Abstract
Background:
Sarcomas are rare and heterogeneous cancers. We assessed the contribution of DICER1 mutations to sarcoma development.
Methods:
The coding region of DICER1 was sequenced in 67 sarcomas using a custom Fluidigm Access Array. The RNase III domains were Sanger sequenced in six additional sarcomas to identify hotspot DICER1 variants.
Results:
The median age of sarcoma diagnosis was 45.7 years (range: 3 months to 87.4 years). A recurrent embryonal rhabdomyosarcoma (ERMS) of the broad ligament, first diagnosed at age 23 years, harboured biallelic pathogenic somatic DICER1 variants (1 truncating and 1 RNase IIIb missense). We identified nine other DICER1 variants. One somatic variant (p.L1070V) identified in a pleomorphic sarcoma and one germline variant (c.2257-7A>G) may be pathogenic, but the others are considered to be benign.
Conclusions:
We show that deleterious DICER1 mutations underlie the genetic basis of only a small fraction of sarcomas, in particular ERMS of the urogenital tract.
Keywords: sarcoma, DICER1, mutations, biallelic, embryonal rhabdomyosarcoma
Sarcomas are a rare group of histologically and genetically heterogeneous tumours of mesenchymal origin (Fletcher et al, 2013). Most sarcomas arise sporadically. However, a small number of cases manifest in individuals with germline mutations in genes associated with cancer predisposition syndromes, such as TP53, NF1, RB1, APC, RECQL4, and HRAS (Fletcher et al, 2013; Ballinger et al, 2016; Thomas and Ballinger, 2016). The age of onset of sarcomas is often lower than that observed for most epithelial cancers and, as such, the heritable proportion of sarcomas is likely to be higher than is currently documented (Fletcher et al, 2013; Thomas and Ballinger, 2016). Along with translocations, intra-exonic somatic mutations may also contribute to sarcoma development. In a heterogeneous series of 811 next-generation-sequenced sarcomas, the Cancer Genome Atlas Research Network identifies TP53, PIK3CA, ATRX, PCLO, and LRP1B to be the five most frequently somatically mutated genes (Supplementary Tables S1a and S1b) (cBioPortal for Cancer Genomics).
There are rare reports of sarcomas arising in the context of the DICER1 syndrome (Foulkes et al, 2011; Rio Frio et al, 2011; Kim et al, 2013; Doros et al, 2014; Schultz et al, 2016), a rare paediatric tumour predisposition syndrome caused by germline mutations in DICER1 (OMIM 601200). Priest et al (1996) noted the occurrence of paediatric-onset sarcomas co-occurring with pleuropulmonary blastoma, a tumour now known to be prototypic of the syndrome. Hill et al (2009) further substantiated the association by reporting sarcomas in germline DICER1 mutation carriers. Subsequent reports of sarcomas in DICER1 germline-mutated patients include a para-spinal rhabdomyosarcoma in a 20-year-old (Rio Frio et al, 2011) and a pleomorphic sarcoma of the thigh (consistent with a leiomyosarcoma) in a 26-year-old (Foulkes et al, 2011). A cervical primitive neuroectodermal tumour (Ewing/cPNET) was also reported in a germline DICER1-mutated patient (Foulkes et al, 2011). However, as testing for characteristic second somatic DICER1 RNase IIIb mutations (Foulkes et al, 2014) was not performed, it is not possible to discern whether the lesions are manifestations of the syndrome or co-incidental occurrences. In contrast, an Askin/Ewing family tumour that arose in a 13-year-old germline DICER1 mutation carrier (for more details, see de Kock et al, 2014b) was not found to harbour a characteristic RNase IIIb hotspot mutation (Foulkes, unpublished data). There are also several reports of somatic DICER1 RNase IIIb hotspot mutations in uterine carcinosarcoma (Table 1 and Supplementary Table S1c).
