Abstract
MicroRNAs (miRNAs) are a series of conserved, short, non-coding RNAs that modulate gene expression in a posttranscriptional manner. miRNAs are involved in almost every physiological and pathological process. Type 1 diabetes (T1D) is an autoimmune disease that is the result of selective destruction of pancreatic β-cells driven by the immune system. miRNAs are also important participants in T1D pathogenesis. Herein, we review the most recent data on the potential involvement of miRNAs in T1D. Specifically, we focus on two aspects: the roles of miRNAs in maintaining immune homeostasis and regulating β-cell survival and/or functions in T1D. We also discuss circulating miRNAs as potent biomarkers for the diagnosis and prediction of T1D and investigate potential therapeutic approaches for this disease.
Keywords: autoimmunity, β-cell destruction, miRNA, type 1 diabetes
INTRODUCTION
Type 1 diabetes (T1D) is an autoimmune disease that results from the selective destruction of pancreatic β-cells driven by autoimmunity. In genetically susceptible individuals, abnormal immune responses induced by ill-defined environmental triggers insult pancreatic islets, subsequently resulting in β-cell loss and insulin deficiency.1, 2, 3, 4 The pathogenic processes of this disease involve autoimmune responses and β-cell destruction.5 A better understanding of the exact mechanisms involved in these pathogenic events would help to not only uncover the pathogenesis of T1D but also hunt for new therapeutic approaches for this disease.
MicroRNAs (miRNAs) are endogenous, evolutionarily conserved and short (21–23 nucleotides) non-coding RNAs that function as key modulators of gene expression in a posttranscriptional manner.6 The majority of miRNA-coding genes are located in the introns of protein-coding genes. However, these genes can also be localized in exons, intergenic regions, or 5′ and 3′ untranslated regions (UTRs). miRNA genes are transcribed in the nucleus primarily by RNA polymerase II to produce primary miRNAs (pri-miRNAs), which have multiple stem-loop structures. Most pri-miRNAs are processed to produce miRNA precursors (pre-miRNAs) by the Drosha–DGCR8 microprocessor complex (canonical pathway). Then, the pre-miRNAs are transported to the cytoplasm and further cleaved by the RNase III endonuclease Dicer to generate mature miRNAs. The mature miRNAs can be loaded into the RNA induced silencing complex (RISC) assembly, which subsequently mediates the regulatory functions of miRNAs. Generally, miRNAs exert their functions via binding with the 3′ UTRs of their target genes, resulting in translational silencing or direct mRNA degradation.6 Interestingly, recent findings have revealed that miRNAs may also modulate gene expression in a positive manner.7, 8 A representative paradigm is that miR-10a can bind to the 5′ UTR of ribosomal protein mRNAs and enhance their translation.7 In addition, miR-21 has been found to directly target mitochondrial cytochrome b (mt-Cytb) and positively regulate mt-Cytb translation.8 miRNAs play important roles in the normal maintenance of cell homeostasis and biological functions. Aberrant miRNA expression is associated with many pathological conditions in humans, including cancers, neurodegenerative disorders, infectious diseases and autoimmune diseases.9, 10, 11, 12, 13 Recently, a growing number of studies have implicated miRNAs in the pathogenesis of autoimmune T1D.
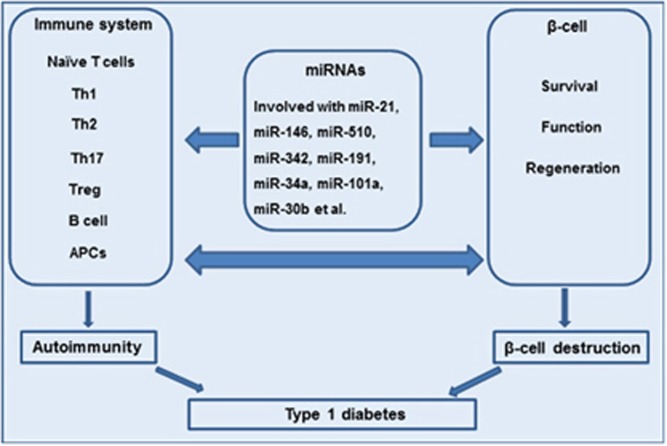
Herein, we focus predominantly on the key roles of miRNAs in the pathogenic events of T1D (Figure 1), including (1) the roles of miRNAs in maintaining immune homeostasis and the effect of aberrant miRNA expression on autoimmunity during the onset and progression of autoimmune T1D and (2) the roles of miRNAs in pancreatic β-cell apoptosis and/or dysfunction in T1D. In addition, we discuss how miRNAs serve as potent biomarkers for the diagnosis and prediction of T1D and investigate their therapeutic potential for this disease.
Figure 1.
miRNAs: new regulators of autoimmunity and pancreatic β-cell destruction in type 1 diabetes. miRNAs may regulate naïve T cell, Th cell (including Th1, Th2, Th17 and Treg cells), B cell and APC differentiation, proliferation, cell death and functions, consequently inducing the appearance of autoimmunity and causing β-cell destruction. These effects lead to the onset and development of type 1 diabetes. In addition, miRNA dysregulation may affect β-cell survival, functions and regeneration, thereby contributing to the pathogenesis of type 1 diabetes
miRNAs: REGULATORS OF IMMUNE PATHOGENESIS IN T1D
The immunopathological mechanisms in T1D
Islet-reactive T cells are excited, expanded and subsequently recruited into islets in both humans and animal models, eventually causing damage to β-cells and accelerating the onset of T1D.14, 15 T cells (including CD4+ and CD8+ T cells), macrophages, dendritic cells (DCs), natural killer (NK) cells, B lymphocytes and a series of chemokines and cytokines released by various immune cells are involved in the autoimmune attack on β-cells.2, 3 Major histocompatibility complex (MHC) class II molecules are not expressed on β-cells.15 Thus, antigen-presenting cells (APCs), such as macrophages and DCs, take up β-cell-related autoantigens and present the respective antigens to naïve T cells with the help of the MHC II molecules expressed on the APCs. Upon excitation, T cells are expanded and differentiated into functional immune cells.15
Naïve CD4+ T cells eventually develop into four major phenotypes (T-helper 1 (Th1), Th2, Th17 and regulatory T (Treg) cells).16, 17 Considering the importance of these cells in autoimmunity, the pathogenic contributions of Th1 cells and protective effects of Th2 cells also play crucial roles in the pathogenesis of T1D.16 The Th1 population mainly produces principle cytokines including interleukin-2 (IL-2), interferon-γ (IFN-γ) and tumor necrosis factor-β (TNF-β), whereas Th2 cells predominantly secrete IL-4, IL-5 and IL-10.16 In T1D patients, β-cell autoantigen-specific CD4+ T cells show a more proinflammatory phenotype with concomitant secretion of IFN-γ.14, 18 Conversely, CD4+ T cells from healthy individuals seem to have similar β-cell specificity but exhibit higher IL-10-secreting activity, and the phenotypes and functions of these cells are more regulatory.14, 18 The transfer of Th1 cells into neonatal non-obese diabetic (NOD) mice, which is an animal model of spontaneous T1D, leads to T1D onset.19 Detailed experiments using mouse models have shown that IFN-γ receptor (IFN-γR) mutations that abrogate the expression of the IFN-γRα chain to render it unresponsive to IFN-γ not only delay the appearance of insulitis in NOD mice but also completely prevent cyclophosphamide-induced diabetes in BCR2.5 transgenic mice.20 Administration of exogenous IL-10 or IL-4 has been demonstrated to halt T1D development in NOD mice.21, 22 Tregs are a heterogeneous subgroup of CD4+ T cells that intervene by averting the proliferation and activation of auto-reactive effector T cells (Teffs) and inducing immune tolerance, thereby leading to a decline in inflammatory responses.23, 24 Tregs exert their immunosuppressive functions mainly via four mechanisms: cell-to-cell contact, secretion of immunosuppressive cytokines, killing or modification of APCs and competition for growth factors.25, 26, 27 Treg deficiency may deprive or blunt their immunosuppressive capabilities, thereby accelerating autoimmunity.28 The transcription factor Foxp3 is believed to be the master key controlling Treg cells. Thus, altered Foxp3 expression in CD4+ T cells will influence Treg development and functions. Deletion of Foxp3 or administration of an anti-CD25 antibody (Ab) can accelerate diabetes