Abstract
Bone is a common site for metastasis in breast cancer patients and is associated with a series of complications that significantly compromise patient survival, partially due to the advanced stage of disease at the time of detection. Currently, no clinically-approved biomarkers can identify or predict the development of bone metastasis. We recently identified a unique peptide fragment of parathyroid hormone-related protein (PTHrP), PTHrP(12-48), as a validated serum biomarker in breast cancer patients that correlates with and predicts the presence of bone metastases. In this study, the biological activity and mode of action of PTHrP(12-48) was investigated. Sequence-based and structure-based bioinformatics techniques predicted that the PTHrP(12-48) fragment formed an alpha helical core followed by an unstructured region after residue 40 or 42. Thereafter, detailed structure alignment and molecular docking simulations predicted a lack of interaction between PTHrP(12-48) and the cognate PTH1 receptor (PTHR1). The in silico prediction was confirmed by the lack of PTHrP(12-48)-stimulated cAMP accumulation in PTHR1-expressing human SaOS2 cells. Using a specific human PTHrP(12-48) antibody that we developed, PTHrP(12-48) was immunolocalized in primary and bone metastatic human breast cancer cells, as well as within human osteoclasts (OCLs) in bone metastasis biopsies, with little or no localization in other resident bone or bone marrow cells. In vitro, PTHrP(12-48) was internalized into cultured primary human OCLs and their precursors within 60 min. Interestingly, PTHrP(12-48) treatment dose-dependently suppressed osteoclastogenesis, via the induction of apoptosis in both OCL precursors as well as in mature OCLs, as measured by the activation of cleaved caspase 3. Collectively, these data suggest that PTHrP(12-48) is a bioactive breast cancer–derived peptide that locally regulates the differentiation of hematopoietic cells and the activity of osteoclasts within the tumor–bone marrow microenvironment, perhaps to facilitate tumor control of bone.
Keywords: OSTEOCLASTS, TUMOR-INDUCED BONE DISEASE, PTH/VIT D/FGF23, CELL/TISSUE SIGNALING, ENDOCRINE PATHWAYS
Introduction
Currently, there are approximately 2 million women in the United States with breast cancer,(1) which is the second leading cause of cancer death in women.(2) Breast cancer has a propensity to metastasize to bone, and bone metastases are frequently associated with complications such as hypercalcemia due to osteolysis, nerve compression, intractable bone pain, and pathological fractures,(3–6) yet no clinically-approved biomarkers that predict the development of bone metastases exist.(5,7)
Since its discovery as the causal agent of the humoral hypercalcemia of malignancy(8) parathyroid hormone-related protein (PTHrP) has been implicated in a plethora of biologic processes,(9,10) which largely require interaction with the cognate PTH receptor (PTHR1).(11,12) Indeed, PTHrP and peptides containing the N terminus are known to preside over endochondral ossification,(13,14) tooth eruption,(15) and calcium release during lactation,(16) as well as in normal breast development(17) and placental calcium transport.(18) These functions provide the rationale for the idea that PTHrP, via activation of receptor activator of NF-κB ligand (RANKL), and other molecules in the bone microenvironment drive the profound osteolysis that is common in breast cancer patients with bone metastasis.(4) However, the identity of specific PTHrP peptides in the circulation of breast cancer patients and thus, the identity of the active PTHrP-derived agents, has been elusive. Therefore, the search for peptide fragments of PTHrP and their biological characterization is an expanding and ongoing endeavor. To this end, we recently reported the discovery and identification of the first circulating fragment of PTHrP, PTHrP(12-48), in the serum of breast cancer patients with bone metastasis.(19)
Studies over many decades have demonstrated that PTHrP is the product of a single copy gene (PTHLH), localized on the p-arm of chromosome 12.(20–22) Unlike the related human PTH gene, the human PTHLH gene is a complex transcriptional unit with nine exons and three distinct promoter regions that drive the transcription of three PTHrP proteins of 139, 141, and 173 amino acids.(20–22) PTHrP is produced by many cancers including breast, and in the case of metastatic breast cancer PTHrP acts to potently induce osteoclastic bone resorption by altering RANKL/osteoprotegerin (OPG) expression, resulting in bone destruction.(23) Importantly, PTHrP(12-48) is not a transcribed product, but the result of proteolytic processing of any of the three human PTHrP isoforms. Thus, characterizing PTHrP(12-48) as a bioactive peptide may provide new insight into breast cancer progression to bone, PTHrP metabolism, and new opportunities for early diagnosis and treatment.
Materials and Methods
Cell lines
Peripheral blood mononuclear cells (PBMCs) (Astarte Biologics, Bothell, WA, USA) stored frozen in liquid nitrogen were thawed quickly at 37°C and resuspended by adding drop-wise to 9 mL of culture media (alpha-MEM [Gibco, Grand Island, NY, USA] + 10% FBS [Hy-Clone Laboratories, Inc., Logan, UT, USA]) + 1× antibiotics (penicillin/streptomycin; Fisher, Pittsburg, PA, USA). The cell suspension was centrifuged at 400g for 10 min at 21°C and a single cell suspension made using culture media. Cells were counted and plated in culture media+ mCSF with or without RANKL (both from R&D Systems, Minneapolis, MN, USA) or other treatments, and incubated at 37°C with 5% CO2. Cells were half-fed on day 3 and day 7 and then treated according to experimental requirements.
SaOS2 cells, human osteosarcoma cells(24,25) were stored in liquid nitrogen, thawed at 37°C, plated in 10-cm culture dishes in RPMI 1640 media (Sigma, St. Louis, MO, USA) + 10% FBS + 1× antibiotics (penicillin/streptomycin; Fisher, Pittsburg, PA, USA) and incubated at 37°C with 5% CO2. The medium was replaced every 3 days. Once cells reached ~90% confluency experiments were initiated.
Modeling the structure of PTHrP(12-48)
Bioinformatic, computational, algorithmic, and performance indices tools were utilized to predict the tertiary structure of PTHrP(12-48). The tools utilized were Fast Interaction REfine-ment in molecular DOCKing (FireDock), a high-throughput web service for flexible refinement and scoring of protein-protein docking solutions generated by rigid-body docking algorithms such as ClusPro 2.0 (available at: http://bioinfo3d.cs.tau.ac.il/FireDock/).(26) ClusPro 2.0 is an automated molecular docking server that uses a combination of grid-based rigid body docking and root-mean-square deviation (RMSD)-based clustering to identify near-native low-energy conformations of protein complexes (available at: http://cluspro.bu.edu/home.php).(27) Iterative Threading ASSEmbly Refinement (I-TASSER) is an automated protein structure prediction server that uses a combination of multiple threading-based template alignments and iterative TASSER Monte Carlo simulations to generate native-like protein models (available at: http://zhanglab.ccmb.med.umich.edu/ITASSER/).(28) QUARK is an ab initio protein structure prediction method that determines the three-dimensional structure of a protein, based solely on its amino acid sequence and no predetermined template information, an important advantage when modeling small targets (<200 amino acids) lacking homologous templates in the protein database, such as PTHrP(12-48).(29)
Human PTHrP(12-48) antibody development and evaluation
A polyclonal antibody (IgY) directed against full-length PTHrP(12-48) was developed in Leghorn White chickens (GenWay Biotech, San Diego, CA, USA). Briefly, chickens were injected subcutaneously with 100 to 250mg of Keyhole Limpet Hemocyanin (KLH)-conjugated PTHrP(12-48) suspended in complete Freund’s adjuvant (CFA) 3 weeks before the egg-laying period, with three booster doses of PTHrP(12-48)-KLH, in incomplete Freund’s adjuvant, administered during the egg laying period at intervals of 2 to 3 weeks. An additional booster dose was administered 1 month after the last booster dose to further increase antibody titer. High IgY titers of 1:100,000 to 1:1,000,000 were achieved. After immunization serum and eggs were collected, yolks harvested, pulverized, and precipitated with polyethylene glycol, and the IgY fraction collected. The IgY fraction was depleted of PTHrP(1-37) by negative selection chromatography and the flow-through positively selected against a PTHrP(12-48) column. Subsequent antibody validation and specificity was determined using indirect ELISA assays, performed against the N-terminal peptide, PTHrP(1-37) and PTHrP(12-48).
