Dear Editor,
Strigolactones (SLs) are a group of carotenoid-derived small molecules synthesized by plants. As a special class of plant hormones, SLs regulate shoot branching1,2,3,4,5,6,7. Intriguingly, SLs are also exuded into the soil as rhizospheric signals for communication with arbuscular mycorrhizal fungi (AMF)8 and seeds of root parasitic weeds9,10,11: SLs facilitate AMF symbiosis with a wide range of land plants, which improves nutrient and water uptake of the host plants1, and stimulate germination of the seeds of root parasitic weeds such as Striga for parasitic growth on host plants, which causes agricultural disaster with the loss of billions of dollars' worth of crops every year9.
Previous studies have identified several key components in the SL signaling pathway. In model plant Arabidopsis, the F-box protein MAX2 (MORE AXILLARY GROWTH2) regulates both SL-repressed shoot branching and karrikin-induced seed germination12,13; however, the α/β hydrolase D14 (DWARF14) regulates shoot branching but not seed germination14, the repressor proteins SMXL6/7/8 (SMAX1-LIKE family) function in shoot branching15,16, while SMAX1 (SUPPRESSOR OF MAX2 1) regulates seed germination17. Most recently, we have defined D14 as a non-canonical hormone receptor that mediates the hydrolysis of SL to a covalently linked intermediate molecule (CLIM), which serves as the active form of SL that covalently engages the interior of D14 thus effecting plant branching suppression18.
Studies on SL perception by parasitic weeds remain challenging due to the genetic intractability of the phenotypes and the obligate parasitic property of the experimental system9. A group of α/β hydrolase ShHTLs/ShKAI2s (Striga hermonthica HYPO-SENSITIVE TO LIGHT/KARRIKIN INSENSITIVE2 proteins), which are paralogs of D14, were uncovered to hydrolyze SL and mediate the SL-induced seed germination9,10,11. These ShHTLs, including the most active ShHTL7, were proposed to act as the SL receptor in Striga hermonthica9,10,11. However, the active form of SL perceived by root parasitic weeds is still unknown. It is also not clear what is the genuine receptor that binds to the active form of SL. Here, we experimentally define ShHTL7 as a non-canonical receptor that generates and perceives the SL hydrolysis intermediate CLIM, which serves as an active form of SL to regulate Striga seed germination.
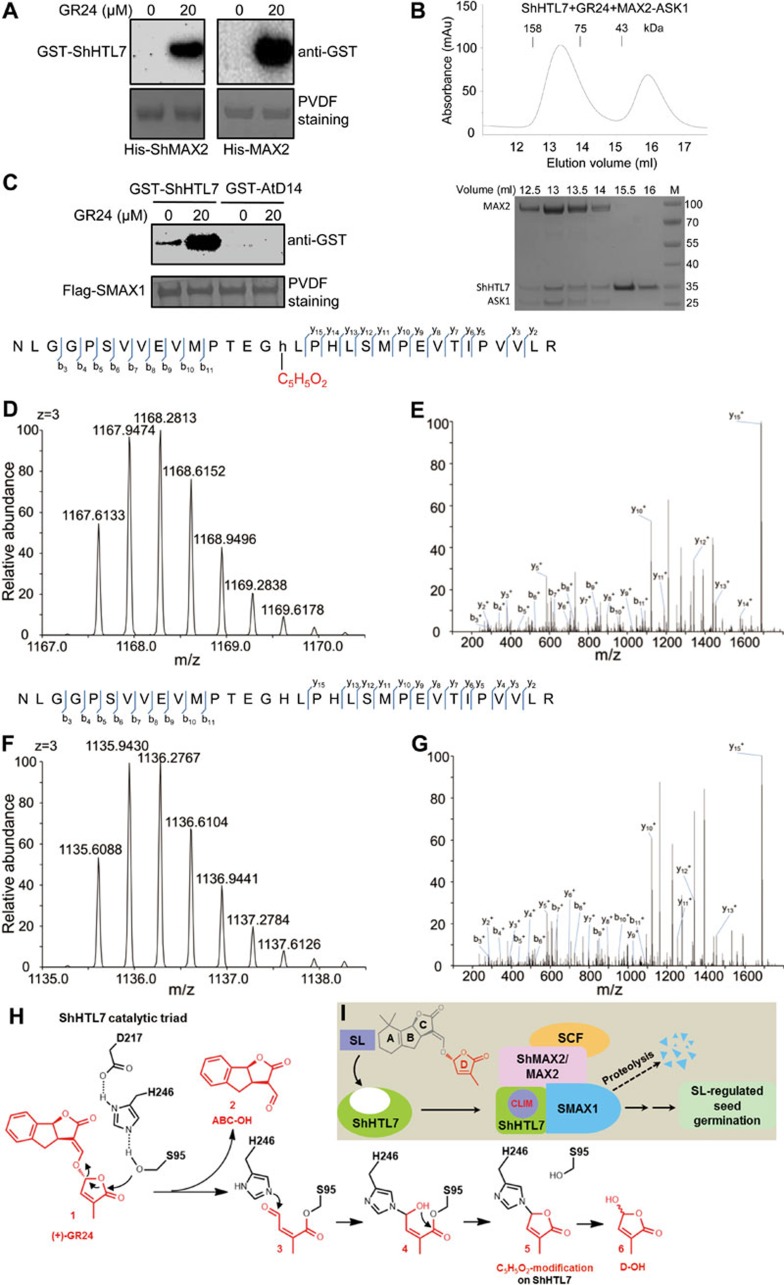
We expressed and purified key signaling components, such as the α/β hydrolases ShHTL7 and AtD14, the repressor proteins SMAX1 and SMXL6, and the F-box proteins ShMAX2 and MAX2 (stabilized by co-expressed ASK1; herein referred to as ShMAX2-ASK1 and MAX2-ASK1), and used these reagents to perform analyses comparing the mechanism of SL-stimulated seed germination with the mechanism of the SL-mediated repression of plant branching. Pull-down assay (Figure 1A and Supplementary information, Figure S1A) and size exclusion chromatography (SEC) (Figure1B and Supplementary information, Figure S1B) showed that ShHTL7 interacted with the F-box protein ShMAX2 or MAX2 in a GR24 (synthetic SL analog)-dependent manner; and this interaction is similar to the SL-dependent D14-D3/MAX2 interaction essential for SL-mediated suppression of plant branching5,6,7,15,18.