onset in NOD mice and enhance its severity.29, 30 At the onset of autoimmune diabetes, a significant enhancement of Foxp3+ Treg cells is observed in the pancreatic lymph node. However, a progressive decrease in Treg cells accompanies the development of T1D. Moreover, intra-islet Foxp3+ Tregs show decreased CD25 and Bcl-2 expression, which probably promotes their sensitivity to apoptosis. Importantly, these abnormalities are ascribed to local IL-2 deficiency.31 Previous studies have shown that low-dose IL-2 therapy promotes Treg survival and leads to a dampening of T1D.31, 32 Th17 is another subset of Th cells that is characterized by secretion of the proinflammatory cytokines IL-17, IL-17F, IL-22 and IL-21. IL-17 is a key cytokine with the ability to induce the synthesis of other inflammatory cytokines and chemokines.33, 34 Recently, the contributions of Th17 cells to T1D pathogenesis have been increasingly recognized.35 Th17 cells exert immunological functions in T1D mainly through the following pathways: (1) Th17 cells counteract Treg cells to expand and disturb the Teff/Treg cell ratio, which allows the development of T1D, (2) Th17 cells may convert to diabetogenic T cells and enhance the islet-destruction effects in T1D and (3) Th17 cells can stimulate CD8+ cytotoxic T lymphocytes (CTLs), thereby contributing to the development of T1D.33 In conclusion, a ‘balanced auto-reactive set-point’ (Th1/Th17 vs Treg/Th2 cells) is a key determinant of the pathological outcome in T1D.
In addition to CD4+ T cells, CD8+ T cells have been implicated in the pathogenesis of T1D. HLA I molecules are primarily expressed on CD8+ T cells and are linked to the risk of the development of autoimmune T1D.36 Islet-specific hyperexpression of the HLA I protein is consistently observed in human islets from post-mortem collections of T1D patients, and CD8+ T cells are predominant in the lymphocyte infiltrates.37 Furthermore, β-cell autoantigen-specific CD8+ T cells have been detected in the peripheral blood (PB) of patients with T1D.38 These data strongly suggested an impact of cytotoxic CTLs on β-cell damage.
The pathogenic effects of B cells in NOD mice have been demonstrated by the observation that their ablation using either a genetic approach or antibody treatments offers strong protection from T1D.39, 40, 41 The unique ability to specifically take up β-cell antigens allows B cells to function as a subset of APCs and improve the expansion of diabetogenic T cells.42, 43
Innate immunity also exhibits vital functions in T1D. For example, APCs present β-cell antigens to T cells, resulting in immunological molecular events and leading to β-cell destruction.44 DCs and other APCs have been observed to infiltrate islets in both human T1D patients and NOD mice.45, 46 The inflammatory cytokines secreted by these immune cells can also destroy β-cells.47
In addition, a series of studies organized by the Network for the Pancreatic Organ Donor with Diabetes (nPOD), which recovers pancreas and lymphoid tissues from organ donors with islet autoimmunity and T1D, have provided new evidence on the etiology and pathogenesis of T1D. For example, C4d deposition has been found to be elevated in the pancreas of T1D patients. Studies have also supported an association of enteroviruses with T1D, including cases with a longer disease duration that might be due to viral persistence or multiple infections.48
The roles of miRNAs in the regulation of T1D immunopathogenesis
miRNAs have been implicated in a wide scope of functions. They play important roles in many physical and pathological processes. Accumulating evidence has shown that miRNAs are important participants in the control of the delicate immune system equilibrium. miRNAs function as key modulators of immune responses mainly through the regulation of immune cell differentiation, development, activation and functions. During T-cell development, deletion of Dicer in immature thymocytes leads to a drastic reduction in total thymocyte numbers, with a significant reduction in the more mature double-positive (CD4+CD8+) and single-positive (CD4+ or CD8+) thymocytes probably as a result of increased cell death and attenuated proliferation.49 In the case of Th cell differentiation, CD4+ T cells lacking Dicer fail to evolve into the Th2 phenotype due to a bias toward Th1 differentiation and their inability to prevent IFN-γ production.50 In addition, the specific overexpression or deletion of individual miRNA genes affects T-cell development. The miR-17-92 family is hyperexpressed in T precursor cells, but the expression levels of these family members decrease concomitant with T-cell maturation. Exogenous overexpression of the miR-17-19 cluster in lymphocytes can result in severe lympho-proliferative disease in mice in which a mass of CD4+ T cells (and CD8+ T cells to a lesser extent) are expanded.51 CD4+ T cells isolated from miR-155-deficient mice are apparently biased toward Th2 differentiation and are accompanied by the secretion of Th2 cytokines. At the same time, the Th1 populations in these mice have altered functions despite normal IFN-γ production.52, 53 Treg cells with specifically deleted miRNAs due to Dicer or Drosha ablation can develop an early fatal onset of autoimmunity.54, 55, 56 Individual miRNA-deficient Tregs are predisposed to have an inadequate ability to underscore their functional relevance. For example, inhibition of miR-155 can disturb Treg development and functions. Tregs with a miR-155 deficiency exhibit upregulated expression of cytokine signaling 1 (Socs1) and a diminished ability to respond to IL-2.57 In addition, miR-21 and miR-31 regulate Treg development by changing Foxp3 expression.58 Several miRNAs have been reported to promote Th17 cell development. For example, miR-326 facilitates Th17 development and accelerates the progression of autoimmunity by directly inhibiting ETS1, which is a negative modulator of Th17 differentiation.59 In addition, the regulatory roles of miRNAs are crucial for early B-cell development. Deletion of Dicer has been demonstrated to halt the pro- to pre-B-cell transition.60 miRNAs also exert important effects on NK, NKT and APC development and functions.61
There is increasing evidence that miRNAs associate with multiple immunity-related genes and pathways, and consequently regulate immune homeostasis; additionally, miRNA alterations can result in autoimmune responses that favor the progression of many types of autoimmune diseases, including systemic lupus erythematosus, multiple sclerosis, rheumatoid arthritis (RA) and T1D.28 For example, dysregulated miRNA expression, such as miR-31, miR-146a and miR-155, in Tregs has been shown to impair Treg functions in RA.28, 62
miR-326 expression is significantly increased in peripheral blood mononuclear cells (PBMCs) from T1D patients; the elevated levels correlate with ongoing islet autoimmunity and the disease severity.63 Several possible target genes of miR-326, including vitamin D receptor and erythroblastosis virus E26 oncogene homolog 1, are vital modulators of the immune system, suggesting that the miRNA probably has important effects on autoimmunity in T1D by targeting these genes.63 Another study performed by Yang et al.64 identified 26 differentially expressed miRNAs in PBMCs from newly diagnosed T1D patients, among which miR-146 was the most downregulated molecule. The study also showed that the miR-146 levels were associated with ongoing islet autoimmunity.64 Moreover, miR-21a and miR-93 are decreased in PBMCs from T1D patients;65 both miRNAs have been reported to participate in inflammatory and apoptosis signaling pathways39 and are not affected by glucose stimuli.65 Thus, the two miRNAs might be involved in the autoimmune responses of T1D.