Dot blots
Dot blots on nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA, USA) were performed using IL-8 (control), PTH (1-34) (control), PTHrP(1-141) (full-length; kind gift of Dr. T.J. Martin, St. Vincent’s Institute for Medical Research, Melbourne, Australia), PTHrP(1-37), and PTHrP(12-48) (New England Peptides Inc., Gardner, MA, USA) at 100 ng, 10 ng, and 1 ng. Blots were incubated with either a specific PTHrP(1-15) monoclonal antibody (kind gift or Dr. J. Chirgwin, Indiana University, Indianapolis, IN, USA) or the PTHrP(12-48) polyclonal antibody and developed by chemiluminescence using a Bio-Rad Chemidoc MP imaging system (Bio-Rad Laboratories).
Western blot analysis
IL-8, PTH1-34 (10 mM), PTHrP(1-37) (1 mM), and PTHrP(12-48) at 1 mM, 100 mM, and 10 mM were resolved on a 12% SDS-PAGE then transferred to a nitrocellulose membrane. The blots were placed in Blokhen (Aves Labs, Inc., Tigard, OR, USA) (diluted 1:10 in water) for 30 min, and washed three times in PBS+ 0.05% Tween-20 before incubating with PTHrP(12-48) antibody and thereafter washed again before incubating with secondary antibody. The primary and secondary antibodies along with antibody dilutions and incubation times are indicated in Supplemental Table 1. Blots were then developed by chemiluminescence using the Bio-Rad Chemidoc MP imaging system. In addition, the blot was stripped for 30 min at room temperature (RT), using Western blot stripping buffer (PBS+ 7ml of β-mercaptoethanol [BME]+ 2% SDS), washed for 30 min and then probed for IL-8 (8 kDa) using a CXCL8/IL-8 specific antibody (Supplemental Table 1) (data not shown).
Immunohistochemistry
Commercially available tissue arrays (Maxivision BioSciences Inc., Kenmore, WA, USA) of a variety of normal human tissues were placed in boiling Citrate Buffer pH 6.0 and allowed to cool for 30min at RT. Endogenous peroxidase was blocked using the peroxidase solution supplied in the anti-chicken horseradish peroxidase (HRP)-diaminobenzidine (DAB) Cell and Tissue Staining Kit (R&D Systems). After 5 min slides were washed with PBS (pH 7.4), blocked with blocking buffer, and incubated overnight with the PTHrP(12-48) polyclonal antibody (dilution 1:200). The next morning, sections were washed in PBS (pH7.4), incubated with secondary antibody for 30 min, washed again, and DAB was applied. After development of DAB, sections were washed with distilled water, counter-stained with Mayer’s Hematoxylin, dehydrated, cleared, and mounted with Permaslip. Archival formalin-fixed paraffin-embedded primary human breast cancer and bone metastasis biopsy specimens were immunostained for PTHrP(12-48). UAMS and Penn State Institutional IRBs approved the studies performed.
Immunofluorescence assays
Human PBMCs were seeded at 500,000 cells/well onto chamber well slides (Lab-Tek, Vernon Hills, IL, USA) using culture media + 1× antibiotics (penicillin/streptomycin; Fisher, Pittsburg, PA, USA) with mCSF (25 ng/mL) alone or with RANKL (50 ng/mL) and allowed to proliferate at 37°C with 5% CO2.
PTHrP(12-48) cell internalization assays
On day 7 (when differentiated osteoclasts are present), PTHrP-(12-48) (100 nM) in the presence of serum was added to cells for 15, 30, 60, and 90 min. Cells were fixed with 4% paraformaldehyde in PBS at 4°C for 15 min and permeabilized with 0.02% PBS-Triton X-100 for 15 min. Primary antibody block was performed using BlokHen (1:10 dilution in water; Aves Labs, Inc.,Tigard, OR, USA) followed by the addition of primary and secondary antibody using dilutions and incubation times described in Supplemental Table 1.
Cleaved caspase 3 assays
Cells were treated for 24 hours with TNFα (200 ng/mL), or PTHrP(12-48) (50 nM) on day 0 and day 7. Cells were fixed, permeabilized, and stained using a specific primary cleaved caspase 3 (Asp175) antibody(30) at 1:400 dilution in PBS. Cells were washed in PBS three times followed by incubation for 1 hour with secondary antibodies (Supplemental Table 1). For both assays, ProLong Gold Antifade reagent with 4,6-diamidino-2-phenylindole (DAPI; Cell Signaling, Danvers, MA, USA), used to stain nuclei (blue), was added. Coverslipped slides were allowed to dry in the dark at RT overnight and then sealed using clear nail polish and kept at 4°C. To visualize cells, either fluorescence microscopy was performed (Evos-FL microscope; Advanced Microscopy group [AMG], Bothell, WA, USA), fitted with light cubes for GFP, DAPI, Texas Red, and red fluorescent protein [RFP]), or confocal fluorescent microscopy was performed (Zeiss LSM 510 confocal microscope; Zeiss, Oberkochen, Germany). Images obtained using DAPI (405 nm), FITC/GFP (488 nm), and Cy5 (561 nm) light cubes were processed using Zen 2009 software (Zeiss, Oberkochen, Germany).
cAMP ELISA
Human SaOS2 cells were plated in 10-cm culture dishes in RPMI 1640 media.(31) Confluent cells were serum starved for 6 hours prior to treatment. Cells were first treated with 3-isobutyl-1-methylxanthine (IBMX) for 15min(32) and then either left untreated, treated with IBMX, or treated with increasing concentrations of PTHrP(12-48) (10 to 1000 nM). PTH(1-34) (50 nM) activates cAMP production via PTHR1,(33) and was used as a positive control. Cells were lysed using lysis buffer and a competitive cAMP ELISA (Enzo Life Sciences, Farmingdale, NY, USA) was performed, according to the manufacturer’s protocol.(34)
Tartrate-resistant acid phosphatase staining
Osteoclast cultures were stained for tartrate-resistant acid phosphatase (TRAP).(35) Briefly, cells were stained for 60 min at 37°C using chemical reagents purchased from Sigma (St. Louis, MO, USA) or Fisher (Pittsburg, PA, USA) and then rinsed using deionized water. Cells containing three or more nuclei (multinucleated [MNC]) and TRAP stained (red) (TRAP+MNC) per well were enumerated.
Statistics
Multigroup data were analyzed by analysis of variance (ANOVA) with Student-Newman-Keul’s post hoc test as appropriate. For differences between only two experimental groups, data were analyzed by Student’s t test. All data are presented as mean ± SE. Values of p <0.05 between groups was considered significant and is reported as such.