Figure 1.
ShHTL7 acts as a non-canonical receptor to generate and perceive CLIM. (A) ShHTL7 interacts with ShMAX2 and MAX2 in a GR24-dependent manner. Pull-down assays using GST-ShHTL7 and His-ShMAX2-ASK1 or His-MAX2-ASK1 in the absence or presence of 20 μM GR24. GST-ShHTL7 was detected by anti-GST antibody and PVDF membrane was stained with Memstain to show equal loading. GR24 used in this work is stereoisomer (+)-GR24. (B) GR24 induced the interaction of ShHTL7 and MAX2 in an SEC assay. SEC analysis (upper panel) of the interaction between ShHTL7 and MAX2-ASK1 in the presence of 200 μM GR24. SDS-PAGE analysis (lower panel) of peak fractions from the upper panel; M, molecular weight ruler (kDa). (C) GR24 enhances ShHTL7-SMAX1 interaction. Pull-down assays using Flag-SMAX1 and GST-ShHTL7 or GST-AtD14 in the absence or presence of 20 μM GR24. GST fusion proteins were detected by anti-GST antibody and PVDF membrane was stained with Memstain to show equal loading. (D-G) ShHTL7 hydrolyzes GR24 to effect a C5H5O2-modification at catalytic residue H246 of ShHTL7. A triply charged peptide (244-NLGGPSVVEVMPTEGHLPHLSMPEVTIPVVLR-262) of ShHTL7 with the GR24-derived C5H5O2-modification on H246 was identified by MS/MS (D and E, m/z = 1167.6133); the modified peptide was isolated from trypsin digestion of ShHTL7 in the GR24-induced ShHTL7-MAX2-ASK1 complex collected in SEC. As control, no modification on H246 of ShHTL7 was detected when ShHTL7 without pre-induction of GR24 was subjected to MS/MS analysis (F and G, m/z = 1135.6088). Labeled peaks in E and G correspond to masses of y and b ions of the peptide displayed on the top. Lowercase 'h' indicates the modified H246. (H) Schematic diagram of the proposed ShHTL7-mediated hydrolysis process of GR24. Hydrolysis of (+)-GR24 (1) involves a nucleophilic attack by S95 which produces ABC-OH (2) and compound 3, and the generation of D-OH (6). The Nε2 of H246 attacks the aldehyde carbon of the S95-linked compound 3 to form the H246- and S95-linked linear compound 4, referred to as the covalently-linked intermediate molecule (CLIM). Compound 4 initiates an intramolecular nucleophilic attack to generate the H246-linked circular compound 5, which appears as C5H5O2-modification on H246 detected by MS/MS and indicates the existence of CLIM inside the MAX2-bound ShHTL7. Compound 5 will be further hydrolyzed from H246 to produce D-OH (6). Similar process of AtD14-mediated GR24 hydrolysis can be found in our recent study18. (I) A simplified model of SL perception by ShHTL7. ShHTL7 docks SL in the catalytic cavity, hydrolyses SL into a D-ring-derived intermediate (CLIM) that covalently binds inside ShHTL7, undergoes conformational changes, and interacts with ShMAX2/MAX2-based SCF complex and SMAX1 to trigger SL signal transduction and regulate seed germination.