In autoimmune T1D, self-reactive T cells in the periphery develop into pancreas-infiltrating T lymphocytes (PILs), which induce insulitis and cause β-cell damage. T1D does not occur in the absence of CD3+ T cells with either a CD4+ or CD8+ phenotype. Furthermore, the development of T1D follows the changing levels of immune-related genes during the evolution of thymocytes into peripheral CD3+ T cells.66 In a comparative analysis of thymocytes derived from pre-diabetic 1-month-old NOD mice (1 mo), CD3+ peripheral T cells from pre-diabetic (1 mo or 7 mo) and diabetic NOD mice (>7 mo), and PILs from pre-diabetic (1 mo) NOD mice, Fornari et al.66 found that the thymocytes exhibited four altered miRNAs (upregulated miR-34a and miR-345-5p, and downregulated miR-29c and miR-101a), the CD3+ peripheral T cells exhibit two altered miRNAs (upregulated miR-378 and downregulated miR-296-5p) and the PILs exhibited seven upregulated miRNAs (miR-10b, miR-202-3p, miR-697, miR-705, miR-709, miR-712 and miR-877). Through the construction of miRNA–mRNA interaction networks, the authors determined the links between miR-202-3p and the CCR7 chemokine receptor and Cd247 (Cd3 zeta chain) mRNAs, which have been proven to participate in the control of autoimmunity in NOD mice.66 A miRNA profile provided by De Jong et al. suggested that some miRNAs predicted to target the apoptotic genes Trail, Trail-R2, Fas and Faslg (that is, miR-23b, miR-98 and miR-590-5p) were simultaneously hyperexpressed in auto-reactive CD8+ T cells derived from T1D patients. The transfection of these miRNAs into primary T cells led to reduced Fas and Trail mRNA levels, which subsequently performed their relevant functions and generated excessive expansion of diabetogenic cytotoxic T cells.67
Different miRNA expression patterns have been observed within specific T-cell subtypes between pre-T1D and healthy donors. Naïve CD4+ T cells derived from pre-T1D patients displayed 32 dysregulated miRNAs compared with the healthy controls, of which 28 were decreased. Naïve natural Tregs (nTregs) in pre-T1D patients displayed two dysregulated miRNAs, and nTregs activated in vivo displayed a large number of dysregulated miRNAs. These differentially expressed miRNAs share a common characteristic: they can alter T-cell functions.68 For instance, both naïve CD4+ and transitional memory cells hyperexpress miR-101 in the pre-T1D stage. One of its targets is the histone 3 methyltransferase Ezh2, which is a catalytic subunit of the polycomb repressive complex 2. Ezh2 deficiency in naïve CD4+ T cells can induce differentiation of these cells with a bias toward a Th1 phenotype in both mice and humans.69, 70 nTregs in pre-T1D display two differentially expressed miRNAs (upregulated miR-15a and downregulated let-7c). In addition, one target of miR-15a is the antiapoptotic gene Bcl2, and increased miR-15a can impair rTreg survival.71 Let-7c is a TGF-β-regulated miRNA, and its decreased level in serum has been found to correlate with rapid progression to end-stage renal disease in T1D.72 Direct comparison of activated nTregs between pre-T1D patients and healthy controls has shown a significant increase in miR-31 and miR-26a in the pre-T1D group.68 miR-31 has been demonstrated to reduce Foxp3 expression.58 Similar to miR-101, miR-26a has also been shown to target Ezh2, which is related to Foxp3 expression and the suppressive functions of nTregs.68 Jeker et al. found that miR-10a expression was lower in Tregs from NOD mice than in Tregs from autoimmunity-resistant C57BL/6 mice.73 miR-10a is preferentially expressed in Treg cells derived from the mouse thymus but cannot directly modulate Foxp3 or other molecules involved in Treg homeostasis.73 A recent study showed that miR-10a attenuated the phenotypic conversion of inducible Treg cells into follicular helper T cells, which was associated with the inhibition of transcriptional repressor Bcl-6 and co-repressor Ncor2.74 Therefore, because miR-10a exerts important functions in the maintenance of a specific Treg phenotype, the dysregulation of miR-10a may contribute to T1D development. Genome-wide miRNA profiling of Treg cells from patients with T1D has shown a remarkable increase in miR-510 and decreases in miR-191 and miR-342.75 miR-510 is involved in irritable bowel syndrome and plays a crucial role in chronic bowel inflammatory processes.76 miR-342 has been reported to be downregulated in human leukocytes after lipopolysaccharide treatment.77 In addition, regulators of cytokine signaling, such as Ep300, Bmpr2 and Pfgfra, are predicted to be possible targets of miR-342.
Berry et al.78 showed a protective effect of miR-34 against T1D. miR-34a is prominently highly expressed in B-cell progenitors and marginal zone B cells from NOD.B10 Idd9.3 mice and can negatively regulate B-cell lymphopoiesis, subsequently leading to an ineffective expansion of β-cell-specific CD4+ T cells and a protective effect against T1D in NOD.B10 Idd9.3 mice.78 Multiple miRNAs within the 14q32 cluster, such as miR-342, have been found to target the mRNAs of some autoantigens (for example, IA-2, IA-2β and GAD65) in T1D.79
miRNAs: PARTICIPANTS IN AUTOIMMUNITY-MEDIATED β-CELL DAMAGE
Pancreatic β-cells have distinct miRNA expression patterns that can regulate β-cell development and/or functions. miRNA dysregulation in β-cells induced by proinflammatory cytokines or infiltrating immune cells in islets is implicated in β-cell dysfunction (for example, impaired insulin synthesis or secretion) and cell apoptosis, which contribute to the development of T1D.
