Abstract
The stalk region of the influenza virus hemagglutinin is relatively well conserved compared with the globular head domain, which makes it a potential target for use as a universal vaccine against influenza. However, the role of CD4 T cells in the hemagglutinin stalk-specific immune response is not clear. Here we identified a mouse CD4 T-cell epitope that encompasses residues HA2113-131 from the hemagglutinin stalk domain after a sub-lethal infection of influenza. In response to stimulation with the identified epitope, splenocytes derived from the infected mice showed significant polyfunctionality as shown by IL-2, TNF-α and IFN-γ production as well as degranulation. Moreover, mice immunized with the peptide corresponding to this CD4 T-cell epitope exhibited interindividual sharing of the CD4 T-cell receptor β sequences, and they had a higher survival rate following a challenge with a lethal dose of pandemic H1N1 influenza virus. Thus, our data demonstrated a crucial role of hemagglutinin stalk-specific CD4 T cells in the host immune response against influenza virus infection.
Keywords: H1N1, hemagglutinin, influenza, T-cell receptor
Introduction
The influenza virus causes respiratory infections in humans, resulting in significant morbidity and mortality every year worldwide. The entry of the influenza virus into host cells is mediated by its surface glycoprotein, hemagglutinin (HA), which has become a major target for neutralizing antibodies and vaccine design. Each monomer of this trimeric protein is post-translationally cleaved by host proteases into two subunits, HA1 and HA2. The membrane-distal globular domain of HA lies in the HA1 subunit and contains the receptor-binding domain as well as most of the antigenic sites, which avoid neutralization by existing host antibodies through the accumulation of mutations, leading to seasonal influenza outbreaks.1, 2 Compared with the globular domain, the membrane-proximal stalk domain of HA is relatively conserved among influenza viruses. The HA stalk domain contains the N- and the C-terminal domains of HA1 and the N-terminal domain of the HA2 subunit, and together, they form a stalk-like structure that anchors the globular domain to the cellular or viral membrane.3, 4 Although in the native virus the epitopes of the HA stalk domain are less accessible to antibodies than those of the globular domain, HA stalk-specific antibodies are made during natural infection in humans as well as in mice.5 Recently, a number of monoclonal antibodies against the stalk domain have been isolated, and many of them show broad neutralizing activity against multiple influenza virus strains.6, 7, 8, 9, 10, 11
In addition to neutralizing antibodies, protective immunity against the influenza virus is also mediated by cellular immune responses. For example, a previous vaccine study in BALB/c mice demonstrated that the induction of CD8 T-cell responses targeting an epitope on the HA stalk domain of the PR8 virus (A/PR/8/34), which spans residues HA533-541 (IYSTVASSL), leads to effective protection against lethal infection by heterologous strains of the influenza virus.12 Thus, antigen-specific CD8 effector T cells are implicated in the control of pulmonary inflammation and viral clearance, whereas CD4 T cells are mostly known to have a role in maintaining CD8 T-cell and B-cell memory responses.13, 14 However, many recent studies have demonstrated that independent of antibodies, influenza-specific CD4 T cells with cytotoxic characteristics closely correlate with viral clearance and limitations on disease severity,15, 16, 17, 18 implying an essential role of CD4 T cell in protection against influenza. It has been shown that influenza-specific CD4 T cells that accumulated in the lungs before viral clearance can secrete a significantly greater amount of IFN-γ,19 suggesting that type 1 T helper cells (Th1) are critical for recovery. Thus, CD4 effector T cells could be a vital component of vaccine-induced protection against influenza infection.
To further dissect the role of influenza-specific CD4 T cells in protective immunity, it is essential to identify influenza HA CD4 T-cell epitopes. We focused on the epitope mapping of the stalk protein because little is known about stalk-specific CD4 T cells. In a BALB/c mouse model, we mapped an epitope to residues HA2 113-131 in the long α-helix region of the HA stalk domain. We showed that immunization with this CD4 T-cell epitope provides partial protection against a lethal pH1N1 challenge and that this was mainly due to cytotoxic polyfunctionality, such as perforin-dependent cytotoxicity and IFN-γ production. Previous studies have demonstrated that the expansion of certain virus-specific T-cell receptors (TCRs) is imperative to confer protection against viral infection.20, 21 Hence, we performed high-throughput sequencing of a TCR repertoire to investigate the vaccine-driven expansion of specific TCR clonotypes. Our data clearly showed an expansion of the shared CD4 TCRβ clonotypes that respond to the identified CD4 T-cell epitope in immunized mice with and without a virus challenge, implying that these shared TCRβ clonotypes may act as reliable predictors of protective CD4 T-cell immune responses. Collectively, the identification of the CD4 T-cell epitope in the conserved stalk domain of the HA protein not only facilitates our understanding of HA stalk-specific CD4 T cells but also represents a tool for evaluating the protective CD4 T-cell responses against the HA stalk following vaccination.
Materials and methods
Mice and infection
We purchased female BALB/c mice from Harlan Laboratories, Inc. (Horst, The Netherlands) and performed all the mouse experiments in accordance with protocols approved by the Animal Welfare Structure of the Luxembourg Institute of Health. For virus infection, 10-week-old BALB/c mice were intranasally infected with a sublethal dose (8 × 104 half maximal tissue culture infectious dose; TCID50) of pandemic H1N1 (pH1N1) influenza strain A/Luxembourg/46/2009 in 50 μl phosphate-buffered saline (PBS) after anesthesia with isoflurane. All experiments using the live influenza virus were conducted in a biosafety level 3 containment facility.
HA stalk peptide library
A panel of 15-mer peptides overlapping by 13 residues and covering the entire sequence of the HA stalk protein from the pH1N1 virus (A/Luxembourg/46/2009) were synthesized by a MultiPep RS peptide synthesizer (Intavis AG, Tuebingen, Germany) using a modified SPOT synthesis protocol. The peptides were neither conjugated nor modified otherwise. They were lyophilized in a 96-well plate and were resuspended in 1 ml ultrapure water to generate a stock solution with concentration of 1 mg/ml. For the in vitro ELISPOT assay, the peptide stocks were diluted 1:100 in RPMI-1640 medium (Gibco-BRL, Gaithersburg, MD, USA) supplemented with 10% FCS, 2 mM l-glutamine, 5 mM HEPES, 50 μg/ml gentamicin and 50 μg/ml penicillin–streptomycin.