Table 1. Literature review—sarcomas with somatic DICER1 mutations.
| Sarcoma type | Site | Sex | Age of Sarc. Dx | Case | Somatic DICER1 mutation(s) | Germline DICER1 mutation | Mutations in cis or in trans? | Clinically suspicious at time of sarcoma Dx?a | Evidence of DICER1 syndrome (age of Dx) | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| ERMS | Ovary | F | 6y | 1 | c.5425G>A; p.G1809R | c.1196_1197dupAG; p.W400Sfs*59 | Not known | Nob | CN (12y); MNG (13y) | de Kock et al, 2015 |
| Uterine cervix | F | 13y | 2 | c.5113G>A; p.E1705K | c.3907_3908delCT; p.L1303Vfs*4 | Not known | Yes | MNG (11y); LC (13y) | Foulkes et al, 2011 and de Kock et al, 2016 | |
| F | 14y | 3 | c.5438A>G; p.E1813G | c.3611_3616delACTACAinsT | Not known | Yes | MNG (14y) | Foulkes et al, 2011 and de Kock et al, 2016 | ||
| F | 53y | 4 | c.5439G>T; p.D1813D | c.2457C>G; p.I813_Y819del | Not known | Yes | MNG (17y) | Rio Frio et al, 2011 and de Kock et al, 2016 | ||
| F | — | 5 | c.5428G>T; p.D1810Y | None identified | NA | No | — | Heravi-Moussavi et al, 2012 | ||
| F | 13y | 6 | c.5437G>A; p.E1813K | c.3535_3538delTCTT; p.S1179Tfs*12 | Not known | No | LC, likely PPB Type Ir | Tomiak et al, 2014 | ||
| F | Adult | 7 | c.5125G>A; p.D1709Nc | Not done | NA | — | — | Conlon et al, 2015 | ||
| F | 44y | 8 | c.2062C>T; p.R688* & c.5438A>G; p.E1813G | None identified | Not known | No | None | de Kock et al, 2016 | ||
| Uterus | F | 12y | 9 | c.5365-1G>T | None identified | NA | No | — | Doros et al, 2012 | |
| Abdomen | — | — | 10 | c.4259_4261delGAG; p.1418_1420delE | Not done | NA | No | — | Doros et al, 2012 | |
| Brain stem | F | 21y | 11 | c.5125G>A (& LOH) | c.4050+1G>A | Not known | Yes | cERMSd | de Kock et al, 2014a | |
| Lower genital tracte | F | 14y | 12 | c.5428G>C; p.D1810H | c.5387C>T, p.Q1783* | In trans | Nob | MNG (20y) | Fernández-Martínez et al, 2017 | |
| Anaplastic sarcoma | Kidney | F | 21y | 1 | c.2233C>T; p.R745* & c.5437G>A; p.E1813K | None identified | Not known | No | — | Doros et al, 2014 |
| F | 1.75y | 2 | c.5425G>A; p.G1809R | None identified | NA | No | — | Doros et al, 2014 | ||
| F | 9y | 3 | c.5425G>A; p.G1809R | c.2062C>T; p.R688* | In trans | Yes | PPB Type I (8mo) | Wu et al, 2016a | ||
| — | 12y | 4 | c.5125G>A; p.D1709N & c.5138A>T; p.D1713V | Negative | Not known | No | — | Wu et al, 2014 meeting | ||
| F | 7mo | 5 | c.5438A>G; p.E1813G | c.2450delC; p.P817Lfs*15 | In transf | Yes | ASK in CN (7mo) | Wu et al, 2016b | ||
| Liposarcoma | Site not stated | — | — | 1 | p.E1797Dg | Not done | NA | No | — | Kim et al, 2013 |
| — | — | 2 | p.E1797Dg | Not done | NA | No | — | Kim et al, 2013 | ||
| Carcinosarcoma | Uterus | F | — | 1 | c.5425G>A; p.G1809R | c.2516C>T; p.S839F | In trans | No | — | Chen et al, 2015 |
| F | Adult | 2 | c.5437G>C; p.E1813Qh | Not done | NA | — | — | Conlon et al, 2015 | ||
| Ovary | F | — | 3 | c.5438A>G; p.E1813G | Suspected inactivation | In trans | No | — | Chen et al, 2015; Heravi-Moussavi et al, 2012 | |
| Undifferentiated sarc. | Ovary | F | 10y | 1 | c.5125G>A; p.D1709N | c.5096-12G>A | Not known | Nob | SLCT (14y) | Schultz et al, 2016 |
| STS (unknown subtype) | Site not stated | F | 30-39y | 1 | p.G1809R | c.3665dupT; p.L1222fs*13 | Not known | — | — | Schrader et al, 2016 |
Abbreviations: ASK=anaplastic sarcoma of kidney; CN=cystic nephroma; ERMS=embryonal rhabdomyosarcoma; F=female; LC=lung cysts; MNG=multinodular goitre; mo=months; NA=not applicable; PPB=pleuropulmonary blastoma; Sarc.=sarcoma; SLCT=Sertoli–Leydig cell tumour; STS=soft tissue sarcoma; y=years.
