Abstract
Studying the early function of essential genes is an important and challenging problem in developmental biology. Here, we established a method for rapidly inducing CRISPR-Cas9-mediated mutations in one blastomere of two-cell stage embryos, termed 2-cell embryo-CRISPR-Cas9 injection (2CC), to study the in vivo function of essential (or unknown) genes in founder chimeric mice. By injecting both Cre mRNA and CRISPR-Cas9 targeting the gene of interest into fluorescent reporter mice, the 2CC method can trace both wild-type and mutant cells at different developmental stages, offering internal control for phenotypic analyses of mutant cells. Using this method, we identified novel functions of the essential gene Tet3 in regulating excitatory and inhibitory synaptic transmission in the developing mouse cerebral cortex. By generating chimeric mutant mice, the 2CC method allows for the rapid screening of gene function in multiple tissues and cell types in founder chimeric mice, significantly expanding the current armamentarium of genetic tools.
Keywords: CRISPR-Cas9, two-cell embryo, gene function, Tet3, cerebral cortex, neocortical development
Introduction
Forward and reverse genetics in animal models have been pivotal for identifying the functions of genes and proteins1. Deleting the genomic sequence encoding a gene of interest using standard “knockout” methods is routinely used for determining in vivo gene function1,2,3. However, for the substantial number of essential genes4, this approach has significant limitations. Furthermore, compensatory mechanisms are sometimes activated in complete gene knockouts, obscuring the true function of the gene5. In rodent studies, these problems have been circumvented by generating conditional knockouts, where a mouse line carrying the floxed allele (sandwiched by LoxP sites) is crossed with a mouse line carrying Cre recombinase under the control of a specific promoter (termed Cre/LoxP system)6, to generate mice carrying homozygous floxed alleles and Cre transgene. This technology allows for precise spatial and temporal control of gene expression7 and has significantly advanced the power of mammalian genetics8. However, generating homozygous mutants with different Cre drivers is very time-consuming even when both floxed alleles and Cre drivers are available, as it takes at least two generations to obtain the progenies of interest. Moreover, for genes with pleotropic functions and/or relatively unknown functions, screening through more than 500 available Cre drivers8 can be a daunting task. Thus, although the Cre/LoxP system can provide cell type and spatial-temporal specificity for studying gene function, there is an unmet need for a method that can rapidly screen through multiple tissues and cell types.
Recently, the CRISPR-Cas9 system from bacteria, consisting of Cas9 nuclease and a single guide RNA (sgRNA) targeting a gene of interest, has been applied to rapid genome editing in different species9,10,11,12,13,14,15,16. This method has the significant advantage that direct injection of CRISPR-Cas9 into zygotes can generate animals carrying mutations in multiple endogenous genes in a single step10,13,14,17. We reasoned that this one-step approach can be adapted from injecting CRISPR-Cas9 at the one-cell stage to injecting one blastomere of a two-cell stage embryo to generate chimeric embryos. At the two-cell stage, both blastomeres are totipotent, and thus can differentiate into all cell types and integrate into all tissues of the future embryo18. For essential genes, this approach can potentially generate surviving individuals for studying their postnatal function in different tissues and cell types. A gene of particular interest is the ten-eleven translocation (Tet) family member Tet3, an enzyme that initiates the active DNA demethylation process by oxidizing 5-methycytosine (5mC) into 5-hydroxymethycytosine (5hmC), and has been shown to play critical roles during multiple steps of development18,19,20. As Tet3-knockout mice are neonatally lethal21, we surmised that the blastomere injection approach we propose may generate some surviving individuals to study Tet3 function in tissues postnatally. Following various optimizations, we established a method of injecting multiple sgRNAs into one blastomere of 2-cell stage embryos, termed 2CC, that effectively generated large deletions or insertions in Tet3. Viable chimeric mice were generated and novel functions of Tet3 in regulating excitatory and inhibitory synaptic transmission in the developing mouse neocortex were revealed.
