Abstract
Under the microscope, eukaryotic animal cells can adopt a variety of different shapes and sizes. These cells also move and deform, and the physical mechanisms driving these movements and shape changes are important in fundamental cell biology, tissue mechanics, as well as disease biology. This article reviews some of the basic mechanical concepts in cells, emphasizing continuum mechanics description of cytoskeletal networks and hydrodynamic flows across the cell membrane. We discuss how cells can generate movement and shape changes by controlling mass fluxes at the cell boundary. These mass fluxes can come from polymerization/depolymerization of actin cytoskeleton, as well as osmotic and hydraulic pressure-driven flow of water across the cell membrane. By combining hydraulic pressure control with force balance conditions at the cell surface, we discuss a quantitative mechanism of cell shape and volume control. The broad consequences of this model on cell mechanosensation and tissue mechanics are outlined.
Keywords: cell mechanics, cell water dynamics, cytoskeleton mechanics
1. Introduction
In a cluttered corridor of a rather drab-looking concrete laboratory tower, a young graduate student, S, is walking briskly, immersed in thought. She is on her way to a meeting with her mentors, still dressed in a stained lab coat and clutching safety glasses. These weekly meetings are a part of her interdisciplinary training program, where researchers from physics and cell biology are brought together to tackle basic biophysics questions. Today, she just completed a round of laboratory experiments, and now her mind is brimming with excitement and questions. The door is open, and in seconds she is in the office of biology professor B.
Her co-mentors, physics professor P together with B, are already seated at the small conference table. S gives a slight nod in their direction and sits down.
S: This week, I performed live cell microscopy experiments on different types of eukaryotic cells. I was able to capture images of several fluorescently labeled cytoplasmic proteins and the cell membrane. What I saw was fascinating! Cells seem to have a mind of their own, and can take on variety of shapes and sizes. Cells can also migrate and change shapes at different stages of their cell cycle (figure 1). What are the molecular components that allow cells to behave this way? Cytoplasmic proteins I observed also can move and form patterns that are correlated with the cell shape and cell movement. Do we understand how cells control their shape and movement?
Figure 1.
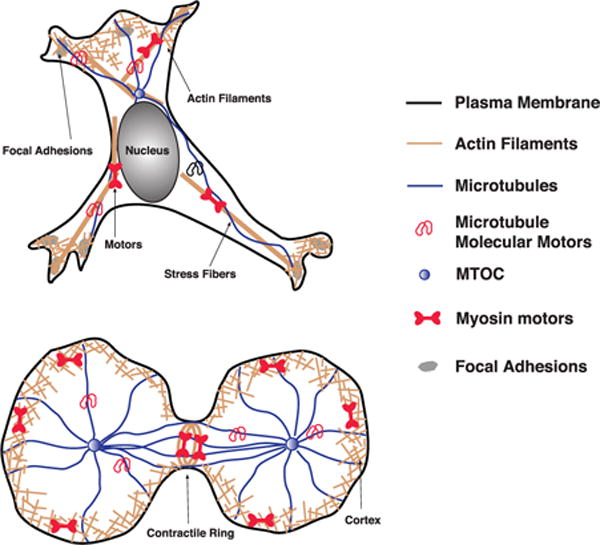
Living animal cells must move and deform to accomplish essential processes such as cell division or cell motility. These active movements are thought to be generated by cytoskeletal filaments and molecular motors within the cell. Motors such as myosin can generate contractile forces within actin networks. Actin filaments can also polymerize, and together with actin binding proteins to form loose open networks such as the lamellipodium, or bundled structures such as stress fibers. These structural elements in the cell can all influence the shape and movement of cell boundaries.
B: These are of course key questions in understanding how cells work. We now know the genetic sequences of many organisms, and it is fairly straightforward to find out the protein content (the proteome) of a given cell type [1]. But how these expressed proteins are organized and controlled to perform essential cell functions like movement and division are still not clear. There is a lack of quantitative models that adequately address phenomena at the cell scale. From experiments, it is also apparent that there are higher-order mechanical structures in the cell. For instance, we know that the cytoplasm of the cell is not a structureless liquid. There are complex micron-sized assemblies such as cross-linked actin filament networks, microtubules and intermediate filaments [2–7]. These filaments are very dynamic, undergoing cycles of polymerization and depolymerization, regulated by other proteins. In addition, organelles such as the nucleus, mitochondria and endoplasmic reticulum have dynamic and distinct shapes as well [8]. Several different types of molecular motors also work together with the cytoskeleton filaments to generate movement and transport within the cell [9–11].
S: So how do cells uses these elements to actively move, divide and change shape?
2. Forces and conservation laws
P: Newton’s laws state that motion is related to forces. In the context of cells, we clearly see a lot of movement, and forces can be generated in several ways. Molecular motors can directly provide forces derived from small molecules such as ATP or GTP. These motor molecules make small (nanometer-sized) conformational changes during the ATP hydrolysis cycle [11–13]. When many of these molecules sum up their forces, large cell-scale forces are developed. But there are other ways of generating forces as well. From fundamental conservation laws, mass transport is connected to momentum flux and therefore movement. For a continuous material with density ρ, fundamental mass conservation is [14, 15]
| (1) |
where g = ρv is the momentum density and v is the spatial velocity field. J is a source (or sink) term coming from an external mass flux. For simplicity, let us consider the cell as a 1D continuum strip of material with length L (figure 2(A)), and the mass flux can come from material outside of the cell or added from the cytoplasmic fluid phase. There is new mass added at the front x = x2, and mass is removed at the back, x = x1. Therefore the source term can be written as J=J2δ(x−x2) − J1δ(x−x1) If the material density ρ is constant, then the mass conservation equation is simply
| (2) |
for x∈(x1,x2) implying that the velocity field inside (x1,x2) is a constant. However, from overall mass conservation (x1,x2) must be changing. Since the overall mass balance within the domain is
| (3) |
where L = x2 − x1 is the length of the strip, this gives a condition for the boundary velocities
| (4) |
The actual constant velocity field within the domain is not known, and must be determined from another condition. In cases where material points are fixed in space such as from adhesions to stationary substate (figure 2(A)), then ν(x,t) = 0 within the domain, and the only points that are moving are the boundaries, giving velocity conditions for individual boundaries: . Thus, from mass fluxes alone, we can get motion!
Figure 2.
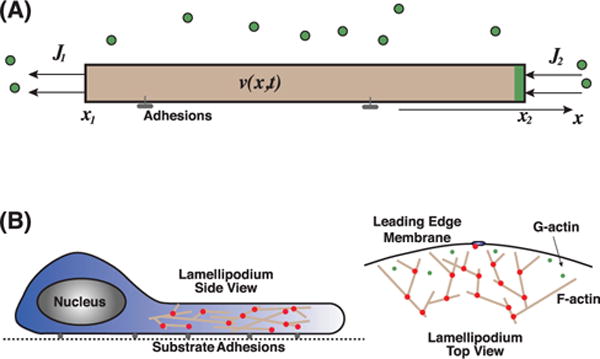
(A) Mass conservation is related to cell boundary movement. There is a velocity field, v(x, t ), within the shaded region representing the material in consideration. The boundary of the material can move from mass fluxes at the boundary J1,2. Cells build actin filaments from actin monomers; therefore the actin network can extend and retracting according monomer fluxes. (B) For a moving cell with a flat and broad lamellipodium, there is a polymerizing actin network at the leading edge. Mass flux of actin monomers (G-actin) increases F-actin network mass and drives membrane protrusion.
Note that this problem becomes more complicated in 2D, when there are two unknown velocity fields v = (vx,vy). In this case, other conservation laws must come into play in order to obtain a solution. The momentum density also satisfy momentum conservation (Newton’s 2nd Law) [14, 15], therefore
| (5) |
where σ is now the momentum current tensor, or the stress tensor. In the low Reynolds number limit that is appropriate for the cellular scale, we can neglect the nonlinear term (2nd term) [16]. The momentum current tensor contains information about mechanical forces on a small volume element in the material. In order to mathematically solve the coupled equations (1) and (5), is it necessary to introduce a constitutive law, i.e. a relationship between σ and the velocity field v. This constitutive relationship depends on the type of material, and the microscopic molecular structure. This relationship can also be measured experimentally.
Without complicating things, in the simplest picture, mass fluxes J are sufficient to push the cell boundary and generate cell shape changes.
B: In the cell, there are multiple sources of mass flux. One possibility is from polymerization and depolymerization of actin cytoskeleton or microtubule cytoskeleton. From experiments, it has been shown that the actin cytoskeletal network is important for cell shape and cell movement [17–19]. New actin networks in the cell are formed from monomers (G-actin) that polymerizes into actin filaments.
S: Ahh, that means by controlling where and when actin polymerization and depolymerization happens (figure 2(B)), the cell can control its motion and generate forces!
B: Precisely. It is known from experiments on cells on 2D substrates that by depolymerizing the actin cytoskeletal network using drugs such as Latrunculin A/B, cells no longer can change shape or move on 2D flat substrates [20, 21]. Therefore, actin is necessary for 2D migration. From extensive studies on the shape of rapidly migrating cells, we now have very sophisticated models of how actin network dynamics generate the shape of moving cells [22–24]. Features such as myosin contraction, diffusion of G-actin in the cytoplasm and detailed branched structure of the actin network has been incorporated [25]. So we know a lot about how actin drives cell movement.
S: In order to predict how cells move and change shape, we need information on the stress tensor in equation (5), right? A constitutive law as you described?
3. Actin cytoskeleton mechanics
P: You are absolutely right. There has been a lot of work in this area, trying to understand properties of the actin cytoskeleton [26–30].
B: From a biological perspective, the actin network plays a crucial role in a cell’ s morphology, migration and force production [31–33]. In mammalian cells, a large portion of actin is located within a cell cortex, which is just below the plasma membrane [33–37]. It is within this actin rich cortex that F-actin interacts with molecular motors and actin crosslinking proteins [38–40].
P: There has been also a lot of work on structural composition of actin networks in cells. For example, actin crosslinking proteins such as α-actinin, cortexilin, and fascin are important proteins in forming actin networks [41–43]. In addition to exerting active forces, myosin also forms mini-filaments that cross-links F-actin [44]. Branch forming proteins such as Arp2/3 generate a mesh-like network [45]. Speckle microscopy has been developed to measure movement of individual actin monomers [46–48]. So far, hundreds of actin-binding proteins have been identified. These proteins seem to regulate all aspects of the actin network, including polymerization, depolymerization, cross-linking and mechanical rigidity. Thus, to by-pass the molecular complexity, a constitutive model is an useful approach to describe the actin network mechanics. In such a constitutive model, molecular components will influence the mechanical moduli, but the mathematical relationship between kinematic variables and stress remains similar.