Results
Predicted tertiary structure and processing of PTHrP(12-48)
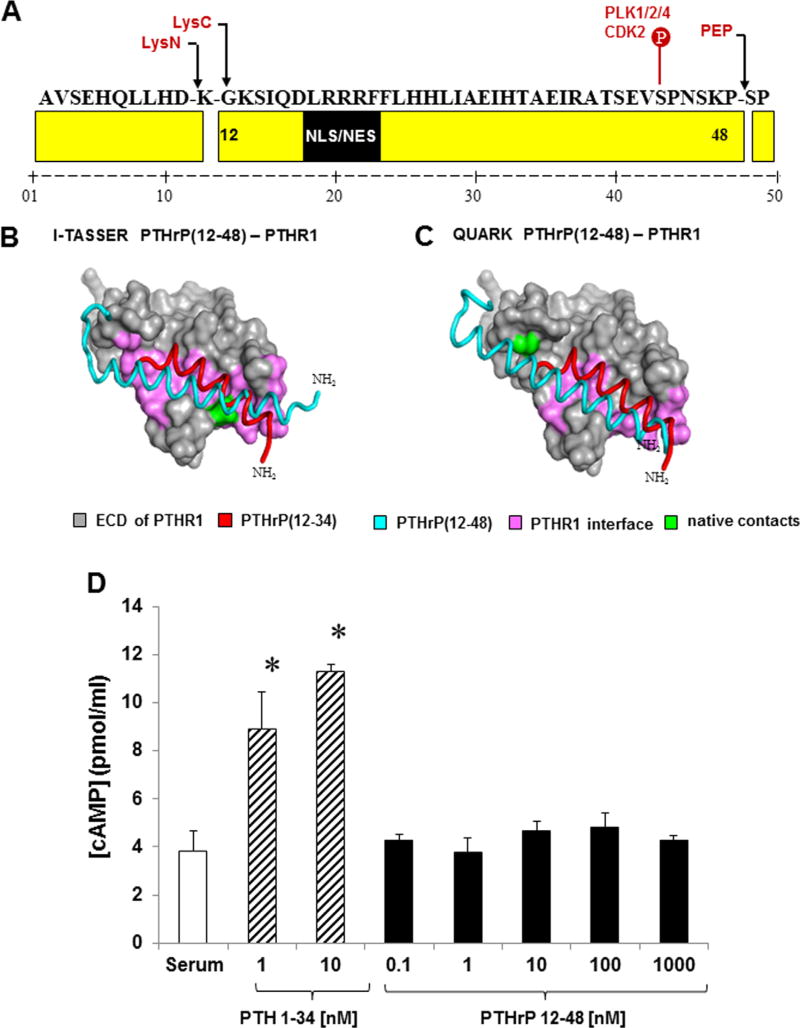
Bioinformatic tools predicted the tertiary structure and cleavage of PTHrP(12-48). C-terminal processing involved the cleavage of Pro48 by a member of the prolyl oligopeptidase family of serine proteases (Fig. 1A). For N-terminal processing, it is unclear whether PTHrP is processed on the amino or carboxyl side of Lys11. However, if the bond between Asp10 and Lys11 is hydrolyzed then the Lys11 residue would be removed by aminopeptidase activity (Fig. 1A). Both proteases are likely to require specific structural cues for the hydrolysis in order to avoid catalysis of nearby lysine and proline residues.
Fig. 1.
(A) Predicted PTHrP(12-48) cleavage sites. Lysine-specific monobasic endoproteases (LysC/LysN) and post prolyl endoprotease family members (PEP) hydrolyze the Lys11 and Pro48 peptide bonds. The PTHrP(12-48) sequence contains a noncanonical monopartite nuclear localization signal (NLS; residues 19-21). Phosphorylation of Ser43 by CDK2 was also predicted. Black arrows indicate potential cleavage sites. (B) I-TASSER, and (C) QUARK PTHrP(12-48)/PTHR1 complexes. Despite obvious similarities to the native complex (red), the extra helical turns in PTHrP(12-48) (blue) confers misorientation in the PTHrP(12-48)/PTHR1 complex and no interaction with PTHR1 (gray). (D) Competitive cAMP ELISA. SaOS2 cells were treated with PTH(1-34) (striped bars) or PTHrP(12-48) (solid black bars). PTH(1-34) stimulated cAMP production and no significant cAMP stimulation was observed with PTHrP(12-48) treatment. *p < 0.05 from other groups as determined by ANOVA and Student-Newman-Keul’s post hoc analysis. Results are representative of three independent experiments.
Both I-TASSER and QUARK predicted that PTHrP(12-48) forms a single alpha helix (Fig. 1B, C). The helix of I-TASSER model spans residues 12 to 41 and similarly, de novo structure prediction by QUARK suggests PTHrP(12-48) forms a slightly bent α-helix (residues 12-42) that unwinds into an unstructured region after residue 42. Because each structure prediction algorithm approaches protein folding differently, the convergence of the independent predictions to a similarly folded PTHrP(12-48) structure is a robust indication of modeling accuracy.
Computational analysis of PTHrP(12-48) bioactivity
The classical biological action of both PTH and PTHrP have been ascribed to the first 34 amino acids that interact with PTHR1.(24) PTHrP(12-48) contains the alpha helical binding domain (residues 12-34) that interacts with the ligand binding domain of the PTHR1; however, PTHrP(12-48) lacks the N-terminal “activation domain” (residues 1-11) (Fig. 1A).(11,36) Thus, it is conceivable that PTHrP(12-48) could interact with the PTHR1, but have no stimulatory effect. To examine any possible PTHrP(12-48) interactions and signaling via PTHR1, molecular docking studies of the PTHrP(12-48) structures with the predicted X-ray crystal structure of the extracellular domain (ECD) of the PTHR1 were performed (ClusPro 2.0) and refined/re-scored (FireDock). I-TASSER and QUARK homology modeling of PTHrP(12-48)/PTHR1 complexes (Fig. 1B, C) demonstrated that PTHrP(12-48) ligand (light blue) is mis-oriented in the hydrophobic pocket(37) of PTHR1 (gray) with respect to the native configuration (red) in the high-resolution crystal structure. I-TASSER and QUARK PTHrP(12-48)/PTHR1 complex models both predicted that PTHrP(12-48) likely interacts with PTHR1 through many of the same residue contacts (identified in green; Fig. 1B, C) as the native PTHrP(12–34) ligand, including several critical contacts (Arg20, Phe23, and Leu24)(37–40) that anchor PTHrP in the hydrophobic pocket, although the extra helical turns in PTHrP(12-48) demonstrate no specific interaction with PTHR1. However, the possibility that PTHrP(12-48) may interact with the receptor through a slightly different binding mode via induced fit accommodations not captured by current docking software, although unlikely, cannot be excluded.
PTHrP(12-48) does not stimulate cAMP in SaOS2 cells
In order to test the negative interaction predictions of the PTHrP(12-48)/PTHR1 models, the activity of PTHrP(12-48) was measured in human SaOS2 cells. We and others have shown that SaOS2 cells respond to both PTH(1-34) and PTHrP(1-36) ligands(11,31,36) and trigger the activation of Gαs.(41,42) SaOS2 cells were treated with PTHrP(12-48) and the activation of PTHR1 measured by cAMP production. The cells were treated with serum alone, PTH(1-34) (1 and 10 nM) and PTHrP(12-48) (0.1, 1, 10, 100, and 1000 nM) for 10 min and cAMP production measured (Fig. 1D). PTH(1-34) treatment stimulated the expected increase in cAMP (Fig. 1D). In contrast, PTHrP(12-48) treatment did not induce cAMP at any concentration tested, confirming the in silico prediction that PTHrP(12-48) does not activate PTHR1 (Fig. 1D), nor does PTHrP(12-48) antagonize agonist activity of PTHrP(1-36) at the PTHR1 (data not shown).
