can be generated with the Cas9/gRNA vector and crossed with the Gal4 line. This method may circumvent problems linked to the strength of the tissue-specific promoters used to drive Cas9 expression.
Finally, the applications of the vector system would be tremendously extended if it also enabled the temporal control of gene silencing. Several options could be explored including coupling the CRISPR/Cas9 technique with the ERT2 inducible system to control the activity of Cre, Gal4, or Cas9 itself.
Owing to the high-throughput nature of the CRISPR/Cas9 technology, the tissue-specific CRISPR system is a tractable tool for precise in vivo reverse genetic screens. It may make it possible to uncover new genes that regulate the biology of a particular tissue, without affecting the rest of the organism. It may also allow researchers to revisit or clarify the role of known genes by inactivating them in different cell types, measuring their respective contributions to the global phenotype.
Acknowledgments
We thank Jonathan E. Henninger for critical reading of the manuscript. This work was supported by NIH grants R01 CA103846, R01 HL04880, R01 DK53298, PO1 HL32262, P30 DK49216, U01 HL10001, and R24 DK092760. In addition, L. I. Z. is a Howard Hughes Medical Institute Investigator. L.I.Z. is a founder and stock holder of Fate, Inc., Scholar Rock, and a scientific advisor for Stemgent.
References
- Ablain J, Durand EM, Yang S, Zhou Y, Zon LI. A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Developmental Cell. 2015;32(6):756–764. doi: 10.1016/j.devcel.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(35):12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Research. 2014;24(1):142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, … Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, 2nd, … Ekker SC. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491(7422):114–118. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, Fletcher CD, … Look AT. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(2):407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, … van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, … Sharp PA. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell. 2015;160(6):1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature Biotechnology. 2013;31(3):230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, … Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, … Amache SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature Biotechnology. 2008;26(6):702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, … Boggs C. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don’t stop making antisense. Development. 2008;135(10):1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nature Biotechnology. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Ahkmetova L, Pauli A, … Schier AF. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9(5):e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, … Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, … Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154(2):442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, … Nüsslein-Volhard C. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Research. 2000;10(11):1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisano Y, Sakuma T, Nakade S, Ohga R, Ota S, Okamoto H, … Kawahara A. Precise in-frame integration of exogenous DNA mediated by CRISPR/Cas9 system in zebrafish. Scientific Reports. 2015;5:8841. doi: 10.1038/srep08841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, … Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang HT, Kathrein KL, Barton A, Gitlin Z, Huang YH, Ward TP, … Zon LI. A network of epigenetic regulators guides developmental haematopoiesis in vivo. Nature Cell Biology. 2013;15(12):1516–1525. doi: 10.1038/ncb2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, … Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nature Biotechnology. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(34):13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature Biotechnology. 2013;31(3):233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Developmental Cell. 2004;7(1):133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Research. 2009;19(7):1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Scientific Reports. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera Mdel C, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nature Biotechnology. 2014;32(3):267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, … Lawson ND. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Developmental Cell. 2015;32(1):97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nature Biotechnology. 2014;32(7):677–683. doi: 10.1038/nbt.2916. [DOI] [PubMed] [Google Scholar]
- Li J, Zhang BB, Ren YG, Gu SY, Xiang YH, Du JL. Intron targeting-mediated and endogenous gene integrity-maintaining knockin in zebrafish using the CRISPR/Cas9 system. Cell Research. 2015;25(5):634–637. doi: 10.1038/cr.2015.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, … Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature Biotechnology. 2008;26(6):695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, … Rebar EJ. ATALE nuclease architecture for efficient genome editing. Nature Biotechnology. 2011;29(2):143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Research. 2014;42(Web Server issue):W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nature Methods. 2015;12(10):982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development. 2011;138(1):169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: in search of genes controlling development in a vertebrate. Current Biology. 1994;4(3):189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nature Genetics. 2000;26(2):216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Parant JM, George SA, Pryor R, Wittwer CT, Yost HJ. A rapid and efficient method of genotyping zebrafish mutants. Developmental Dynamics. 2009;238(12):3168–3174. doi: 10.1002/dvdy.22143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Shandilya H, D’Alessio JM, O’Connor K, Durocher J, Gerard GF. Mutation detection using Surveyor nuclease. Biotechniques. 2004;36(4):702–707. doi: 10.2144/04364PF01. [DOI] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DYR. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015;524(7564):230–233. doi: 10.1038/nature14580. [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JR. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnology. 2011;29(8):697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB. Rapid reverse genetic screening using CRISPR in zebrafish. Nature Methods. 2015;12(6):535–540. doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Zhang X, Chai Y, Zhu Z, Yi P, Feng G, … Ou G. Conditional knockouts generated by engineered CRISPR-Cas9 endonuclease reveal the roles of coronin in C. elegans neural development. Developmental Cell. 2014;30(5):625–636. doi: 10.1016/j.devcel.2014.07.017. [DOI] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507(7490):62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summerton J, Weller D. Morpholino antisense oligomers: design, preparation, and properties. Antisense and Nucleic Acid Drug Development. 1997;7(3):187–195. doi: 10.1089/oli.1.1997.7.187. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Research. 2003;13(3):382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, … Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, Wang J, Qiu Z, Chang T, … Yee JK. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nature Biotechnology. 2015;33(2):175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- Xue Z, Wu M, Wen K, Ren M, Long L, Zhang X, Gao G. CRISPR/Cas9 mediates efficient conditional mutagenesis in Drosophila. G3 (Bethesda) 2014;4(11):2167–2173. doi: 10.1534/g3.114.014159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509(7501):487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]