Autoantibodies (aAbs) are generated by the immune system in response to a number of stimuli, one of which is believed to be aberrant protein targeting to the plasma surface. In this study, the presence of human cytochrome P450 enzyme CYP4Z1 on the outer surface of the plasma membrane of MCF-7 breast cancer cells is demonstrated by immunofluorescence. Moreover, anti-CYP4Z1 aAbs could clearly be detected in breast cancer patient sera by both immunoprecipitation and western blot analysis, while weak or no signals were obtained when testing controls. In an indirect CYP4Z1 ELISA, reactive aAb titers were significantly higher in breast cancer patient samples than in controls. High-resolution epitope mapping of five breast cancer patient sera revealed strong recognition of an epitope that is located on the surface of the enzyme.
Cytochrome P450 enzymes (CYPs or P450s) are a superfamily of heme-containing monooxygenases that are widely distributed in every domain of life. Many of the 57 human CYP enzymes are involved in the bioconversion of xenobiotics, which includes hepatic drug metabolism and the bioactivation of chemical carcinogens, but some family members have their predominant role in the biosynthesis of physiologically important compounds such as steroids and fatty acids. However, these two groups are not strictly separated from each other and overlaps exist; for instance, some CYPs involved in steroid hydroxylation are less specific than once thought and can also oxidize foreign compounds with related structures. Human CYPs are membrane-bound enzymes that are either located on the cytoplasmic side of the endoplasmic reticulum or on the matrix side of the inner mitochondrial membrane; for their activity they typically depend on specific electron transfer proteins that are co-localized with them.1 Interestingly, several studies have demonstrated that some microsomal CYPs (such as CYP1A2, CYP2D6 and CYP2E1) as well as cytochrome P450 reductase are transported through secretory vesicles from the endoplasmic reticulum to the outer surface of rodent or human hepatocytes where they face the extracellular space and are catalytically active. Such plasma membrane-localized CYPs are probably one cause for the emergence of anti-CYP aAbs found in patients with a number of liver diseases as well as in patients with some endocrine or autoimmune disorders; for instance, CYP21A2 is the major adrenal cortex autoantigen in idiopathic Addison’s disease,2 and CYP2D6 is the molecular target of anti-LKM1 (liver kidney microsomal type 1) Abs, which are detectable in type II autoimmune hepatitis and in some patients with HCV hepatitis.3, 4 All CYPs for which plasma membrane localization has been shown are also known antigens. However, anti-CYP aAbs in cancer patients have not been reported to date, despite the fact that a number of human CYPs (including members of the families CYP1-4 as well as CYP19A1) are known to be overexpressed in various types of malignancies, such as breast, colorectal, lung or ovarian cancer. One of these enzymes is CYP4Z1, which is selectively expressed in mammary tissue and almost undetectable in other healthy human tissues, but displays a strong overexpression in breast cancer and ovarian cancer cells.5, 6 We have previously shown that CYP4Z1 catalyzes the monohydroxylation of lauric and myristic acid, although it is not known whether this activity is its major physiological function.7
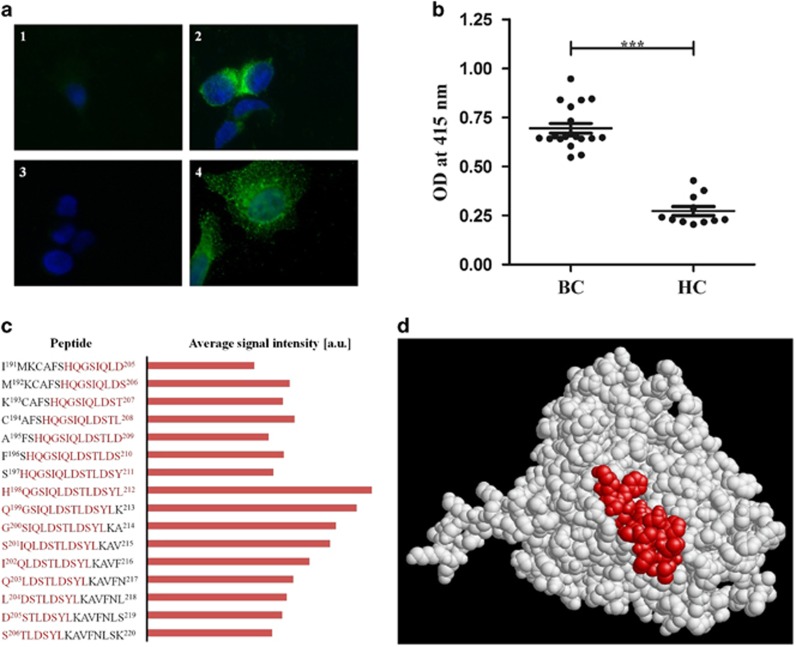
In this study, we investigated by indirect immunofluorescence microscopy whether CYP4Z1 might also be detected on the plasma membrane of cells of the human epithelial breast cancer cell line MCF-7. Immunostaining of non-permeabilized MCF-7 cells with an anti-CYP4Z1 polyclonal antibody and a fluorescein-conjugated secondary antibody resulted in strong fluorescence labeling on the outer surface of the cells, while no labeling was observed in the control (Figure 1a). This result constitutes the first proof of CYP4Z1 expression on the plasma membrane of breast cancer cells; to the best of our knowledge, this also is the first evidence of a plasma membrane localization of a human CYP in non-hepatic cells. Next, we wanted to test whether there is recognition of membrane targets on MCF-7 cells by circulating auto-aAbs of a breast cancer patient. Immunostaining of MCF-7 with a breast cancer patient serum and a fluorescein-conjugated anti-human IgG secondary antibody again resulted in extensive fluorescence labeling on the surface of the cells, while no signal was found in the control. It is tempting to speculate that other human CYPs might also be found on the plasma membrane of non-hepatic cells; obvious candidates are those CYPs that are known antigens, such as CYP21A2.