Results
A two-step injection protocol (termed 2CC method) for mutating genes of interest in blastomeres
To test the feasibility of injection into one blastomere of a two-cell stage embryos, we used a double-fluorescent Cre reporter mouse line that expressed membrane-targeted tdTomato (mT) prior to Cre-mediated excision and membrane-targeted green fluorescent protein (mG) post excision (termed mT/mG)22 to respectively label the descendent cells of the untreated and manipulated blastomeres23. We first tested whether injection of Cre mRNA randomly into one blastomere of two-cell stage embryos generated chimeric embryos containing red and green cells in all tissues (Figure 1A). Two-cell stage embryos were collected from superovulated females (mated with heterozygous mT/mG male mice) 46-48 h after human chorionic gonadotrophin (hCG) injection, as previously reported, to ensure that the cytoplasms of the two blastomeres were completely separated24. To determine the optimal concentration of Cre mRNA required for in vivo excision, we microinjected mRNA encoding Cre recombinase at different concentrations into the cytoplasm of one blastomere of two-cell stage embryos and assessed the developmental potential of injected embryos. Almost all embryos injected with low concentrations of Cre mRNA (0.5 or 2.5 ng/μl) developed to the blastocyst stage in vitro, indicating that these mRNA concentrations did not affect early embryonic development (Supplementary information, Table S1 and Figure S1A). The blastocysts showed significant mG expression, demonstrating successful excision caused by Cre mRNA injection.
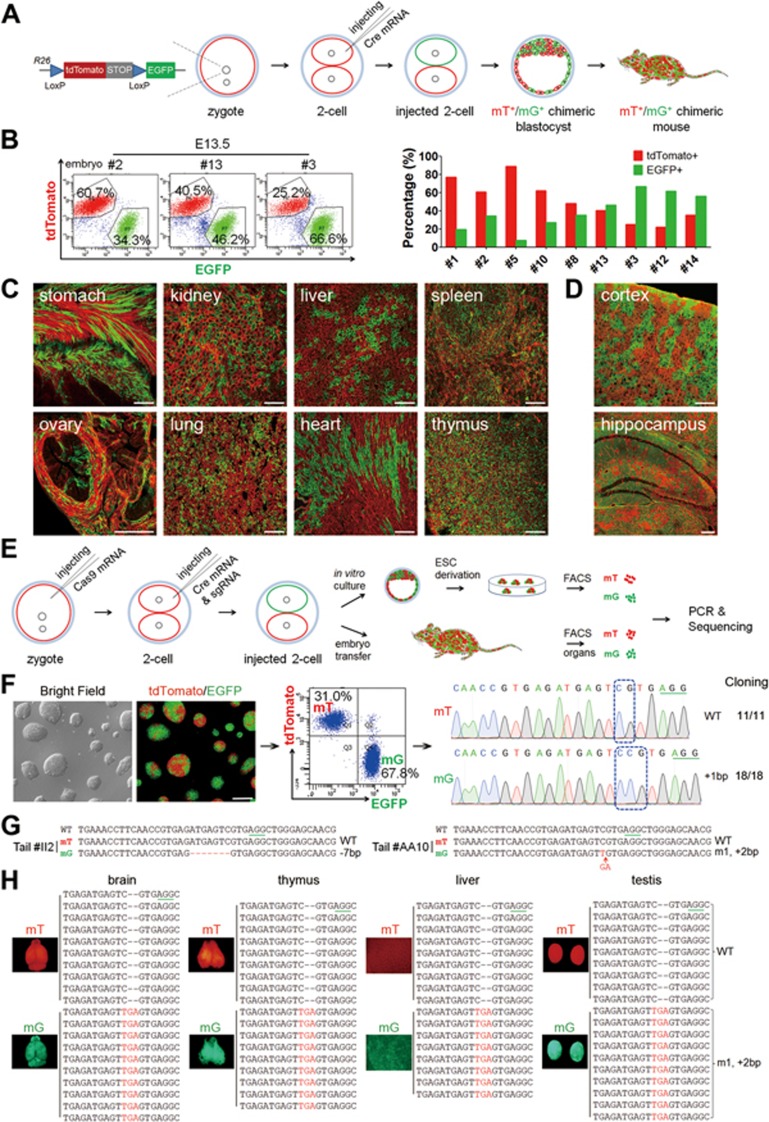
Figure 1.