There are several broad classes of constitutive models. Of course, solids and fluids are already familiar, the primary difference being that solids can sustain shear stress at static mechanical equilibrium while fluids cannot. Actin networks are examples of semi flexible polymer networks [49–52]. These networks are viscoelastic, meaning that they retain some properties of solids and fluids. However, it is important to keep in mind that actin networks in cells are viscoelastic fluids. Therefore, the stress at long times is only a function of deformation rate:
| (6) |
where I is the identity tensor and vn is the spatial velocity field of the actin network. μ is the shear viscosity of the network. The diagonal part of the stress tensor acts like a pressure, but the shear part (off-diagnol) vanishes when vn = 0 or when vn is spatially constant. This is the long time behavior of actin networks.
The dynamic nature of the cross-linking proteins and accessory proteins that depolymerize actin allow the cell to tune the mechanical properties of its actin network [53–55]. When cross-linking proteins bind to filaments they contribute to the elasticity within the meshwork and when they unbind they allow the filaments to slide past one another, thus contributing to the development of shear stress (figure 3). The elastic behavior of the network is modeled by a Young’s modulus, E. When it is combined with the viscosity μ, a relaxation time scale τ=E/μ emerges. After an initial step perturbation or deformation, τ is the time scale which the material still retains some elastic characteristics. After τ, the material behaves as a fluid and flows. For actin in the cytoplasm, τ is about 10 s [56, 57].
Figure 3.
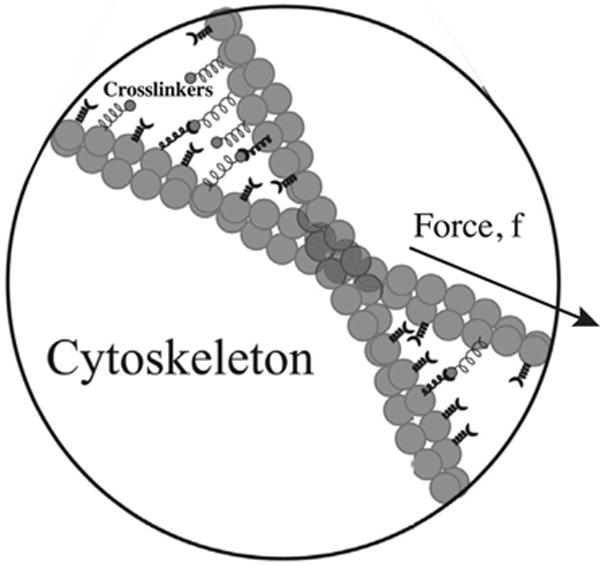
Actin cytoskeletal network behaves as a viscoelastic fluid, primarily due to force-dependent binding and unbinding of cross-linker proteins and filament depolymerization. When forces are applied to filaments, cross-linker dynamics acts as a viscous friction between filaments at time scales longer than cross-linker binding rates. The network flows under force, giving a constitutive relationship as in equation (6).
S: Okay, so measurements show that the viscosity of actin network, μ, is about 1000 times higher than that of water [26, 58, 59].
P: Correct. It is also important to note that a single time scale as described above is a simplification. Rheological properties of actin network also depend on other factors like filament stiffness, filament bending and entropic fluctuations. There are multiple time scales in the viscous behavior [60, 61]. In the simplified case of rigid filaments with dynamic cross-links, the force-velocity behavior of filaments sliding with respect to each other resembles viscous friction, and the friction coefficient can be explicitly related to cross-linker binding kinetics and linker stiffness [62–64]. Furthermore, when forces are applied to the network, the mutual sliding friction tend to align the filaments and generate large actin bundles, so-called stress fibers [65, 66]. These stress fibers tend to attach to substrates through integrin focal adhesions [67–69]. The movement of adhesions depend on substrate stiffness [62].
B: Myosin molecular motors also add to the fluidity of the actin network. The heads of myosin motors bind F-actin and utilize ATP hydrolysis to generate force to move toward the positive end of actin filaments [39, 44]. This movement also slides filaments with respect to each other and further fluidizing the network [39, 52]. Indeed, activities of small myosin assemblies is responsible for the contractile nature of actin networks in cells. If the contraction is isotropic, the contractile stress can be modeled as an active part of σ0 in equation (6). The contraction acts as a negative pressure, i.e.
| (7) |
where σp is the passive pressure in the actin network and σa is the negative pressure from active contraction of myosin. σa depends on energy input, such as ATP hydrolysis [32, 70], and signaling pathways that controls concentrations of myosin [71–73]. In order for sustained network contraction by myosin to occur, it is not sufficient to only have filament sliding, since the sliding movement does not decrease overall network volume. It is shown that buckling of actin is also necessary for contraction [74, 75]. In addition, filaments under mechanical compression should also depolymerize faster [76]. This elevated depolymerization in compressed networks allows recycling of G-actin and sustained contractile stress generation by myosin. Finally, network contraction can result not just from myosin motor activity, but also from passive crosslinking. Actin cross-linkers such as α-actinin can directly cause network contraction [64, 77], simply from favoring closer filament-filament interaction.
P: There is a growing body of literature on the active behavior of the actin cytoskeleton, including non-isotropic networks and active chiral behavior [78–81]. In these more complex scenarios, the stress depends on possible nematic order in the network and active stresses are not necessarily isotropic [70]. As one can imagine, studying the actin network within a cell is difficult due to the presence of other structures within the cell. Researchers have therefore been using reconstituted actin works in vitro to further investigate the behavior of the actin cytoskeleton [54, 61, 62, 82–86].
S: Ok, but the cytoplasm is more complex than just the cytoskeleton right? What about the water phase? I recently read that water is apparently 70% of the cell mass content [87], and the cytoplasm is a porous material where water can flow [88]. Are there other mass fluxes that are important for cell shape and mechanics?
4. Water dynamics in cells
P: Indeed, in addition to cytoskeletal polymerization, the cell can passage water through the cell membrane directly, or through the membrane water channel aquaporin [89, 90]. The direction and magnitude of the water flux depend on the water chemical potential difference, Δμ, across the cell membrane, which is influenced by both the hydrostatic pressure difference, ΔP, and the osmotic pressure difference, ΔΠ [91]:
| (8) |
Here α is a permeation constant proportional to aquaporin density. Since this flux depends on two kinds of pressures, mechanical forces that influence ΔP and solute concentration changes that influence ΔΠ will both drive water flow across the membrane (figure 4). Therefore, mechanics and chemical contents inside and outside of the cell determine water flow.
Figure 4.
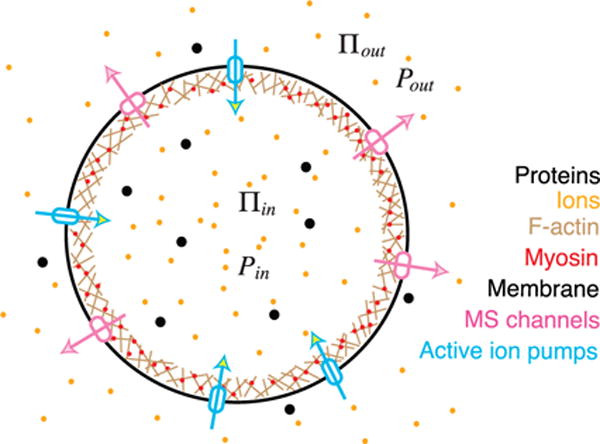
Cell pressure and volume regulation. There is typically an elevated cytoplasmic osmotic pressure (Πin) as compared to the extracellular environment (Πout). Therefore, hydrostatic pressure inside the cell is higher: Pin > Pout. The hydrostatic pressure difference is largely sustained by tension in the actin cortex underneath the membrane. When the osmotic pressure difference changes, this causes a change in the hydrostatic pressure difference and a change in cortical/membrane tension. Cells will re-adjust its osmotic content by opening and closing ion channels and pumps, and re-adjust cortical tension. When the cell experiences mechanical forces, the hydrostatic pressure difference changes, and cause the same chain of osmolyte flux and actomyosin tension change. It is unclear whether the cell maintains a constant osmotic pressure difference in all conditions.
B: This water flux has been studied in various physiological contexts, including coupled transport of water and salt across epithelia [92]. Indeed, aquaporin was discovered in kidney epithelium cells and every day your kidney re-absorbs 180 liters of water from blood that leaks into the glomerulus [93]. For cells, water flux also plays a role in membrane blebbing [94, 95], and the ameboid mode of cell migration [96, 97], where extruded blebs is refilled with cytoskeleton, and the cell can move by repeated development of blebs [36, 98–100]. Outside of physiological situations, osmotic effects can drive microfluidic pumping [101] and has been proposed as a way to generate energy [102, 103].
S: So water can flow in and out of the cytoplasm. Does this mean that the cell size increases when there is net water flux into the cell? How fast can the cell size change? What are the factors influencing the cell size?
4.1. Cellular pressure/volume regulation: a qualitative description
P: Yes, water permeation is an important element in the regulation of cell volume [104]. Mathematically, on the time scale of water flow, we have [105],
| (9) |
where V and A are the volume and surface area of the cell, respectively. Here, the hydrostatic pressure difference ΔP is mechanically balanced by cell surface tension and surface curvature. We will discuss the precise force balance conditions a little later. The osmotic pressure difference, ΔΠ=Πin − Πout depends on concentration of solutes inside and outside of the cell. Potentially there is larger concentrations of solutes in the cell, therefore Πin > Πout. Note that the osmotic pressure is an entropic force, arising from entropy of mixing [106], and therefore does not depend on molecular details. Πin ≈ RATcin=RAT(n+Np)/V, where n is the number of small transportable ions and molecules such as Na+, K+, Cl− and in the cytoplasm. Np is the number of untransportable proteins and macromolecules in the cytoplasm, and Ra is the gas constant and T is the temperature. Note this estimate of the osmotic pressure is valid for simple solutes in dilute solutions, and additional corrections could be necessary for complex cytoplasmic environments.
B: The cell of course actively controls both the protein content Np, and the ion content, n. For ions, there are many ionic species to consider. Na+, K+, and Cl− are the most abundant ones, each with concentrations in the 100 mM range. But Ca2+, H+, , and other small organic molecules are also involved. Transport of sugars and other small nutrients are obviously needed for increasing Np. Ions flow across the cell membrane both passively and actively. Passive ion channels allow ions to flow according to the chemical potential difference of each ionic species. Active ion pumps can transport ions against the chemical potential gradient; and the pumping process requires energy input. Some common pumps and co-transportors include Na/K pump [107], Na-K-Cl cotransport [108], and Na/H exchanger [109, 110]. These channels and pumps are essential for cells to maintain intracellular osmolyte concentrations.
P: These ion channels and pumps work on relatively fast time scale (~s). We can describe cell volume regulation on this time scale by considering water and ion fluxes in mathematical models. If osmolarity and cell mechanical behavior are the main concerns, we can consider simplified models with neutral osmolytes [105]. Equation (9) implies that at equilibrium, the hydrostatic pressure difference is equal to the osmotic pressure difference: ΔP = ΔΠ, and the water content does not change. One can induce water flow and cell volume change by applying osmotic shock [111], i.e. changing ∏out, or by changing the hydrostatic pressure difference either by changing Pout [112] or applying mechanical forces to the cell. When the extracellular osmolarity varies, the ion flux across the membrane changes, leading to an increase or decrease of intracellular ions within a few minutes. This process changes the volume of the cell, primarily from ion fluxes driving water flow. From experiments, it is known that cells can adapt to osmotic shocks, and the cell volume can recover within tens of minutes [111]. The cell volume change also depends on the mechanical stress in the actomyosin cortex, which directly influences ΔP. The coupled water and ion fluxes together with cortical contraction are responsible for the volume recovery [105, 111]. Note that water fluxes are equally influenced by both hydrostatic pressure and osmotic pressure (equation (9)). Therefore, mechanical response of the cell on the time scale of minutes is also influenced by water and ion dynamics as well as actomyosin dynamics.