Furthermore, we found that GR24 strongly enhanced the interaction between ShHTL7 and SMAX1 (Figure 1C), which is similar to the SL-induced AtD14-SMXL6 interaction15. Interestingly, GR24 did not enhance the interaction between ShHTL7 and SMXL6, and the interaction between AtD14 and SMAX1 (Supplementary information, Figure S1C and Figure 1C), which provides a mechanistic explanation for the distinctive functions of ShHTL7-SMAX1 interaction, which specifically controls seed germination, and AtD14-SMXL6 interaction, which specifically regulates shoot branching. Given that SL-induced AtD14-SMXL6 interaction is essential for SMXL6 ubiquitination and degradation15, SL-enhanced ShHTL7-SMAX1 interaction may have a similar effect on SMAX1.
Since both AtD14 and ShHTL7 possess enzymatic activity that hydrolyzes SL molecules9,18, we investigated whether ShHTL7, like AtD14, can catalyze the production of active SL molecule CLIM and covalently bind it. The GR24-induced ShHTL7-MAX2-ASK1 complex was collected in SEC assay (Figure 1B) and run on SDS-PAGE to separate ShHTL7 for trypsin digestion followed by mass spectrometric analysis. We identified a chemical modification on the catalytic residue H246 in peptide 231-NLGGPSVVEVMPTEGhLPHLSMPEVTIPVVLR-262 of ShHTL7, which caused a shift in molecular weight by 96.0211 Da (Figure 1D and 1E); and this change is identical to the increase in molecular weight of AtD14 resulted from chemical modification of the corresponding His residue (H247)18. As the control, the unmodified peptide was identified when ShHTL7 without GR24 induction was subjected to MS/MS analysis.
Our results have revealed similarities between the ShHTL7-mediated SL signaling for induction of seed germination and the D14-triggered SL signaling for repression of plant branching. Taking advantage of the comprehensive analyses on D14-mediated SL signaling from our recent work18, we are able to conclude that the shift in molecular weight by 96.0211 Da (Figure 1D and 1E) corresponds to a covalent addition of C 5H 4O 2 on H246 of ShHTL7 (Figure 1H, compound 5) and compound 4 (Figure 1H) is the active form of SL in the SL-induced ShHTL7-MAX2 complex. The same modification was readily detected when a natural SL, 5-deoxystrigol (5DS), was used in MS/MS assay (Supplementary information, Figure S1D).
We further generated structural models for the ShHTL7-MAX2 complex and the ShHTL7-ShMAX2 complex using crystal structure of AtD14-D3 complex (PDB code: 5HZG)18 as the template. We found that the overall topologies of corresponding proteins, such as α/β hydrolases (ShHTL7 and AtD14) and F-box proteins (ShMAX2, MAX2 and D3), and the interfaces are almost identical (Supplementary information, Figure S1E), implying that, during the conversion of SL into CLIM, ShHTL7 also undergoes conformational changes similar to AtD14 and generate a conserved surface for the interaction with MAX2/ShMAX2 to initiate SL signaling in Striga.