miRNAs control pancreatic β-cell fate and pancreas formation
Dicer1 or Ago2 deficiency can affect the valuable formation of overall functional miRNA patterns in pancreatic β-cells. A study has shown that specific ablation of Dicer1 in the developing endocrine pancreas in mice at embryonic day 10.5 causes a prominent loss of the β-cell mass.80 However, Dicer deficiency at a late stage of β-cell development does not have an obvious impact on the proliferation and function of mature β-cells but instead affects β-cell apoptosis.80 Moreover, mice lacking Ago2 are unable to achieve compensatory expansion of β-cells in response to insulin resistance.81 During early endocrine pancreatic development, miR-7 negatively regulates α- and β-cell differentiation via direct inhibition of Pax6, which is essential for endocrine pancreatic differentiation.82 Moreover, miR-124a directly targets the transcription factor Foxa2, which is expressed during endocrine pancreatic development and plays an important role in β-cell differentiation.83 Ngn3 is a marker of endocrine progenitor cells and is indispensable for endocrine pancreatic development. Inhibition of miR-15 during pancreatic regeneration leads to increased Ngn3 expression and initiates the regeneration process.84 miR-199b-5p enhances β-cell proliferation at least partially through Mlk3 downregulation.85 miR-375 has also been reported to be essential for the formation and maintenance of the β-cell mass.86
miRNAs control insulin synthesis and secretion
β-cell-specific ablation of Dicer1 can result in the onset of diabetes as a consequence of impaired insulin biosynthesis and glucose-stimulated insulin secretion (GSIS).87, 88, 89 Recent studies have indicated that Ago2 is also critical for the insulin-secreting capacity and compensatory expansion of β-cells.90 Deletion of Ago2 in insulin-producing β-cells (the MIN6 cell line) enhances insulin production.90 miR-375, which is abundantly expressed in islets, reduces glucose-induced insulin synthesis by targeting 3′-phosphoinositide-dependent protein kinase-1 (Pdk1).91 miR-375 overexpression has also been proven to regulate GSIS through directly targeting some genes relevant for insulin exocytosis, such as Aifm1, Gephyrin, Ywhaz and Mtpn.90, 92 miR-30d facilitates insulin gene expression in β-cells by negatively regulating Map4k4, which is a negative modulator of the insulin transcription factor Mafa.93 Moreover, miR-9 has been demonstrated to exert its suppressive effects on insulin secretion via cleavage of the transcription factor Onecut2 and an increase in Granuphilin expression, which accelerate exocytosis processing.94
miRNA dysfunction mediated by autoimmunity contributes to β-cell damage
In 2010, Roggli et al.95 determined the influences of proinflammatory cytokines on miRNA expression in β-cells and investigated their presumable relationship with β-cell survival and/or functions. In their study, Roggli et al. exposed MIN6 cells to the proinflammatory cytokines typically produced by infiltrating immune cells (IL-1β or a cytokine mixture of IL-1β, TNF-α and IFN-γ). The global microarray profiles revealed three significantly upregulated miRNAs (miR-21, miR-34a and miR-146a). The three miRNAs were enhanced upon treatment with IL-1β or the cytokine mixture. IL-1β alone also strongly increased miR-21 and miR-146a expression, although the two miRNAs were also increased in the presence of TNF-α but not IFN-γ. miR-34a is equally induced by stimulation with IL-1β and TNF-α, whereas IFN-γ does not have any impact on its expression. Similar results were observed following exposure of cultured human islets to IL-1β. Interestingly, miRNA expression patterns were obtained from pancreatic islets derived from NOD mice at 8 and 13 weeks of age compared to the patterns obtained from 4-week-old mice. The pancreatic islets of NOD mice at 4 weeks of age did not display any signs of insulitis. Further experiments found that miRNA dysregulation induced by inflammatory cytokines could affect β-cell survival and/or functions. Overexpression of miR-34a led to a small reduction in the insulin content together with a decreased proinsulin mRNA level. In contrast, overexpression of miR-21 or miR-146a did not exert a significant effect on insulin promoter activity, proinsulin mRNA levels, or the insulin content. In addition, miR-34a or miR-21 overexpression did not have any effect on basal insulin secretion but reduced GSIS, which was associated with decreased Vamp2 expression (a soluble N-ethylmaleimide-sensitive factor attachment protein receptor (Snare) protein required for β-cell exocytosis). In addition, inhibition of miR-34a and miR-146a can protect MIN6 cells from cytokine-induced cell apoptosis.95 These findings have indicated a crucial role for these miRNAs in cytokine-mediated β-cell dysfunction. In Ruan’s study, he and his colleagues described a particular β-cell death regulatory pathway involving miR-21, its target gene Pdcd4 and the upstream transcriptional factor nuclear factor-κB (NF-κB). C-Rel and p65 are important members of the NF-κB family. In β-cells, they can activate the promoter of the miR-21 gene, thereby upregulating miR-21 expression; miR-21 sequentially inhibits Pdcd4 expression,96 which is involved in the progression to cell death. Previous studies have shown that Pdcd4-deficient β-cells are resistant to death. In vivo, Pdcd4 deficiency has been found to protect NOD mice against spontaneous diabetes and renders C57BL/6 mice resistant to streptozotocin (STZ)-induced diabetes.96
Another study also performed by Roggli et al.97 showed that changes in the levels of the miR-29 family contributed to the β-cell dysfunction induced by proinflammatory cytokines during the initial phases of T1D. miR-29a/b/c is upregulated in islets isolated from NOD mice during the phases preceding diabetes manifestation. These miRNAs are also augmented in islets isolated from mice and humans after exposure to these cytokines. Transfection of miR-29a/b/c mimics into MIN6 and dissociated islet cells leads to impaired GSIS. In these cells, defective insulin production is associated with a decreased Onecut2 level and a subsequent increase in granuphilin. miR-29a/b/c overexpression also promotes apoptosis through suppressing the expression of the antiapoptotic protein Mcl1.97
In our previous study, we found that IL-1β treatment induced increased miR-101a and miR-30b expression in β-cells and that the two miRNAs contributed to cytokine-mediated β-cell apoptosis and dysfunction.98 miR-101a and miR-30b downregulate proinsulin expression and reduce the insulin content via direct suppression of the transcriptional factor Neurod1. Furthermore, miR-101a/miR-30b-mediated β-cell apoptosis is related to decreased Bcl2 expression. We also showed that miR-101a impaired GSIS by inhibiting Onecut2.98
Recently, Grieco et al. reported that miR-23a, miR-23b and miR-149 were downregulated upon exposure of β-cells to the proinflammatory cytokines IL-1β+IFN-γ. These downregulated miRNAs were further demonstrated to modulate the expression levels of the pro-apoptotic Bh3-only proteins Dp5 and Puma, consequently affecting β-cell apoptosis.99
Multiple low-dose streptozotocin (MLDS)-induced diabetes is an autoimmune disease with clinicopathological features similar to those of human T1D. MLDS can induce marked production of inflammatory cytokines, such as IFN-γ, IL-1β and TNF-α, which mediates β-cell loss. By crossing Dicer loxp mice with mice carrying a Cre allele driven by rat insulin promoter (RIP) II, Mi et al.100 specifically removed Dicer from differentiated β-cells. In their study, the Dicer.RIP cre knockout mice exhibited a significantly accelerated appearance and higher prevalence of MLDS-induced autoimmune diabetes.100 This result highly suggested that miRNA deficiency in β-cells promoted susceptibility to MLDS-induced autoimmune diabetes.