Specificity of PTHrP(12-48) antibody
To determine sites of physiologic PTHrP(12-48) expression, a polyclonal antibody specifically directed against human PTHrP(12-48) was developed, and the sensitivity and specificity of the PTHrP(12-48) polyclonal antibody (Ab) was determined by immunoblot analyses. PTH(1-34), recombinant PTHrP (1-141), PTHrP(1-37), PTHrP(12-48) (100 ng, 10 ng, and 1 ng) and interleukin 8 (IL-8), were blotted onto nitrocellulose membranes and incubated with either a PTHrP(1-15)-mAb antibody (Fig. 2A) or with the PTHrP(12-48) polyclonal Ab (Fig. 2B). Both IL-8 and PTH(1-34) were not recognized by either PTHrP Ab. The N-terminal PTHrP(1-15) antibody recognized an antigen present in full-length PTHrP(1-141) as well as PTHrP(1-37) but not PTHrP(12-48). Similarly and most importantly, the PTHrP(12-48)-specific Ab recognized PTHrP(12-48), but not PTHrP(1-141), likely because amino acids 12 to 48 are not uncovered in the PTHrP(1-141) protein in the nondenaturing dot blot. PTH(1-34) was not detected by either antibody. These data suggest the PTHrP(12-48) polyclonal antibody is specific for PTHrP(12-48) and that the active epitope resides within amino acids 12 to 37 of PTHrP. Next, PTH(1-34) (10 µM), PTHrP(1-37) (1mM), PTHrP(12-48) (1 mM, 100 µM, 10µM), and IL-8 were resolved by SDS-PAGE (Fig. 2C), blotted to nitrocellulose, and incubated with a PTHrP(12-48) Ab. The PTHrP(12-48) Ab specifically recognized the 4-kDa PTHrP(12-48) confirming PTHrP(12-48) antibody specificity.
Fig. 2.
Human PTH(1-34), PTHrP(1-141), PTHrP(1-37), PTHrP(12-48), and IL-8 were transferred to nitrocellulose and incubated with (A) PTHrP(1-15)-mAb and (B) PTHrP(12-48) Ab. PTHrP(12-48) is recognized by the PTHrP(12-48) Ab and not by the N-terminal Ab. (C) Immunoblot analysis of IL-8 (1 mM), PTH (1-34) (10 µM, 1.6 µg), PTHrP(1-37) (1000 µM, 219 µg), and PTHrP(12-48) (1000, 100, 10 µM; 170.4, 17, 1.7 µg). PTHrP(12-48) was specifically recognized by PTHrP(12-48) Ab. Results are representative of two independent series of experiments. (D, brown stain) IHC detects PTHrP(1-15) expression and (E) not PTHrP(12-48) expression in human breast duct. (F, G, brown stain) IHC detects PTHrP(12-48) expression in metastatic tumor cells (F) and OCLs (G). (H) Immunofluorescent detection of PTHrP(12-48) in human OCLs. PBMCs were treated with mCSF (i-iv) or mCSF+ RANKL (v-viii). Following serum starvation, day 7 cells were treated with PTHrP(12-48) for 30, 60, and 90 min. Panels (i) and (v): No primary Ab (control). Panels (ii-iv) and (vi-viii): intracellular detection of PTHrP(12-48) using PTHrP(12-48) Ab. PTHrP(12-48) was localized in OCLs (yellow arrowheads) at 60 and 90 min (Bar = 100 µm). DAPI (blue) was used to identify the nuclei of individual cells. Results are representative of three independent experiments.
Because the PTHrP(12-48) Ab specifically recognized PTHrP-(12-48), we next performed immunohistochemical (IHC) staining of a variety of normal human tissues, using commercially available tissue arrays (Maxivision BioSciences Inc., Kenmore, WA, USA). IHC staining was performed on tissue sections with either the PTHrP(1-15)-mAb or the PTHrP(12-48) Ab. As expected, normal breast tissue stained positive for PTHrP (1–15) (Fig. 2D), because breast is a well-recognized site for PTHrP expression, but intriguingly the same tissue appeared negative for PTHrP(12-48) (Fig. 2E). In addition, other classic PTHrP target tissues, such as placenta and cartilage, appeared negative for PTHrP(12-48) expression, suggesting that PTHrP(12-48) is not proteolytically processed in normal PTHrP-expressing tissues (Supplemental Fig. 1). Next, archival bone metastasis specimens were immunostained for PTHrP(12-48) (Fig. 2F, G). Immunoreactive and positive PTHrP(12-48) staining of metastatic tumor cells was observed (Fig. 2F), while the majority of surrounding normal cells, including osteoblasts and osteocytes, appeared negative. Surprisingly, OCLs that are also present in the bone microenvironment stained positive for PTHrP(12-48) (Fig. 2G). In fact, osteoclasts showed positive, cytosolic as well as what appeared to be nuclear staining. These data indicate that PTHrP(12-48) not only circulates at measureable levels in breast cancer patient serum,(19) but is detectable in metastatic tumor cells. In addition, the IHC staining suggested that PTHrP(12-48) may be taken up by OCLs. This latter idea was tested in the next series of experiments.
PTHrP(12-48) translocates into OCLs and inhibits OCL precursor survival and differentiation
Next, immunofluorescence assays were performed to determine whether PTHrP(12-48) entered human OCLs and if so, determine the time course of PTHrP(12-48) internalization in human OCLs. Human PBMCs were cultured in the presence of: mCSF (25 ng/mL) alone or mCSF+ RANKL (50 ng/mL). On day 7 (when some differentiated OCLs are present), PTHrP(12-48) (100 nM) was added to cells in the presence of serum for 15, 30, 60, and 90 min prior to fixation and immunofluorescence. Specific PTHrP(12-48) staining was observed in OCLs 60 min after the addition of PTHrP(12-48) (Fig. 2G, yellow arrows). Interestingly, similar treatment of day 7 cells with PTHrP(1-36) showed no internalization, whereas in agreement with others PTHrP(1-141) and PTHrP(12-48) was internalized into SaOS2 cells (Supplemental Fig. 2).(43) Specific PTHrP(12-48) staining was observed in mononuclear cells cultured in the presence of mCSF alone (Fig. 2H, panel iv). These data showed that PTHrP(12-48) was able to efficiently enter differentiated OCLs as well as potential precursors in vitro, raising the intriguing possibility that PTHrP(12-48) may have some specific biologic activity in OCLs. This interesting idea was tested in the experiments in the next section.
PTHrP(12-48) inhibits OCL proliferation and survival
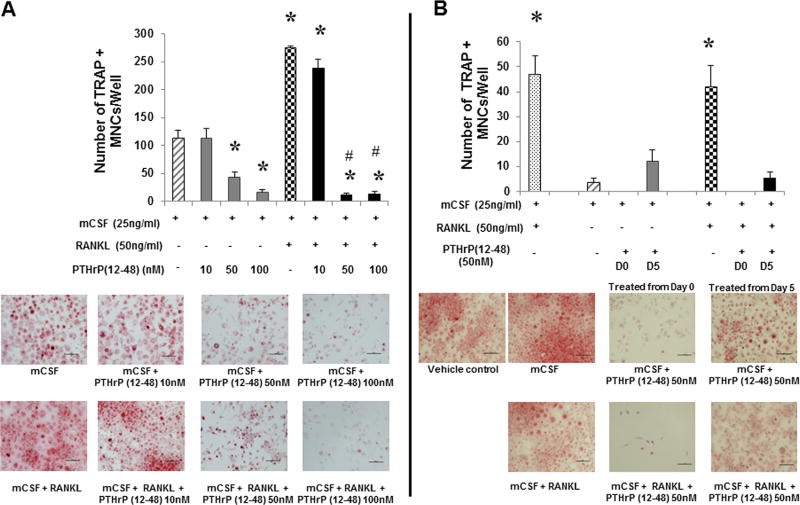
Although OCLs do not express any demonstrable PTHR1,(44) OCL nuclei immunostained positive for PTHrP(12-48) (Fig. 2F). Thus, the effect of PTHrP(12-48) on osteoclastogenesis was tested. To determine whether PTHrP(12-48) treatment altered survival, proliferation, and/or differentiation of OCLs and their precursors in vitro, human OCL precursors (Fig. 3) were treated with mCSF (25 ng/mL) alone or in the presence of RANKL (50 ng/mL), or in combination with increasing concentrations of PTHrP(12-48) (10 nM, 50 nM, and 100 nM) on day 0 and cultured toward osteoclastogenesis. On day 10, fixed OCLs were TRAP stained and the TRAP+ MNCs enumerated (Fig. 3A). RANKL significantly increased the number of TRAP+ MNCs. Interestingly, PTHrP (12-48) treatment significantly and dose-dependently decreased the number of TRAP+ MNCs in cultures treated with mCSF alone (Fig. 3A), showing that PTHrP(12-48) significantly inhibits the survival of OCL precursors. In addition, PTHrP(12-48) also significantly decreased the number of TRAP+ MNCs in RANKL-treated cultures (Fig. 3A). Collectively, these data support the novel conclusion that PTHrP(12-48) inhibits both the survival and RANKL-dependent differentiation of human OCLs and their precursors.
