Abstract
Cells require one-carbon units for nucleotide synthesis, methylation and reductive metabolism, and these pathways support the high proliferative rate of cancer cells. As such, anti-folates, drugs that target one-carbon metabolism, have long been used in the treatment of cancer. Amino acids, such as serine are a major one-carbon source, and cancer cells are particularly susceptible to deprivation of one-carbon units by serine restriction or inhibition of de novo serine synthesis. Recent work has also begun to decipher the specific pathways and sub-cellular compartments that are important for one-carbon metabolism in cancer cells. In this review we summarise the historical understanding of one-carbon metabolism in cancer, describe the recent findings regarding the generation and usage of one-carbon units and explore possible future therapeutics that could exploit the dependency of cancer cells on one-carbon metabolism.
Keywords: cancer metabolism, one-carbon metabolism, serine metabolism, folate cycle, nucleotide synthesis, methylation
Introduction and historical overview
Cancer cells adapt their metabolism in order to support enhanced proliferation and survival. The initial evidence for this observation can be attributed to Otto Warburg, who, in the late 1920s reported increased aerobic glycolysis in cancer cells, the eponymous ‘Warburg effect’. Many hypotheses as to the exact causes/metabolic advantages conferred by aerobic glycolysis have been presented: Defects in mitochondria/OXPHOS, enhanced ATP turnover, increased use of glycolytic intermediates as anabolic precursors, avoidance of ROS production, and spatial and energetic constraints have all been suggested to contribute. Although specific targeting of this pathway by chemotherapeutics remains elusive, the increased uptake of glucose by tumours has been exploited for imaging purposes via positron emission topography. Despite this historic appreciation of altered metabolism in cancer cells, it was not included in the initial hallmarks of cancer, published in 2000 (Hanahan and Weinberg, 2000). However, interest in this subject has grown dramatically, culminating in the inclusion of reprogrammed energy metabolism in the 2011 updated hallmarks of cancer (Hanahan and Weinberg, 2011).
Although the importance and function of one-carbon metabolism in cancer has been of intense interest in recent years (Locasale, 2013; Ducker and Rabinowitz, 2017; Mattaini et al, 2016; Yang and Vousden, 2016), the involvement of this metabolic pathway in cancer is long established. One-carbon metabolism encompasses both the folate and methionine cycles and allows cells to generate one-carbon units (also referred to as methyl groups) and utilise them for the biosynthesis of important anabolic precursors and for methylation reactions. Tetrahydrofolate (THF) is synthesised from dietary folic acid and functions as a universal one-carbon acceptor. THF accepts one-carbon units derived from amino acids such as serine and glycine and the resulting ‘methylated-THF’ exists in several interchangeable forms with varying chemical structure. These include formyl-THF, methyl-THF and methylene-THF, which, respectively, donate their one-carbon units to purine synthesis, the methionine recycling pathway (via homocysteine methylation) and thymidylate synthesis (Figure 1) (Tibbetts and Appling, 2010).
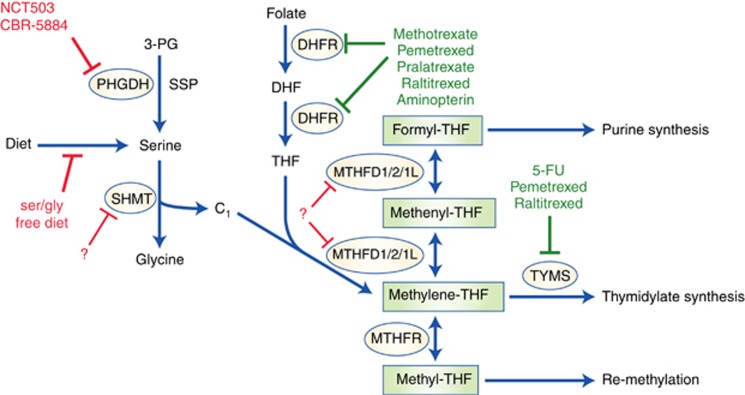
Figure 1.
An overview of one-carbon metabolism and the established/future therapeutics that target this pathway. One-carbon metabolic pathways with established chemotherapeutics highlighted in green and possible targets for future interventions highlighted in red. Solid red lines indicate interventions that are currently in development. Dashed lines indicate possible targets that may be subject to further investigation. Enzymes that regulate these pathways are circled. Serine can be obtained from the diet, or synthesised de novo from the glycolytic intermediate, 3-PG by the SSP, of which PHGDH is a key enzyme. Dietary folate is converted by DHFR first to DHF and then to THF, a one-carbon unit acceptor. Serine is catabolised to glycine by SHMT1/2, which yields a one-carbon unit (C1) that is accepted by THF to form methylene-THF. Methylene-THF can then be converted to formyl-THF via the intermediate methenyl-THF by the action of MTHFD1/2/1L. Formyl-THF donates its one-carbon unit to purine synthesis. Methylene-THF can either donate its one-carbon unit to thymidylate synthesis or be converted by MTHFR to methyl-THF, which supplies one-carbon units for methionine recycling.