Clinically suspicious for DICER1 syndrome.
DICER1 syndrome was not clinically suspected at time of sarcoma diagnosis, but later development of DICER1-associated lesion with or without germline mutation identification led to identification of the syndrome in these patients.
Mutation identified in a metastasis from a primary cervical RMS.
cERMS from this patient not included in the table as no somatic testing was performed.
The site of the ERMS was not reported in the publication; personal communication with the authors revealed the site to be in the lower genital tract.
Not definitive.
Mutation found in tumour, but not confirmed to be somatic.
Mutation identified within a rhabdomyosarcomatous component of a uterine carcinosarcoma.
—No data.
The asterisks shown at the end of certain mutations is HGVS nomenclature indicating a termination codon.
More recently, DICER1 mutations have been strongly implicated in the pathogenesis of embryonal rhabdomyosarcoma (ERMS) of the uterine cervix (cERMS) (Tomiak et al, 2014; de Kock et al, 2016) the ovary (de Kock et al, 2015), and anaplastic sarcoma of the kidney (D1ASK) (Doros et al, 2014; Wu et al, 2016). Characteristic hotspot DICER1 RNase IIIb mutations were identified in the three aforementioned lesions. Biallelic somatic DICER1 mutations were similarly detected in a case of adult-onset cERMS (de Kock et al, 2016).
Despite the above evidence, the true contribution of DICER1 mutations to sarcomas is not yet known. In this study, we aimed to uncover the contribution of DICER1 mutations to a convenience sample of 61 predominantly adult-onset sarcomas of various subtypes. We recruited an additional 12 Ewing sarcomas consequent to the observation of a cPNET/Ewing and Askin/Ewing family tumour in DICER1 kindred, as described above, for a total of 73 sarcomas.
Materials and methods
Patients and samples
We collected 73 sarcomas of 24 different subtypes, as detailed in the Supplementary Materials and Methods. Age of diagnosis ranged from ages 3 months to 87.4 years (median age 45.7 years), and 38 of the patients were female and 35 were male. This study was approved by the Institutional Review Board (IRB) of the Faculty of Medicine of McGill University, Montreal, Quebec, Canada, number A12-M117-11A, and patients signed consent forms in accordance with the IRB approval.
DICER1 screening
Fluidigm access array
We screened the full DICER1 coding region and exon–intron boundaries in tumour gDNA from 67 (of 73) sarcomas (Supplementary Tables S2a and S2b) using a custom Fluidigm Access Array, which targets all exons and exon–intron boundaries of DICER1, followed by next-generation sequencing on an Illumina (San Diego, CA, USA) MiSeq, as previously described (de Kock et al, 2014a). All identified mutations were validated by Sanger sequencing and matched-normal gDNA, if available, was used to determine whether mutations were germline or somatic in origin.
Sanger sequencing
For the six remaining sarcomas (all FFPE-derived) (Supplementary Tables S2a and S2b), we focused our investigation on the RNase domains of DICER1 to identify known hotspot mutations (Foulkes et al, 2014). The regions encoding the RNase III domains were PCR amplified and Sanger sequenced (de Kock et al, 2014b). Other regions of DICER1 were not sequenced in these six samples.
MLPA assay
We screened for deletions or duplications of DICER1 in the germline of 53 patients from whom good quality non-tumour DNA was available (cases 1–52 and 56) using an in-house multiplex ligation-dependent probe amplification (MLPA) assay, as described previously (Sabbaghian et al, 2014).
Details of bioinformatics methods, cloning experiments, mosaicism experiments, TruSight Tumour 15 panel sequencing of case 1 (including TP53 gene, Illumina), and DICER1 copy number variation (CNV) experiments are provided in the supplement (Materials and Methods section).