Antibodies and flow cytometric analysis
Mice were killed by intraperitoneal injection of 200 μg/mg of natrium pentobarbital, and the spleens were then excised. The splenocytes were incubated with anti-Fcγ receptor (2.4G2) followed by surface staining with anti-CD49b (DX5; BioLegend, San Diego, CA, USA), anti-CD3e (142-2C11), anti-CD4 (RM4-5), anti-CD8 (53-6.7) and anti-CD14 (mC5-3; all from BD Biosciences, San Jose, CA, USA) for phenotypic analyses and sorting. We excluded dead cells by using the APC-Cy7 Live/Dead stain kit (Invitrogen, Carlsbad, CA, USA).
The magnitude and polyfunctionality of the HA stalk-specific T-cell responses were determined using intracellular cytokine staining. In brief, 4 × 106 splenocytes were cultured in 96-well plates and stimulated for 6 h in 10% FCS-supplemented RPMI-1640 medium (Gibco-BRL) containing 10 μg/ml of Brefeldin A (Sigma-Aldrich, St Louis, MO, USA), anti-CD107a (1D4B; BioLegend) and 10 μg/ml of the HA stalk peptides. Following the stimulation and surface staining, the splenocytes were then fixed and permeabilized using the Cytofix/Cytoperm kit (BD Biosciences). Then, the cells were intracellularly stained with the following antibodies: anti-IFNγ (XMG1.2, BD Biosciences), anti-IL-2 (JES6-5H4), anti-IL-21 (mhalx21), anti-IL-4 (11B11), and anti-TNFα (MP6-XT22; all from eBioscience, San Diego, CA, USA). A FACSAria SORP flow cytometer (BD Biosciences) was used, and the data analysis was performed with the FlowJo software (version 8.8.6, Tree Star, Inc., Ashland, OR, USA).
Cell depletion
CD8+ T cells and CD49b+ NK cells were depleted from the splenocytes using CD8a and CD49b magnetic micro-beads according to the manufacturer’s instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). The in vivo depletion of CD4+ T cells was performed using an intraperitoneal injection of 0.3 mg of the monoclonal antibody GK1.5 in 0.2 ml of sterile PBS 3, 2, and 1 day(s) before the challenge experiment, as suggested in the manufacturer’s instructions. The depletion of the cells ranged between 90% and 99%, as confirmed by flow cytometry (Supplementary Figure 1).
ELISPOT
The number of cells secreting HA stalk-specific IFN-γ was determined using a mouse IFN-γ ELISPOT kit (BD Biosciences) following the manufacturer’s instruction. In short, ELISPOT plates were coated with the capturing antibodies at 4 °C overnight, followed by one wash and 2 h of blocking with 10% FBS supplemented RPMI 1640 (Gibco-BRL). The freshly prepared cell suspensions (5 × 105) were added to every well and stimulated with the HA stalk peptides (10 μg/ml). After incubation at 37 °C, 5% CO2, and 99% humidity, the plates were washed twice with deionized water and three times with PBS containing 0.05% Tween-20. Following incubation with the detection antibodies for 2 h at room temperature and three more washes with PBS containing 0.05% Tween-20, streptavidin-horseradish peroxidase was added to each well and left to incubate for 1 h at room temperature. The colored spots were then developed by incubating the samples with the final substrate solution for 15–30 min in the dark, and the reaction was terminated by a wash with deionized water. The quantification of the spots was performed using the ImageQuant software (Molecular Dynamics, Sunnyvale, CA, USA).
Mouse immunization and challenge experiment
As shown in Figure 2a, 8-week-old BALB/c mice (n=12 mice/group) were intraperitoneally immunized three times with a mixture of the peptides 105, 106 and 107, or PBS in aluminum hydroxide gel adjuvant (Brenntag Biosector, Denmark). Two weeks after the final boost, mice were killed for the antigen-specific detection of CD154 and for CD4 T-cell repertoire analysis.22, 23 In the challenge-protection assay, all mice received a 50% Mouse Lethal Dose (5 × MLD50) of A/Luxembourg/46/2009 (pH1N1) intranasally five times after anesthesia with isoflurane. Mice were weighed daily post challenge and killed if their weight decreased by more than 20% of their pre-challenge weight.
Restimulation culture conditions
Splenocytes from mice immunized with the mixture of peptides or PBS were harvested to characterize their antigen-reactive CD4 T cells using surface CD154 expression. Approximately 1 × 107 splenocytes were restimulated on a 48-well plate in the presence of anti-CD154 (MR1, 5 μg/ml), anti-CD40 (HM40-35, 2 μg/ml; both from eBioscience), anti-Fcγ receptor (50 μg/ml), and antigen (HA stalk peptides, 100 μg/ml) for 12 h. Cells were washed twice and stained with anti-CD3e and anti-CD4 for 15 min on ice.
High-throughput next generation sequencing
We isolated untouched mouse CD4+ T cells by negative selection using a commercial isolation kit (Miltenyi Biotec) according to the manufacturer’s instructions. Flow cytometry analysis confirmed that the cells were over 90% pure. The TCRβ CDR3 regions were amplified and sequenced from 300 ng of extracted DNA from each sample on the ImmunoSEQ platform (Adaptive Biotechnologies, Seattle, WA, USA). An ImmunoSEQ Analyser completed the subsequent processing and analysis of the data. Error corrections of the sequencing results24 were automatically made by the analysis platform for the precise quantification of rare T-cell clones.25 The resulting data were normalized for PCR bias, and the detailed properties of all samples are shown in Table 1. For each sample, we had on average ~1.4e5 productive reads that are in-frame and fully annotated (V, J segments assigned).
Table 1. Sample properties.
| Sample Name | Organ | Raw reads | Productive total reads | Productive unique reads |
|---|---|---|---|---|
| M1 | Spleen | 110 858 | 80 440 | 11 119 |
| M2 | Spleen | 476 609 | 347 647 | 34 962 |
| M3 | Spleen | 132 505 | 94 063 | 12 532 |
| M4 | Spleen | 174 175 | 128 509 | 19 325 |
| P1 | Spleen | 120 695 | 81 974 | 11 296 |
| P2 | Spleen | 190 061 | 168 220 | 9740 |
| P3 | Spleen | 122 833 | 90 635 | 6497 |
| P4 | Spleen | 160 565 | 118 258 | 14 892 |
| PC1 | Spleen | 155 199 | 140 766 | 11 507 |
| PC2 | Spleen | 132 270 | 124 145 | 12 125 |
Software and statistical analysis
PESTLE and SPICE, software provided by the NIH (http://exon.niaid.nih.gov), were used to visualize the multifunctional T-cell responses. Statistical analyses were performed using the GraphPad Prism 5 software (GraphPad, San Diego, CA, USA). Significant differences in animal survival were analyzed using the log-rank test and pairwise comparisons. Student’s t-test (unpaired, two-tailed) was used for comparisons between data from specified different conditions. P<0.05 was considered significant (*).