The importance of one-carbon metabolism in cancer was initially recognised >60 years ago; in 1948, Sydney Farber observed that dietary folate deficiency in children with acute leukaemia reduced their leukaemic cell number. Specific targeting of folate metabolism in these patients with the folic acid antagonist aminopterin indeed produced a temporary remission (Farber and Diamond, 1948). These discoveries led to the development of the class of drugs known as antifolates, the most well known being methotrexate, which continues to be used as a treatment for many types of cancer. Further insight into the mechanistic action of antifolates was provided in 1958, when it was reported that these drugs bind to and inhibit dihydrofolate reductase (DHFR), the enzyme responsible for the production of THF from folate. Antifolates therefore prevent the formation of THF and thus block one-carbon metabolism (Osborn et al, 1958). However, these drugs have many deleterious side effects due to the importance of THF in healthy tissues. The recent revitalisation in cancer metabolism research is starting to provide further mechanistic insight into the functions and workings of these pathways specifically in the context of cancer. In this review, we will describe the metabolic pathways through which cancer cells obtain one-carbon units and the ways in which these are utilised to support cancer cell proliferation.
One-carbon metabolic pathways
There are several pathways through which cells generate one-carbon units. This includes serine metabolism to glycine, the glycine cleavage system (GCS) as well as metabolism of choline and other amino acids. Recent evidence suggests that cancer cells can alter or become more reliant upon these pathways in order to sustain one-carbon supply for proliferation. Serine can be converted to glycine by the methyltrasferases SHMT1 (cytoplasmic) and SHMT2 (mitochondrial). During this reaction, the one-carbon unit that is cleaved from serine is transferred to THF, generating methylene-THF (Tibbetts and Appling, 2010). This reaction can also run in the opposite direction, producing serine from glycine at the cost of one-carbon units (Labuschagne et al, 2014; Ducker et al, 2016). Serine is a non-essential amino acid and, as such, can be sourced from the intracellular de novo serine synthesis pathway (SSP) that makes serine from the glycolytic intermediate, 3-phosphoglycerate (3-PG). Serine can modulate the availability of 3-PG for the SSP as it is an activator of the glycolytic enzyme, PKM2. During serine starvation, the activity of PKM2 is decreased leading to an increase in 3-PG to supply the SSP (Chaneton et al, 2012). However, cells can readily take up extracellular serine, and cancer cells frequently augment their serine supply by uptake. Generation of serine through lysosomal processes, which can break down proteins from cell intrinsic (e.g., autophagy; Galluzzi et al, 2015) or extrinsic sources (e.g., macropinocytosis; Commisso et al, 2013; Kamphorst et al, 2015), also have the potential to contribute intracellular serine, though the relative importance for these pathways in cancer-specific serine metabolism remains unexplored.
Interest in the role of de novo serine synthesis in cancer was initiated as early as 1955, when it was found that tumours could generate serine from extracellular glucose (Kit, 1955). Further research in the 1980s identified upregulated activity of enzymes of the SSP, as well as SHMT, in cancer cells (Snell, 1985; Snell and Weber, 1986). The incorporation of a one-carbon unit from radiolabelled serine was found in nucleotides, a known output for one-carbon units, suggesting a role for serine in this pathway (Snell et al, 1987). Indeed, more recent studies have found that cancer cells require SHMT (particularly SHMT2) for optimal proliferation and tumorigenicity, indicating the importance of serine catabolism in cancer (Jain et al, 2012; Ye et al, 2014). Additionally, upregulation of SSP genes occurs in several types of cancer, including breast cancer and melanoma (Locasale et al, 2011; Pollari et al, 2011; Possemato et al, 2011). Inhibition of the SSP by small-molecule inhibitors or RNAi targeting core SSP genes reduces xenograft tumour growth (Possemato et al, 2011; Pacold et al, 2016). Extracellular serine also has an important role in supporting cancer cell proliferation through the provision of one-carbon units. Cancer cells that do not exhibit upregulated SSP genes rely upon extracellular serine for survival (Possemato et al, 2011; Maddocks et al, 2013; DeNicola et al, 2015). Serine starvation reduces cancer cell proliferation and xenograft growth by depriving them of the necessary one-carbon units to support anabolism (Maddocks et al, 2013; Labuschagne et al, 2014; Maddocks et al, 2016). There is therefore clear evidence that cancer cells require serine-derived one-carbon units for optimal growth, some support this using exogenous serine, while others increase de novo serine synthesis through genetic upregulation of SSP genes.