Another study by Bravo-Egana et al.101 estimated the effects of inflammation on miRNA expression in transplanted rat islets. A series of islet miRNAs was influenced by inflammation, of which 26 were upregulated and 5 downregulated. The induction of miRNAs is dependent on NF-κB, which is an important pathway involved in inflammatory responses. Using miRNA and mRNA target-predicting algorithms, a bioinformatics analysis of genome-wide RNA studies of β-cells identified 238 potential miRNA target genes, several of which were theoretically involved in insulin signaling, islet physiology and diabetes pathogenesis.101
Thioredoxin-interacting protein (Txnip) is a key regulator of β-cell loss in diabetes. Txnip expression is increased in diabetes and causes β-cell apoptosis, whereas Txnip deficiency is protective against both T1D and type 2 diabetes. The proapoptotic and diabetogenic functions of Txnip are associated with the Txnip-mediated changes in miRNAs, such as miR-204 and miR-200.102, 103 Txnip can induce miR-204 expression, which in turn regulates Mafa expression and thus modulates β-cell functions, such as insulin production.102 In addition, Txnip impairs β-cell functions by inducing miR-200.103 miR-200 has been demonstrated to target and decrease Zeb1 and promote β-cell apoptosis.103 A recent study showed that IFN-γ increased Txnip expression in β-cells via inhibition of miR-17, whereas overexpression of miR-17 alleviated the cytokine effect.104 IFN-γ has been reported to lead to miR-17 degradation, probably due to the effective activation of Ire1α.105
POTENTIAL APPLICATIONS OF miRNAs: NEW BIOMARKERS AND POTENTIAL THERAPEUTIC TARGETS FOR T1D
miRNAs act as potent biomarkers for T1D
miRNAs in blood cells can be used as potential biomarkers for many autoimmune diseases, including T1D. As reviewed above, miR-326 expression is upregulated in Ab-positive T1D patients compared with Ab-negative T1D subjects. The increase in miR-326 expression correlates with ongoing islet autoimmunity and may serve as a candidate biomarker for the autoimmune process in T1D.63 Yang et al. identified 26 miRNAs and 1218 genes that were differentially expressed in PBMCs from newly diagnosed T1D patients. miR-146 was one of the most downregulated molecules and was linked to ongoing islet autoimmunity in T1D patients.64 In addition, miR-21a and miR-93 are downregulated in the PBMCs of T1D patients compared with healthy individuals.65
In addition to controlling gene expression in blood cells, many miRNAs are also detected in serum or plasma and other body fluids (that is, urine, saliva and breast milk) in association with microparticles, such as microvesicles or exosomes, or with proteins (for example, Argo-2). Although the functions of these miRNAs remain obscure, circulating miRNAs can serve as potential biomarkers for the detection of various forms of cancers and autoimmune diseases. The analysis of miRNA expression patterns in serum, plasma or other body fluids also aids in the development of new approaches for the prediction of T1D. The miR-21 and miR-210 levels are markedly upregulated in the plasma and urine of T1D patients. Urinary miR-126 levels in diabetic patients are markedly lower than the levels in age- or gender-matched controls and are negatively associated with the glycated hemoglobin levels of the patients. In contrast, the plasma miR-126 levels are comparable in both cohorts. T1D patients exhibit dysregulated miR-21, miR-126 and miR-210 levels in plasma or urinary samples, which may indicate an early onset of diabetes-associated diseases.106 In a comprehensive sequencing analysis of serum from new-onset T1D patients, Nielsen et al.107 identified 12 upregulated serum miRNAs (miR-24, miR-25, miR-26a, miR-27a, miR-27b, miR-29a, miR-30a-5p, miR-148a, miR-152, miR-181a, miR-200a and miR-210) in T1D patients, several of which might regulate the expression of genes related to β-cell survival and functions. Notably, miR-25 has been demonstrated to be negatively associated with β-cell functions and positively associated with glycemic control.107 Recently, plasma miRNAs were prospectively obtained from a group of 40 children 1, 3, 6, 12 and 60 months after diagnosis and evaluated for miRNA expression. Six miRNAs (miR-24, miR-146a, miR-194, miR-197, miR-301a and miR-375) at 3 months were associated with residual β-cell functions 6–12 months after the diagnosis.108 miR-375 is highly expressed in islets and is essential for pancreas development, insulin gene expression and insulin secretion. miR-375 can serve as a biomarker for the detection of β-cell death and the prediction of diabetes. Circulating miR-375 is markedly augmented in STZ-injected mice prior to the occurrence of hyperglycemia or in NOD mice 2 weeks before the onset of diabetes, whereas its levels are decreased 1 week post diabetes.109 Interestingly, another study showed that the serum miR-375 level was lower in children with newly diagnosed T1D than in the age-matched control individuals.110 Therefore, we speculated that a sudden increase in circulating miR-375 prior to T1D onset was likely to result from a pool of miR-375 immediately released during the process of β-cell death; in turn, this level will decrease after T1D onset, which may reflect β-cell residual functions.
miRNAs may be potential therapeutic targets for T1D
Abnormal miRNA expression patterns can lead to the dysfunction of immune responses and insulin secretion. Restoration of miRNA expression to normal levels may have therapeutic potential for the control of immune homeostasis and the maintenance of sufficient insulin release. The potential of miRNA gene therapy strategies has been tested in several studies. Anti-miRNA oligonucleotides are one of the most common strategies in miRNA gene therapy; in this strategy, anti-miRNA oligonucleotides specifically bind to miRNA molecules and subsequently prevent the binding of miRNAs to their target genes. Other oligonucleotide-based strategies involve miRNA mimics, which have the same nucleotide sequences as endogenous miRNAs. In addition, the regulation of miRNAs by viral-based or reagent-based transfection has been successfully applied in vivo and in vitro, and has exhibited therapeutic potential for T1D. As discussed above, mice with overexpression of the miR-17-19 cluster in lymphocytes tend to develop autoimmunity.51 The transfection of miR-23b, miR-98 or miR-590-5p mimics into primary T cells leads to reduced Fas and Trail mRNA levels and consequently excessive expansion of diabetogenic cytotoxic T cells.67 Blocking miR-21, miR-34a or miR-146a activity with anti-miRNA oligonucleotides prevents impairment of GSIS in MIN6 cells with IL-1β stimulation.95 However, Lovis et al. showed that blocking miR-34a or miR-146 using oligonucleotides partially prevented palmitate-treated MIN6 cells from undergoing apoptosis but was insufficient to restore normal insulin release.111 In our previous study, we found that transfection of a miR-101a or miR-30b mimic caused β-cell dysfunction, whereas inhibition of miR-101a and miR-30b with antisense molecules alleviated inflammatory cytokine-induced β-cell dysfunction.98
CONCLUSIONS
Accumulating evidence has identified roles for miRNAs in autoimmune T1D. miRNAs act as novel players in the modulation of the immune system and β-cells through regulating their survival and functions. Interestingly, circulating miRNAs also serve as important biomarkers for T1D. Considering the regulatory roles of miRNAs in immune cells and β-cells, we believe that miRNAs can be used as an effective strategy for the treatment of T1D. Although to the best of our knowledge studies on the roles of miRNAs in T1D are in their infancy, progress in our understanding of the roles of miRNA in this disease may help advance its clinical management in the future.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81502359, 81300648 and 81170725) and the Hunan Province Natural Sciences Foundation of China (15JJ3132).
Footnotes
The authors declare no conflict of interest.