Results
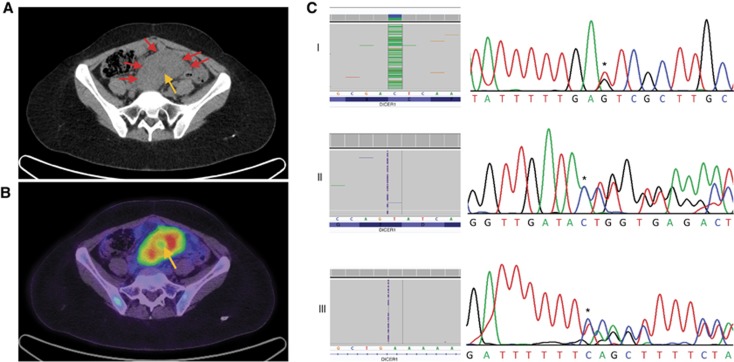
We identified multiple DICER1 variants in an ultimately fatal case of abdominal ERMS that arose in a 23-year-old female following a short history of abdominal pain (case 1) (Figure 1 and Supplementary Tables S2a and S3). Two of these variants are likely to be pathogenic (discussed below). The ERMS was detected on ultrasound as a mixed solid and cystic pelvic mass in the broad ligament, measuring ∼20 cm in its longest diameter with a 10–11 cm solid component (Figure 1). The ERMS, obtained following chemo- and radiotherapy (see Figure 1), harboured a DICER1 RNase IIIb hotspot mutation in exon 25 (c.5439G>T; p.E1813D), which co-occurred with a predicted-truncating DICER1 mutation in exon 11 (c.1785_1786insA; p.T596Nfs*3), both of which were not detected by regular sequencing techniques in the patient’s germline. The patient carried an additional germline insertion (c.2040+53_2040+54insT) in intron 12 of DICER1 (Figure 1C). Experiments to investigate a potential mosaic origin of the exon 25 and exon 11 mutations suggest that neither are likely to be mosaic in nature (Supplementary Table S4). Given the young age of sarcoma onset, we also screened the patient’s germline and tumour samples for TP53 mutations and did not identify any pathogenic TP53 alterations (Supplementary Table S5). Further characterisation of the DICER1 mutations revealed that the exon 11 mutation was in trans with both the intron 12 and exon 25 mutations. The latter two were therefore present in cis (Figure 2 and Supplementary Figure S1). Cloning of a cDNA fragment encompassing all three mutations revealed that the transcript bearing the exon 11, c.1785_1786insA insertion was almost always degraded by nonsense-mediated decay as only 3 of 48 sequenced clones expressed the mutation. No cDNA clones were found to exhibit aberrant splicing as a consequence of the intron 12, c.2040+53_2040+54insT variant, indicating that this variant is most likely to be non-contributory (Supplementary Figure S1).
Figure 1.
Diagnostic images and mutations for case 1. (A) Axial computed tomography (CT) of pelvis demonstrating a solid pre-sacral soft tissue mass (arrows) with low attenuation signal suggesting central cystic/necrotic change (bottom right arrow). (B) Fused positron emission tomographic and CT image of pre-sacral mass demonstrating high metabolic activity as reflected by F-18 flourodeoxyglucose avidity. Central area of reduced activity coincides with area of central tumor necrosis (arrow). Following surgical resection of the tumour, the patient underwent chemotherapy (vincristine, doxorubicin, and cyclophosphamide) for 4 months and radiotherapy of the abdomen and pelvis (24 Gy). Recurrent pelvic disease was detected after an 18-month disease-free interval. Surgical resection was attempted, but complications were incurred. Three months later, recurrent disease was again noted on positron emission tomography imaging. Two cycles of irinotecan/temozolamide chemotherapy were administered. Fifty-two months after initial diagnosis, the patient succumbed to her disease. (C) The exon 25 c.5439G>T somatic mutation (Panel I), exon 11 c.1785_1786insA somatic mutation (Panel II) and intron 12 c.2040+53_2040+54insT germline mutation (Panel III) as seen in Fluidigm-derived data (left) and chromatogram (right). The mutations are indicated by an asterisk and the wild-type sequence is provided below each chromatogram. A full color version of this figure is available at the British Journal of Cancer journal online.