In addition to the production of one-carbon units from the conversion of serine to glycine, glycine itself is also a potential source of one-carbon units via the GCS. This process is localised to mitochondria and functions to catabolise glycine oxidatively (Kikuchi et al, 2008). With regard to one-carbon metabolism, the GCS cleaves a methylene group from glycine, which is accepted by THF to produce methylene-THF for use in downstream reactions requiring one-carbon units. This pathway also regenerates NADH from NAD+ and results in the release of CO2 and ammonia (Kikuchi et al, 2008). As a source of one-carbon units, it has been suggested that glycine, and thereby the GCS, promote cancer cell growth. This is supported by the upregulation of and dependency upon a core enzyme of the GCS, glycine decarboxylase, in lung tumour-initiating cells (Zhang et al, 2012) and glioblastoma-derived cells (Kim et al, 2015). Inhibition of the GCS in cancer cell lines with high GCS activity reduces xenograft growth (Zhang et al, 2012). However, further studies have shown that the GCS does not universally support the proliferation of cancer cells or necessarily contribute one-carbon units to nucleotide synthesis (Labuschagne et al, 2014). Excess glycine is in fact detrimental to cancer cell proliferation and tumour growth (Rose et al, 1999a; Rose et al, 1999b). Under conditions of excess glycine and low serine in colorectal cancer cells, the dominant SHMT reaction is the conversion of glycine to serine, which further depletes one-carbon units and inhibits nucleotide synthesis and proliferation (Labuschagne et al, 2014). In glioblastoma cells with elevated SHMT2 activity, increased glycine production can lead to toxicity via accumulation of aldehydes if the glycine is not degraded by the GCS (Kim et al, 2015). Taken together, these studies suggest that, while the GCS can support tumorigenesis, its activity seems to be more dependent on glycine breakdown/detoxification than generation of one-carbon units for nucleotide synthesis. The directionality of serine/glycine conversion is also an important factor in cancer cell metabolism and evidence suggests that mitochondrial SHMT2 is the major serine-to-glycine conversion enzyme in this context (Lewis et al, 2014).
Choline, a vitamin obtained from the diet, can also be a source of one-carbon units through its metabolism to betaine. Betaine is a cofactor that supports the regeneration of methionine from homocysteine. During this process, betaine itself is converted to dimethylglycine. Subsequently, a one-carbon unit is cleaved from dimethylglycine, which is accepted by THF to form methenyl-THF (Ueland, 2011). One-carbon units may also be derived from histidine. Histidine is metabolised to the intermediate, formiminoglutamate. Formiminoglutamante and THF are then converted to glutamate and formimino-THF. Upon removal of an ammonium ion, formimino-THF becomes methenyl-THF and enters the folate cycle. One-carbon units may also be obtained from tryptophan. Formate is produced from tryptophan metabolism and forms formyl-THF. Although these less well-known pathways can theoretically contribute one-carbon units, their importance for one-carbon metabolism in cancer cells is yet to be fully described.
Utilisation of one-carbon units in cancer cells
Nucleotide synthesis
One-carbon units are necessary for the biosynthesis of both purine and pyrimidine nucleotides, both of which are essential for DNA and RNA synthesis. Given the high proliferation rate of cancer cells and the requirement of nucleotides for proliferation, cancer cells have a large demand for one-carbon units for nucleotide synthesis.
Purine nucleotides are synthesised from ribose-5-phosphate, which is generated by the pentose phosphate pathway. Through a series of steps, which require the incorporation of two one-carbon units and one molecule of glycine, inosine monophosphate (IMP) is produced. This is the common precursor to all purine nucleotides. Depletion of serine in cancer cells inhibits proliferation and reduces the level of purine nucleotides (Maddocks et al, 2013; Labuschagne et al, 2014; Maddocks et al, 2016). Inhibition of serine metabolism by serine starvation, deletion, or RNAi-mediated knockdown of SHMT2, causes a build-up in precursors upstream of IMP prior to one-carbon unit incorporation (Labuschagne et al, 2014; Kim et al, 2015; Ducker et al, 2016). Deprivation of serine therefore inhibits cancer cell proliferation by depleting cells of one-carbon units for purine biosynthesis.
One-carbon units are also required for the production of the pyrimidine nucleotide, thymidylate. Specifically, this occurs during the methylation of dUMP to form dTMP, a reaction catalysed by thymidylate synthase (TYMS) and using methylene-THF as the methyl donor. Methylene-THF is converted to DHF during this reaction and is reduced back to THF by DHFR. Both folate deficiency and methotrexate treatment inhibit dTMP synthesis to such a degree that uracil is incorporated into DNA in its place (Goulian et al, 1980; Blount et al, 1997). Methylene-THF production and TYMS activity are coupled to DNA synthesis by the localisation of SHMT1 and TYMS to the nucleus during replication (Woeller et al, 2007; MacFarlane et al, 2011). SHMT1 functions as a scaffold protein to target a thymidylate-producing enzyme complex, including TYMS and DHFR, to DNA. This complex is enriched at replication forks in order to supply thymidylate for DNA synthesis (Anderson et al, 2012). 5-Fluoruoracil (5-FU) is one of the most commonly used chemotherapeutics. It affects many pathways, and one of its functions is as an inhibitor of TYMS. It has been suggested that pyrimidine metabolism is the most affected pathway during 5-FU treatment of colorectal cancer cells (Ser et al, 2016). The success of 5-FU in inhibiting cancer cell proliferation by blocking TYMS demonstrates that one-carbon units, which TYMS also requires, are critical.