B: On longer time scales, tens of minutes to hours, cell volume is determined by changes in Np, or the total protein content of the cell. Cells actively import molecules and nutrients to grow. And over the cell cycle, the cell volume roughly doubles [113–115]. Indeed, one possibility is that the osmotic regulation system roughly keeps a constant osmolyte concentration in the cell, and therefore Np and volume should increase at the same rate [116]. The regulation of Np is an important aspect of the cell cycle and signaling pathways are involved in this regulation [117–119]. A full quantitative picture is still not clear. It is evident that these pathways must also couple with ionic fluxes, actin cytoskeleton and myosin contraction. More work is needed to understand this area.
S: Huh, cell volume regulation is quite complex! I guess it makes sense since the cell volume determines the concentration of everything in the cell. The cell has to get that right. Coupling with water dynamics is also interesting. Water fluxes are clearly important in the kidney. Are there other situations where water fluxes has an important role?
4.2. Water-driven cell migration—an osmotic engine
P: It’s a good question. So far, we have discussed how isotropic (i.e. spatially uniform) water flux can lead to cell volume increase and decrease. If the water flux is spatially varying, this mass flux can lead to cell movement, exactly in the same manner as depicted in figure 2(A). For cells confined in a 1D narrow channel, it was discovered experimentally that cells can move without actin or myosin [21, 120]. Therefore, we can neglect their contribution to mass fluxes. In this case, the cell moves by permeating water through the cell: the so-called osmotic engine model (OEM, figure 5) [121]. This concept has been proposed sometime ago [122–124], and it is possible because, for a polarized cell, the intracellular osmolyte concentration is non-uniform. This allows an influx of water at the leading edge of the cell and an efflux at the trailing edge. When the intracellular fluid is stationary with respect to a fixed frame, the polarized water flux drives cell migration through the extension of cell membrane (figure 5(A)).
Figure 5.
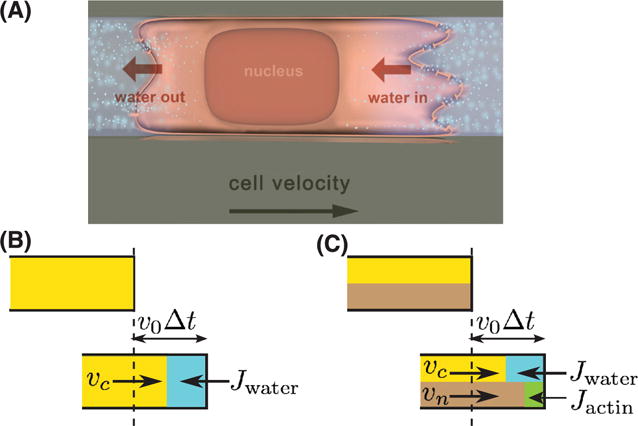
Directional water flux, like actin polymerization flux, can drive cell movement. (A) Cells in confined environments can generate water influx at the leading edge and water efflux at the trailing edge. The water flux is connect with directional ionic flows. (B) Fluid flux gives a quantitative kinematic condition for the cell boundary: equation (10). Here vc is the fluid velocity in the cytoplasm and v0 is the velocity of the cell leading edge. In time Δt, the new volume element is v0Δt which is equal to the sum of vcΔt and JwaterΔt. (C) If the cytoplasm is considered as a 2-phase fluid, the fluid phase only occupies a fraction of the cell volume θc. In this case, the new volume is given by a combination of vc, actin velocity vn, and Jwater and Jactin (equation 11).
B: In order to develop the gradient of osmolytes in the cell, it is important to have a polarized distribution of ion channels and pumps in the cell. In the experiments of [121], they studied cancer cells migrating in 1D confined channels, a situation mimicking cancer metastasis in tissues. They observed that the cell has an elevated concentration of Na/H exchanger at the leading edge [121].
B: The interesting aspect is that the cell appears to be able to move with and without actin [120, 121]. The cell speed in actin-dependent motility appears to be faster, and the osmotic engine appears to be slower. Therefore, the cell seems to have two different gears, possibly for different environmental conditions. Water fluxes can also come from asymmetric external hydrostatic pressures, which also can drive cell migration [125].
S: Ions have electrical charges. Therefore, is it correct to consider neutral osmolytes in the OEM?
P: Only in a simplified phenomenological manner. In general we need to consider charged ionic species and the associated electric fields and voltage gradients [107, 126, 127]. This is true for both cell volume regulation and OEM. When transmembrane voltages are introduced, there is then a natural coupling between the osmotic engine model and cell migration under external electric fields (electro-motility) [128].
B: Electro-motility has been experimentally explored in 2D cells [129, 130]. Cells can go to either the cathode or the anode, and this ‘galvanotaxis’ appears to be cell-type dependent. Again, just as in the osmotic engine case, polarization of the cell plays an important role. It is not clear how cells turn these external electrical signals into spatial arrangements of various proteins in the cell. In general, actin and myosin are perhaps involved in both polarization dynamics and driving motility, and in the case of galvanotaxis, cell polarization maybe organized by downstream signal pathway from the electric signals [129].
P: In 2D the story could be more complicated. The water flux across the membrane now can come from any direction; it is the spatial distribution of water flux that determines cell shape and movement. In addition, we also need to consider the intracellular fluid dynamics [94, 131]. But in 1D, the story is simpler. We can think of 1D cells with non-zero intracellular fluid velocity. In this case, as shown in figure 5(B), both the intracellular fluid velocity, vc, and water flux, Jwater, contribute to the cell migration. At the leading edge of the cell, we have a simple relation for the leading edge velocity
| (10) |
This velocity condition is again a result of mass conservation.
B: Absolutely. Experimental observation shows that knocking out aquaporins in cells reduces cell migration speed [121, 132]. In many cases water flux and actin polymerization probably work together to facilitate cell migration. Although the exact details require further elucidation.
4.3. Actin and water together? A two-phase model of the cytoplasm
P: Yes. Indeed, for the general cell migration case, we can consider a two-fluid-phase model to understand physical elements giving rise to cell velocity. Let’s still consider a 1D cell. Let the volume fraction of the fluid (actin network) phase be θc (θn).θc+θn= 1 by volume conservation. Since now the fluid only occupies a fraction of the total mass of the cell, as illustrated in figure 5(C), then the velocity of the cell leading edge is related to the velocity of the fluid and fluxes by
| (11) |
whereJactin is the mass flux of new actin polymerizing at the cell boundary.Jactin depends on the G-actin concentration and the density of polymerizing actin ends. Now we observe that the water flux and actin polymerization add constructively to the velocity of the leading edge, as seen in the experiments [133]. Note that all the variables in entire system are potentially coupled,Jwater,Jactin, vc, and vn could all depend on each other.
S: That’s interesting! The actin phase and the water phase are coupled. Does this mean that water flux is always necessary during cell migration?
B: The kinematic relationship in equation (11) is clear, but how the actin and water velocities are regulated is unclear. The regulation of these kinematic variables has to do with mechanisms of cell polarization, and it is known that in addition to chemical factors, mechanical factors such as the cell membrane tension are involved [25, 134]. Actin perhaps is involved in setting up a polarized cell as well as contributing to membrane velocity. The answer is still out there.
5. Cell volume and cell shape dynamics: a quantitative view
P: In order to examine the combined contributions of actomyosin dynamics and osmotic flow in cell mechanics, it is useful to develop a quantitative description of forces and flows in cell volume regulation and cell shape change. To start we have to consider how mechanical forces are balanced at the cell surface for different cell geometries. This force balance requires us to consider the cell membrane, the actomyosin cortex, active contraction and polymerization, and potential water flows across the membrane. Using a continuum mechanics approach, it is possible to get an order of magnitude estimates of these contributions to the steady state cell shape.
5.1. Cell surface force balance
P: To see how the details work, we can focus on a small section of the cell surface which includes the membrane and the actomyosin cortex underneath (figure 6). Since the cortex is permeable to water, at steady state the hydrostatic pressure within the cortex is relatively uniform. At mechanical equilibrium, the total force acting on the membrane surface element S enclosed by C is zero (figure 6(A)), and we have
| (12) |
where σ · n is the total force acting on the surface element S from the actomyosin network. σ is the mechanical stress in the cortex. T is the membrane tension, and the last term can be shown to be:
| (13) |
where the right hand side is the total force from membrane tension. Therefore, within the continuum framework, force balance gives an explicit relationship between actomyosin activity, cell hydrostatic pressure and membrane tension.
Figure 6.
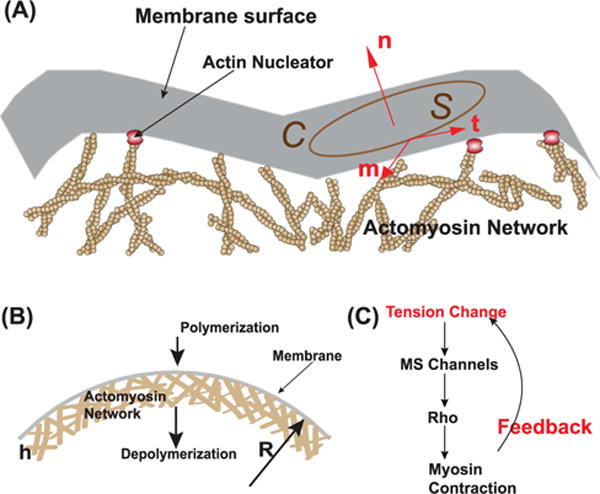
(A) Schematic illustration of mechanical forces on a membrane element S. This element experiences forces from stress in the actin cortex below, membrane tension in plane of the membrane, and hydrostatic pressure that acts normal to the membrane. (B) When actin polymerization and myosin contraction are consider, we can estimate the stress in the cortex. The sum of mechanical forces yields force balance condition equation (14). (C) Myosin contraction in the cortex is regulated by signaling networks. Mechanosensitive channels can sense membrane tension change, and activation myosin contraction relatively quickly.
S: Ok, what is the stress in the actomyosin cortex, σ? How do we compute it?
B: Within the cytoskeleton network, actin actively polymerizes near the cell membrane and depolymerizes some distance from the membrane. This continuous mass flux generates an actin flow, and maintains a relatively constant cortical thickness, h ~ 200–500 nm [135]. As discussed earlier, myosin actively applies a contractile stress tangential to the membrane surface. Therefore, the total stress is σ=σp+σactive, and σactive = −σa(t ⊗ t+m ⊗ m), where we have assumed that the myosin-generated stress is isotropic: equal in both t and m directions. The negative sign in front of σa indicates that it is contractile. Since signaling networks can control the activation of myosin (figure 6(C)), σa is proportional to the amount of activated myosin, M: σa = KmaxM, in which Kmax is the maximum contractile stress when 100% of myosin is activated. Kmax is a function of protein expression, and therefore can be assumed to be a constant over a short period of time (<hours).