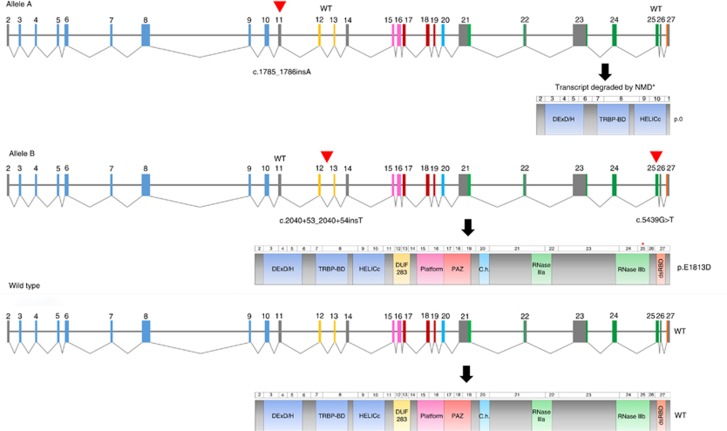
Figure 2.
Graphic depiction of biallelic nature of DICER1 mutations identified in the recurrent ERMS of case 1. The exon 11 c.1785_1786insA mutation is in trans (Allele A, top panel) with the intron 12 c.2040+53_2040+54insT and exon 25 c.5439G>T mutations (Allele B, middle panel). The mutations are indicated by a triangle. Only 3 of 48 clones were found to express the exon 11 mutation, suggesting that the mutated transcript is almost always degraded by nonsense mediated decay (NMD) and thus, no protein is likely to be produced from this allele (p.0). No clones were found to exhibit aberrant splicing as a consequence of the intronic c.2040+53_2040_54insT mutation. As such, the resulting protein is predicted to be normal, except for the single amino acid substitution at position p.E1813 (asterisk, Allele B, middle panel). The wild-type (WT) scenario is depicted in the bottom panel. A full color version of this figure is available at the British Journal of Cancer journal online.
Because of DICER1’s involvement in the above-mentioned ERMS, we sequenced a further 72 sarcomas (60 sarcomas of various subtypes and 12 Ewing sarcomas; Supplementary Tables S2a and S2b) and an additional 9 DICER1 variants were identified (Supplementary Table S3). Of the nine variants, six were established to be germline in origin, two were somatic, and for one variant, the germline vs somatic origin remains undetermined (no germline DNA sample available). One somatic variant, c.3208C>G (p.L1070V), identified in a pleomorphic sarcoma with giant cells (case 46), is predicted to be damaging by both PolyPhen2 and SIFT with a score of 1 and 0.01, respectively. However, no additional characteristic RNase IIIb hotspot mutation was found within this sarcoma and therefore its causal role remains speculative. An intronic DICER1 variant, c.2257-7A>G, had previously been identified in the germline of patient 73. However, no RNase IIIb mutation was identified in the Ewing sarcoma. Based on mutation frequency data and in silico effect predictions, the remaining seven variants are unlikely to be involved in the pathogenesis of the sarcomas in question. Germline deletions in DICER1 have also been found to predispose to the DICER1 syndrome (Sabbaghian et al, 2014). We therefore screened for deletions or duplications in the germline of 53 patients from whom good quality non-tumour DNA was available (cases 1–52 and 56) and no such alterations were identified (Supplementary Figure S3). Copy number alterations of DICER1 have been identified in various cancers including breast cancer, ovarian cancer and melanoma (Zhang et al, 2006; Pugh et al, 2014). We screened for CNVs of DICER1 in 59 sarcomas using a ddPCR experiment (chosen due to low DNA input requirement) and detected copy number changes involving the DICER1 locus in 5 cases (8.5%), each of which was a unique subtype (Supplementary Table S6). However, the extent of the CNVs is not accurately definable using the ddPCR system.
Discussion
DICER1 is an RNase III endoribonuclease responsible for processing hairpin precursor microRNAs (miRNAs) into mature miRNAs, which in turn, regulate the expression of messenger RNAs (Foulkes et al, 2014). Germline mutations in DICER1 predispose to several early childhood or adolescent-onset phenotypes, including pleuroplumonary blastoma, Sertoli-Leydig cell tumour and paediatric cystic nephroma (Foulkes et al, 2014). Genetically, DICER1 syndrome-associated tumours are most often characterised by a predisposing germline DICER1 mutation that inactivates one allele, coupled with a highly distinctive second somatic missense mutation affecting one of the RNase IIIb metal ion-binding sites on the other allele (Foulkes et al, 2014). The biallelically mutated recurrent ERMS in our study (case 1) demonstrates that such mutations may contribute to the development of ERMS, even if both mutations are acquired somatically. Although most DICER1-related lesions manifest in early childhood (Foulkes et al, 2014), it is becoming increasingly evident that the acquisition of two somatic DICER1 mutations can lead to a later-onset of neoplasia (de Kock et al, 2016), as was observed in case 1.