Methylation pathways
Tumours often display altered patterns of DNA methylation. DNA methylation regulates gene expression, and of particular interest in cancer, hypermethylation of tumour-suppressor gene promoters can reduce their expression (Kulis and Esteller, 2010). RNA is also subjected to methylation that can regulate translation (Fu et al, 2014). Proteins themselves can be posttranslationally modified by methylation, which can alter function and protein–protein interactions. S-adenosylmethionine (SAM) is a universal methyl donor and is generated by the addition of adenosine from ATP to methionine (Figure 2). Upon the transfer of its methyl group to an acceptor such as DNA, SAM becomes S-adenosylhomocysteine, which is converted to homocysteine. Homocysteine can be recycled back to methionine by the contribution of a methyl group from methyl-THF (Figure 2). Although the primary role of serine in supporting the methionine cycle was thought to be by providing one-carbons for methionine recycling, the activity of this pathway in cancer cells has been found to be low (Shlomi et al, 2014; Mehrmohamadi et al 2014; Maddocks et al, 2016). Recent work shows that serine and glycine metabolism can support the methionine cycle by an alternate mechanism, that of de novo ATP synthesis. Conversion of methionine into SAM requires ATP-derived adenosine, and serine helps provide this adenosine by supplying precursors for de novo purine synthesis, which makes new molecules of ATP. (i.e., ATP created by de novo synthesis, rather than energetic ATP turnover). Hence, serine restriction results in decreased transfer of methyl units to DNA and RNA in cancer cells by decreasing de novo ATP synthesis (Maddocks et al, 2016).
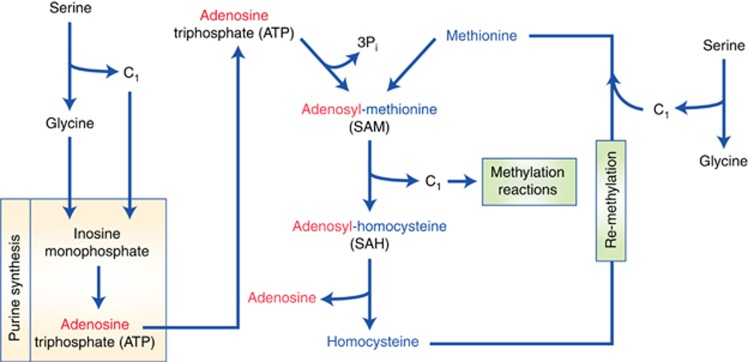
Figure 2.
The contribution of one-carbon metabolism to methylation. Serine-dependent one-carbon metabolism supports the methionine cycle and methylation reactions via two distinct pathways. Serine catabolism provides one-carbon units (C1) and glycine, which are both required for de novo ATP synthesised via purine synthesis. ATP synthesised in this way contributes adenosine for the production of SAM from methionine. After participating in a methylation reaction, SAM becomes S-adenosylhomocysteine (SAH) and then homocysteine. Homocysteine can be re-methylated and recycled back to methionine, which requires a one-carbon unit that can also be sourced from serine catabolism to glycine.
Recent work has highlighted the cross-talk between metabolism and the epigenome. Metabolites such as actyl-coA, AMP and SAM are required for histone acetylation, phosphorylation and methylation of DNA and histones, respectively. The metabolic pathways and enzymes that supply these key compounds are therefore critical for the maintenance and adaptation of the epigenome. Indeed, dietary reduction of methionine decreases SAM levels, leading to diminished histone methylation with significant effects upon gene expression (Mentch et al, 2015). The specific contribution of one-carbon metabolism-dependent DNA methylation in pancreatic cancer has recently been explored. Loss of the serine–threonine kinase, LKB1, promotes tumorigenesis in KRas-mutant pancreatic cancer, and in this context, loss of LKB1 promotes increased expression of SSP enzymes, leading to increased de novo serine synthesis. Serine sourced through this pathway contributes one-carbon units that support SAM synthesis, through either ATP production or methionine recycling. In concordance with this, global DNA methylation was increased in KRas-mutant LKB1-deleted cells as well as the levels of several DNA methyltransferases for which SAM is a critical cofactor. This serine-dependent DNA methylation upon the loss of LKB1 in KRas-mutant cells contributes to tumour growth, presumably through alterations in gene expression (Kottakis et al, 2016). As alterations to DNA methylation are known to be important in cancer, the wider epigenetic implications of one-carbon metabolism inhibition in cancer cells is an important topic for further research.