Results
Identification of a CD4 T-cell epitope on the HA stalk domain
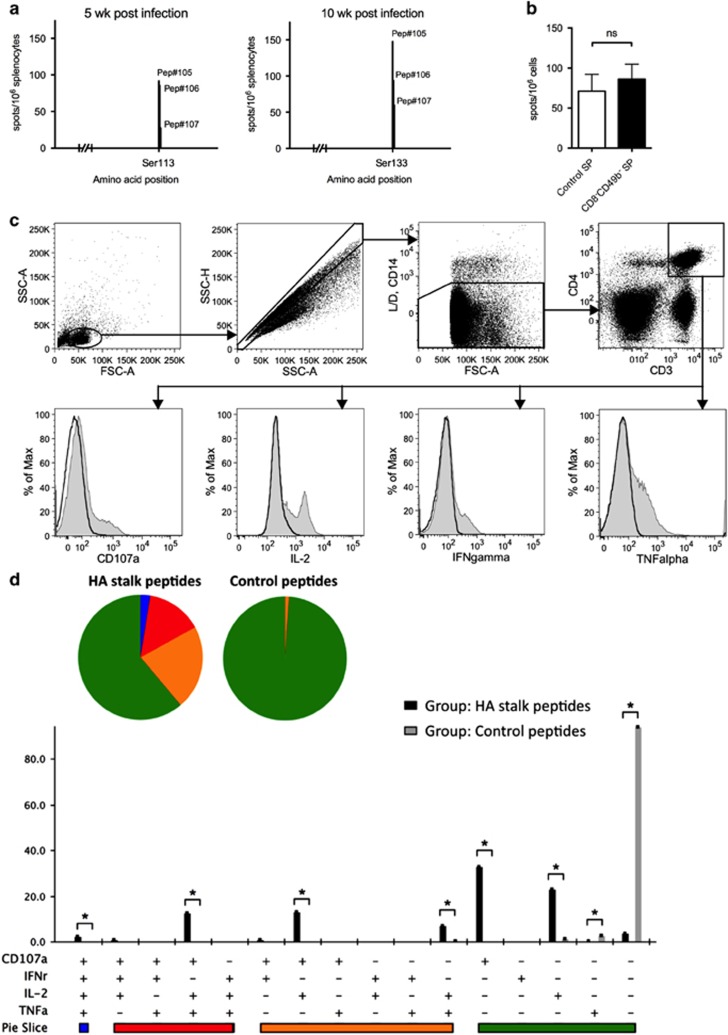
To determine the role of CD4 T-cell responses in the cross-protective immunity against influenza infection, we initially identified the HA stalk-specific CD4 T-cell epitopes. A series of 96 overlapping 15-mer peptides spanning the entire HA stalk domain (332 amino acids in length) of the pH1N1 virus was used for the stimulation of splenocytes obtained from BALB/c mice after 5 and 10 weeks of pH1N1 sub-lethal infection. As shown in Figure 1a, a peptide spanning residues HA2 113-131 (SNVKNLYEKVRSQLKNNAK) was identified by positive IFN-γ ELISPOT responses. The position of the identified CD4 T-cell epitope is located on the membrane-proximal region of the HA stalk domain (Supplementary Figure 2A). Alignments of the amino-acid sequences and a phylogenetic analysis were performed to examine the conservation of this epitope with pH1N1 A/Luxembourg/46/2009 with the corresponding regions of other influenza subtypes (Supplementary Figures 2B and C). Our results showed that the identified CD4 T cell epitope is not only highly conserved among the H1N1 strains tested (with 94.7% sequence identity) but also more closely clustered with those of the group 1 subtypes. The mean values of the protein sequence identities were 69.6% and 48.8% among the group 1 and group 2 subtypes, respectively.
Figure 1.
Identification of an HA stalk-specific CD4 T-cell epitope and memory HA stalk-specific CD4 T-cell immune responses. (a) IFN-γ ELISPOT analysis of splenocytes obtained 5 and 10 weeks after pH1N1 sublethal infection (n=8 and n=12, respectively). Cells were stimulated with a set of overlapping 15-mer peptides spanning the HA stalk protein. Stimulations were performed with individual peptides. (b) IFN-γ ELISPOT analysis of splenocytes obtained 10 weeks post pH1N1 sublethal infection (n=4) with the depletion of CD8± T cells and CD49b± NK cells (CD8−49b− SP) or without depletion (control SP). ns, non-significant. (c) Splenocytes were harvested 10 weeks after pH1N1 sublethal infection and stimulated with a mixture of peptides 105, 106 and 107 or were stimulated with negative control peptides. Responses were visualized using an ICS assay. Representative flow cytometry profiles are shown. (d) The magnitude and polyfunctionality of CD4 T-cell responses. All of the possible combinations of the responses are shown on the X-axis, whereas the percentages of the functionally distinct cell populations within the total CD4 T-cell population are shown on the Y-axis. Rows: C, CD107a; I, IFN- γ 2, IL-2; T, TNF-α. Responses are grouped and color coded on the basis of the number of functions. *P<0.05. Error bars represent the s.d.
Functional characterization of HA stalk-induced CD4 T-cell responses
To confirm the ELISPOT data and to ensure the peptide HA2 113-131 indeed harbors a CD4 T-cell epitope, we harvested mouse splenocytes 10 weeks post sub-lethal pH1N1 infection and used the peptide to stimulate the splenocytes with the depletion of CD8+ T cells and CD49b+ NK cells (CD8−49b− SP) or without depletion (control SP). The results showed that there is no difference in the peptide-induced IFN-γ responses with or without the presence of CD8+ T cells and NK cells (Figure 1b). In addition, we compared splenic lymphocytes stimulated with the peptide HA2 113-131 or an irrelevant control peptide (IKRFAKREEFMOKHIE). Figure 1c shows responses in the ICS assay using specific antibodies to determine T-cell lineage (CD3 and CD4), degranulation (CD107a) and the cytokine-producing cells (TNF-α, IL-2 and IFN-γ). The quality of the anti-viral CD4 T-cell response was examined by analyzing TNF-α, IL-2 and IFN-γ secretion and examining the surface expression of CD107a as an indirect marker for cytotoxicity. In total, 16 different antigen-reactive CD4 T-cell populations were characterized (Figure 1d). The elicited antigen-reactive CD4 T-cell responses were polyfunctional, with 38% of the CD4 T cells exhibiting two, three or four functions. Compared with the negative control, the stronger antigen-specific response obtained in the infected mice re-stimulated with peptides containing the HA stalk epitope ex vivo was due to an enhancement in the absolute frequencies of the CD4 T-cell population producing IL-2, TNF-α and CD107a or IL-2 and CD107a.