Most ERMS that arise in the context of DICER1 mutations involve the urogenital system and interestingly, the biallelically mutated ERMS from our study arose in the broad ligament, which is the peritoneal fold that attaches the uterus, fallopian tubes and ovaries to the pelvis. Although a limited number of other sarcoma subtypes have been found to carry both truncating and/or RNase IIIb hotspot somatic DICER1 mutations (Table 1 and Supplementary Table S1c) (de Kock and Foulkes, 2016), ERMS appears to be the subtype that is most commonly DICER1 mutated. Clinicians should be mindful of the association between ERMS and DICER1 syndrome. Genetic testing should be performed particularly if ERMS are seen to arise in constellation with one or more known DICER1 syndrome phenotypes, as the identification of germline DICER1 mutations has important implications for the screening and counselling of patients and their families.
In summary, our study demonstrates that likely-pathogenic DICER1 mutations underlie the genetic basis of only a small fraction of sarcomas, with ERMS the most likely sarcoma subtype to harbour such mutations. Conversely, the occurrence of a sarcoma at a site other than the genito-urinary system and of a type other than an ERMS (with the exception of anaplastic sarcoma of the kidney) is not suggestive of the DICER1 syndrome.
Acknowledgments
We thank the patients for their participation in the study, Ms Leanne Taylor, Ms Brooke Backman, and Mr HJ Dubbink for providing pathology material, and Dr Leon van Kempen, Dr George Chong, and Karin Delorme of the Molecular Diagnostics Laboratory, Jewish General Hospital, for assistance with the TruSight Tumor 15 capture and sequencing of case 1. We also thank Nelly Sabbaghian for assistance with the MLPA assay, and Dr Tara Paton and Guillermo Casallo of The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada, for assistance with the ddPCR experiments. A portion of biospecimens and data used in this research were obtained from the Victorian Cancer Biobank, Victoria, Australia, with appropriate ethics approval. The Victorian Cancer Biobank is supported by the Victorian Government. This work was supported by Alex’s Lemonade Stand Foundation and by C17, the latter with funding from Childhood Cancer Canada Foundation (to WDF), the Vanier Canada Graduate Scholarship (to LdK), and the Kate McGarrigle Fellowship in Sarcoma Research of the Cedars Cancer Centre (to BR).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Ballinger ML, Goode DL, Ray-Coquard I, James PA, Mitchell G, Niedermayr E, Puri A, Schiffman JD, Dite GS, Cipponi A, Maki RG, Brohl AS, Myklebost O, Stratford EW, Lorenz S, Ahn SM, Ahn JH, Kim JE, Shanley S, Beshay V, Randall RL, Judson I, Seddon B, Campbell IG, Young MA, Sarin R, Blay JY, O'Donoghue SI, Thomas DM (2016) Monogenic and polygenic determinants of sarcoma risk: an international genetic study. Lancet Oncol 17(9): 1261–1271. [DOI] [PubMed] [Google Scholar]
- cBioPortal for Cancer Genomics The Cancer Genome Atlas (TCGA) Research Network: Sarcoma. Available at: http://www.cbioportal.org/index.do (accessed 23 August 2016).