NADH/NADPH production
NADH and NADPH are important cofactors that provide electrons for redox reactions. These molecules can be produced by one-carbon metabolism and are critical for multiple metabolic and biosynthetic pathways.
One-carbon units derived from formate are accepted by THF to produce formyl-THF. This is an ATP-dependent reaction that is catalysed, in mitochondria, by MTHFD1L. Alternatively, serine-derived one-carbon units form methylene-THF. Formyl-THF and methylene-THF donate their one-carbon units specifically to purine or thymidylate synthesis, respectively. During anabolism, the enzyme MTHFD allows for conversion of methylene-THF to formyl-THF for use in purine biosynthesis. NAD(P)+ is used as a cofactor in this reaction and is reduced to NAD(P)H. The mitochondrial forms of this enzyme, MTHFD2 and MTHFD2L, can use either NAD+ or NADP+ as a cofactor, whereas the cytosolic form, MTHFD1, specifically uses NAD+. During catabolism, the MTHFD2 reaction is run at a much higher rate than the one-carbon unit demands for purine synthesis. This enables cells to increase NADH production, which is subjected to mitochondrial oxidative phosphorylation to turnover ATP (Ducker et al, 2016; Meiser et al, 2016).
Mitochondrial MTHFD2/MTHFD2L can also use NADP+ as a cofactor to convert serine-derived methylene-THF to formyl-THF, this results in the concomitant production of NADPH (Lewis et al, 2014). Mitochondrial NADPH can also be generated by the oxidation of formyl-THF to CO2 and THF by the ALDH1L2 enzyme. Mitochondrial NADPH produced by such pathways provides reducing power for proline synthesis (Ducker et al, 2016). Although predominantly performed in the mitochondria, methylene-THF can be oxidised by MTHFD1 in the cytoplasm and form NADPH, which can support fatty acid synthesis (Ducker et al, 2016). Fatty acids are required for the production of lipid signalling molecules and membranes, and both of these processes are critical for maintaining cancer cell proliferation (Currie et al, 2013).
Future therapeutic strategies
One-carbon units contribute to multiple downstream pathways that are known to or are likely to benefit cancer cell survival. Further detailed understanding of these may allow for more precise targeting of the specific pathways that are most important for cancer cell survival. Traditional antifolate chemotherapeutics such as methotrexate and 5-FU already target one-carbon metabolic pathways (Table 1). However, these drugs have many deleterious side effects due to the importance of folate pathways in healthy proliferating cells and resistance of cancer cells to antifolates is a common problem. Future therapeutics may better target one-carbon metabolism in cancer cells by more selectively inhibiting individual one-carbon pathway enzymes (Figure 1).
Table 1. Established antifolate antineoplastic agents.
| Drug name | Targets | Therapeutic uses |
|---|---|---|
| Aminopterin | DHFR | Initially found to reduce leukaemic cells in children; no longer in use |
| Methotrexate | DHFR | Used to treat a wide range of neoplastic disease |
| Pemetrexed | DHFR, TYMS (and SHMT; Daidone et al, 2011) | Non-small cell lung carcinoma; pleural mesothelioma |
| Pralatrexate | DHFR | Peripheral T-cell lymphoma |
| Raltitrexed | DHFR and TYMS | Metastatic colorectal cancer |
| 5-Fluoruoracil | TYMS | Used to treat a wide range of neoplastic disease |
Abbreviations: DHFR=dihydrofolate reductase; SHMT=serine hydroxymethyltransferase; TYMS=thymidylate synthase.
Small-molecule inhibitors of the SSP targeting PHGDH have been recently developed (Mullarky et al, 2016; Pacold et al, 2016). These have been successful in vitro, reducing cancer cell proliferation (Mullarky et al, 2016; Pacold et al, 2016) and inhibiting xenograft growth specifically in PHGDH-dependent cell lines (Pacold et al, 2016). In order for this approach to be successful clinically, tumours that are PHGDH dependent must be identifiable. Importantly, PHGDH may have other functions that must be considered if PHGDH is to be targeted therapeutically (Liu et al, 2013). For example, PHGDH is overexpressed in glioma and stabilises the transcription factor, FOXM1, leading to increased expression in genes that promote tumour invasion, angiogenesis, and regulate the cell cycle (Liu et al, 2013). Although targeting of the SSP may be a promising approach for tumours that are dependent upon this pathway, this is less likely to be successful in cancer cells that depend upon other sources of serine/one-carbon units.