HA stalk-reactive CD4 T cells are induced by peptide immunization
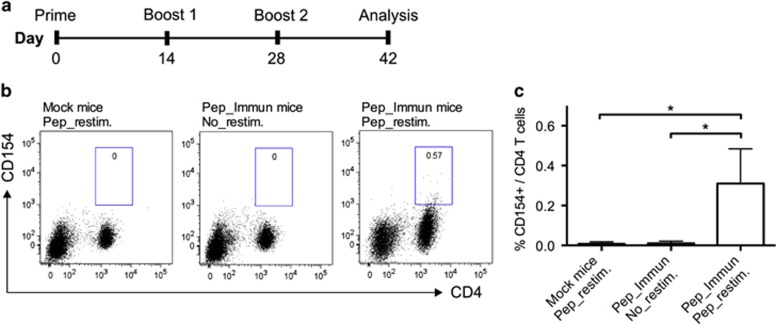
To induce HA stalk-specific CD4 T cells, mice were immunized three times (2 week intervals) with the peptide HA2 113-131 (Pep_Immun group) or PBS (Mock group). Both were emulsified in Alum. Two weeks following the last boost, mouse splenocytes were re-stimulated ex vivo by the same peptide or an irrelevant control peptide. The cells were subsequently stained for CD3, CD4 and CD154 surface markers to identify antigen-reactive CD4 T cells by FACS analysis (Figure 2b). A small but distinct population of CD4 T cells expressing CD154 (0.32±0.17%) was detected in the immunized group but not in the Mock group (P<0.05, Figure 2c). Moreover, no spontaneous upregulation of CD154 by CD4 T cells could be detected in the immunized mice that were not re-stimulated with the peptides (P<0.05, Figure 2c). However, no IFN-γ-producing CD4 T cells were detected by ICS or the ELISPOT assay following peptide immunization (data not shown).
Figure 2.
Detection of CD154-expressing CD4 T cells following HA stalk peptide immunization. (a) Scheme of peptide immunization. Mice were primed and boosted with peptides containing an HA stalk epitope (Pep_Immun group) or PBS (Mock group) in Alum. Two weeks after the final boost, splenocytes from mice were harvested for antigen-specific detection. (b) Splenocytes were re-stimulated for 12 h with or without the presence of peptides (Pep_restim. or No_restim.) and then stained for CD4 and CD154. One representative flow cytometry profile of four experiments is shown. (c) The frequency of CD154 expression in CD4 T-cell populations was measured. *P<0.05. Error bars represent the s.d.
HA stalk-specific CD4 TCRβ signatures
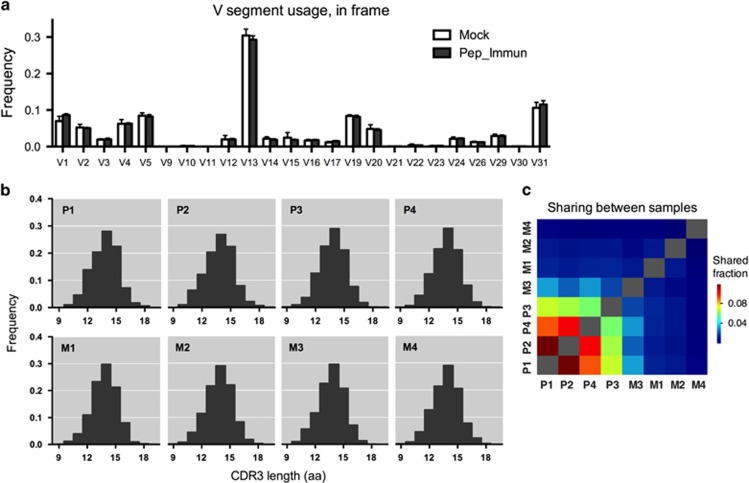
To explore the alterations in the CD4 T-cell repertoire landscape following peptide immunization, we analyzed the TCRβ repertoire in the spleens of mice from the Pep_Immun group (P1–P4; n=4) and the Mock group (M1–M4; n=4) using high-throughput sequencing. First, we examined the gene segment usages (Figure 3a). Qualitatively, similar Vβ distributions were observed in all samples, with only a few segments that differed in frequency between the two groups. For example, Vβ1 and Vβ31 were overexpressed in the Pep_Immune group compared with the Mock group, whereas Vβ13 was under-expressed in the Pep_Immune group. As another measure to examine repertoire evenness, the spectratyping analysis of the CDR3 length distributions was applied for the evaluation of clonal dominance in the CD4 T-cell repertoires (see Figures 3b for representative plots). The analysis revealed that the length distributions were highly homogenous among the samples, without any skewness of a specific CDR3 length. Hence, when comparing both the Vβ usages and spectratyping analysis, no significant change was observed in splenic CD4 T cells of the two groups, suggesting the maintenance of the phenotypic balance after peptide immunization.
Figure 3.
Comparison of the TCRβ repertoires of CD4 T cells. (a) Vβ usage analysis. Each bar represents the mean frequency of a Vβ gene segment in the spleens of mice from the Pep_Immun group or the Mock group. (b) CDR3 spectratyping analysis. Each bar represents the relative frequency of a particular CDR3 length (in amino acids) measured across each individual spleen sample. Upper panel: Pep_Immun group (P1-P4); lower panel: Mock group (M1-M4). (c) Sharing of clones between each sample computed by averaging across the two ratios of shared reads over total reads for each sample.