P: σpassive can be estimated using the constitutive relation in equation (6). The results are straightforward [116], and we find that the passive stress within the network is relatively small when compared with the contractile stress and hydrostatic pressure, and the normal component of the force balance obtained from dotting equation (12) with n becomes
| (14) |
The tangential component is also interesting, and depending on the spatial variations in σ, there could be membrane flow and tension gradients. Geometrically, ∇ · n = 2H where H is the cell surface mean curvature [136]. Therefore we see that the active stress in the cortex and the membrane tension both balance the hydrostatic pressure difference. Also, from our discussions earlier, if the cell is steady and there is no net water flow across the membrane, i.e. ΔP = ΔΠ, which means that osmotic pressure is related to membrane and cortical tension.
B: Another good thing about equation (14) is that it is essentially exact for rapid cortical turnover. Other cortical components like actin cross-linkers, Arp2/3 and bundling proteins may change the viscosity of the actin network, and could introduce a passive stress contribution in equation (14). Proteins like spectrin may further help to stiffen the cortex and generate elastic behavior [137]. But the general form of equation (14) still remains the same, only the passive stress contribution may change.
S: So we have local force balance for a stationary cell. What does this say about cell volume and cell shape?
P: The force balance equation is sufficient to determine the overall cell shape! Since ∇ · n is related to the mean curvature, we can relate the cell geometry to forces at the cell surface. For a spherical cell, ∇ · n = 2/R, where R is the cell radius. For other geometries, computation of the complete cell shape is possible from equation (14). We can discuss this a little later.
B: Moreover, equation (14) gives us a good view on how active regulation of myosin activity contributes to cell volume adjustment (figure 6(C)). σa depends on active chemical regulation of myosin. Some details on this are fuzzy, but studies suggest that mechanosensitive transmembrane protein is responsible for Rho activation, a crucial signal upstream for activating myosin. For example, mechanosensitive Ca2+ channel, such as TRPV4, can directly activate Rho [138, 139]. These mechanosensitive ion channels are membrane tension sensitive. Estimates have shown that membrane tension at steady state is low, in the order of 0.01 pN nm−1 [140, 141]. However, if there is a sudden changes in cell mechanical and chemical environment, such as during a osmotic shock or changes in mechanical force, the membrane tension can increase dramatically. The jump in membrane tension opens these mechanosensitive channels, and signals to the cell to activate Rho and myosin contraction. Since equation (14) is correct at times scales of tens of seconds, changes in σa re-adjusts ΔP and T, so that overall cell volume can adapt to change.
5.2. Dynamic control of cell volume and pressure
P: The sequence of events described by B can be put in mathematical form. We will limit our discussion to spherical cells where everything is spatially uniform. The first equation describes the total water content of the cell,
| (15) |
Here ΔΠ = RAT(n + Np)/V − RATcout where cout is the extracellular concentration. From what we know about ion channels, n changes much faster than Np, therefore we have
| (16) |
where Iactive could be the flux through the active ion pumps and Ipassive is the flux through passive ion channels. The functional forms of Ipssive,active will depend on specific channels, and could depend on T and membrane electrical potential ϕm, and (cin, cout) where cin,out are concentrations inside and outside of the cell. For example, for the Na/K exchanger, which is an ATP-driven ion pump that exchanges 3Na+ with 2K+ ions, the flux depends on both intracellular and extracellular Na+ and K+ concentrations. The flux expression has been proposed, and is dependent on transmembrane voltage, ϕm [126]. Na+ flux is coupled to Cl− flux through Na-K-Cl co-transporters [108].
The remaining quantity to define is ΔP which according to equation (14) is
| (17) |
This introduce unknowns T and σa If σa is governed by a feedback signaling mechanism then
| (18) |
where K1,2 are phosphorylation/dephosphorylation rate constants. From properties of the signaling network, K1,2 can depend on membrane tension and ion concentrations inside the cell. It is also possible that shear stress within the cortex could signal to myosin, and K1 depend on σp, or myosin intrinsic mechanosensitivity [142]. Finally, membrane tension can be described by another constitutive relation, e.g.
| (19) |
where A0 is the unstretched membrane area, and A would be the current membrane area. A would be a function of R. Note that in cells, the membrane is likely highly folded, therefore the actual constitutive law must incorporate entropic elasticity, and the relationship between A and R maybe complicated [143, 144].
Equations (15)–(19) are a closed set of dynamic equations that describe changes in (R, n, σa, T) for given parameters (Np,A0, Kmax, cout) and associated flux constants in α and Ipassive, active. This system of equations has a steady state (Rss, nss, σa,ss, Tss), which are the homeostatic values of these quantities in the cell. This steady state should be stable against perturbations, and the phase space region around the steady state will describe how cells responds to changes in variables such as Np and cout.
S: Does this mean that we can predict cell volume for a given condition?
P: Precisely.
B: The details are significantly more complicated than described here. First, there are multiple types of permeable ions, at least Na+, K+ and Cl− are involved. Ca2+ is another secondary messenger. Furthermore, flux equations for Ipassive.active are generally not known. Since ions are charged, membrane potential ϕm also comes into play [128]. Since ionic species are coupled through co-transporters, flux couplings become very complicated. It is likely that cells have compensatory mechanisms when fluxes are disrupted. Currently there are no predictive models of ionic flux balance in cells.
S: So this problem is not completely understood at all!
5.3. Adherent cells: a test of the model
P: Spherical cells are not very common. But further verification of these ideas can come from examining adherent cells: cells that are growing on flat substrates. Let us consider a static axisymmetric cell confined on a circular substrate (figure 7). At steady state, T ≪ σah. Equation (14) then becomes: (∇ · n)−1 = hσa./ΔP For the coordinate system defined in figure 7(A), D, which describes the shape of the adherent cell, is a function of θ. ∇ · n can be expressed as a function of D(θ), we therefore have
| (20) |
where D′ = ∂D/∂θ and D″=∂2D/∂θ2. This is a nonlinear 2nd order ODE, therefore two boundary conditions (BCs) are needed to determine the cell shape. One of the BC is the adhered size, R0, which sets D(θ = 0) = R0. The other BC can be obtained by assuming the cell surface is smooth at θ = π/2. Notice σa, h all can be functions of θ, but if they are constants, then there is a single size parameter r0 = 2hσa/ΔP. In this case, if R0 = r0, the cell is a prefect hemisphere. If R0 > r0, the cell will be stretched at the top, with a flatter region near the substrate. Figure 7 shows some computed cell shape for different adhesion size R0. The result predicts that for the same r0, cells with larger spread area tend to have larger volume (figure 7(B)).
Figure 7.
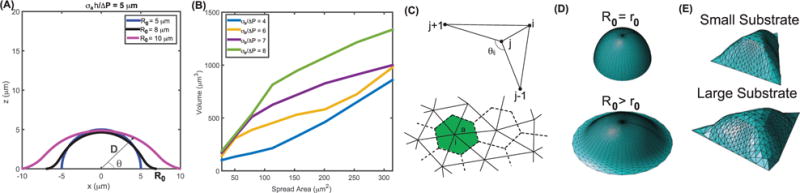
(A) Computed 3D shapes of adherent cells for different substrate adhesion size, R0. The cell surface is described by the function D(θ) (inset). Equation (20) predicts that the cell shape varies with increasing R0. (B) Cell volume increases as substrate size increases. The critical parameter governing the volume-area correlation is σa/ΔP. (C) Mean curvature can be approximately computed using triangular surfaces using equation (21). The triangulated surface can describe cell shape in arbitrary geometry. ((D), (E)) Computed 3D cell shapes using triangulation of cell surface for cells on circular and triangular substrates. Here r0 = 2σah/ΔP.
S: It seems that equation (14) can predict at lot of features of the cell. But you only mentioned about suspended or axisymmetric cell shape. What about non-axisymmetric cells?
P: Of course, equation (14) is applicable for all types of cells under any condition. But the distribution of σa is in general a function of space, and depends on local signaling activity of Rho, Rac GTPases. If we can measure hσa/ΔP as a function of space, then we can compute the cell shape for any adhesion geometry. For example, by triangulating the cell surface, we can compute the local mean curvature around the ith vertex as (figure 7(C)) [146, 147]
| (21) |
where ai is the area around the vertex, and the sum is over j vertices connected to the i-th vertex. lij and θij are the length of the connecting edge and the angle between triangles sharing the edge, respectively. Using a weak form of equation (14), we can compute the shape of the cell surface for a given hσa/ΔP for any substrate geometry (figure 7(C)).
The other unknown is ΔP, which if there are no protein and ionic gradients, ΔP should be spatially uniform. But we do not know whether it remains constant in all conditions. There has been efforts to measure intracellular ΔP [145]. In general, the right hand side of equation (20) is not clear, and generally depends on θ.
S: Ok, but why are we interested in cell shape dependence on substrate geometry? How would this influence important biological functions?
B: We know that substrate properties can influence many aspects of cell function [68, 148, 149]. For example, cell cycle progression seems to depend on substrate stiffness [150]. R0 seems to change depending on cell substrate properties. These changes also seem to influence mesenchyemal stem cell differentiation [151]. Therefore, many essential cell functions depend on substrate stiffness and substrate adhesion [67]. Extracellular matrix stiffness also influences cell migration and cancer metastasis [152], the list goes on… Indeed, gene expression profiles are changed depending on substrate adhesion and substrate stiffness [153]. From mechanics alone, we see that cell volume and cell shape depend on substrate adhesion and geometry. And since cell volume will influence all kinds of protein concentrations, there could be a physical basis to these biological changes [154]. There are lots of questions in this direction and further work is needed
5.4. Higher order models: cell and tissue mechanics
S: We see that water flux is influenced by hydrostatic and osmotic pressures equally. Therefore, it should be possible to drive water flow from externally applied mechanical forces too, right?
P: This is correct! In equation (14), if there is an externally applied mechanical force to the cell Fext, it simply adds to the force balance equation and modifies it to
| (22) |
This says that the hydrostatic pressure difference can be changed by externally applied mechanical force. A change in ΔP leads to water flux, and changes in osmotic pressure as well. Therefore, a complex response is initiated when an external force is applied to the cell [105, 155]. The response depends on the rate of deformation: since water flows across the membrane slowly, a rapidly applied force will generate deformations that essentially preserves the cell volume [105]. Alternatively, if the deformation is slow, then water will flow, leading to cell volume change. It appears that the relevant deformation time scale is μm s−1 [105, 116]. With this time scale or slower, cells can change volume significantly.
B: This time scale of deformation is important. During organismal development, most shape changes occur slowly. Therefore mechanical forces in developing tissues must influence water dynamics in cells and tissues [156–159].