Fig. 3.
PBMCs were treated with mCSF (25 ng/mL) ± RANKL (50 ng/mL) (striped/checkered bars). (A) Cells were treated with PTHrP(12-48) (10 to 100nM) (solid gray/black) or (B) PTHrP(12-48) (50 nM) (solid gray/black) either on day 0 (D0) or day 5 (D5). On day 10, OCLs were stained for TRAP. The mean TRAP+MNCs/well ±SE is reported. In A: *= significantly different from mCSF control (p < 0.001); # = significantly different from mCSF+RANKL (p < 0.001). In B: * = significantly different from other groups (p < 0.05). Results are representative of four independent experiments.
Therefore, we next examined the time-course of the PTHrP-(12-48)-mediated inhibition of survival in mature OCLs and their precursors. OCL precursors were treated with mCSF (25 ng/mL) alone or in combination with RANKL (50 ng/mL) and allowed to differentiate into OCLs (day 10). Cells were treated with 50nM PTHrP(12-48) (effective dose from Fig. 3A) from day 0 or from day 5. On day 10 of culture, cells were fixed and stained for TRAP and TRAP+ MNCs enumerated (Fig. 3B). RANKL significantly increased osteoclastogenesis in both cases. However, PTHrP (12-48) treatment from either day 0 or day 5 significantly inhibited osteoclastogenesis induced by RANKL. These data suggest that PTHrP(12-48) potently inhibits not only the survival of mCSF-dependent OCL precursors but also RANKL-induced OCL survival and differentiation.
PTHrP(12-48) induces apoptosis in OCLs and their precursors
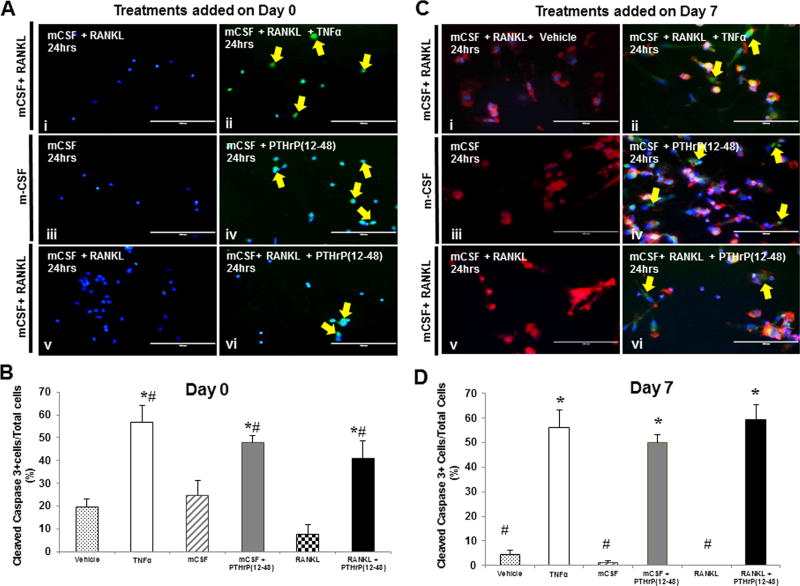
To determine whether the inhibition of OCL survival and proliferation induced by PTHrP(12-48) treatment was apoptosis-related, the effect of PTHrP(12-48) on OCL apoptosis was measured. OCL precursors were treated for 24 hours with the known apoptosis-inducing agent TNFα (200 ng/mL) or with PTHrP(12-48) (50 nM) on day 0 and day 7. The treatment-induced activation of caspase 3 was determined by immunofluorescence, using a specific cleaved caspase 3 (Asp175) antibody.(30) The cells positive for the expression of cleaved caspase 3, a measure of apoptosis, were enumerated (Fig. 4B, D).
Fig. 4.
Human PBMCs were treated with mCSF alone or mCSF+RANKL up to day 7. At (A) day 0 or (C) day 7, cells were treated with either vehicle, TNFα, mCSF±PTHrP(12-48), or mCSF+RANKL+PTHrP(12-48) for 24 hours. Panels: (i) vehicle control; (ii) TNFα (positive control); (iv-vi) PTHrP(12-48). Intracellular immunodetection of cleaved caspase-3 is observed (yellow arrows). Cells were also stained with phalloidin (red staining) to visualize multinucleated cells. (Bar = 100 µm). (B, D) The percent of cleaved caspase 3-positive cells per total cells was enumerated from 10 independent fields of view. TNFα induced ~55% apoptosis and PTHrP(12-48) induced >55% apoptosis. In B: * = significantly different from vehicle control; # = significantly different from RANKL treated. In D: * = significantly different from #, as determined by ANOVA and post hoc analysis, p < 0.05. Results are representative of three independent experiments.
In OCL precursors treated with vehicle or mCSF without or with RANKL on day 0 (Fig. 4A, B) for 24 hours, less than 25% of the total cells were positive for cleaved caspase-3. Treatment with PTHrP(12-48) (50 nM) in the presence of mCSF alone or mCSF+ RANKL significantly increased the number of caspase 3–positive cells compared to control, in OCL precursors as well as in differentiated OCLs. Similarly, in cells cultured for 7 days (Figure 4C, D), treatment with PTHrP(12-48) (50 nM) significantly increased the number of cleaved caspase 3-positive cells. The increase was similar to that seen following treatment with the potent apoptotic agent TNFa (200 ng/ml). These data provide convincing in vitro evidence that PTHrP(12-48) is a potent inhibitor of OCL proliferation and survival via the robust stimulation of cell apoptosis, as measured by the induction of cleaved caspase 3.
Summary and Discussion
This study provides the first insight into the biology of PTHrP (12-48). We hypothesized that PTHrP(12-48) circulated as a biomarker and possessed biological activity. Through a series of in silico, biochemical, and IHC approaches, the biologic activity of PTHrP(12-48) was shown. PTHrP(12-48) is the first peptide fragment of PTHrP confirmed to circulate in vivo and it is more than likely the product of enzymatic processing of the three known full-length PTHrP isoforms.(8,20,21,45) However, no data exists regarding the processing/metabolism of any PTHrP isoform or peptide. In silico cleavage site analyses of full-length PTHrP isoforms identified that endoproteases, which utilize specific structural cues to selectively hydrolyze the peptide bonds at Lys11 and Pro48,(46) are potentially involved in the processing of the isoforms, resulting in PTHrP(12-48). Although the processing enzymes are not directly identified here, data strongly suggests that both enzymes are endoproteases involved in proprotein processing and secretion, raising the exciting possibility that PTHrP(12-48) may be actively secreted by breast cancer cells. The identification and characterization of these specific endoproteases is currently the focus of intensive ongoing investigations.
Indeed, the physiological implications regarding the extent of posttranslational processing of PTHrP as well as the identity of the functional peptides are still poorly understood. Data do exist that support the idea that secreted forms of N-terminal, mid-regional, and C-terminal regions of PTHrP mediate the spectrum of PTHrP-mediated local and endocrine effects,(47,48) although none are confirmed in the systemic circulation. However, the discovery of PTHrP (12-48) as a circulating biomarker significantly correlated with breast cancer bone metastasis(19) uniquely positions PTHrP(12-48) as a validated, circulating bioactive PTHrP peptide.