High-throughput sequencing of the TCR repertoire enables us to compare repertoires at a higher resolution beyond the CDR3 length distribution or the gene segment usage using an analysis at the level of individual TCR sequences. Thus, the similarity between samples was subsequently examined by a comparison of the overlapping clonotypes. Productive sequences that shared identical Vβ and Jβ gene segments and the CDR3 amino-acid sequence were used for the analysis. The percentage of sample overlap was computed by averaging the two ratios of the shared reads over the total reads for each sample using the ImmunoSEQ Analyser (Materials and methods). The results indicated that the fraction of shared clones in the Pep_Immun group is higher than those in the Mock group (Figure 3c). An average sharing of 0.086% was found between the Pep_Immun mice, whereas only 0.006% was shared between the Mock mice. To further estimate the overall similarity across samples from the same group, the average of all the pairwise correlation coefficients for each group comparison was calculated. We found that the Pep_Immun group had a higher within-group correlation compared with the Mock group (r2=0.1420 vs r2=0.0225). Thus, these results suggest that there might be convergent HA stalk-specific TCR repertoire signatures derived from peptide immunization.
HA stalk-reactive CD4 T cells and TCRβ signatures in protective immunity
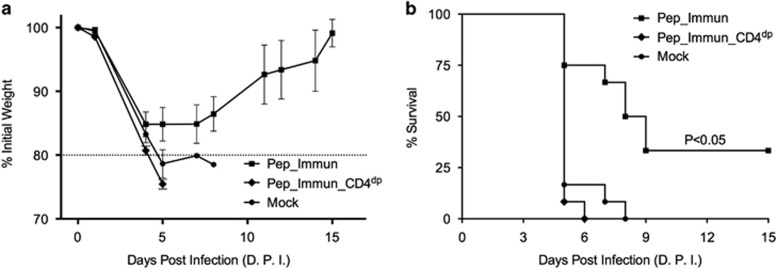
As HA stalk-specific CD4 T cells are induced in vivo, we evaluated their role in protection. We challenged the mice with the pH1N1 virus after peptide immunization as described above. As the primary read-outs for protective immunity, we monitored the weight loss and the survival time of the mice (Figure 4). Our results showed that the immunization of the mice with the peptide emulsified in alum showed partial protection (33%) against lethal infection by the pH1N1 virus. This result was significantly different from the Pep_Immun_CD4dp group (P<0.05) and the Mock group (P<0.05), in which no mouse survived. We next examined the TCRβ clonotypes in the CD4 T-cell populations of two immunized mice that survived the virus challenge (analyzed on day 15 post challenge, PC1 and PC2) in order to search for the identical Vβ and Jβ gene usage and the CDR3 amino-acid sequences. The identified 36 overlapping clonotypes and their frequencies are listed in Table 2. These clonotypes were then tracked in the CD4 T-cell populations of mice in the Pep_Immun group (P1–P4) and the Mock group (M1–M4). Three clones (numbered 1, 12 and 25) were found in the CD4 subsets of the majority of the mice in the Pep_Immun group and not present in any mouse of the Mock group (Table 2). The higher frequencies of these three TCRβ clonotypes after the virus challenge may imply the expansion of the HA stalk-specific clones in order to protect the mice from the viral challenge.
Figure 4.
Protective immunity. Mice immunized with HA stalk peptides (Pep_Immun), mice immunized with HA stalk peptides and depleted of CD4 T cells (Pep_Immun_CD4dp), and mice immunized with PBS in alum (Mock) were challenged by 5 × MLD50 of mouse-adapted pH1N1 virus (A/Luxembourg/46/2009) two weeks after the final boost (n=12 mice/group, representative of two distinct experiments). (a) Body weight was monitored daily. (b) The percent survival rate after the virus challenge is shown by a Kaplan–Meier survival plot and was analyzed with the log-rank test and pairwise comparisons.
Table 2. HA-stalk reactive CD4 T-cell clones identified in immunized mice that survived after virus challenge.
| no. | Vβ gene | CDR3 (amino acid) | Jβ gene |
Frequency of the overlapped clone (%) in Pep_Immun. group after virus challenge |
Frequency of the tracked clone (%) in Pep_Immun. group |
Frequency of the tracked clone (%) in Mock group |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | P1 | P2 | P3 | P4 | M1 | M2 | M3 | M4 | ||||
| 1 | TCRβV01 | CTCSAGQYEQYF | TCRβJ02-07 | 0.050725557 | 0.158041525 | 0.020669 | 0.018674 | 0.036635 | 0.006851 | 0 | 0 | 0 | 0 |
| 2 | TCRβV13 | CASGDLGSGNTLYF | TCRβJ01-03 | 0.068841827 | 0.089866749 | 0 | 0 | 0 | 0 | 0 | 0.099327 | 0 | 0 |
| 3 | TCRβV31 | CAWSLGGSGNTLYF | TCRβJ01-03 | 0.052537184 | 0.086767896 | 0.001148 | 0 | 0 | 0.017438 | 0 | 0 | 0.030764 | 0 |
| 4 | TCRβV31 | CAWRGTANSDYTF | TCRβJ01-02 | 0.0670302 | 0.030988534 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5 | TCRβV13 | CASGETGYEQYF | TCRβJ02-07 | 0.03623254 | 0.058878215 | 0 | 0 | 0 | 0.001868 | 0 | 0 | 0 | 0 |
| 6 | TCRβV13 | CASSDSSGNTLYF | TCRβJ01-03 | 0.014493016 | 0.080570189 | 0 | 0.010281 | 0 | 0.023044 | 0 | 0 | 0 | 0 |
| 7 | TCRβV01 | CTCSASSGNTLYF | TCRβJ01-03 | 0.043479049 | 0.027889681 | 0.005741 | 0.001259 | 0 | 0 | 0 | 0 | 0 | 0 |
| 8 | TCRβV29 | CASRDNYEQYF | TCRβJ02-07 | 0.025362778 | 0.055779362 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 9 | TCRβV13 | CASGDRGPGNTLYF | TCRβJ01-03 | 0.023551151 | 0.052680508 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10 | TCRβV05 | CASSQDLQANTGQLYF | TCRβJ02-02 | 0.003623254 | 0.086767896 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11 | TCRβV13 | CASSDRSNSDYTF | TCRβJ01-02 | 0.03623254 | 0.027889681 | 0.003445 | 0 | 0 | 0 | 0 | 0 | 0.027688 | 0 |