S: In some of my readings, there are already proposed models for tissue mechanics. For example, an approach that relates cell volume and surface area with a ‘mechanical energy’ has been popular [160–166] (figure 8):
| (23) |
where the first sum is over cells, and the second sum is over cell-cell interfaces. The first term models cell volume change with respect to a reference V0. The second term is a contractility term, describing the propensity to shrink the cell surface area from myosin activity. The last term is a surface energy term. The surface energy term may model cell-cell contact energy arising from chemical bonds such as E/N Cadherins [167]. This model has used to describe cell motility in the confluent epithelium [163, 168, 169], and it has been shown that different combinations of surface energy term and contractility can lead to a jamming transition, i.e. a possible lost of global cell mobility within the epithelium [169]. How do we interpret this model, since the cell is an active object that does not obey energy conservation principles?
Figure 8.
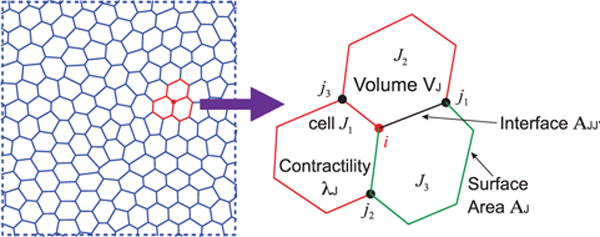
Epithelial cell layer mechanics. When cells form a confluent layer, the shape and forces within the tissue can be described by an energy formulation of the vertex model (equation (23)). The energy function describes cell-cell adhesion, cell volume change and cell surface contractility, and can be understood as a Lyapunov function of the shape and volume dynamical system described by equations (15)–(19).
P: Well, this is a very interesting point. This is a phenomenological model that roughly describes cell shape and volume dynamics using an energy. It could be interpreted as a Lyapunov function of the dynamical systems model, equations (15)–(19). Lyapunov function is an energy-like function that describes phase space flow near the steady state fixed point [170]. This function is generally difficult to find. For cells in normal physiological situations, since it actively controls its cell shape and volume, the steady state fixed point must be stable, and a Lyapunov function likely exists. In other contexts, the system may exhibit interesting limit cycle dynamics [171, 172] Therefore it would be interesting to connect the cell volume and cell shape model at the single cell level to the multi-cellular model in equation (23).
In addition, a continuum model for the tissue layer can be built from a discrete model in equation (23). Indeed, the material described by equation (23) can behave as a jammed solid or a fluid [166]. At long times, cell division and apoptosis relieves shear stress and overall tissue again behaves as a fluid [173].
6. Conclusions and perspectives
S: So mechanics has a lot to do with how cells move and control their size and shape. How cells generate force and respond to applied force depends on functions of cytoskeleton as well as water flow.
B: This is good, but the next level of question for biology is how these mechanical forces and cell shape information are integrated to gene expression and epigenetic modification. For example, we know that when mechanical forces are applied to the cell, the cell responds at the seconds time scale by opening and closing ion channels and pumps. At the minute time scale, signaling molecules such as Rho, Rac and ROCK integrates this information to regulate cytoskeleton activity and motor function [71, 174]. At time scales longer than minutes, however, we observe transcription factor movement across the nuclear envelope, and gene expression profiles of the cell starts to change [153, 175, 176]. Ultimately, these changes will modify cell cycle dynamics, and perhaps even the cell phenotype. Therefore, the connection between cell mechanics and genetic regulation is the critical step in advancing biology.
P: This is absolutely right. The connection between mechanics, forces and genes ultimately underlies organismal development, tissue morphogenesis and disease biology [151, 177]. Often, there is a circular loop between mechanics and genes. Transcriptional changes alter cell force generation; this then of course changes the cell organization, shape and motility, which further alter gene activity (figure 9(A)). The opposite loop can also occur: when forces and mechanics experienced by cells change, this could alter signaling pathways and modify gene activity, which then leads to protein expression change and alterations in cell force production and shape. Biology is probably best explained when we consider forces and mechanics, together with genes as a combined system.
Figure 9.
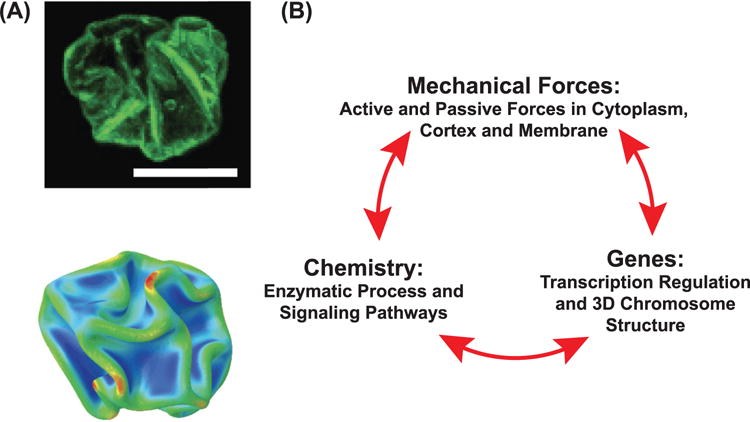
(A) Nuclear shape and volume is directly impacted by cell mechanics. The upper image is of a crumpled nucleus when a tissue cell is detaching from the substrate. The lower image is a computational analysis of an elastic shell undergoing crumpling transition when the pressure difference across the shell is increasing. (B) Mechanistic models that describe cell dynamics must incorporate cell mechanics, biochemistry and gene expression together in a single picture. These systems interact with each other and give rise to complex cell behavior in biological contexts.
B: P makes an excellent point. Some preliminary evidence could link cell mechanics with gene expression. For example, by changing the substrate size, the variable R0 in figure 7, an important transcription factor can change its localization between the cytoplasm and the nucleus [175]. We also know that the genome of the cell is organized and folded in a reasonably well defined 3D structure [178, 179]. The topology of this 3D structure seems to determine the epigenetic state of the cell. Indeed, genetic material itself seems to be a mechanically active material [180]. Mechanical forces of course, can alter the structure of biological molecules as well as chromatin [181–184]. It is reasonable to consider how forces might directly alter 3D chromatin organization, and some studies have demonstrated that there is a connection between nuclear mechanics and cytoskeletal mechanics [185–188]
P: Indeed, there seems to be experimental evidence that links cell volume with cell nuclear volume and shape (figure 9(B)). When cell volume decreases, the nuclear volume also shrinks [189]. Cytoskeletal networks are also physically connected to the nuclear envelop, and actively exert forces to shape the nucleus [185, 186, 188, 190–192]. This would compact the chromatin loops and change transcriptional program. Of course, mechanosensory molecules such as membrane channels, Rho, myosin and actin network itself may also directly communicate with nuclear transport and transcriptional factor localization, leading to direct connection between genes and cell mechanics.
S: Ahh, so many things to consider in order to understand biological organization. Where can physical models help?
B: From a biologist perspective, mathematical modeling provides a framework to think about functional relationship between measurable quantities. For example, a relationship between cell size and cell cortical tension is predicted by equation (14). We can then check this by measuring cell size and myosin activity. It is not necessarily possible to achieve quantitative prediction, but it is absolutely necessary to make predictions about how things maybe related to each other. These functional relationships are also opportunities for us to test the model.
P: From the modeling perspective, models are only as good as their assumptions! Therefore, experimental work that test assumptions are valuable. Some of the modeling assumptions are not easily understood, or even noticed! Therefore, in addition to testing functional relationships provided by the models, another way to test models is to simply test assumptions of the model, implicit or otherwise!
S: Great, let me get back to the lab and checkout what we talked about today. Next week, I will come back and show you what I find, and I’m sure I will have more questions!
Acknowledgments
The work is made possible by funding from National Institutes of Health: R01GM114675, U54CA210173 and T32CA153952.
Footnotes
(Some figures may appear in colour only in the online journal)
References
- 1.Blackstock WP, Weir M. Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol. 1999;17:121–7. doi: 10.1016/s0167-7799(98)01245-1. [DOI] [PubMed] [Google Scholar]
- 2.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin: dnase i complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 3.Bindschadler M, Osborn EA, Dewey CF, McGrath JL. A mechanistic model of the actin cycle. Biophys J. 2004;86:2720–39. doi: 10.1016/S0006-3495(04)74326-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–42. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 5.Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–76. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 6.Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann H, Bär H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–73. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- 8.Voeltz GK, Prinz WA. Sheets, ribbons and tubuleshow organelles get their shape. Nat Rev Mol Cell Biol. 2007;8:258–64. doi: 10.1038/nrm2119. [DOI] [PubMed] [Google Scholar]
- 9.Schliwa M, Woehlke G. Molecular motors. Nature. 2003;422:759–65. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- 10.Jülicher F, Ajdari A, Prost J. Modeling molecular motors. Rev Mod Phys. 1997;69:1269. [Google Scholar]
- 11.Kolomeisky AB, Fisher ME. Molecular motors: a theorist’s perspective. Annu Rev Phys Chem. 2007;58:675–95. doi: 10.1146/annurev.physchem.58.032806.104532. [DOI] [PubMed] [Google Scholar]
- 12.Goldman YE, Brenner B. Special topic: molecular mechanism of muscle contraction. Annu Rev Physiol. 1987;49:629–36. doi: 10.1146/annurev.ph.49.030187.003213. [DOI] [PubMed] [Google Scholar]
- 13.Walcott S, Sun SX. Hysteresis in cross-bridge models of muscle. Phys Chem Chem Phys. 2009;11:4871–81. doi: 10.1039/b900551j. [DOI] [PubMed] [Google Scholar]
- 14.Chaikin PM, Lubensky TC. Principles of Condensed Matter Physics. Cambridge: Cambridge University Press; 2000. [Google Scholar]
- 15.Gurtin ME. An Introduction to Continuum Mechanics. New York: Academic; 1981. [Google Scholar]
- 16.Pozrikidis C. Introduction to Theoretical and Computational Fluid Dynamics. Oxford; Oxford University Press; 2011. [Google Scholar]