Next, de novo structural modeling predicted PTHrP(12-48) to be an alpha helix followed by an unstructured region after residue 42. Molecular docking simulations with the PTHR1 in the ligand bound conformation produced a PTHrP(12-48)/PTHR1 interaction model predicting that PTHrP(12-48) is mis-oriented in the hydrophobic pocket of PTHR1, perhaps due to its extended continuous helix,(49) although many of the residue contacts (Arg20, Phe23, Leu24) critical for binding and anchoring of PTHrP in the hydrophobic pocket of PTHR1 are maintained.(37) Collectively, the molecular modeling suggested that PTHrP(12-48) may be capable of weak interactions with PTHR1. However, biochemical, conformational, pharmacologic, and biophysical evidence strongly indicate that PTHrP(12-48) is not a PTHR1 agonist/antagonist, because it lacks the critical N-terminal amino acids (residues 1-11) that activate G-protein– coupled signaling systems that increase cAMP.(11,36) In addition, the overall shift of PTHrP(12-48) in the PTHR1 binding pocket identified during molecular docking strongly suggests PTHrP-(12-48) is unlikely to have any functional biological interaction with the PTHR1. Supporting the model’s prediction, PTHrP (12-48) treatment did not induce cAMP accumulation in PTHR1-expressing SaOS2 cells. Furthermore, we have been unable to show any significant antagonist or partial antagonist actions of PTHrP(12-48) at the PTHR1 in SaOS2 or any other PTHR1-expressing cell (data not shown).
Interestingly, using a specific polyclonal PTHrP(12-48) antibody we developed, specific PTHrP(12-48) immunostaining was observed. Human breast, a classic N-terminal PTHrP target tissue (Fig. 2) was positive for N-terminal PTHrP(1-15), but negative for PTHrP(12-48). Human breast cancer bone metastasis specimens convincingly showed that tumor cells and even OCLs appeared PTHrP(12-48)-positive, with little observable staining in other resident bone marrow cells, including osteocytes and osteoblasts. These data strongly suggest that PTHrP(12-48) is expressed by both primary(19) and metastatic breast tumors.
PTHrP(12-48) is translocated into OCLs and their precursors. PTHrP(1-108), PTHrP(67-93), and PTHrP(94-108) have all been shown to translocate into cells in a PTHR1-dependent fashion.(50–52) PTHrP nuclear localization in chondrocytes has been shown to involve a canonical nuclear localization sequence (NLS) in the region spanning amino acids 67 to 93.(53) Interestingly the sequence of PTHrP(12-48) contains a putative noncanonical intracellular translocation sequence at position 19 to 21,(54) which potentially mediates the uptake and intracellular localization of PTHrP(12-48).
Because PTHrP(12-48) was actively translocated into OCLs, the effect of PTHrP(12-48) treatment on PBMCs and differentiation to OCLs in vitro was examined. PTHrP(12-48) dose- and time-dependently inhibited not only the survival of OCL precursors, but also subsequent differentiation into OCLs. These data have led us to further posit that the inhibition of mCSF-responsive cells in PBMC cultures (some of which can become OCLs) may be a mechanism by which tumor cells remodel the local bone marrow microenvironment, and potentially anywhere that PTHrP(12-48) is released. To date we have been unable to show an effect of PTHrP(12-48) on mesenchymal lineage cells or observe any apparent uptake in other resident bone and bone marrow cells in vitro or in vivo. These data currently support the notion that in vivo, PTHrP(12-48) may alter lineage determination and differentiation of other cells of hematopoietic origin. This and other intriguing ideas are currently being investigated to more precisely define the role of PTHrP(12-48) in tumor biology.
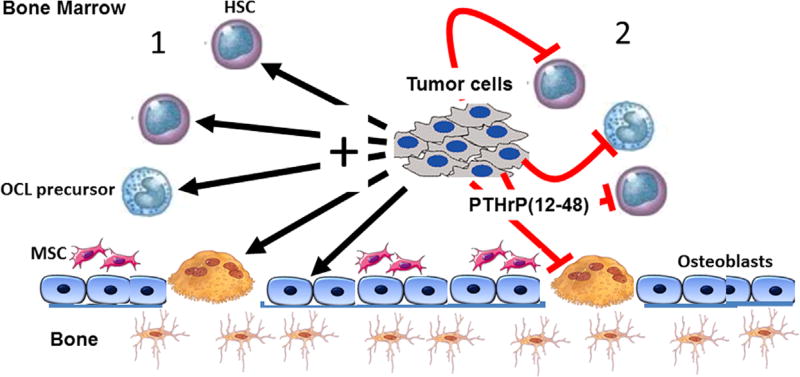
Because PTHrP(12-48) was able to effectively inhibit osteoclastogenesis, we next sought to ascertain a potential mechanism. We and others have shown that the survival of OCLs and their precursors can be modulated by changes in apoptosis.(30,55) PTHrP(12-48) significantly stimulated the expression of cleaved caspase 3 in OCLs and their precursors. The stimulation of this well-documented measure of cell apoptosis was comparable to the effects observed following treatment with TNFα. It is important to recognize that these data support the novel idea that PTHrP(12-48) is released as a paracrine inhibitor of hematopoietic cell survival (Fig. 5). It is important to recognize that this PTHrP(12-48) activity is occurring in the face of the release by colonizing tumor cells of other tumor-derived agents that act in an endocrine, paracrine, and/or autocrine fashion in the tumor-bearing bone marrow(4,6) (Fig. 5).
Fig. 5.
Schematic of the local actions of PTHrP(12-48) in the tumor–bone marrow microenvironment. Metastatic tumors in the bone marrow niche drive bone marrow and bone cell activity and differentiation. (1) Tumor cells release molecules that activate osteoclast differentiation and osteolysis, increase mesenchymal cell activity and other hematopoietic cells (including osteoclast precursors) locally or distantly (IL-8, PTHrP, TGF-β). (2) The same tumor also releases inhibitory molecules PTHrP(12-48) that locally inhibit monocyte/macrophage osteoclast precursor cell survival, differentiation, and osteolysis, as well other hematopoietic cells. The local PTHrP(12-48) action remodels the local microenvironment, permitting expansion of mesenchymal cells known to be important for the tumor niche, while modifying other hematopoietic elements. Validating this model phenomenon in humans is crucial to understanding the physiological roles of PTHrP(12-48). HSC = hematopoietic stem cell; MSC = mesenchymal stromal cell.
Indeed, the bone marrow consists of a wide array of progenitor cells destined to become mature cells such as osteoblasts and adipocytes as well as hematopoietic elements.(56) Marrow cells are profoundly influenced by a plethora of endocrine-derived, paracrine-derived, and autocrine-derived factors that ultimately function to contribute to the regulation of bone remodeling, as well as other actions.(3,56) In this context, the release of many tumor-derived factors (N-terminal PTHrP, IL-8, TGFβ)(3) as well as the paracrine release of PTHrP(12-48) in the bone marrow by colonizing tumor cells collectively initiates a cellular crosstalk that on one hand activates osteolysis (IL-8, N-terminal PTHrP) to support tumor growth, while PTHrP(12-48) remodels local hematopoietic elements, including osteoclast precursors in favor of tumor growth and potentially the evasion of immune surveillance (Fig. 5).
Collectively, these data suggest that in addition to being a circulating breast cancer bone metastatic biomarker,(19) PTHrP-(12-48) has specific and potent biological activity. It is produced by breast cancer cells at both primary and bone metastasis sites and is taken up by local hematopoietic lineage cells (OCLs and their precursors), independent of the expression of PTHR1. The result of this is the potent inhibition of cell proliferation and survival. These intriguing observations support the idea that via this previously unrecognized mechanism, tumor cells remodel the local hematopoietic bone marrow niche. Such a strategy could provide an important survival advantage for disseminated tumor cells and identifies potential new therapeutic opportunities that are worthy of further interrogation.