Cancer cells that do not upregulate SSP activity are instead dependent upon the uptake of extracellular serine for survival (Pollari et al, 2011; Possemato et al, 2011; Maddocks et al, 2013). These may be targeted by lowering the availability of exogenous serine. Indeed, serine and glycine starvation is successful in reducing xenograft and autochtonous tumour growth and significantly improving survival in multiple mouse models of cancer (Maddocks et al, 2013; Maddocks et al, 2017). As such, a dietary intervention to reduce sources of one-carbon units such as serine and glycine may be appropriate to diminish the raw materials from this pathway, rather than inhibiting it with small molecules.
A more universal approach could be taken by targeting enzymes required for one-carbon metabolism regardless of serine origin, such as SHMT1/2 and MTHF enzymes. However, when targeting specific components of one-carbon metabolism, it is possible that tumours could re-wire their metabolism to compensate. MTHF enzymes are responsible for the conversion of methylene-THF to formyl-THF and methyl-THF for use in nucleotide synthesis and methionine recycling. There are several forms of MTHF: cytoplasmic MTHFD1 and mitochondrial MTHFD1L, MTHFD2 and MTHFD2L. MTHFD2 is only expressed in embryonic, tumour, and non-differentiated tissue, whereas MTHFD2L is much more widely expressed (Bolusani et al, 2011; Nilsson et al, 2014). Cells predominantly use the mitochondrial enzymes for one-carbon metabolism; however, if this is inhibited, cells can compensate by using cytoplasmic MTHFD1 (Ducker et al, 2016), which may prove problematic when designing inhibitors. MTHFD1 is critical for cell survival, making it a potentially viable target. This is because THF conjugates cannot be transported across the mitochondrial membrane. Instead, mitochondrial one-carbon units are converted to formate, which can be transported into the cytoplasm and is reconstituted by MTHFD1 to THF forms for cytoplasmic nucleotide synthesis (Ducker et al, 2016).
Further understanding of one-carbon metabolic pathways will allow for the development of new inhibitors and provide better understanding of the contribution of diet to cancer progression. Both of these goals have the potential to improve cancer treatment and, importantly, to inform on which combinations of treatments will achieve the greatest benefit in specific patient populations.
Acknowledgments
We acknowledge funding from Cancer Research UK (CRUK) Career Development Fellowship (to ODKM) C53309/A19702. We thank K Vousden, C Labuschagne and C Winchester for reading the manuscript.
Footnotes
ODKM contributed to CRUK Cancer Research Technology filing of UK Patent Application No. 1609441.9.
References
- Anderson DD, Woeller CF, Chiang EP, Shane B, Stover PJ (2012) Serine hydroxymethyltransferase anchors de novo thymidylate synthesis pathway to nuclear lamina for DNA synthesis. J Biol Chem 287(10): 7051–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolusani S, Young BA, Cole NA, Tibbetts AS, Momb J, Bryant JD, Solmonson A, Appling DR (2011) Mammalian MTHFD2L encodes a mitochondrial methylenetetrahydrofolate dehydrogenase isozyme expressed in adult tissues. J Biol Chem286(7): 5166–5174. [DOI] [PMC free article] [PubMed]
- Blount BC, Mack MM, Wehr CM, MacGregor JT, Hiatt RA, Wang G, Wickramasinghe SN, Everson RB, Ames BN (1997) Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc Natl Acad Sci USA 94(7): 3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaneton B, Hillmann P, Zheng L, Martin AC, Maddocks OD, Chokkathukalam A, Coyle JE, Jankevics A, Holding FP, Vousden KH, Frezza C, O'Reilly M, Gottlieb E (2012) Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491(7424): 458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, Grabocka E, Nofal M, Drebin JA, Thompson CB, Rabinowitz JD, Metallo CM, Vander Heiden MG, Bar-Sagi D (2013) Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497(7451): 633–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie E, Schulze A, Zechner R, Walther TC, Farese RV Jr. (2013) Cellular fatty acid metabolism and cancer. Cell Metab 18(2): 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daidone F, Florio R, Rinaldo S, Contestabile R, di Salvo ML, Cutruzzolà F, Bossa F, Paiardini A (2011) In silico and in vitro validation of serine hydroxymethyltransferase as a chemotherapeutic target of the antifolate drug pemetrexed. Eur J Med Chem 46(5): 1616–1621. [DOI] [PubMed] [Google Scholar]