- Chen J, Wang Y, McMonechy MK, Anglesio MS, Yang W, Senz J, Maines-Bandiera S, Rosner J, Trigo-Gonzalez G, Grace Cheng SW, Kim J, Matzuk MM, Morin GB, Huntsman DG (2015) Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis. J Pathol 237(2): 215–225. [DOI] [PubMed] [Google Scholar]
- Conlon N, Schultheis AM, Piscuoglio S, Silva A, Guerra E, Tornos C, Reuter VE, Soslow RA, Young RH, Oliva E, Weigelt B (2015) A survey of DICER1 hotspot mutations in ovarian and testicular sex cord-stromal tumors. Mod Pathol 28(12): 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kock L, Boshari T, Martinelli F, Wojcik E, Niedziela M, Foulkes WD (2016) Adult-onset cervical embryonal rhabdomyosarcoma and DICER1 mutations. J Low Genit Tract Dis 20(1): e8–e10. [DOI] [PubMed] [Google Scholar]
- de Kock L, Druker H, Weber E, Hamel N, Traubici J, Malkin D, Arseneau J, Stewart CJ, Bouron-Dal Soglio D, Priest JR, Foulkes WD (2015) Ovarian embryonal rhabdomyosarcoma is a rare manifestation of the DICER1 syndrome. Hum Pathol 46(6): 917–922. [DOI] [PubMed] [Google Scholar]
- de Kock L, Foulkes WD (2016) Sarcoma and germ-line DICER1 mutations. Lancet Oncol 17(11): e470. [DOI] [PubMed] [Google Scholar]
- de Kock L, Sabbaghian N, Plourde F, Srivastava A, Weber E, Bouron-Dal Soglio D, Hamel N, Choi JH, Park SH, Deal CL, Kelsey MM, Dishop MK, Esbenshade A, Kuttesch JF, Jacques TS, Perry A, Leichter H, Maeder P, Brundler MA, Warner J, Neal J, Zacharin M, Korbonits M, Cole T, Traunecker H, McLean TW, Rotondo F, Lepage P, Albrecht S, Horvath E, Kovacs K, Priest JR, Foulkes WD (2014. a) Pituitary blastoma: a pathognomonic feature of germ-line DICER1 mutations. Acta Neuropathol 128(1): 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kock L, Sabbaghian N, Soglio DB, Guillerman RP, Park BK, Chami R, Deal CL, Priest JR, Foulkes WD (2014. b) Exploring the association between DICER1 mutations and differentiated thyroid carcinoma. J Clin Endocrinol Metab 99(6): E1072–E1077. [DOI] [PubMed] [Google Scholar]
- Doros L, Yang J, Dehner L, Rossi CT, Skiver K, Jarzembowski JA, Messinger Y, Schultz KA, Williams G, Andre N, Hill DA (2012) DICER1 mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr Blood Cancer 59(3): 558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doros LA, Rossi CT, Yang J, Field A, Williams GM, Messinger Y, Cajaiba MM, Perlman EJ, A Schultz K, Cathro HP, Legallo RD, LaFortune KA, Chikwava KR, Faria P, Geller JI, Dome JS, Mullen EA, Gratias EJ, Dehner LP, Hill DA (2014) DICER1 mutations in childhood cystic nephroma and its relationship to DICER1-renal sarcoma. Mod Pathol 27(9): 1267–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez L, Villegas JA, Santamaria I, Pitiot AS, Alvarado MG, Fernandez S, Torres H, Paredes A, Blay P, Balbin M (2017) Identification of somatic and germ-line DICER1 mutations in pleuropulmonary blastoma, cystic nephroma and rhabdomyosarcoma tumors within a DICER1 syndrome pedigree. BMC Cancer 17(1): 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher C, Hogendoorn P, Merterns F, Bridge J (2013) (eds). World Health Organisation Classification of Tumours of Soft Tissue and Bone. International Agency for Research on Cancer Press: Lyon, France. [Google Scholar]
- Foulkes WD, Bahubeshi A, Hamel N, Pasini B, Asioli S, Baynam G, Choong CS, Charles A, Frieder RP, Dishop MK, Graf N, Ekim M, Bouron-Dal Soglio D, Arseneau J, Young RH, Sabbaghian N, Srivastava A, Tischkowitz MD, Priest JR (2011) Extending the phenotypes associated with DICER1 mutations. Hum Mutat 32(12): 1381–1384. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Priest JR, Duchaine TF (2014) DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer 14(10): 662–672. [DOI] [PubMed] [Google Scholar]
- Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, Fejes AP, Chow C, Tone A, Kalloger SE, Hamel N, Roth A, Ha G, Wan AN, Maines-Bandiera S, Salamanca C, Pasini B, Clarke BA, Lee AF, Lee CH, Zhao C, Young RH, Aparicio SA, Sorensen PH, Woo MM, Boyd N, Jones SJ, Hirst M, Marra MA, Gilks B, Shah SP, Foulkes WD, Morin GB, Huntsman DG (2012) Recurrent DICER1 hotspot mutations in endometrial tumours and their impact on microRNA biogenesis. New Engl J Med 366(3): 234–42.22187960 [Google Scholar]