Supplementary Material
Acknowledgments
This manuscript is dedicated to the fond memory of the late Dr. Marco G. Cecchini. His intellect, humor, and understanding of bone physiology are deeply missed.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
Authors’ roles: LJS, DG, NSA, BJA, FLS, DKW, and AK designed and performed the research, analyzed the data and wrote and revised the manuscript. CLW, KL, and LJS analyzed data and edited the manuscript. AL and KL contributed to research design. All authors contributed to writing and editing of the manuscript.
References
- 1.Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–89. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics. 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Makhoul I, Montgomery CO, Gaddy D, Suva LJ. The best of both worlds - managing the cancer, saving the bone. Nat Rev Endocrinol. 2016;12(1):29–42. doi: 10.1038/nrendo.2015.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev Cancer. 2002;2(8):584–93. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 5.Suva LJ. Adjuvant bisphosphonates in breast cancer: the ABCSG-12 study. Curr Osteoporos Rep. 2010;8(2):57–9. doi: 10.1007/s11914-010-0012-5. [DOI] [PubMed] [Google Scholar]
- 6.Weilbaecher KN, Guise TA, McCauley LK. Cancer to bone: a fatal attraction. Nat Rev Cancer. 2011;11(6):411–25. doi: 10.1038/nrc3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coleman R, Cameron D, Dodwell D, et al. Adjuvant zoledronic acid in patients with early breast cancer: final efficacy analysis of the AZURE (BIG 01/04) randomised open-label phase 3 trial. Lancet Oncol. 2014;15(9):997–1006. doi: 10.1016/S1470-2045(14)70302-X. [DOI] [PubMed] [Google Scholar]
- 8.Suva LJ, Winslow GA, Wettenhall RE, et al. A parathyroid hormone-related protein implicated in malignant hypercalcemia: cloning and expression. Science. 1987;237(4817):893–6. doi: 10.1126/science.3616618. [DOI] [PubMed] [Google Scholar]
- 9.Foley J, Longely BJ, Wysolmerski JJ, Dreyer BE, Broadus AE, Philbrick WM. PTHrP regulates epidermal differentiation in adult mice. J Invest Dermatol. 1998;111(6):1122–8. doi: 10.1046/j.1523-1747.1998.00428.x. [DOI] [PubMed] [Google Scholar]
- 10.Wysolmerski JJ, Stewart AF. The physiology of parathyroid hormone-related protein: an emerging role as a developmental factor. Annu Rev Physiol. 1998;60:431–60. doi: 10.1146/annurev.physiol.60.1.431. [DOI] [PubMed] [Google Scholar]
- 11.Adams AE, Pines M, Nakamoto C, et al. Probing the bimolecular interactions of parathyroid hormone and the human parathyroid hormone/parathyroid hormone-related protein receptor. 2. Cloning, characterization, and photoaffinity labeling of the recombinant human receptor. Biochemistry. 1995;34(33):10553–9. doi: 10.1021/bi00033a030. [DOI] [PubMed] [Google Scholar]
- 12.Juppner H, Abou-Samra AB, Freeman M, et al. A G protein-linked receptor for parathyroid hormone and parathyroid hormone-related peptide. Science. 1991;254(5034):1024–6. doi: 10.1126/science.1658941. [DOI] [PubMed] [Google Scholar]
- 13.Huch K, Kleffner S, Stove J, Puhl W, Gunther KP, Brenner RE. PTHrP, PTHr, and FGFR3 are involved in the process of endochondral ossification in human osteophytes. Histochem Cell Biol. 2003;119(4):281–7. doi: 10.1007/s00418-003-0519-2. [DOI] [PubMed] [Google Scholar]
- 14.Chung UI, Lanske B, Lee K, Li E, Kronenberg H. The parathyroid hormone/parathyroid hormone-related peptide receptor coordinates endochondral bone development by directly controlling chondrocyte differentiation. Proc Natl Acad Sci U S A. 1998;95(22):13030–5. doi: 10.1073/pnas.95.22.13030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boabaid F, Berry JE, Koh AJ, Somerman MJ, McCcauley LK. The role of parathyroid hormone-related protein in the regulation of osteo-clastogenesis by cementoblasts. J Periodontol. 2004;75(9):1247–54. doi: 10.1902/jop.2004.75.9.1247. [DOI] [PubMed] [Google Scholar]
- 16.Abbas SK, Pickard DW, Rodda CP, et al. Stimulation of ovine placental calcium transport by purified natural and recombinant parathyroid hormone-related protein (PTHrP) preparations. Q J Exp Physiol. 1989;74(4):549–52. doi: 10.1113/expphysiol.1989.sp003303. [DOI] [PubMed] [Google Scholar]
- 17.Wysolmerski JJ, Dann PR, Zelazny E, et al. Overexpression of parathyroid hormone-related protein causes hypercalcemia but not bone metastases in a murine model of mammary tumorigenesis. J Bone Miner Res. 2002;17(7):1164–70. doi: 10.1359/jbmr.2002.17.7.1164. [DOI] [PubMed] [Google Scholar]
- 18.MacIsaac RJ, Heath JA, Rodda CP, et al. Role of the fetal parathyroid glands and parathyroid hormone-related protein in the regulation of placental transport of calcium, magnesium and inorganic phosphate. Reprod Fertil Dev. 1991;3(4):447–57. doi: 10.1071/rd9910447. [DOI] [PubMed] [Google Scholar]
- 19.Washam CL, Byrum SD, Leitzel K, et al. Identification of PTHrP(12-48) as a plasma biomarker associated with breast cancer bone metastasis. Cancer Epidemiol Biomarkers Prev. 2013;22(5):972–83. doi: 10.1158/1055-9965.EPI-12-1318-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangin M, Ikeda K, Dreyer BE, Broadus AE. Isolation and characterization of the human parathyroid hormone-like peptide gene. Proc Natl Acad Sci U S A. 1989;86(7):2408–12. doi: 10.1073/pnas.86.7.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suva LJ, Mather KA, Gillespie MT, et al. Structure of the 5′ flanking region of the gene encoding human parathyroid-hormone-related protein (PTHrP) Gene. 1989;77(1):95–105. doi: 10.1016/0378-1119(89)90363-6. [DOI] [PubMed] [Google Scholar]
- 22.Yasuda T, Banville D, Hendy GN, Goltzman D. Characterization of the human parathyroid hormone-like peptide gene Functional and evolutionary aspects. J Biol Chem. 1989;264(13):7720–5. [PubMed] [Google Scholar]
- 23.Henderson MA, Danks JA, Slavin JL, et al. Parathyroid hormone-related protein localization in breast cancers predict improved prognosis. Cancer Res. 2006;66(4):2250–6. doi: 10.1158/0008-5472.CAN-05-2814. [DOI] [PubMed] [Google Scholar]
- 24.Araki S, Mezawa M, Sasaki Y, et al. Parathyroid hormone regulation of the human bone sialoprotein gene transcription is mediated through two cAMP response elements. J Cell Biochem. 2009;106(4):618–25. doi: 10.1002/jcb.22039. [DOI] [PubMed] [Google Scholar]
- 25.Wang B, Yang Y, Abou-Samra AB, Friedman PA. NHERF1 regulates parathyroid hormone receptor desensitization: interference with beta-arrestin binding. Mol Pharmacol. 2009;75(5):1189–97. doi: 10.1124/mol.108.054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–32. doi: 10.1093/nar/gkn186. (Web Server issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozakov D, Beglov D, Bohnuud T, et al. How good is automated protein docking? Proteins. 2013;81(12):2159–66. doi: 10.1002/prot.24403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5(4):725–38. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu D, Zhang Y. Ab initio protein structure assembly using continuous structure fragments and optimized knowledge-based force field. Proteins. 2012;80(7):1715–35. doi: 10.1002/prot.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fowler TW, Kamalakar A, Akel NS, Kurten RC, Suva LJ, Gaddy D. Activin A inhibits RANKL-mediated osteoclast formation, movement and function in murine bone marrow macrophage cultures. J Cell Sci. 2016;128(4):683–94. doi: 10.1242/jcs.157834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakamoto C, Behar V, Chin KR, et al. Probing the bimolecular interactions of parathyroid hormone with the human parathyroid hormone/parathyroid hormone-related protein receptor. 1. Design, synthesis and characterization of photoreactive benzophenone-containing analogs of parathyroid hormone. Biochemistry. 1995;34(33):10546–52. doi: 10.1021/bi00033a029. [DOI] [PubMed] [Google Scholar]