References
- Anaya JM. The diagnosis and clinical significance of polyautoimmunity. Autoimmun Rev 2014; 13: 423–426. [DOI] [PubMed] [Google Scholar]
- Kahaly GJ, Hansen MP. Type 1 diabetes associated autoimmunity. Autoimmun Rev 2016; 15: 644–648. [DOI] [PubMed] [Google Scholar]
- Morran MP, Omenn GS, Pietropaolo M. Immunology and genetics of type 1 diabetes. Mt Sinai J Med 2008; 75: 314–327. [DOI] [PubMed] [Google Scholar]
- Stankov K, Benc D, Draskovic D. Genetic and epigenetic factors in etiology of diabetes mellitus type 1. Pediatrics 2013; 132: 1112–1122. [DOI] [PubMed] [Google Scholar]
- Simmons KM, Gottlieb PA, Michels AW. Immune intervention and preservation of pancreatic beta cell function in type 1 diabetes. Curr Diab Rep 2016; 16: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JS, Carlson JA, Brock G. miRNA: the new gene silencer. Am J Clin Pathol 2007; 128: 830–836. [DOI] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5'UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell 2008; 30: 460–471. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang X, Wang F, Zhou L, Yin Z, Fan J et al. MicroRNA-21 lowers blood pressure in spontaneous hypertensive rats by upregulating mitochondrial translation. Circulation 2016; 134: 734–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garo LP, Murugaiyan G. Contribution of MicroRNAs to autoimmune diseases. Cell Mol Life Sci 2016; 73: 2041–2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res 2016; 57 (Suppl 1): i106–i111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstrom K. Micro-RNA in disease and gene therapy. Curr Drug Discov Technol 2011; 8: 76–86. [DOI] [PubMed] [Google Scholar]
- Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol 2015; 888: 85–105. [DOI] [PubMed] [Google Scholar]
- Verma P, Pandey RK, Prajapati P, Prajapati VK. Circulating microRNAs: potential and emerging biomarkers for diagnosis of human infectious diseases. Front Microbiol 2016; 7: 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Tourino I, Arif S, Eichmann M, Peakman M. T cells in type 1 diabetes: instructors, regulators and effectors: a comprehensive review. J Autoimmun 2016; 66: 7–16. [DOI] [PubMed] [Google Scholar]
- Yadav D, Judkowski V, Flodstrom-Tullberg M, Sterling L, Redmond WL, Sherman L et al. B7-2 (CD86) controls the priming of autoreactive CD4 T cell response against pancreatic islets. J Immunol 2004; 173: 3631–3639. [DOI] [PubMed] [Google Scholar]
- Sia C. Imbalance in Th cell polarization and its relevance in type 1 diabetes mellitus. Rev Diabet Stud 2005; 2: 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard CJ, Krakauer M, Bendtzen K, Lund H, Sellebjerg F, Nielsen CH. T helper cell type 1 (Th1), Th2 and Th17 responses to myelin basic protein and disease activity in multiple sclerosis. Immunology 2008; 125: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arif S, Tree TI, Astill TP, Tremble JM, Bishop AJ, Dayan CM et al. Autoreactive T cell responses show proinflammatory polarization in diabetes but a regulatory phenotype in health. J Clin Invest 2004; 113: 451–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science 1995; 268: 1185–1188. [DOI] [PubMed] [Google Scholar]
- Wang B, Andre I, Gonzalez A, Katz JD, Aguet M, Benoist C et al. Interferon-gamma impacts at multiple points during the progression of autoimmune diabetes. Proc Natl Acad Sci USA 1997; 94: 13844–13849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapoport MJ, Jaramillo A, Zipris D, Lazarus AH, Serreze DV, Leiter EH et al. Interleukin 4 reverses T cell proliferative unresponsiveness and prevents the onset of diabetes in nonobese diabetic mice. J Exp Med 1993; 178: 87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennline KJ, Roque-Gaffney E, Monahan M. Recombinant human IL-10 prevents the onset of diabetes in the nonobese diabetic mouse. Clin Immunol Immunopathol 1994; 71: 169–175. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell 2008; 133: 775–787. [DOI] [PubMed] [Google Scholar]
- Jaberi-Douraki M, Pietropaolo M, Khadra A. Continuum model of T-cell avidity: understanding autoreactive and regulatory T-cell responses in type 1 diabetes. J Theor Biol 2015; 383: 93–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T, Xiang Y, Chang C, Zhou Z. Alteration of regulatory T cells in type 1 diabetes mellitus: a comprehensive review. Clin Rev Allergy Immunol 2014; 47: 234–243. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S, Miyara M, Costantino CM, Hafler DA. FOXP3+ regulatory T cells in the human immune system. Nat Rev Immunol 2010; 10: 490–500. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Roncarolo MG. Immune intervention with T regulatory cells: past lessons and future perspectives for type 1 diabetes. Semin Immunol 2011; 23: 182–194. [DOI] [PubMed] [Google Scholar]
- Sharma VK. Kaveri SV, Bayry J. Impaired regulatory T cell function in autoimmune diseases: are microRNAs the culprits? Cell Mol Immunol 2016; 13: 135–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellanby RJ, Thomas D, Phillips JM, Cooke A. Diabetes in non-obese diabetic mice is not associated with quantitative changes in CD4+ CD25+ Foxp3+ regulatory T cells. Immunology 2007; 121: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Herman AE, Matos M, Mathis D, Benoist C. Where CD4+CD25+ T reg cells impinge on autoimmune diabetes. J Exp Med 2005; 202: 1387–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Q, Adams JY, Penaranda C, Melli K, Piaggio E, Sgouroudis E et al. Central role of defective interleukin-2 production in the triggering of islet autoimmune destruction. Immunity 2008; 28: 687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg-Bleyer Y, Baeyens A, You S, Elhage R, Fourcade G, Gregoire S et al. IL-2 reverses established type 1 diabetes in NOD mice by a local effect on pancreatic regulatory T cells. J Exp Med 2010; 207: 1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, He F, Yang Y, Yuan G, Zhang M, Yu X. Th17 cells in type 1 diabetes. Cell Immunol 2012; 280: 16–21. [DOI] [PubMed] [Google Scholar]
- Aggarwal S, Gurney AL. IL-17: prototype member of an emerging cytokine family. J Leukoc Biol 2002; 71: 1–8. [PubMed] [Google Scholar]
- Marwaha AK, Leung NJ, McMurchy AN, Levings MK. TH17 cells in autoimmunity and immunodeficiency: protective or pathogenic? Front Immunol 2012; 3: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton-Williams EE, Palmer SE, Charlton B, Slattery RM. Beta cell MHC class I is a late requirement for diabetes. Proc Natl Acad Sci USA 2003; 100: 6688–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askenasy N. Mechanisms of diabetic autoimmunity: I—the inductive interface between islets and the immune system at onset of inflammation. Immunol Res 2016; 64: 360–368. [DOI] [PubMed] [Google Scholar]
- Di Lorenzo TP, Peakman M, Roep BO. Translational mini-review series on type 1 diabetes: systematic analysis of T cell epitopes in autoimmune diabetes. Clin Exp Immunol 2007; 148: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CY, Rodriguez-Pinto D, Du W, Ahuja A, Henegariu O, Wong FS et al. Treatment with CD20-specific antibody prevents and reverses autoimmune diabetes in mice. J Clin Invest 2007; 117: 3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Nagafuchi S, Anzai K, Kondo S, Kitamura D, Wakana S et al. Direct evidence for the contribution of B cells to the progression of insulitis and the development of diabetes in non-obese diabetic mice. Int Immunol 1997; 9: 1159–1164. [DOI] [PubMed] [Google Scholar]