Figure 1.
(a) Detection of CYP4Z1 on the outer surface of non-permeabilized MCF-7 cells by rabbit anti-CYP4Z1 IgG and by aAbs from a breast cancer patient serum. Non-permeabilized MCF-7 cells were probed with antibodies as indicated below and stained with DAPI. Fluorescence was detected on a Eclipse E600 microscope (Nikon) using exposure times of 80 ms for DAPI and 2 s for FITC, respectively. Magnification is × 1000 for all images. The images correspond to: (1) MCF-7 cells exposed to secondary anti-rabbit Ab (goat anti-rabbit IgG conjugated to Alexa fluor 488; 1:2000) only; (2) MCF-7 cells probed with rabbit anti-CYP4Z1 Ab (1:1000) and secondary anti-rabbit Ab (as in 1); (3) MCF-7 cells exposed to secondary anti-human Ab (goat anti-human IgG conjugated to Alexa fluor 488; 1:2000) only; (4) MCF-7 cells probed with human breast cancer serum (1:500) and secondary anti-human Ab (as in 3). Breast cancer serum samples used in this study were from female breast cancer patients of the Tianjin Medical University Cancer Institute and Hospital, while control serum samples were from female patients of the Tianjin University Hospital with no malignancies. All patients gave written, informed consent under a study protocol approved by the institutional ethics committees. (b) ELISA assay detected higher titers of anti-CYP4Z1 aAbs in breast cancer patients’ sera. Recombinant CYP4Z1 full-length protein was used to coat an ELISA plate. Wells were incubated with sera from breast cancer patients (BC; n=19) or healthy controls (HC; n=11), respectively. The average of all samples in each group is displayed by the dark line, with error bars representing the s.e.m. for each group (P<0.0001). (c) Comparison of aAb binding to peptides in the vicinity of amino acids 198–212 of CYP4Z1. High-resolution epitope mapping of five patient sera was done by PreMed Biotech Co. Ltd. (Beijing, China) using 15-mer peptides covering the entire CYP4Z1 sequence with an offset of one amino acid. For each patient sera, signal intensities were normalized with respect to the peptide giving the highest intensity; from these, data averages were calculated and plotted in arbitrary units. Shown are the peptide sequences and average signal intensities for peptides number 198–213. Amino acids His-198 to Leu-212 (which constitute peptide number 205) are highlighted in red. (d) Localization of amino acids His-198 to Leu-212 on the surface of CYP4Z1. We recently published a homology model of CYP4Z1 on the basis of the X-ray structure of human CYP3A4 as a template.8 The graph shows this structure with amino acids His-198 to Leu-212 highlighted in red.
In order to test for the presence of anti-CYP4Z1 aAbs, sera from breast cancer patients were incubated with recombinant CYP4Z1 protein and protein A-sepharose beads. The resulting immune complexes were precipitated, washed and subjected to SDS-PAGE and western blot transfer. Western blot development was done using a rabbit polyclonal anti-CYP4Z1 antibody and a corresponding secondary antibody coupled to horseradish peroxidase (HRP), while signals were monitored using enhanced chemiluminescence. Strong signals for CYP4Z1 could be detected in all breast cancer samples while only very weak or no signals were found in control sera (data not shown). This result shows for the first time that anti-CYP4Z1 aAbs are present in breast cancer patients’ sera. Since in this experiment, the aAbs reacted with the full-length folded CYP4Z1 protein, they could in principle be directed against either linear or conformational epitopes. In order to test whether linear epitopes are involved, we performed western blot analysis of recombinant CYP4Z1 protein using the patient sera as primary antibody and an anti-human secondary antibody. Again, we found strong signals in all breast cancer samples while only very weak or no signals were observed in control sera (data not shown). This experiment proves that at least one linear epitope within CYP4Z1 is recognized by anti-CYP4Z1 aAbs in breast cancer sera. The next aim of this study was to quantify the anti-CYP4Z1 aAb signals in breast cancer sera and to compare them to controls. For this purpose, we set up an indirect ELISA using human recombinant CYP4Z1 full-length protein as the target, breast cancer patient sera or controls, respectively, as probes and a HRP-coupled anti-human secondary antibody. Signal measurement was done by monitoring HRP-dependent conversion of 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) at 415 nm. Under these conditions, all breast cancer sera gave signals between OD415 0.5 and 1.0, while control signals were all significantly lower (Figure 1b).