| 12 | TCRβV01 | CTCSADSGNTLYF | TCRβJ01-03 | 0.016304643 | 0.058878215 | 0.014928 | 0.001888 | 0 | 0.016816 | 0 | 0 | 0 | 0 |
| 13 | TCRβV13 | CASRDNYAEQFF | TCRβJ02-01 | 0.014493016 | 0.058878215 | 0 | 0.001679 | 0 | 0 | 0 | 0 | 0 | 0 |
| 14 | TCRβV13 | CASSETGREQYF | TCRβJ02-07 | 0.023551151 | 0.043383948 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 15 | TCRβV05 | CASSQDLGGEQYF | TCRβJ02-07 | 0.030797659 | 0.030988534 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | TCRβV13 | CASSDSAETLYF | TCRβJ02-03 | 0.021739524 | 0.040285095 | 0.002297 | 0 | 0 | 0.001868 | 0 | 0 | 0 | 0 |
| 17 | TCRβV04 | CASSYSGNTLYF | TCRβJ01-03 | 0.007246508 | 0.065075922 | 0 | 0.001049 | 0.012212 | 0.002491 | 0.046049 | 0 | 0 | 0 |
| 18 | TCRβV13 | CASGGTDSGNTLYF | TCRβJ01-03 | 0.010869762 | 0.052680508 | 0 | 0 | 0 | 0.008096 | 0 | 0 | 0 | 0 |
| 19 | TCRβV04 | CASSRDWGYEQYF | TCRβJ02-07 | 0.032609286 | 0.015494267 | 0.010909 | 0 | 0 | 0 | 0.01842 | 0 | 0 | 0 |
| 20 | TCRβV05 | CASSQSSYEQYF | TCRβJ02-07 | 0.021739524 | 0.027889681 | 0 | 0.001259 | 0 | 0 | 0 | 0 | 0 | 0 |
| 21 | TCRβV05 | CASSQQISNERLFF | TCRβJ01-04 | 0.016304643 | 0.034087388 | 0.000419631186150492 | 4.20E-04 | 0 | 0 | 0 | 0 | 0 | 0 |
| 22 | TCRβV13 | CASRQISNERLFF | TCRβJ01-04 | 0.023551151 | 0.018593121 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 23 | TCRβV04 | CASSLRLISNERLFF | TCRβJ01-04 | 0.014493016 | 0.034087388 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 24 | TCRβV04 | CASSYRGAETLYF | TCRβJ02-03 | 0.005434881 | 0.049581655 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 25 | TCRβV20 | CGARGQSGNTLYF | TCRβJ01-03 | 0.009058135 | 0.043383948 | 0.003445 | 0.003147 | 0 | 0.004982 | 0 | 0 | 0 | 0 |
| 26 | TCRβV04 | CASSLGASQNTLYF | TCRβJ02-04 | 0.01811627 | 0.024790827 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 27 | TCRβV05 | CASSQDWGGAEQFF | TCRβJ02-01 | 0.009058135 | 0.037186241 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 28 | TCRβV20 | CGASRDTSNTEVFF | TCRβJ01-01 | 0.012681389 | 0.030988534 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 29 | TCRβV13 | CASGDATNSDYTF | TCRβJ01-02 | 0.025362778 | 0.006197707 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 30 | TCRβV05 | CASSQELGGREQYF | TCRβJ02-07 | 0.023551151 | 0.006197707 | 0 | 0.003357 | 0 | 0 | 0 | 0 | 0 | 0 |
| 31 | TCRβV13 | CASSGTGQDTQYF | TCRβJ02-05 | 0.010869762 | 0.018593121 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 32 | TCRβV01 | CTCSAGGAETLYF | TCRβJ02-03 | 0.007246508 | 0.024790827 | 0.001722 | 0.005875 | 0 | 0 | 0 | 0 | 0 | 0 |
| 33 | TCRβV19 | CASSNERLFF | TCRβJ01-04 | 0.010869762 | 0.012395414 | 0 | 0 | 0 | 0.00436 | 0 | 0 | 0 | 0 |
| 34 | TCRβV05 | CASSPGTGYAEQFF | TCRβJ02-01 | 0.010869762 | 0.012395414 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 35 | TCRβV31 | CAWSLGGNQDTQYF | TCRβJ02-05 | 0.009058135 | 0.015494267 | 0 | 0.002518 | 0 | 0.007474 | 0 | 0 | 0.009229 | 0 |
| 36 | TCRβV31 | CAWSLRGANTGQLYF | TCRβJ02-02 | 0.007246508 | 0.00929656 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
TCRβ sequences with convergent features are denoted in bold.
Discussion
In the present study, we report the mapping of a CD4 T-cell epitope at the stalk region of the influenza HA protein. Ex vivo stimulation of splenocytes from infected BALB/c mice with the HA stalk peptide, which spanned residues HA2 113-131 (SNVKNLYEKVRSQLKNNAK), resulted in polyfunctional CD4 T-effector responses, including IL-2, IFN-γ and TNF-α secretion and degranulation. Our results showed that immunization with this CD4 T-cell epitope peptide provides partial protection against a challenge of a lethal dose of the pH1N1 virus. The identification of this HA stalk-specific CD4 T-cell epitope allows us to characterize and determine the requirements for protective cellular immune responses against the influenza virus.
In addition to our findings in BALB/c mice, Yang et al.26 also reported that the same CD4 T-cell epitope (SNVKNLYEKVRSQLKNNAKE) within the pH1N1 HA stalk protein in DRB5-restricted humans using an ex vivo tetramer staining approach, implying a future application by exploiting the identified epitope in human vaccine development. However, our analyses suggested that the identified CD4 T-cell epitope is restricted to the HA stalk of H1 subtype viruses. Our sequence alignment showed that the CD4 T-cell epitope of the pH1N1 virus shares 47.37% amino-acid sequence identity with that of the H3N2 (A/Aichi/2/1968) virus (Supplementary Figure 2B). There was no immune response detectable in the splenocytes from H3N2 (A/Aichi/2/1968) sub-lethally infected mice after re-stimulation with the identified HA stalk peptide. In addition, the splenocytes from the pH1N1 sub-lethally infected mice did not react to the CD4 T-cell epitope peptides that were previously identified in the H3 HA stalk protein.27 These results demonstrate that H1 and H3 influenza viruses harbor distinct CD4 T-cell epitopes on their respective HA stalk proteins, implying that there might be no cross-reactive CD4 T cell toward both the H1 and H3 HA stalk proteins in BALB/c mice.