- Serreze DV, Chapman HD, Varnum DS, Hanson MS, Reifsnyder PC, Richard SD et al. B lymphocytes are essential for the initiation of T cell-mediated autoimmune diabetes: analysis of a new "speed congenic" stock of NOD.Ig mu null mice. J Exp Med 1996; 184: 2049–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert C, Riseili B, Rojas M, Thomas JW. B cell specificity contributes to the outcome of diabetes in nonobese diabetic mice. J Immunol 2001; 167: 5535–5538. [DOI] [PubMed] [Google Scholar]
- Silveira PA, Johnson E, Chapman HD, Bui T, Tisch RM, Serreze DV. The preferential ability of B lymphocytes to act as diabetogenic APC in NOD mice depends on expression of self-antigen-specific immunoglobulin receptors. Eur J Immunol 2002; 32: 3657–3666. [DOI] [PubMed] [Google Scholar]
- Ferris ST, Carrero JA, Unanue ER. Antigen presentation events during the initiation of autoimmune diabetes in the NOD mouse. J Autoimmun 2016; 71: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox A, Richardson SJ, Bone AJ, Foulis AK, Morgan NG. Analysis of islet inflammation in human type 1 diabetes. Clin Exp Immunol 2009; 155: 173–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MS, Bluestone JA. The NOD mouse: a model of immune dysregulation. Annu Rev Immunol 2005; 23: 447–485. [DOI] [PubMed] [Google Scholar]
- Cieslak M, Wojtczak A. Role of pro-inflammatory cytokines of pancreatic islets and prospects of elaboration of new methods for the diabetes treatment. Acta Biochim Pol 2015; 62: 15–21. [DOI] [PubMed] [Google Scholar]
- Pugliese A, Yang M, Kusmarteva I, Heiple T, Vendrame F, Wasserfall C et al. The Juvenile Diabetes Research Foundation Network for Pancreatic Organ Donors with Diabetes (nPOD) Program: goals, operational model and emerging findings. Pediatr Diabetes 2014; 15: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J et al. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med 2005; 201: 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med 2005; 202: 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 2008; 132: 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y et al. Regulation of the germinal center response by microRNA-155. Science 2007; 316: 604–608. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR et al. Requirement of bic/microRNA-155 for normal immune function. Science 2007; 316: 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med 2008; 205: 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med 2008; 205: 1993–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med 2008; 205: 1983–1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu LF, Thai TH, Calado DP, Chaudhry A, Kubo M, Tanaka K et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 2009; 30: 80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouas R, Fayyad-Kazan H, El Zein N, Lewalle P, Rothe F, Simion A et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol 2009; 39: 1608–1618. [DOI] [PubMed] [Google Scholar]
- Du C, Liu C, Kang J, Zhao G, Ye Z, Huang S et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat Immunol 2009; 10: 1252–1259. [DOI] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C et al. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 2008; 132: 860–874. [DOI] [PubMed] [Google Scholar]
- Zhu S, Pan W, Qian Y. MicroRNA in immunity and autoimmunity. J Mol Med 2013; 91: 1039–1050. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Haupt S, Kreuzer JT, Hammitzsch A, Proft F, Neumann C et al. Decreased expression of miR-146a and miR-155 contributes to an abnormal Treg phenotype in patients with rheumatoid arthritis. Ann Rheum Dis 2015; 74: 1265–1274. [DOI] [PubMed] [Google Scholar]
- Sebastiani G, Grieco FA, Spagnuolo I, Galleri L, Cataldo D, Dotta F. Increased expression of microRNA miR-326 in type 1 diabetic patients with ongoing islet autoimmunity. Diabetes Metab Res Rev 2011; 27: 862–866. [DOI] [PubMed] [Google Scholar]
- Yang M, Ye L, Wang B, Gao J, Liu R, Hong J et al. Decreased miR-146 expression in peripheral blood mononuclear cells is correlated with ongoing islet autoimmunity in type 1 diabetes patients 1miR-146. J Diabetes 2015; 7: 158–165. [DOI] [PubMed] [Google Scholar]
- Salas-Perez F, Codner E, Valencia E, Pizarro C, Carrasco E, Perez-Bravo F. MicroRNAs miR-21a and miR-93 are down regulated in peripheral blood mononuclear cells (PBMCs) from patients with type 1 diabetes. Immunobiology 2013; 218: 733–737. [DOI] [PubMed] [Google Scholar]
- Fornari TA, Donate PB, Assis AF, Macedo C, Sakamoto-Hojo ET, Donadi EA et al. Comprehensive survey of miRNA-mRNA interactions reveals that Ccr7 and Cd247 (CD3 zeta) are posttranscriptionally controlled in pancreas infiltrating T lymphocytes of non-obese diabetic (NOD) mice. PLoS ONE 2015; 10: e0142688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong VM, van der Slik AR, Laban S, van 't Slot R, Koeleman BP, Zaldumbide A et al. Survival of autoreactive T lymphocytes by microRNA-mediated regulation of apoptosis through TRAIL and Fas in type 1 diabetes. Genes Immun 2016; 17: 342–348. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng ZP, Naselli G, Bell F, Wettenhall J, Auyeung P et al. MicroRNAs in CD4(+) T cell subsets are markers of disease risk and T cell dysfunction in individuals at risk for type 1 diabetes. J Autoimmun 2016; 68: 52–61. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Kinkel S, Maksimovic J, Bandala-Sanchez E, Tanzer MC, Naselli G et al. The polycomb repressive complex 2 governs life and death of peripheral T cells. Blood 2014; 124: 737–749. [DOI] [PubMed] [Google Scholar]
- Tumes DJ, Onodera A, Suzuki A, Shinoda K, Endo Y, Iwamura C et al. The polycomb protein Ezh2 regulates differentiation and plasticity of CD4(+) T helper type 1 and type 2 cells. Immunity 2013; 39: 819–832. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 2005; 102: 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pezzolesi MG, Satake E, McDonnell KP, Major M, Smiles AM, Krolewski AS. Circulating TGF-beta1-regulated miRNAs and the risk of rapid progression to ESRD in type 1 diabetes. Diabetes 2015; 64: 3285–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeker LT, Zhou X, Gershberg K, de Kouchkovsky D, Morar MM, Stadthagen G et al. MicroRNA 10a marks regulatory T cells. PLoS One 2012; 7: e36684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Kanno T, Nakayamada S, Hirahara K, Sciume G, Muljo SA et al. TGF-beta and retinoic acid induce the microRNA miR-10a, which targets Bcl-6 and constrains the plasticity of helper T cells. Nat Immunol 2012; 13: 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezova RSO, Faltejskova P, Mikulkova Z, Buresova I, Raja KR, Hodek J et al. microRNA-342, microRNA-191 and microRNA-510 are differentially expressed in T regulatory cells of type 1 diabetic patients. Cell Immunol 2010; 260: 70–74. [DOI] [PubMed] [Google Scholar]
- Kapeller J, Houghton LA, Monnikes H, Walstab J, Moller D, Bonisch H et al. First evidence for an association of a functional variant in the microRNA-510 target site of the serotonin receptor-type 3E gene with diarrhea predominant irritable bowel syndrome. Hum Mol Genet 2008; 17: 2967–2977. [DOI] [PubMed] [Google Scholar]
- Schmidt WM, Spiel AO, Jilma B, Wolzt M, Muller M. In vivo profile of the human leukocyte microRNA response to endotoxemia. Biochem Biophys Res Commun 2009; 380: 437–441. [DOI] [PubMed] [Google Scholar]
- Berry GJ, Budgeon LR, Cooper TK, Christensen ND, Waldner H. The type 1 diabetes resistance locus B10 Idd9.3 mediates impaired B-cell lymphopoiesis and implicates microRNA-34a in diabetes protection. Eur J Immunol 2014; 44: 1716–1727. [DOI] [PubMed] [Google Scholar]