- 17.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–65. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 18.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–12. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mogilner A, Oster G. Cell motility driven by actin polymerization. Biophys J. 1996;71:3030–45. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayot C, Debeir O, Van Ham P, Van Damme M, Kiss R, Decaestecker C. Characterization of the activities of actin-affecting drugs on tumor cell migration. Toxicol Appl Pharmacol. 2006;211:30–40. doi: 10.1016/j.taap.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Balzer EM, Tong Z, Paul CD, Hung W-C, Stroka KM, Boggs AE, Martin SS, Konstantopoulos K. Physical confinement alters tumor cell adhesion and migration phenotypes. FASEB J. 2012;26:4045–56. doi: 10.1096/fj.12-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theriot JA, Mitchison TJ. Actin microfilament dynamics in locomoting cells. Nature. 1991;352:126–31. doi: 10.1038/352126a0. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Ishihara A, Theriot JA, Jacobson K. Principles of locomotion for simple-shaped cells. Nature. 1993;362:167–71. doi: 10.1038/362167a0. [DOI] [PubMed] [Google Scholar]
- 24.Raynaud F, Ambühl ME, Gabella C, Bornert A, Sbalzarini IF, Meister J-J, Verkhovsky AB. Minimal model for spontaneous cell polarization and edge activity in oscillating, rotating and migrating cells. Nat Phys. 2016;12:367–73. [Google Scholar]
- 25.Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA. Mechanism of shape determination in motile cells. Nature. 2008;453:475–80. doi: 10.1038/nature06952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wirtz D. Particle-tracking microrheology of living cells: principles and applications. Annu Rev Biophys. 38:301–26. doi: 10.1146/annurev.biophys.050708.133724. [DOI] [PubMed] [Google Scholar]
- 27.Mason TG, Ganesan K, Van Zanten JH, Wirtz D, Kuo SC. Particle tracking microrheology of complex fluids. Phys Rev Lett. 1997;79:3282. [Google Scholar]
- 28.MacKintosh FC, Schmidt CF. Microrheology. Curr Opin Colloid Interface Sci. 1999;4:300–7. [Google Scholar]
- 29.Gardel ML, Shin JH, MacKintosh FC, Mahadevan L, Matsudaira P, Weitz DA. Elastic behavior of cross-linked and bundled actin networks. Science. 2004;304:1301–5. doi: 10.1126/science.1095087. [DOI] [PubMed] [Google Scholar]
- 30.Yamada S, Wirtz D, Kuo SC. Mechanics of living cells measured by laser tracking microrheology. Biophys J. 2000;78:1736–47. doi: 10.1016/S0006-3495(00)76725-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrlicher AJ, Krishnan R, Guo M, Bidan CM, Weitz DA, Pollak MR. Alpha-actinin binding kinetics modulate cellular dynamics and force generation. Proc Natl Acad Sci USA. 2015;112:6619–24. doi: 10.1073/pnas.1505652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jülicher F, Kruse K, Prost J, Joanny J-F. Active behavior of the cytoskeleton. Phys Rep. 2007;449:3–28. [Google Scholar]
- 33.Joanny J-F, Prost J. Active gels as a description of the actin-myosin cytoskeleton. HFSP J. 2009;3:94–104. doi: 10.2976/1.3054712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bray D, White JG. Cortical flow in animal cells. Science. 1988;239:883–8. doi: 10.1126/science.3277283. [DOI] [PubMed] [Google Scholar]
- 35.Tinevez J-Y, Schulze U, Salbreux G, Roensch J, Joanny J-F, Paluch E. Role of cortical tension in bleb growth. Proc Natl Acad Sci USA. 2009;106:18581–6. doi: 10.1073/pnas.0903353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Charras G, Paluch E. Blebs lead the way: how to migrate without lamellipodia. Nat Rev Mol Cell Biol. 2008;9:730–6. doi: 10.1038/nrm2453. [DOI] [PubMed] [Google Scholar]
- 37.Salbreux G, Charras G, Paluch E. Actin cortex mechanics and cellular morphogenesis. Trends Cell Biol. 2012;22:536–45. doi: 10.1016/j.tcb.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 38.Janmey PA, Hvidt S, Lamb J, Stossel T. Resemblance of actin-binding protein/actin gels to covalently crosslinked networks. Nature. 1990;345:89–92. doi: 10.1038/345089a0. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey D, Duggan C, Saha D, Smith D, Käs J. Active fluidization of polymer networks through molecular motors. Nature. 2002;416:413–6. doi: 10.1038/416413a. [DOI] [PubMed] [Google Scholar]
- 40.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science; 1994. [Google Scholar]
- 41.Wagner B, Tharmann R, Haase I, Fischer M, Bausch AR. Cytoskeletal polymer networks: the molecular structure of cross-linkers determines macroscopic properties. Proc Natl Acad Sci USA. 2006;103:13974–8. doi: 10.1073/pnas.0510190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ren Y, Effler JC, Norstrom M, Luo T, Firtel RA, Iglesias PA, Rock RS, Robinson D. Mechanosensing through cooperative interactions between myosin ii and the actin crosslinker cortexillin i. Curr Biol. 2009;19:1421–8. doi: 10.1016/j.cub.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vignjevic D, Kojima S-I, Aratyn Y, Danciu O, Svitkina T, Borisy GG. Role of fascin in filopodial protrusion. J Cell Biol. 2006;174:863–75. doi: 10.1083/jcb.200603013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–90. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mullins RD, Heuser JA, Pollard TD. The interaction of arp2/3 complex with actin: nucleation, high affinity pointed end capping, and formation of branching networks of filaments. Proc Natl Acad Sci USA. 1998;95:6181–6. doi: 10.1073/pnas.95.11.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–6. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 47.Danuser G, Waterman-Storer CM. Quantitative fluorescent speckle microscopy of cytoskeleton dynamics. Annu Rev Biophys Biomol Struct. 2006;35:361–87. doi: 10.1146/annurev.biophys.35.040405.102114. [DOI] [PubMed] [Google Scholar]
- 48.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–5. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 49.Rubinstein M, Colby RH. Polymer Physics. Oxford; Oxford University Press; 2003. [Google Scholar]
- 50.MacKintosh FC, Käs J, Janmey PA. Elasticity of semiflexible biopolymer networks. Phys Rev Lett. 1995;75:4425. doi: 10.1103/PhysRevLett.75.4425. [DOI] [PubMed] [Google Scholar]
- 51.Broedersz CP, MacKintosh FC. Modeling semiflexible polymer networks. Rev Mod Phys. 2014;86:995. [Google Scholar]
- 52.Marchetti MC, Joanny JF, Ramaswamy S, Liverpool TB, Prost J, Rao M, Simha RA. Hydrodynamics of soft active matter. Rev Mod Phys. 2013;85:1143. [Google Scholar]
- 53.Wottawah F, Schinkinger S, Lincoln B, Ananthakrishnan R, Romeyke M, Guck J, Käs J. Optical rheology of biological cells. Phys Rev Lett. 2005;94:098103. doi: 10.1103/PhysRevLett.94.098103. [DOI] [PubMed] [Google Scholar]
- 54.Kim T, Gardel ML, Munro E. Determinants of fluidlike behavior and effective viscosity in cross-linked actin networks. Biophys J. 2014;106:526–34. doi: 10.1016/j.bpj.2013.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bausch AR, Ziemann F, Boulbitch AA, Jacobson K, Sackmann E. Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry. Biophys J. 1998;75:2038–49. doi: 10.1016/S0006-3495(98)77646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10:1393–400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ponti A, Vallotton P, Salmon WC, Waterman-Storer CM, Danuser G. Computational analysis of F-actin turnover in cortical actin meshworks using fluorescent speckle microscopy. Biophys J. 2003:3336–52. doi: 10.1016/S0006-3495(03)70058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dix JA, Verkman AS. Mapping of fluorescence anisotropy in living cells by ratio imaging. Application to cytoplasmic viscosity. Biophys J. 1990;57:231. doi: 10.1016/S0006-3495(90)82526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gardel ML, Kasza KE, Brangwynne CP, Liu J, Weitz DA. Mechanical response of cytoskeletal networks. Meth Cell Biol. 2008;89:487–519. doi: 10.1016/S0091-679X(08)00619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature. 2005;435:191–4. doi: 10.1038/nature03521. [DOI] [PubMed] [Google Scholar]
- 61.Mizuno D, Tardin C, Schmidt CF, MacKintosh FC. Nonequilibrium mechanics of active cytoskeletal networks. Science. 2007;315:370–3. doi: 10.1126/science.1134404. [DOI] [PubMed] [Google Scholar]
- 62.Walcott S, Sun SX. A mechanical model of actin stress fiber formation and substrate elasticity sensing in adherent cells. Proc Natl Acad Sci USA. 2010;107:7757–62. doi: 10.1073/pnas.0912739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srinivasan M, Walcott S. Binding site models of friction due to the formation and rupture of bonds: state-function formalism, force-velocity relations, response to slip velocity transients, and slip stability. Phys Rev E. 2009;80:046124. doi: 10.1103/PhysRevE.80.046124. [DOI] [PubMed] [Google Scholar]
- 64.Walcott S, Sun SX. Active force generation in cross-linked filament bundles without motor proteins. Phys Rev E. 2010;82:050901. doi: 10.1103/PhysRevE.82.050901. [DOI] [PubMed] [Google Scholar]
- 65.Amano M, Chihara K, Kimura K, Fukata Y, Nakamura N, Matsuura Y, Kaibuchi K. Formation of actin stress fibers and focal adhesions enhanced by Rho-Kinase. Science. 1997;275:1308–11. doi: 10.1126/science.275.5304.1308. [DOI] [PubMed] [Google Scholar]
- 66.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol. 2006;173:383–94. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parsons JT, Horwitz AR, Schwartz MA. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat Rev Mol Cell Biol. 2010;11:633–43. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geiger B, Spatz JP, Bershadsky AD. Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol. 2009;10:21–33. doi: 10.1038/nrm2593. [DOI] [PubMed] [Google Scholar]
- 69.Chan CE, Odde DJ. Traction dynamics of filopodia on compliant substrates. Science. 2008;322:1687–91. doi: 10.1126/science.1163595. [DOI] [PubMed] [Google Scholar]
- 70.Prost J, Jülicher F, Joanny JF. Active gel physics. Nat Phys. 2015;11:111–7. [Google Scholar]
- 71.Machacek M, Hodgson L, Welch C, Elliot H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Met Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 73.Murrell M, Oakes PW, Lenz M, Gardel ML. Forcing cells into shape: the mechanics of actomyosin contractility. Nat Rev Mol Cell Biol. 2015;16:486–98. doi: 10.1038/nrm4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lenz M, Gardel ML, Dinner AR. Requirements for contractility in disordered cytoskeletal bundles. New J Phys. 2012;14:033037. doi: 10.1088/1367-2630/14/3/033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lenz M, Thoresen T, Gardel ML, Dinner AR. Contractile units in disordered actomyosin bundles arise from F-actin buckling. Phys Rev Lett. 2012;108:238107. doi: 10.1103/PhysRevLett.108.238107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yogurtcu ON, Kim J-S, Sun SX. A mechanochemical model of actin filaments. Biophys J. 2012;103:719–27. doi: 10.1016/j.bpj.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun SX, Walcott S, Wolgemuth CW. Cytoskeletal cross-linking and bundling in motor-independent contraction. Curr Biol. 2010;20:R649–54. doi: 10.1016/j.cub.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Naganathan SR, Fürthauer S, Nishikawa M, Jülicher F, Grill SW. Active torque generation by the actomyosin cell cortex drives left-right symmetry breaking. Elife. 2014;3:e04165. doi: 10.7554/eLife.04165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fürthauer S, Strempel M, Grill SW, Jülicher F. Active chiral processes in thin films. Phys Rev Lett. 2013;110:048103. doi: 10.1103/PhysRevLett.110.048103. [DOI] [PubMed] [Google Scholar]