Hormone perception is essential for a hormone to exert its effect. Enormous efforts made in the last one hundred years have established a general mechanism underlying the perception for all hormones: the active form of hormone, which is generated through sequential actions of biosynthesis- and modification-related enzymes, non-covalently binds its receptor to trigger hormone signal transduction, and reversibly dissociates from its receptor without being altered. This feature of reversibility distinguishes hormone-receptor interaction from substrate-enzyme interaction. This work uncovers a signal perception mechanism that integrates both substrate-enzyme interaction and hormone-receptor interaction utilized by ShHTL7, which acts as a non-canonical receptor of SLs that generates and perceives the SL active form CLIM. Similar to D1418, ShHTL7 also hydrolyzes SLs into an active intermediate CLIM and covalently binds CLIM to interact with ShMAX2/MAX2 to stimulate seed germination (Figure 1I). As analyzed in our recent work18, both compound 4 and 5 are the covalently linked SL hydrolysis intermediate that binds SL receptor, whereas density maps of SL-induced AtD14-D3-ASK1 complex crystal structure suggest compound 4 as the active form CLIM18 (Figure 1H). Additional structural analysis is needed to test if compound 5 exists in SL-induced complex of ShHTL7-ShMAX2/MAX2 or D14-D3/MAX2, thus serving as an active form of SL.
The infestation of arable land by parasitic weeds including Striga threatens to spread in the developing countries endangering food security around the world. This study extends our understanding of how parasitic plants perceive and transduce SL signal to stimulate seed germination, thus providing tangible targets that new technology can aim to interfere for the purpose of effectively combating parasitic plants.
Acknowledgments
We thank Steven M Smith at University of Tasmania for useful suggestions, and Zhongyuan Sun at Tsinghua University (Center of Biomedical Analysis) for help with MS/MS analysis. We apologize that we are unable to cite many excellent original works due to limits on the number of references. This work was supported by the National Natural Science Foundation of China (31230008, 31421001, 31500228 and 91417302) and the Ministry of Science and Technology of China (2016YFA0500500).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Supplementary Figures S1A–1E
References
- Al-Babili S, Bouwmeester HJ. Annu Rev Plant Biol 2015; 66:161–186. [DOI] [PubMed]
- Gomez-Roldan V, Fermas S, Brewer PB, et al. Nature 2008; 455:189–194. [DOI] [PubMed]
- Umehara M, Hanada A, Yoshida S, et al. Nature 2008; 455:195–200. [DOI] [PubMed]
- Kretzschmar T, Kohlen W, Sasse J, et al. Nature 2012; 483:341–344. [DOI] [PubMed]
- Hamiaux C, Drummond RS, Janssen BJ, et al. Curr Biol 2012; 22:2032–2036. [DOI] [PubMed]
- Jiang L, Liu X, Xiong G, et al. Nature 2013; 504:401–405. [DOI] [PMC free article] [PubMed]
- Zhou F, Lin Q, Zhu L, et al. Nature 2013; 504:406–410. [DOI] [PMC free article] [PubMed]
- Akiyama K, Matsuzaki K, Hayashi H. Nature 2005; 435:824–827. [DOI] [PubMed]
- Tsuchiya Y, Yoshimura M, Sato Y, et al. Science 2015; 349:864–868. [DOI] [PubMed]
- Toh S, Holbrook-Smith D, Stogios PJ, et al. Science 2015; 350:203–207. [DOI] [PubMed]
- Conn CE, Bythell-Douglas R, Neumann D, et al. Science 2015; 349:540–543. [DOI] [PubMed]
- Stirnberg P, Furner IJ, Ottoline Leyser HM. Plant J 2007; 50:80–94. [DOI] [PubMed]
- Nelson DC, Scaffidi A, Dun EA, et al. Proc Natl Acad Sci USA 2011; 108:8897–8902. [DOI] [PMC free article] [PubMed]
- Waters MT, Nelson DC, Scaffidi A, et al. Development 2012; 139:1285–1295. [DOI] [PubMed]
- Wang L, Wang B, Jiang L, et al. Plant Cell 2015; 27:3128–3142. [DOI] [PMC free article] [PubMed]
- Soundappan I, Bennett T, Morffy N, et al. Plant Cell 2015; 27:3143–3159. [DOI] [PMC free article] [PubMed]
- Stanga JP, Smith SM, Briggs WR, et al. Plant Physiol 2013; 163:318–330. [DOI] [PMC free article] [PubMed]
- Yao R, Ming Z, Yan L, et al. Nature 2016; 536:469–473. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures S1A–1E