- Abuhatzira L, Xu H, Tahhan G, Boulougoura A, Schaffer AA, Notkins AL. Multiple microRNAs within the 14q32 cluster target the mRNAs of major type 1 diabetes autoantigens IA-2, IA-2beta, and GAD65. FASEB J 2015; 29: 4374–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn FC, Skewes-Cox P, Kosaka Y, McManus MT, Harfe BD, German MS. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes 2007; 56: 2938–2945. [DOI] [PubMed] [Google Scholar]
- Tattikota SG, Rathjen T, McAnulty SJ, Wessels HH, Akerman I, van de Bunt M et al. Argonaute2 mediates compensatory expansion of the pancreatic beta cell. Cell Metab 2014; 19: 122–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kredo-Russo S, Mandelbaum AD, Ness A, Alon I, Lennox KA, Behlke MA et al. Pancreas-enriched miRNA refines endocrine cell differentiation. Development 2012; 139: 3021–3031. [DOI] [PubMed] [Google Scholar]
- Baroukh N, Ravier MA, Loder MK, Hill EV, Bounacer A, Scharfmann R et al. MicroRNA-124a regulates Foxa2 expression and intracellular signaling in pancreatic beta-cell lines. J Biol Chem 2007; 282: 19575–19588. [DOI] [PubMed] [Google Scholar]
- Joglekar MV, Parekh VS, Mehta S, Bhonde RR, Hardikar AA. MicroRNA profiling of developing and regenerating pancreas reveal post-transcriptional regulation of neurogenin3. Dev Biol 2007; 311: 603–612. [DOI] [PubMed] [Google Scholar]
- Kunisada R, Yoshida N, Nakamura S, Uchiyama H, Matsumoto H. Enhanced expression of miR-199b-5p promotes proliferation of pancreatic beta-cells by down-regulation of MLK3. Microrna 2016; 5: 57–65. [DOI] [PubMed] [Google Scholar]
- Poy MN, Hausser J, Trajkovski M, Braun M, Collins S, Rorsman P et al. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc Natl Acad Sci USA 2009; 106: 5813–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalis M, Bolmeson C, Esguerra JL, Gupta S, Edlund A, Tormo-Badia N et al. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS ONE 2011; 6: e29166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelbaum AD, Melkman-Zehavi T, Oren R, Kredo-Russo S, Nir T, Dor Y et al. Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. Exp Diabetes Res 2012; 2012: 470302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkman-Zehavi T, Oren R, Kredo-Russo S, Shapira T, Mandelbaum AD, Rivkin N et al. miRNAs control insulin content in pancreatic beta-cells via downregulation of transcriptional repressors. EMBO J 2011; 30: 835–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tattikota SG, Sury MD, Rathjen T, Wessels HH, Pandey AK, You X et al. Argonaute2 regulates the pancreatic beta-cell secretome. Mol Cell Proteomics 2013; 12: 1214–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ouaamari A, Baroukh N, Martens GA, Lebrun P, Pipeleers D, van Obberghen E. miR-375 targets 3'-phosphoinositide-dependent protein kinase-1 and regulates glucose-induced biological responses in pancreatic beta-cells. Diabetes 2008; 57: 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature 2004; 432: 226–230. [DOI] [PubMed] [Google Scholar]
- Zhao X, Mohan R, Ozcan S, Tang X. MicroRNA-30d induces insulin transcription factor MafA and insulin production by targeting mitogen-activated protein 4 kinase 4 (MAP4K4) in pancreatic beta-cells. J Biol Chem 2012; 287: 31155–31164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisance V, Abderrahmani A, Perret-Menoud V, Jacquemin P, Lemaigre F, Regazzi R. MicroRNA-9 controls the expression of Granuphilin/Slp4 and the secretory response of insulin-producing cells. J Biol Chem 2006; 281: 26932–26942. [DOI] [PubMed] [Google Scholar]
- Roggli E, Britan A, Gattesco S, Lin-Marq N, Abderrahmani A, Meda P et al. Involvement of microRNAs in the cytotoxic effects exerted by proinflammatory cytokines on pancreatic beta-cells. Diabetes 2010; 59: 978–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Wang T, Kameswaran V, Wei Q, Johnson DS, Matschinsky F et al. The microRNA-21-PDCD4 axis prevents type 1 diabetes by blocking pancreatic beta cell death. Proc Natl Acad Sci USA 2011; 108: 12030–12035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roggli E, Gattesco S, Caille D, Briet C, Boitard C, Meda P et al. Changes in microRNA expression contribute to pancreatic beta-cell dysfunction in prediabetic NOD mice. Diabetes 2012; 61: 1742–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Wang Z, Tu Y, Shen H, Dai Z, Lin J et al. miR-101a and miR-30b contribute to inflammatory cytokine-mediated beta-cell dysfunction. Lab Invest 2015; 95: 1387–1397. [DOI] [PubMed] [Google Scholar]
- Grieco FA, Sebastiani G, Juan-Mateu J, Villate O, Marroqui L, Ladriere L et al. MicroRNAs miR-23a-3p, miR-23b-3p and miR-149-5p regulate the expression of pro-apoptotic BH3-only proteins DP5 and PUMA in human pancreatic beta cells. Diabetes 2016; 66: 100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi QS, He HZ, Dong Z, Isales C, Zhou L. microRNA deficiency in pancreatic islet cells exacerbates streptozotocin-induced murine autoimmune diabetes. Cell Cycle 2010; 9: 3127–3129. [DOI] [PubMed] [Google Scholar]
- Bravo-Egana V, Rosero S, Klein D, Jiang Z, Vargas N, Tsinoremas N et al. Inflammation-mediated regulation of microRNA expression in transplanted pancreatic islets. J Transplant 2012; 2012: 723614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Chen J, Jing G, Shalev A. Thioredoxin-interacting protein regulates insulin transcription through microRNA-204. Nat Med 2013; 19: 1141–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filios SR, Xu G, Chen J, Hong K, Jing G, Shalev A. MicroRNA-200 is induced by thioredoxin-interacting protein and regulates Zeb1 protein signaling and beta cell apoptosis. J Biol Chem 2014; 289: 36275–36283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K, Xu G, Grayson TB, Shalev A. Cytokines regulate beta-cell thioredoxin-interacting protein (TXNIP) via distinct mechanisms and pathways. J Biol Chem 2016; 291: 8428–8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JP, Wang L, Han D, Wang ES, Huskey NE, Lim L et al. IRE1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic Caspase-2. Science 2012; 338: 818–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipova J, Fischer DC, Dangwal S, Volkmann I, Widera C, Schwarz K et al. Diabetes-associated microRNAs in pediatric patients with type 1 diabetes mellitus: a cross-sectional cohort study. J Clin Endocrinol Metab 2014; 99: E1661–E1665. [DOI] [PubMed] [Google Scholar]
- Nielsen LB, Wang C, Sorensen K, Bang-Berthelsen CH, Hansen L, Andersen ML et al. Circulating levels of microRNA from children with newly diagnosed type 1 diabetes and healthy controls: evidence that miR-25 associates to residual beta-cell function and glycaemic control during disease progression. Exp Diabetes Res 2012; 2012: 896362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samandari N, Mirza AH, Nielsen LB, Kaur S, Hougaard P, Fredheim S et al. Circulating microRNA levels predict residual beta cell function and glycaemic control in children with type 1 diabetes mellitus. Diabetologia 2016; 60: 354–363. [DOI] [PubMed] [Google Scholar]
- Erener S, Mojibian M, Fox JK, Denroche HC, Kieffer TJ. Circulating miR-375 as a biomarker of beta-cell death and diabetes in mice. Endocrinology 2013; 154: 603–608. [DOI] [PubMed] [Google Scholar]
- Marchand L, Jalabert A, Meugnier E, Van den Hende K, Fabien N, Nicolino M et al. miRNA-375 a sensor of glucotoxicity is altered in the serum of children with newly diagnosed type 1 diabetes. J Diabetes Res 2016; 2016: 1869082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovis P, Roggli E, Laybutt DR, Gattesco S, Yang JY, Widmann C et al. Alterations in microRNA expression contribute to fatty acid-induced pancreatic beta-cell dysfunction. Diabetes 2008; 57: 2728–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]