- 32.Moorthy BS, Gao Y, Anand GS. Phosphodiesterases catalyze hydrolysis of cAMP-bound to regulatory subunit of protein kinase A and mediate signal termination. Mol Cell Proteomics. 2011 Feb;10(2) doi: 10.1074/mcp.M110.002295. M110.002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pines M, Adams AE, Stueckle S, et al. Generation and characterization of human kidney cell lines stably expressing recombinant human PTH/PTHrP receptor: lack of interaction with a C-terminal human PTH peptide. Endocrinology. 1994;135(4):1713–6. doi: 10.1210/endo.135.4.7925136. [DOI] [PubMed] [Google Scholar]
- 34.Chang CI, Zoghi B, Liao JC, Kuo L. The involvement of tyrosine kinases, cyclic AMP/protein kinase A, and p38 mitogen-activated protein kinase in IL-13-mediated arginase I induction in macrophages: its implications in IL-13-inhibited nitric oxide production. J Immunol. 2000;165(4):2134–41. doi: 10.4049/jimmunol.165.4.2134. [DOI] [PubMed] [Google Scholar]
- 35.Suva LJ, Hartman E, Dilley JD, et al. Platelet dysfunction and a high bone mass phenotype in a murine model of platelet-type von Willebrand disease. Am J Pathol. 2008;172(2):430–9. doi: 10.2353/ajpath.2008.070417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaich G, Orloff JJ, Atillasoy EJ, Burtis WJ, Ganz MB, Stewart AF. Amino-terminal parathyroid hormone-related protein: specific binding and cytosolic calcium responses in rat insulinoma cells. Endocrinology. 1993;132(3):1402–9. doi: 10.1210/endo.132.3.8382601. [DOI] [PubMed] [Google Scholar]
- 37.Pioszak AA, Parker NR, Gardella TJ, Xu HE. Structural basis for parathyroid hormone-related protein binding to the parathyroid hormone receptor and design of conformation-selective peptides. J Biol Chem. 2009;284(41):28382–91. doi: 10.1074/jbc.M109.022905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gensure RC, Shimizu N, Tsang J, Gardella TJ. Identification of a contact site for residue 19 of parathyroid hormone (PTH) and PTH-related protein analogs in transmembrane domain two of the type 1 PTH receptor. Mol Endocrinol. 2003;17(12):2647–58. doi: 10.1210/me.2003-0275. [DOI] [PubMed] [Google Scholar]
- 39.Wittelsberger A, Corich M, Thomas BE, et al. The mid-region of parathyroid hormone (1-34) serves as a functional docking domain in receptor activation. Biochemistry. 2006;45(7):2027–34. doi: 10.1021/bi051833a. [DOI] [PubMed] [Google Scholar]
- 40.Zhou AT, Bessalle R, Bisello A, et al. Direct mapping of an agonist-binding domain within the parathyroid hormone/parathyroid hormone-related protein receptor by photoaffinity crosslinking. Proc Natl Acad Sci U S A. 1997;94(8):3644–9. doi: 10.1073/pnas.94.8.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blind E, Raue F, Knappe V, Schroth J, Ziegler R. Cyclic AMP formation in rat bone and kidney cells is stimulated equally by parathyroid hormone-related protein (PTHrP) 1-34 and PTH 1-34. Exp Clin Endocrinol. 1993;101(3):150–5. doi: 10.1055/s-0029-1211222. [DOI] [PubMed] [Google Scholar]
- 42.Kukreja SC, Shemerdiak WP, Lad TE, Johnson PA. Elevated nephrogenous cyclic AMP with normal serum parathyroid hormone levels in patients with lung cancer. J Clin Endocrinol Metab. 1980;51(1):167–9. doi: 10.1210/jcem-51-1-167. [DOI] [PubMed] [Google Scholar]
- 43.Aarts MM, Rix A, Guo J, Bringhurst R, Henderson JE. The nucleolar targeting signal (NTS) of parathyroid hormone related protein mediates endocytosis and nucleolar translocation. J Bone Miner Res. 1999;14(9):1493–503. doi: 10.1359/jbmr.1999.14.9.1493. [DOI] [PubMed] [Google Scholar]
- 44.Nakchbandi IA, Weir EE, Insogna KL, Philbrick WM, Broadus AE. Parathyroid hormone-related protein induces spontaneous osteoclast formation via a paracrine cascade. Proc Natl Acad Sci U S A. 2000;97(13):7296–300. doi: 10.1073/pnas.110553397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangin M, Ikeda K, Dreyer BE, Milstone L, Broadus AE. Two distinct tumor-derived, parathyroid hormone-like peptides result from alternative ribonucleic acid splicing. Mol Endocrinol. 1988;2(11):1049–55. doi: 10.1210/mend-2-11-1049. [DOI] [PubMed] [Google Scholar]
- 46.Fulop V, Bocskei Z, Polgar L. Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis. Cell. 1998;94(2):161–70. doi: 10.1016/s0092-8674(00)81416-6. [DOI] [PubMed] [Google Scholar]
- 47.Rholam M, Fahy C. Processing of peptide and hormone precursors at the dibasic cleavage sites. Cell Mol Life Sci. 2009;66(13):2075–91. doi: 10.1007/s00018-009-0007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, Goltzman D, Rabbani SA. Processing of pro-PTHRP by the prohormone convertase, furin: effect on biological activity. Am J Physiol. 1995;268(5 Pt 1):E832–8. doi: 10.1152/ajpendo.1995.268.5.E832. [DOI] [PubMed] [Google Scholar]
- 49.Huang SY. Search strategies and evaluation in protein-protein docking: principles, advances and challenges. Drug Discov Today. 2014;19(8):1081–96. doi: 10.1016/j.drudis.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 50.Lam MH, Briggs LJ, Hu W, Martin TJ, Gillespie MT, Jans DA. Importin beta recognizes parathyroid hormone-related protein with high affinity and mediates its nuclear import in the absence of importin alpha. J Biol Chem. 1999;274(11):7391–8. doi: 10.1074/jbc.274.11.7391. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen M, He B, Karaplis A. Nuclear forms of parathyroid hormone-related peptide are translated from non-AUG start sites downstream from the initiator methionine. Endocrinology. 2001;142(2):694–703. doi: 10.1210/endo.142.2.7944. [DOI] [PubMed] [Google Scholar]
- 52.Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol Cell Biol. 1999;19(2):1210–7. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lam MH, Thomas RJ, Loveland KL, et al. Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol Endocrinol. 2002;16(2):390–401. doi: 10.1210/mend.16.2.0775. [DOI] [PubMed] [Google Scholar]
- 54.Gu Z, Liu Y, Zhang Y, et al. Absence of PTHrP nuclear localization and carboxyl terminus sequences leads to abnormal brain development and function. PLoS One. 2012;7(7):e41542. doi: 10.1371/journal.pone.0041542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vaira S, Alhawagri M, Anwisye I, Kitaura H, Faccio R, Novack DV. RelA/ p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J Clin Invest. 2008;118(6):2088–97. doi: 10.1172/JCI33392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reagan MR, Rosen CJ. Navigating the bone marrow niche: translational insights and cancer-driven dysfunction. Nat Rev Rheumatol. 2016;12(3):154–68. doi: 10.1038/nrrheum.2015.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.