- DeNicola GM, Chen PH, Mullarky E, Sudderth JA, Hu Z, Wu D, Tang H, Xie Y, Asara JM, Huffman KE, Wistuba II, Minna JD, DeBerardinis RJ, Cantley LC (2015) NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat Genet 47(12): 1475–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Rabinowitz JD (2017) One-Carbon Metabolism in Health and Disease. Cell Metab25: 27–42. [DOI] [PMC free article] [PubMed]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, Rabinowitz JD (2016) Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab 23(6): 1140–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber S, Diamond LK (1948) Temporary remissions in acute leukemia in children produced by folic acid antagonist, 4-aminopteroyl-glutamic acid. N Engl J Med 238(23): 787–793. [DOI] [PubMed] [Google Scholar]
- Fu Y, Dominissini D, Rechavi G, He C (2014) Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat Rev Genet 15(5): 293–306. [DOI] [PubMed] [Google Scholar]
- Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, Kimmelman A, Kumar S, Levine B, Maiuri MC, Martin SJ, Penninger J, Piacentini M, Rubinsztein DC, Simon HU, Simonsen A, Thorburn AM, Velasco G, Ryan KM, Kroemer G (2015) Autophagy in malignant transformation and cancer progression. EMBO J 34(7): 856–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M, Bleile B, Tseng BY (1980) Methotrexate-induced misincorporation of uracil into DNA. Proc Natl Acad Sci USA 77(4): 1956–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100(1): 57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5): 646–674. [DOI] [PubMed] [Google Scholar]
- Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK (2012) Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 336(6084): 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, Vander Heiden MG, Miller G, Drebin JA, Bar-Sagi D, Thompson CB, Rabinowitz JD (2015) Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res 75(3): 544–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi G, Motokawa Y, Yoshida T, Hiraga K (2008) Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci 84(7): 246–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Fiske BP, Birsoy K, Freinkman E, Kami K, Possemato RL, Chudnovsky Y, Pacold ME, Chen WW, Cantor JR, Shelton LM, Gui DY, Kwon M, Ramkissoon SH, Ligon KL, Kang SW, Snuderl M, Vander Heiden MG, Sabatini DM (2015) SHMT2 drives glioma cell survival in ischaemia but imposes a dependence on glycine clearance. Nature 520(7547): 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kit S (1955) The biosynthesis of free glycine and serine by tumors. Cancer Res 15(11): 715–718. [PubMed] [Google Scholar]
- Kottakis F, Nicolay BN, Roumane A, Karnik R, Gu H, Nagle JM, Boukhali M, Hayward MC, Li YY, Chen T, Liesa M, Hammerman PS, Wong KK, Hayes DN, Shirihai OS, Dyson NJ, Haas W, Meissner A, Bardeesy N (2016) LKB1 loss links serine metabolism to DNA methylation and tumorigenesis. Nature 539(7629): 390–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulis M, Esteller M (2010) DNA methylation and cancer. Adv Genet 70: 27–56. [DOI] [PubMed] [Google Scholar]
- Labuschagne CF, van den Broek NJ, Mackay GM, Vousden KH, Maddocks OD (2014) Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep 7(4): 1248–1258. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, Metallo CM (2014) Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol Cell 55(2): 253–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Guo S, Li Q, Yang L, Xia Z, Zhang L, Huang Z, Zhang N (2013) Phosphoglycerate dehydrogenase induces glioma cells proliferation and invasion by stabilizing forkhead box M1. J Neurooncol 111(3): 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW (2013) Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer13: 572–583. [DOI] [PMC free article] [PubMed]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG (2011) Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nat Genet 43(9): 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacFarlane AJ, Anderson DD, Flodby P, Perry CA, Allen RH, Stabler SP, Stover PJ (2011) Nuclear localization of de novo thymidylate biosynthesis pathway is required to prevent uracil accumulation in DNA. J Biol Chem 286(51): 44015–44022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH (2013) Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493(7433): 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks OD, Labuschagne CF, Adams PD, Vousden KH (2016) Serine metabolism supports the methionine cycle and DNA/RNA Methylation through de novo ATP synthesis in cancer cells. Mol Cell 61(2): 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks ODK, Athineos D, Cheung EC, Lee P, Zhang T, van den Broek NJF, Mackay GM, Labuschagne CF, Gay D, Kruiswijk F, Blagih J, Vincent DF, Campbell KJ, Ceteci F, Sansom OJ, Blyth K, Vousden KH (2017) Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature544: 372–376. [DOI] [PubMed]
- Mattaini KR, Sullivan MR, Vander Heiden MG (2016) The importance of serine metabolism in cancer. J Cell Biology214: 249–257. [DOI] [PMC free article] [PubMed]