In accordance with previous observations, in vivo exposure to stimulation with the peptide antigen is sufficient to prompt T-cell proliferation, whereas the possession of effector functions is contingent on stricter antigen presentation requirements.28, 29 For example, a previous report by Storni et al.30 showed that the injection of the virus-derived peptide alone was enough to stimulate virus-specific T cells to proliferate extensively in mice, but not sufficient to induce IFN-γ secretion. Harari et al.31 further reported that the differentiation of Th1 cell-polarized T cells, which have an important role in the clearance of infectious pathogens, requires prolonged exposure to antigens. In agreement with these findings, Jelley-Gibbs et al.32 demonstrated that antigen presentation by highly activated antigen-presenting cells is essential to induce highly differentiated CD4 T-effector cells, which are able to produce high levels of the cytokine IFN-γ. Our own results add support to these observations that HA stalk-specific CD4 T cells capable of IFN-γ secretion were only found in mice after pH1N1 sub-lethal infection but not in peptide-immunized mice. In addition, these HA stalk-specific CD4 T effectors induced by sub-lethal virus infection could persist efficiently as long-term memory cells for more than 10 weeks after infection.
The reason that we could not detect any cytotoxic characteristics in the HA stalk-specific CD4 T cells derived from peptide immunization may be that certain inflammation-induced co-stimulatory signals derived from the virus, such as pattern recognition molecules, are too few to induce Th1-cell polarization and the subsequent IFN-γ secretion. McKinstry et al.33 have demonstrated that adaptive transfer of Th1-polarized memory CD4 T cells in mice provides full protection against lethal influenza infections, whereas adaptive transfer of unpolarized (Th0) memory CD4 T cells gives only partial protection. Similarly, we observed that the induction of unpolarized HA stalk-specific CD4 T cells by peptide immunization protected one-third of the mice from lethal influenza virus infection. Collectively, these results emphasize the importance of eliciting Th1 responses for better protection against virus infection. Indeed, in the vaccination approach that utilizes a recombinant Adenovirus vector encoding HA from the pH1N1 virus, which provides complete protection against the lethal pH1N1 virus challenge, we observed a significant Th1-polarized memory CD4 T-cell population targeting the identified epitope of the HA stalk. These IFN-γ-producing and HA stalk-specific CD4 T cells expanded significantly following further lethal virus challenge, indicating their vital role in fighting against the viral infection. Thus, for the future optimization of the influenza vaccine, the selection of adjuvant candidates, such as the cytosolic RIG-like helicases, which can orient the Th1 response,34 should be taken into consideration in order to provide better cellular immune protection.
Previous experiments by Swain et al.35 presented evidence that although influenza-specific memory T cells were readily found in the periphery, few memory CD4 T cells could be recovered from the lungs. This is compatible with the finding that only the most highly differentiated and activated CD4 T-effector cells acquire the proper adhesion molecules for tissue homing and for trafficking to inflammatory sites. After the development of a resting memory state, these CD4 effectors lose the expression of tissue homing molecules and migrate more restrictedly to lymphoid sites.36 In addition, access to a high level of influenza antigens as well as the time duration available for naive CD4 T cells to undergo multiple thresholds of differentiation and proliferation could both be important factors for the induction of protective memory CD4 T cells.37 In this report, we observed the occurrence of antigen-specific memory CD4 T-cell clones in mouse spleens by applying a vaccination strategy in which BALB/c mice were intraperitoneally immunized with 60 μg of HA stalk peptide every 2 weeks for a total of three times. These peptide-induced stalk-specific CD4 T cells expanded significantly in mice after they survived an intranasal challenge with a lethal dose of the virus, implying their vital role in resolution of infection.
Finally, using high-throughput sequencing, we conducted a comprehensive analysis of the mouse CD4 TCRβ repertoire in order to investigate whether immunization with the identified HA stalk peptide gave rise to predictable CD4 T-cell repertoires with clonotypes shared across individuals. As in other similar studies,21, 24 we investigated only the mouse TCRβ because of the limitations of the current next generation sequencing techniques to analyze TCRα chain complexity.38 The emergence of shared TCR sequences can be derived by selective advantages of TCR clonotypes that harbor structural features optimal for peptide-MHC recognition.39 For example, a previous study in rhesus macaques with simian immunodeficiency virus infection showed that the occurrence of shared virus-specific TCR clones correlates with protection and inversely correlates with viral load.20 Likewise, several human studies have shown that control of HIV replication by elite controllers without any antiretroviral therapy is mediated by some protective TCR clonotypes with certain protective MHC alleles, such as HLA-B*27 and HLA-B*57.40, 41 Consistent with these findings, mice immunized with the HA stalk peptide exhibited interindividual sharing of convergent TCRβ sequences, which was not found in the non-immunized mice. After the lethal virus challenge, the expansion of certain shared TCRβ clonotypes was observed in the surviving mice. Therefore, the association found between shared TCR clonotypes among different individuals with protective immune responses implies the potential of antigen-specific TCR clones as surrogate predictors of disease outcome.21
Taken together, it is critical to identify which antigens or epitopes derived from the influenza virus can be targeted and recognized by the immune system in order to design optimized vaccination systems. Here, our findings in BALB/c mice not only contribute to identify a novel epitope on the HA stalk domain for mouse CD4 T cells but also provide knowledge of such antigen-specific CD4 T-cell responses against lethal infection by the pH1N1 influenza virus and its antigen-driven protective TCRβ usage signature that can be used to monitor and improve our understanding of the cellular immune responses following vaccination.
Acknowledgments
We thank members of the Department of Infection and Immunity for giving their advice and sharing their experiences concerning high-throughput sequencing and immunoassays, especially René Brons, who helped with the FACS analysis. This work was financed by the European research project FLUTCORE, which is supported by the European Union 7th Framework Program (FP7).
Author contributions
I-NL and CPM designed the research; I-NL, SF and AS performed the experiments; I-NL analyzed the results, prepared the figures and wrote the paper.
Footnotes
Supplementary Information for this article can be found on the Cellular & Molecular Immunology website (http://www.nature.com/cmi)
The authors declare no conflict of interest.