- 80.Mayer M, Depken M, Bois JS, Jülicher F, Grill SW. Anisotropies in cortical tension reveal the physical basis of polarizing cortical flows. Nature. 2010;467:617–21. doi: 10.1038/nature09376. [DOI] [PubMed] [Google Scholar]
- 81.Fürthauer S, Strempel M, Grill SW, Jülicher F. Active chiral fluids. Eur Phys J E. 2012;35:1–13. doi: 10.1140/epje/i2012-12089-6. [DOI] [PubMed] [Google Scholar]
- 82.Shin JH, Gardel ML, Mahadevan L, Matsudaira P, Weitz DA. Relating microstructure to rheology of a bundled and cross-linked F-actin network. in vitro Proc Natl Acad Sci USA. 2004;101:9636–41. doi: 10.1073/pnas.0308733101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc Natl Acad Sci USA. 2006;103:1762–7. doi: 10.1073/pnas.0504777103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bendix PM, Koenderink GH, Cuvelier D, Dogic Z, Koeleman BN, Brieher WM, Field CM, Mahadevan L, Weitz DA. A quantitative analysis of contractility in active cytoskeletal protein networks. Biophys J. 2008;94:3126–36. doi: 10.1529/biophysj.107.117960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Silva MS, Depken M, Stuhrmann B, Korsten M, MacKintosh FC, Koenderink GH. Active multistage coarsening of actin networks driven by myosin motors. Proc Natl Acad Sci USA. 2011;108:9408–13. doi: 10.1073/pnas.1016616108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alvarado J, Sheinman M, Sharma A, MacKintosh FC, Koenderink GH. Molecular motors robustly drive active gels to a critically connected state. Nat Phys. 2013;9:591–7. [Google Scholar]
- 87.Lodish H, Berk A, Matsudaira P, Kaiser CA, Krieger M, Scott MP, Zipursky L, Darnell J. Molecular Cell Biology. 5th. New York: Garland Science; 2004. [Google Scholar]
- 88.Moeendarbary E, Valon L, Fritzsche M, Harris AR, Moulding DA, Thrasher AJ, Stride E, Mahadevan L, Charras GT. The cytoplasm of living cells behaves as a poroelastic material. Nat Mater. 2013;12:253–61. doi: 10.1038/nmat3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agre P. The aquaporin water channels. Proc Am Thorac Soc. 2006;3:5–13. doi: 10.1513/pats.200510-109JH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kruse E, Uehlein N, Kaldenhoff R. The aquaporins. Genome Biol. 2006;7:1. doi: 10.1186/gb-2006-7-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Katchalsky K, Curran PF. Nonequilibrium Thermodynamics in Biophysics. Cambridge, MA: Harvard University Press; 1965. [Google Scholar]
- 92.Weinstein AM, Stephenson JL. Models of coupled salt and water transport across leaky epithelia. J Membr Biol. 1981;60:1–20. doi: 10.1007/BF01870828. [DOI] [PubMed] [Google Scholar]
- 93.Stevens LA, Coresh J, Greene T, Levey AS. Assessing kidney functionmeasured and estimated glomerular filtration rate. New Engl J Med. 2006;354:2473–83. doi: 10.1056/NEJMra054415. [DOI] [PubMed] [Google Scholar]
- 94.Charras GT, Yarrow JC, Horton MA, Mahadevan L, Mitchison TJ. Non-equilibration of hydrostatic pressure in blebbing cells. Nature. 2005;435:365–9. doi: 10.1038/nature03550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taloni A, Kardash E, Salman OU, Truskinovsky L, Zapperi S, La Porta CAM. Phys Rev Lett. 2015;114:208101. doi: 10.1103/PhysRevLett.114.208101. [DOI] [PubMed] [Google Scholar]
- 96.Lämmermann T, Sixt M. Mechanical modes of amoeboidcell migration. Curr Opin Cell Biol. 2009;21:636–44. doi: 10.1016/j.ceb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 97.Ruprecht V, et al. Cortical contractility triggers a stochastic switch to fast amoeboid cell motility. Cell. 2015;160:673–85. doi: 10.1016/j.cell.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Petrie RJ, Yamada KM. At the leading edge of three-dimensional cell migration. J Cell Sci. 2012;125:5917–26. doi: 10.1242/jcs.093732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Paluch EK, Raz E. The role and regulation of blebs in cell migration. Curr Opin Cell Biol. 2013;25:582–90. doi: 10.1016/j.ceb.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zatulovskiy E, Tyson R, Bretschneider T, Kay RR. Bleb-driven chemotaxis of dictyostelium cells. J Cell Biol. 2014;204:1027–44. doi: 10.1083/jcb.201306147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Atzberger PJ, Isaacson S, Peskin CS. A microfluidic pumping mechanism driven by non-equilibrium osmotic effects. Physica D. 2009;238:1168–79. [Google Scholar]
- 102.Yip NY, Elimelech M. Thermodynamic and energy efficiency analysis of power generation from natural salinity gradients by pressure retarded osmosis. Environ Sci Technol. 2012;46:5230–9. doi: 10.1021/es300060m. [DOI] [PubMed] [Google Scholar]
- 103.Feng J, Graf M, Liu K, Ovchinnikov D, Dumcenco D, Heiranian M, Nandigana V, Aluru NR, Kis A, Radenovic A. Single-layer mos2 nanopores as nanopower generators. Nature. 2016;536:197–200. doi: 10.1038/nature18593. [DOI] [PubMed] [Google Scholar]
- 104.Hoffmann EK, Lambert IH, Pedersen SF. Physiology of cell volume regulation in vertebrates. Physiol Rev. 2009;89:193–277. doi: 10.1152/physrev.00037.2007. [DOI] [PubMed] [Google Scholar]
- 105.Jiang H, Sun SX. Cellular pressure and volume regulation and implications for cell mechanics. Biophys J. 2013;105:609–19. doi: 10.1016/j.bpj.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jaynes ET. The gibbs paradox Maximum Entropy and Bayesian Methods. New York: Springer; 1992. pp. 1–21. [Google Scholar]
- 107.Bueno-Orovio A, Sánchez C, Pueyo E, Rodriguez B. Na/K pump regulation of cardiac repolarization: insights from a systems biology approach. Pflugers Arch. 2014;466:183–93. doi: 10.1007/s00424-013-1293-1. [DOI] [PubMed] [Google Scholar]
- 108.Russell JM. Sodium-potassium-chloride cotransport. Physiol Rev. 2000;80:211–76. doi: 10.1152/physrev.2000.80.1.211. [DOI] [PubMed] [Google Scholar]
- 109.Demaurex N, Grinstein S. Na + /H + antiport: modulation by ATP and role in cell volume regulation. J Exp Biol. 1994;196:389–404. doi: 10.1242/jeb.196.1.389. [DOI] [PubMed] [Google Scholar]
- 110.Bianchini L, Kapus A, Lukacs G, Wasan S, Wakabayashi S, Pouyssegur J, Yu FH, Orlowski J, Grinstein S. Responsiveness of mutants of NHE1 isoform of Na + /H + isoform of Na + /H + antiport to osmotic stress. Am J Physiol. 1995;269:C998–1007. doi: 10.1152/ajpcell.1995.269.4.C998. [DOI] [PubMed] [Google Scholar]
- 111.Stewart MP, Helenius J, Toyoda Y, Ramanathan SP, Muller DJ, Hyman AA. Hydrostatic pressure and the actomyosin cortex drive mitotic cell rounding. Nature. 2011;469:226–30. doi: 10.1038/nature09642. [DOI] [PubMed] [Google Scholar]
- 112.Hui TH, Zhou ZL, Qian J, Lin Y, Ngan AHW, Gao H. Volumetric deformation of live cells induced by pressure-activated cross-membrane ion transport. Phys Rev Lett. 2014;113:118101. doi: 10.1103/PhysRevLett.113.118101. [DOI] [PubMed] [Google Scholar]
- 113.Zlotek-Zlotkiewicz E, Monnier S, Cappello G, Le Berre M, Piel M. Optical volume and mass measurements show that mammalian cells swell during mitosis. J Cell Biol. 2015;211:765–74. doi: 10.1083/jcb.201505056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Grover WH, Bryan AK, Diez-Silva M, Suresh S, Higgins JM, Manalis SR. Measuring single-cell density. Proc Natl Acad Sci USA. 2011;108:10992–6. doi: 10.1073/pnas.1104651108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Son S, Kang JH, Oh S, Kirchner MW, Mitchison TJ, Manalis S. Resonant microchannel volume and mass measurements show that suspended cells swell during mitosis. J Cell Biol. 2015;211:757–63. doi: 10.1083/jcb.201505058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tao J, Sun SX. Active biochemical regulation of cell volume and a simple model of cell tension response. Bio phys J. 2015;109:1541–50. doi: 10.1016/j.bpj.2015.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pan D. The hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yu F, Guan K. The Hippo pathway: regulators and regulations. Gene Dev. 2013;27:355–71. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–93. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Panopoulos A, Howell M, Fotedar R, Margolis RL. Glioblastoma motility occurs in the absence of actin polymer. Mol Biol Cell. 2011;22:2212–20. doi: 10.1091/mbc.E10-10-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stroka KM, Jiang H, Chen S-H, Tong Z, Wirtz D, Sun SX, Konstantopoulos K. Water permeation drives tumor cell migration in confined microenvironments. Cell. 2014;157:611–23. doi: 10.1016/j.cell.2014.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mitchell P. Hypothetical thermokinetic and electrokinetic mechanisms of locomotion in micro-organisms. Proc R Phys Soc. 1956;25:32–4. (Edinburgh) [Google Scholar]
- 123.Harold FM. Pumps and currents: a biological perspective. In: Slayman CL, editor. Current Topics in Membranes and Transport. Vol. 16. New York: Academic; 1982. [Google Scholar]
- 124.Lammert PE, Prost J, Bruinsma R. Ion drive for vesicles and cells. J Theor Biol. 1996;178:387–91. [Google Scholar]
- 125.Prentice-Mott HV, Chang C-H, Mahadevan L, Mitchison TJ, Irimia D, Shah J. Biased migration of confined neutrophil-like cells in asymmetric hydraulic environments. Proc Natl Acad Sci USA. 2013;110:21006–11. doi: 10.1073/pnas.1317441110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Armstrong CM. The Na/K pump, clion, and osmotic stabilization of cells. Proc Natl Acad Sci USA. 2003;100:6257–62. doi: 10.1073/pnas.0931278100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chang H, Fujita T. A kinetic model of the thiazide-sensitive Na–Cl cotransporter. Am J Physiol. 1999;276:F952–9. doi: 10.1152/ajprenal.1999.276.6.F952. [DOI] [PubMed] [Google Scholar]
- 128.Li Y, Mori Y, Sun SX. Flow-driven cell migration under external electric fields. Phys Rev Lett. 2015;115:268101. doi: 10.1103/PhysRevLett.115.268101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Allen GM, Mogilner A, Theriot JA. Electrophoresis of cellular membrane components creates the directional cue guiding keratocyte galvanotaxis. Curr Biol. 2013;23:560–8. doi: 10.1016/j.cub.2013.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cohen DJ, Nelson WJ, Maharbiz MM. Galvanotactic control of collective cell migration in epithelial monolayers. Nat Mater. 2014;13:409–17. doi: 10.1038/nmat3891. [DOI] [PubMed] [Google Scholar]
- 131.Keren K, Yam PT, Kinkhabwala A, Mogilner A, Theriot JA. Intracellular fluid flow in rapidly moving cells. Nat Cell Biol. 2009;11:1219–24. doi: 10.1038/ncb1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schwab A, Nechyporuk-Zloy V, Fabian A, Stock C. Cells move when ions and water flow. Pflugers Arch. 2007;453:421–32. doi: 10.1007/s00424-006-0138-6. [DOI] [PubMed] [Google Scholar]
- 133.Hu J, Verkman AS. Increased migration and metastatic potential of tumor cells expressing aquaporin water channels. FASEB J. 2006;20:1892–4. doi: 10.1096/fj.06-5930fje. [DOI] [PubMed] [Google Scholar]