High-resolution mapping of anti-CYP4Z1 aAb epitopes was performed to specifically determine which amino acids were critical for anti-CYP4Z1 antibody binding, using the PEPperCHIP peptide microarray printing technique9 on standard functionalized glass slides. An overlapping peptide library was synthesized that consisted of 15-mer peptides with an offset of one amino acid covering the entire CYP4Z1 amino-acid sequence. Signals detected after incubation with secondary antibody goat anti-human IgG only were subtracted from the signal obtained with the patient sera and the relative signal intensity was measured for all overlapping of CYP4Z1 peptides. It was found that a peptide encompassing residues 198–212 was strongly recognized by all patients’ sera. As expected, the neighboring peptides also display strong signals, with sequences downstream of residues 198–212 giving higher values than those upstream of it (Figure 1c). This suggests that His-198 plays a significant role in this epitope. As of now, an X-ray crystal structure of CYP4Z1 has not been reported; however, we recently published a homology model of this enzyme on the basis of the X-ray structure of human CYP3A4.8 According to this model, residues His-198 to Leu-212 are all on the protein surface (Figure 1d), which supports the notion that this area is a target of anti-CYP4Z1 aAbs. There is still an urgent need to develop new diagnostic tools that allow for a sufficiently early diagnosis of breast cancer before it progresses to an often-incurable metastatic stage. Serum aAbs against tumor-associated antigens are a group of biomarkers, which are of great interest in this respect, since they are easily accessible in blood samples and have a long half-life. Here we show for the first time that CYP4Z1 is a tumor-associated antigen in breast cancer patients. However, at the moment, we can only speculate about the molecular, diagnostic and therapeutic potentials of anti-CYP4Z1 aAbs. If larger patient groups are studied, it is possible that a correlation between tumor progression and anti-CYP4Z1 aAb levels might be found.
Acknowledgments
We are grateful to Feng Yu-Mei from the Department of Biochemistry and Molecular Biology at Tianjin Medical University Cancer Institute and Hospital (Tianjin, China) for providing cancer patient sera and to Zhang Weiying from the Institute of Molecular Biology at Nankai University (Tianjin, China) for providing MCF-7 cells.
Footnotes
The authors declare no conflict of interest.
References
- Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems-biological variations of electron transport chains. Biochim Biophys Acta 2007; 1770: 330–344. [DOI] [PubMed] [Google Scholar]
- Winqvist O, Karlsson FA, Kampe O. 21-Hydroxylase, a major autoantigen in idiopathic Addison's disease. Lancet 1992; 339: 1559–1562. [DOI] [PubMed] [Google Scholar]
- Bogdanos DP, Invernizzi P, Mackay IR, Vergani D. Autoimmune liver serology: current diagnostic and clinical challenges. World J Gastroenterol 2008; 14: 3374–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalekos GN, Zachou K, Liaskos C, Gatselis N. Autoantibodies and defined target autoantigens in autoimmune hepatitis: an overview. Eur J Int Med 2002; 13: 293–303. [DOI] [PubMed] [Google Scholar]
- Downie D, McFadyen MC, Rooney PH, Cruickshank ME, Parkin DE, Miller ID et al. Profiling cytochrome P450 expression in ovarian cancer: identification of prognostic markers. Clin Cancer Res 2005; 11: 7369–7375. [DOI] [PubMed] [Google Scholar]
- Rieger MA, Ebner R, Bell DR, Kiessling A, Rohayem J, Schmitz M et al. Identification of a novel mammary-restricted cytochrome P450, CYP4Z1, with overexpression in breast carcinoma. Cancer Res 2004; 64: 2357–2364. [DOI] [PubMed] [Google Scholar]
- Zöllner A, Dragan CA, Pistorius D, Muller R, Bode HB, Peters FT et al. Human CYP4Z1 catalyzes the in-chain hydroxylation of lauric acid and myristic acid. Biol Chem 2009; 390: 313–317. [DOI] [PubMed] [Google Scholar]
- Yang X, Hutter M, Goh WW, Bureik M. CYP4Z1—A human cytochrome P450 enzyme that might hold the key to curing breast cancer. Curr Pharm Des; e-pub ahead of print 7 February 2017; doi:10.2174/1381612823666170207150156. [DOI] [PubMed]
- Stadler V, Felgenhauer T, Beyer M, Fernandez S, Leibe K, Guttler S et al. Combinatorial synthesis of peptide arrays with a laser printer. Angew Chem Int Ed Engl 2008; 47: 7132–7135. [DOI] [PubMed] [Google Scholar]