Supplementary Material
References
- Krystal M, Elliott RM, Benz Jr EW, Young JF, Palese P. Evolution of influenza A and B viruses: conservation of structural features in the hemagglutinin genes. Proc Natl Acad Sci USA 1982; 79: 4800–4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 2000; 69: 531–569. [DOI] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 2010; 1: pii: e00018–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 2013; 3: 521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Ekiert DC, Wilson IA. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol 2013; 87: 7149–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333: 850–856. [DOI] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC et al. Highly conserved protective epitopes on influenza B viruses. Science 2012; 337: 1343–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M et al. Antibody recognition of a highly conserved influenza virus epitope. Science 2009; 324: 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011; 333: 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 2009; 16: 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap AK, Steel J, Rubrum A, Estelles A, Briante R, Ilyushina NA et al. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog 2010; 6: e1000990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann EA, Kang SM, Legge KL. Protective CD8 T cell-mediated immunity against influenza A virus infection following influenza virus-like particle vaccination. J Immunol 2013; 191: 2486–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stambas J, Guillonneau C, Kedzierska K, Mintern JD, Doherty PC, La Gruta NL. Killer T cells in influenza. Pharmacol Ther 2008; 120: 186–196. [DOI] [PubMed] [Google Scholar]
- Sun J, Madan R, Karp CL, Braciale TJ. Effector T cells control lung inflammation during acute influenza virus infection by producing IL-10. Nat Med 2009; 15: 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson TM, Li CK, Chui CS, Huang AK, Perkins M, Liebner JC et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat Med 2012; 18: 274–280. [DOI] [PubMed] [Google Scholar]
- Zens KD, Farber DL. Memory CD4 T cells in influenza. Curr Topics Microbiol Immunol 2015; 386: 399–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan RJ, Zhong W, Usherwood EJ, Cookenham T, Roberts AD, Woodland DL. Protection from respiratory virus infections can be mediated by antigen-specific CD4(+) T cells that persist in the lungs. J Exp Med 2001; 193: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Roberts AD, Woodland DL. Antibody-independent antiviral function of memory CD4+ T cells in vivo requires regulatory signals from CD8+ effector T cells. J Immunol 2001; 167: 1379–1386. [DOI] [PubMed] [Google Scholar]
- Roman E, Miller E, Harmsen A, Wiley J, Von Andrian UH, Huston G et al. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med 2002; 196: 957–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DA, Asher TE, Wilson NA, Nason MC, Brenchley JM, Metzler IS et al. Public clonotype usage identifies protective Gag-specific CD8+ T cell responses in SIV infection. J Exp Med 2009; 206: 923–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attaf M, Huseby E, Sewell A. K. alphabeta T cell receptors as predictors of health and disease. Cell Mol Immunol 2015; 12: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhoff D, Frentsch M, Leclerk P, Bumann D, Rausch S, Hartmann S et al. Identification and isolation of murine antigen-reactive T cells according to CD154 expression. Eur J Immunol 2007; 37: 2370–2377. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med 2005; 11: 1113–1117. [DOI] [PubMed] [Google Scholar]
- Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O et al. Comprehensive assessment of T-cell receptor beta-chain diversity in alphabeta T cells. Blood 2009; 114: 4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H, Desmarais C, Matthis J, Livingston R, Andriesen J, Reijonen H et al. Ultra-sensitive detection of rare T cell clones. J Immunol Methods 2012; 375: 14–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, James E, Gates TJ, DeLong JH, LaFond RE, Malhotra U et al. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol 2013; 25: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanekova Z, Adkins I, Kosová M, Janulíková J, Sebo P, Varečková E. Heterosubtypic protection against influenza A induced by adenylate cyclase toxoids delivering conserved HA2 subunit of hemagglutinin. Antiviral Res 2013; 97: 24–35. [DOI] [PubMed] [Google Scholar]
- Rogers PR, Huston G, Swain SL. High antigen density and IL-2 are required for generation of CD4 effectors secreting Th1 rather than Th0 cytokines. J Immunol 1998; 161: 3844–3852. [PubMed] [Google Scholar]
- Bird JJ, Brown DR, Mullen AC, Moskowitz NH, Mahowald MA, Sider JR et al. Helper T cell differentiation is controlled by the cell cycle. Immunity 1998; 9: 229–237. [DOI] [PubMed] [Google Scholar]
- Storni T, Ruedl C, Renner WA, Bachmann MF. Innate immunity together with duration of antigen persistence regulate effector T cell induction. J Immunol 2003; 171: 795–801. [DOI] [PubMed] [Google Scholar]
- Harari A, Vallelian F, Meylan PR, Pantaleo G. Functional heterogeneity of memory CD4 T cell responses in different conditions of antigen exposure and persistence. J Immunol 2005; 174: 1037–1045. [DOI] [PubMed] [Google Scholar]
- Jelley-Gibbs DM, Brown DM, Dibble JP, Haynes L, Eaton SM, Swain SL. Unexpected prolonged presentation of influenza antigens promotes CD4 T cell memory generation. J Exp Med 2005; 202: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinstry KK, Strutt TM, Kuang Y, Brown DM, Sell S, Dutton RW et al. Memory CD4+ T cells protect against influenza through multiple synergizing mechanisms. J Clin Invest 2012; 122: 2847–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy B. The perfect mix: recent progress in adjuvant research. Nat Rev Microbiol 2007; 5: 505–517. [DOI] [PubMed] [Google Scholar]
- Swain SL, Dutton RW, Woodland DL. T cell responses to influenza virus infection: effector and memory cells. Viral Immunol 2004; 17: 197–209. [DOI] [PubMed] [Google Scholar]
- Swain SL, Agrewala JN, Brown DM, Roman E. Regulation of memory CD4 T cells: generation, localization and persistence. Adv Exp Med Biol 2002; 512: 113–120. [PubMed] [Google Scholar]
- Swain SL, Agrewala JN, Brown DM, Jelley-Gibbs DM, Golech S, Huston G et al. CD4+ T-cell memory: generation and multi-faceted roles for CD4+ T cells in protective immunity to influenza. Immunol Rev 2006; 211: 8–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HY, Hao B, Krangel MS. Orchestrating T-cell receptor alpha gene assembly through changes in chromatin structure and organization. Immunol Res 2011; 49: 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles JJ, Bulek AM, Cole DK, Gostick E, Schauenburg AJ, Dolton G et al. Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection. PLoS Pathog 2010; 6: e1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol 2012; 13: 691–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu I, Eberhard J, Ahmad F, Bhatnagar N, Behrens G, Jacobs R et al. Elevated CD57 and CD95 expressions are associated with lower numbers of CD4(+) recent thymic emigrants in HIV-1 infected immune responders following antiretroviral treatment. Immunol Lett 2014; 158: 1–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.