- 134.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–88. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clark AG, Dierkes K, Paluch EK. Monitoring actin cortex thickness in live cells. Biophys J. 2013;105:570–80. doi: 10.1016/j.bpj.2013.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spivak M. Calculus on Manifolds a Modern Approach to Classical Theorems of Advanced Calcu. Oxford; Oxford University Press; 1966. [Google Scholar]
- 137.Baines AJ. Evolution of spectrin function in cytoskeletal and membrane networks. Biochem Soc Trans. 2009;37:796–803. doi: 10.1042/BST0370796. [DOI] [PubMed] [Google Scholar]
- 138.Sokabe T, Fukumi-Tominaga T, Yonemura S, Mizuno A, Tominaga M. The trpv4 channel contributes to intercellular junction formation in keratinocytes. J Biol Chem. 2010;285:18749–58. doi: 10.1074/jbc.M110.103606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seminario-Vidal L, et al. Rho signaling regulates pannexin 1-mediated atp release from airway epithelia. J Biol Chem. 2011;286:26277–86. doi: 10.1074/jbc.M111.260562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gauthier NC, Masters TA, Sheetz M. Mechanical feedback between membrane tension and dynamics. Trends Cell Biol. 2012;22:527–35. doi: 10.1016/j.tcb.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 141.Dai J, Sheetz M. Membrane tether formation from blebbing cells. Biophys J. 1999;77:3363–70. doi: 10.1016/S0006-3495(99)77168-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Luo T, Mohan K, Iglesias PA, Robinson D. Molecular mechanisms of cellular mechanosensing. Nat Mater. 12(2013):1064–71. doi: 10.1038/nmat3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Peliti L, Leibler S. Effects of thermal fluctuations on systems with small surface tension. Phys Rev Lett. 1985;54:1690. doi: 10.1103/PhysRevLett.54.1690. [DOI] [PubMed] [Google Scholar]
- 144.Barbetta C, Imparato A, Fournier J-B. On the surface tension of fluctuating quasi-spherical vesicles. Eur Phys J E. 2010;31:333. doi: 10.1140/epje/i2010-10579-1. [DOI] [PubMed] [Google Scholar]
- 145.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–5. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Noguchi H, Gompper G. Shape transitions of fluid vesicles and red blood cells in capillary flows. Proc Natl Acad Sci USA. 2005;102:14159–64. doi: 10.1073/pnas.0504243102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Atilgan E, Sun SX. Shape transitions in lipid membranes and protein mediated vesicle fusion and fission. J Chem Phys. 2007;126:095102. doi: 10.1063/1.2483862. [DOI] [PubMed] [Google Scholar]
- 148.Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 149.Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci. 2006;119:508–18. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- 150.Klein EA, Yin L, Kothapalli D, Castagnino P, Byfield FJ, Xu T, Levental I, Hawthorne E, Janmey PA, Assoian RK. Cell-cycle control by physiological matrix elasticity and in vivo tissue stiffening. Curr Biol. 2009;19:1511–8. doi: 10.1016/j.cub.2009.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 152.Levental KR, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell. 2009;139:891–906. doi: 10.1016/j.cell.2009.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Provenzano PP, Inman DR, Eliceiri KW, Keely PJ. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a fak–erk linkage. Oncogene. 2009;28:4326–43. doi: 10.1038/onc.2009.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Meyers J, Craig J, Odde DJ. Potential for control of signaling pathways via cell size and shape. Curr Biol. 2006;16:1685–93. doi: 10.1016/j.cub.2006.07.056. [DOI] [PubMed] [Google Scholar]
- 155.Fischer-Friedrich E, Hyman AA, Jülicher F, Müller DJ, Helenius J. Quantification of surface tension and internal pressure generated by single mitotic cells. Sci Rep. 2014;4:6213. doi: 10.1038/srep06213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Osterfield M, Du X, Sch T, Wieschaus E, Shvartsman SY. Three-dimensional epithelial morphogenesis in the developing Drosophila egg. Dev Cell. 2013;24:400–10. doi: 10.1016/j.devcel.2013.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shraiman BI. Mechanical feedback as a possible regulator of tissue growth. Proc Natl Acad Sci USA. 2005;102:3318–23. doi: 10.1073/pnas.0404782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gelbart MA, He B, Martin AC, Thiberge SY, Wieschaus EF, Kaschube M. Volume conservation principle involved in cell lengthening and nucleus movement during tissue morphogenesis. Proc Natl Acad Sci. 2012;109:19298–303. doi: 10.1073/pnas.1205258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saias L, Swoger J, D’Angelo A, Colombelli J, Sharpe J, Salbreux G, Solon J. Decrease in cell volume generates contractile forces driving dorsal closure. Dev Cell. 2015;33:611–21. doi: 10.1016/j.devcel.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 160.Marée AFM, Grieneisen VA, Hogeweg P. Single-Cell-Based Models in Biology and Medicine. New York: Springer; 2007. The cellular potts model and biophysical properties of cells, tissues and morphogenesis; pp. 107–36. [Google Scholar]
- 161.Graner F, Glazier JA. Simulation of biological cell sorting using a two-dimensional extended potts model. Phys Rev Lett. 1992;69:2013. doi: 10.1103/PhysRevLett.69.2013. [DOI] [PubMed] [Google Scholar]
- 162.Honda H. Description of cellular patterns by dirichlet domains: the two-dimensional case. J Theor Biol. 1978;72:523–43. doi: 10.1016/0022-5193(78)90315-6. [DOI] [PubMed] [Google Scholar]
- 163.Li B, Sun SX. Coherent motions in confluent cell monolayer sheets. Biophys J. 2014;107:1532–41. doi: 10.1016/j.bpj.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Farhadifar R, Röper J-C, Aigouy B, Eaton S, Jülicher F. The influence of cell mechanics, cell-cell interactions, and proliferation on epithelial packing. Curr Biol. 2007;17:2095–104. doi: 10.1016/j.cub.2007.11.049. [DOI] [PubMed] [Google Scholar]
- 165.Du X, Osterfield M, Shvartsman SY. Computational analysis of three-dimensional epithelial morphogenesis using vertex models. Phys Biol. 2014;11:06607. doi: 10.1088/1478-3975/11/6/066007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bi D, Lopez JH, Schwarz JM, Manning ML. A density-independent rigidity transition in biological tissues. Nat Phys. 2015;11:1074–9. [Google Scholar]
- 167.Borghi B, Sorohina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at the cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci. 2012;109:12568–73. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Loza AJ, Koride S, Schimizzi GV, Li B, Sun SX, Longmore GD. Cell density and actomyosin contractility control the organization of migrating collectives within an epithelium. Mol Biol Cell. 2016;27:3459–70. doi: 10.1091/mbc.E16-05-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bi D, Yang X, Marchetti MC, Manning ML. Motility-driven glass and jamming transitions in biological tissues. Phys Rev X. 2016;6:021011. doi: 10.1103/PhysRevX.6.021011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Strogatz SH. Nonlinear Dynamics and Chaos: with Applications to Physics, Biology, Chemistry, and Eng ineering. Cambridge, MA: Westview Press; 2014. [Google Scholar]
- 171.Salbreux G, Joanny JF, Prost J, Pullarkat P. Shape oscillations of non-adhering fibroblast cells. Phys Biol. 2007;4:268–84. doi: 10.1088/1478-3975/4/4/004. [DOI] [PubMed] [Google Scholar]
- 172.Kücken M, Soriano J, Pullarkat PA, Ott A, Nicola EM. An osmoregulatory basis for shape oscillations in regenerating hydra. Biophys J. 2008;95:978–85. doi: 10.1529/biophysj.107.117655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ranft J, Basan M, Elgeti J, Joanny JF, Prost J, Julicher F. Fluidization of tissues by cell division apoptosis. Proc Natl Acad Sci USA. 2010;107:20863–8. doi: 10.1073/pnas.1011086107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Amano M, Nakayama M, Kaibuchi K. Rho-kinase/rock: a key regulator of the cytoskeleton and cell polarity. Cytoskeleton. 2010;67:545–54. doi: 10.1002/cm.20472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dupont S, et al. Role of yap/taz in mechanotransduction. Nature. 2011;474:179–83. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 176.Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by yap and taz. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- 177.Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–2. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- 178.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rao SSP, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–80. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ganai N, Sengupta S, Menon GI. Chromosome positioning from activity-based segregation. Nucl Acids Res. 2014;42:4145–59. doi: 10.1093/nar/gkt1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosome fluctuations govern the transcriptional dynamics of RNA polymerase II. Science. 2009;325:626–9. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fazal FM, Block SM. Optical tweezers study life under tension. Nat Photon. 2011;5:318–21. doi: 10.1038/nphoton.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Smith SB, Finzi L, Bustamante C. Direct mechanical measurement of the elasticity of single DNA-molecules by using magnetic beads. Science. 1992;258:1122–6. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 184.Celedon A, Nodelman IM, Wildt B, Dewan R, Searson P, Wirtz D, Bowman GD, Sun SX. Magnetic tweezers measurement of single molecule torque. Nano Lett. 2009;9:1720–5. doi: 10.1021/nl900631w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Khatau SB, et al. Sci Rep. 2012;2:488. doi: 10.1038/srep00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Qingsen L, Kumar A, Makhija E, Shivashankar G. The regulation of dynamic mechanical coupling between actin cytoskeleton and nucleus by matrix geometry. Biomaterials. 2014;35:961–69. doi: 10.1016/j.biomaterials.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 187.Jain N, Iyer KV, Kumar A, Shivashankar G. Cell geometric constraints induce modular gene-expression patterns via redistribution of HDAC3 regulated by actomyosin contractility. Proc Natl Acad Sci USA. 2013;110:11349–54. doi: 10.1073/pnas.1300801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fabrikant G, Gupta S, Shivashankar GV, Kozlov M. Model of T-cell nuclear deformation by the cortical actin layer. Biophys J. 2013;105:1316–23. doi: 10.1016/j.bpj.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Kim D-H, Li B, Si F, Phillip JM, Wirtz D, Sun SX. Volume regulation and shape bifurcation in the cell nucleus. J Cell Sci. 2015;128:3375–85. doi: 10.1242/jcs.166330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lammderding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest. 2004;113:370–8. doi: 10.1172/JCI19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Swift J, et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear lamin A/C deficiency induces defects in cell mechanics, polarization, and migration. Biophys J. 2007;93:2542–52. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]