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, Williams G, Bracamontes D, Messinger Y, Goodfellow PJ (2009) DICER1 mutations in familial pleuropulmonary blastoma. Science 325(5943): 965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Lee SH, Yoo NJ, Lee SH (2013) DICER1 exons 25 and 26 mutations are rare in common human tumours besides Sertoli-Leydig cell tumour. Histopathology 63(3): 436–438. [DOI] [PubMed] [Google Scholar]
- Priest JR, Watterson J, Strong L, Huff V, Woods WG, Byrd RL, Friend SH, Newsham I, Amylon MD, Pappo A, Mahoney DH, Langston C, Heyn R, Kohut G, Freyer DR, Bostrom B, Richardson MS, Barredo J, Dehner LP (1996) Pleuropulmonary blastoma: a marker for familial disease. J Pediatr 128(2): 220–224. [DOI] [PubMed] [Google Scholar]
- Pugh TJ, Yu W, Yang J, Field AL, Ambrogio L, Carter SL, Cibulskis K, Giannikopoulos P, Kiezun A, Kim J, McKenna A, Nickerson E, Getz G, Hoffher S, Messinger YH, Dehner LP, Roberts CWM, Rodriguez-Galindo C, Williams GM, Rossi CT, Meyerson M, Hill DA (2014) Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene 33(45): 5295–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio Frio T, Bahubeshi A, Kanellopoulou C, Hamel N, Niedziela M, Sabbaghian N, Pouchet C, Gilbert L, O'Brien PK, Serfas K, Broderick P, Houlston RS, Lesueur F, Bonora E, Muljo S, Schimke RN, Bouron-Dal Soglio D, Arseneau J, Schultz KA, Priest JR, Nguyen VH, Harach HR, Livingston DM, Foulkes WD, Tischkowitz M (2011) DICER1 mutations in familial multinodular goiter with and without ovarian Sertoli-Leydig cell tumors. JAMA 305(1): 68–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbaghian N, Srivastava A, Hamel N, Plourde F, Gajtko-Metera M, Niedziela M, Foulkes WD (2014) Germ-line deletion in DICER1 revealed by a novel MLPA assay using synthetic oligonucleotides. Eur J Hum Genet 22(4): 564–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader KA, Cheng DT, Joseph V, Prasad M, Walsh M, Zehir A, Ni A, Thomas T, Benayed R, Ashraf A, Lincoln A, Arcila M, Stadler Z, Solit D, Hyman DM, Zhang L, Klimstra D, Ladanyi M, Offit K, Berger M, Robson M (2016) Germline variants in targeted tumor sequencing using matched normal DNA. JAMA Oncol 2(1): 104–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz KA, Harris A, Messinger Y, Sencer S, Baldinger S, Dehner LP, Hill DA (2016) Ovarian tumors related to intronic mutations in DICER1: a report from the international ovarian and testicular stromal tumor registry. Fam Cancer 15(1): 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Ballinger ML (2016) Diagnosis and management of hereditary sarcoma. In: Pichert G, Jacobs C (eds) Rare Hereditary Cancers: Diagnosis and Management. Springer International Publishing: Cham, Switzerland, pp. 169–189. [Google Scholar]
- Tomiak E, de Kock L, Grynspan D, Ramphal R, Foulkes WD (2014) DICER1 mutations in an adolescent with cervical embryonal rhabdomyosarcoma (cERMS). Pediatr Blood Cancer 61(3): 568–569. [DOI] [PubMed] [Google Scholar]
- Wu M, Druker H, Thorner P, Traubici J, Grant R, Albrecht S, Weber E, Charles A, Priest JR, Fabian MR, Watanabe N, Vujanic G, Foulkes WD (2014) DICER1 mutations occurring in childhood anaplastic sarcoma of kidney (Abstract/Program No. 3383S W). Presented at the 64th Annual Meeting of The American Society of Human Genetics, 19 October 2014, San Diego, CA, USA.
- Wu MK, Cotter MB, Pears J, McDermott MB, Fabian MR, Foulkes WD, O'Sullivan MJ (2016. a) Tumor progression in DICER1-mutated cystic nephroma-witnessing the genesis of anaplastic sarcoma of the kidney. Hum Pathol 53: 114–120. [DOI] [PubMed] [Google Scholar]
- Wu MK, Goudie C, Druker H, Thorner P, Traubici J, Grant R, Albrecht S, Weber E, Charles A, Priest JR, Fabian MR, Foulkes WD (2016. b) Evolution of renal cysts to anaplastic sarcoma of kidney in a child with DICER1 syndrome. Pediatr Blood Cancer 63(7): 1272–1275. [DOI] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O'Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G (2006) microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci USA 103(24): 9136–9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.