- Mehrmohamadi M, Liu X, Shestov AA, Locasale JW (2014) Characterization of the usage of the serine metabolic network in human cancer. Cell Reports9: 1507–1519. [DOI] [PMC free article] [PubMed]
- Meiser J, Tumanov S, Maddocks O, Labuschagne CF, Athineos D, Van Den Broek N, Mackay GM, Gottlieb E, Blyth K, Vousden K, Kamphorst JJ, Vazquez A (2016) Serine one-carbon catabolism with formate overflow. Sci Adv 2(10): e1601273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, Nichenametla SN, Locasale JW (2015) Histone methylation dynamics and gene regulation occur through the sensing of one-carbon metabolism. Cell Metab 22(5): 861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullarky E, Lucki NC, Beheshti Zavareh R, Anglin JL, Gomes AP, Nicolay BN, Wong JC, Christen S, Takahashi H, Singh PK, Blenis J, Warren JD, Fendt SM, Asara JM, DeNicola GM, Lyssiotis CA, Lairson LL, Cantley LC (2016) Identification of a small molecule inhibitor of 3-phosphoglycerate dehydrogenase to target serine biosynthesis in cancers. Proc Natl Acad Sci USA 113(7): 1778–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R, Jain M, Madhusudhan N, Sheppard NG, Strittmatter L, Kampf C, Huang J, Asplund A, Mootha VK (2014) Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat Commun5: 3128. [DOI] [PMC free article] [PubMed]
- Osborn MJ, Freeman M, Huennekens FM (1958) Inhibition of dihydrofolic reductase by aminopterin and amethopterin. Proc Soc Exp Biol Med 97(2): 429–431. [DOI] [PubMed] [Google Scholar]
- Pacold ME, Brimacombe KR, Chan SH, Rohde JM, Lewis CA, Swier LJ, Possemato R, Chen WW, Sullivan LB, Fiske BP, Cho S, Freinkman E, Birsoy K, Abu-Remaileh M, Shaul YD, Liu CM, Zhou M, Koh MJ, Chung H, Davidson SM, Luengo A, Wang AQ, Xu X, Yasgar A, Liu L, Rai G, Westover KD, Vander Heiden MG, Shen M, Gray NS, Boxer MB, Sabatini DM (2016) A PHGDH inhibitor reveals coordination of serine synthesis and one-carbon unit fate. Nat Chem Biol 12(6): 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollari S, Kakonen SM, Edgren H, Wolf M, Kohonen P, Sara H, Guise T, Nees M, Kallioniemi O (2011) Enhanced serine production by bone metastatic breast cancer cells stimulates osteoclastogenesis. Breast Cancer Res Treat 125(2): 421–430. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM (2011) Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature 476(7360): 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose ML, Cattley RC, Dunn C, Wong V, Li X, Thurman RG (1999. a) Dietary glycine prevents the development of liver tumors caused by the peroxisome proliferator WY-14,643. Carcinogenesis 20(11): 2075–2081. [DOI] [PubMed] [Google Scholar]
- Rose ML, Madren J, Bunzendahl H, Thurman RG (1999. b) Dietary glycine inhibits the growth of B16 melanoma tumors in mice. Carcinogenesis 20(5): 793–798. [DOI] [PubMed] [Google Scholar]
- Ser Z, Gao X, Johnson C, Mehrmohamadi M, Liu X, Li S, Locasale JW (2016) Targeting one carbon metabolism with an antimetabolite disrupts pyrimidine homeostasis and induces nucleotide overflow. Cell Rep 15(11): 2367–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomi T, Fan J, Tang B, Kruger WD, Rabinowitz JD (2014) Quantitation of cellular metabolic fluxes of methionine. Anal Chem 86(3): 1583–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K (1985) Enzymes of serine metabolism in normal and neoplastic rat tissues. Biochim Biophys Acta 843(3): 276–281. [DOI] [PubMed] [Google Scholar]
- Snell K, Natsumeda Y, Weber G (1987) The modulation of serine metabolism in hepatoma 3924A during different phases of cellular proliferation in culture. Biochem J 245(2): 609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell K, Weber G (1986) Enzymic imbalance in serine metabolism in rat hepatomas. Biochem J 233(2): 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibbetts AS, Appling DR (2010) Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu Rev Nutr 30: 57–81. [DOI] [PubMed] [Google Scholar]
- Ueland PM (2011) Choline and betaine in health and disease. J Inherit Metab Dis 34(1): 3–15. [DOI] [PubMed] [Google Scholar]
- Woeller CF, Anderson DD, Szebenyi DM, Stover PJ (2007) Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J Biol Chem 282(24): 17623–17631. [DOI] [PubMed] [Google Scholar]
- Yang M, Vousden KH (2016) Serine and one-carbon metabolism in cancer. Nat Rev Cancer16: 650–652. [DOI] [PubMed]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, Qing G, Liu Z, Simon MC, Rabinowitz JD, Thompson CB (2014) Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer Discov 4(12): 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, Soh BS, Sun LL, Tai BC, Nga ME, Bhakoo KK, Jayapal SR, Nichane M, Yu Q, Ahmed DA, Tan C, Sing WP, Tam J, Thirugananam A, Noghabi MS, Pang YH, Ang HS, Mitchell W, Robson P, Kaldis P, Soo RA, Swarup S, Lim EH, Lim B (2012) Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 148(1–2): 259–272. [DOI] [PubMed] [Google Scholar]