A two-step injection method (termed 2CC) for rapid mutation of genes of interest in mouse chimeras. (A) Experimental procedure for Cre mRNA injection into one blastomere of mouse two-cell stage embryos to generate tdTomato+/EGFP+ (mT+/mG+) chimeras. (B) FACS analysis and quantification of mT+ and mG+ cells in dissociated whole-embryo suspensions of E13.5 embryos (left); in the quantitation, each red or green bar pair represents one mouse embryo (right). (C) Cryosection of tissues from a newborn mT+/mG+ chimera (P17). All tested tissues showed mT+ and mG+ fluorescence. Scale bar, 100 μm. (D) Cryosection of cortex and hippocampus of an adult chimera (2 months old) showing chimeric distribution of mT+ and mG+ cells. Scale bar, 100 μm. (E) Experimental procedure for the two-step injection method: Cas9 mRNA was injected into the zygotes first, followed by coinjection of Cre mRNA and sgRNA into one blastomere of two-cell stage embryos on the following day. Injected two-cell embryos were cultured in vitro to blastocysts to derive ESC lines. To generate chimeric mice, injected two-cell embryos were transplanted into oviducts of pseudopregnant females. (F) A representative Tet3-targeted ESC line derived from a Tet3-targeted mT+/mG+ blastocyst, and its corresponding FACS and sequencing results. mT+ cells carried WT alleles (11 in 11 sequenced alleles), whereas mG+ cells carried mutant alleles (18 in 18 sequenced alleles). Scale bar, 100 μm. (G) Sequencing results of FACS-sorted mT+ and mG+ cells from Tet3-targeted chimeric mouse tails. Tet3-targeted mG+ cells exhibited only one mutant peak, whereas the mT+ cells from the same mouse showed only WT peak. (H) Whole-mount fluorescence analysis of organs from one Tet3-targeted chimeric mouse (#AA10) and the corresponding sequencing results of mT+ and mG+ cells from these organs. All tested organs exhibited the same genotype as the corresponding tail shown in G.
Embryos injected with 2.5 ng/μl Cre mRNA were transferred into pseudopregnant females, and examined at different time points after transplantation (Supplementary information, Table S1 and Figure S1B). Whole-mount fluorescence analysis showed that a significant number of mT-positive (mT+) and mG-positive (mG+) cells were observed in all tissues of the embryos (Supplementary information, Figure S1B). Assessment of the ratios of mT+ and mG+ cells in dissociated single cells from 13.5-day embryos by fluorescence-activated cell sorting (FACS) showed that the average proportion of mG+ cells was ∼39.41% ± 6.62% (mean ± SEM; Figure 1B). We further analyzed the distribution of mG+ cells in newborn and adult mice by analyzing whole mount and cryosection of different organs, and found strong red and green fluorescence in all tissues examined (Figure 1C and 1D; Supplementary information, Figure S1C). Moreover, the ratios of mT+ and mG+ cells in dissociated single cells from tissues of 1-week-old pups were similar to those of 13.5-day embryos (Supplementary information, Figure S1D). The chimerism also extended to the germline, as when a female chimeric mouse was crossed with a wild-type (WT) male mouse, both mT+ and mG+ offspring were generated (Supplementary information, Figure S1E). Taken together, these results demonstrate that injection of Cre mRNA into one blastomere of two-cell stage embryos of mT/mG mice can generate chimeric mice with mosaic distribution of labeled cells in all tissues (hereafter termed mT+/mG+ mice).
We next asked whether coinjection of Cre mRNA and CRISPR-Cas9 targeting a gene of interest could generate chimeric mice, with mT+ cells carrying the WT gene and mG+ cells from the targeted blastomere carrying the mutated gene. To ensure high level and homogenous Cas9 protein expression at the time of sgRNA injection, we employed a two-step injection protocol (termed 2CC method), first injecting Cas9 mRNA (50 ng/μl, as previously reported9) into the zygotic cytoplasm (23-26 h after hCG injection) prior to the first cell division25 (Figure 1E). In the second step, Cre mRNA and sgRNAs corresponding to Tet family members Tet110 or Tet326 (Supplementary information, Figure S2A) were coinjected into the cytoplasm of one blastomere of late two-cell stage embryos. The dually injected embryos developed efficiently into blastocysts, indicating low toxicity of the injected RNAs. To determine the mutagenesis rate, we derived embryonic stem cell (ESC) lines from Tet1-targeted or Tet3-targeted blastocysts, sorted mT+ and mG+ cells by FACS, and independently sequenced them. The Sanger sequencing results showed that mT+ cells carried WT Tet1 or Tet3, whereas mG+ cells carried non-homologous end joining (NHEJ)-mediated mutations of Tet1 or Tet3 (6 of 21 Tet1-targeted and 8 of 21 Tet3-targeted ESC lines carrying one or two mutant sequence peaks). To further confirm the mutant genotypes, we performed TA cloning and sequencing analysis of PCR products from two cell lines. The results showed that all tested clones exhibited the same mutant sequence as that of direct PCR product sequencing (Figure 1F and Supplementary information, Figure S2B), demonstrating successful genome editing in Tet1 or Tet3 through 2CC.
We next transplanted injected embryos into oviducts of pseudopregnant females to obtain chimeric mice. From 563 transferred embryos, a total of 192 mice were obtained (34.1% survival rate). As heterozygous mT/mG male mice were used, only 50% of mice were expected to be transgenic. Of these, 42 mice showing both red and green fluorescence (mT+/mG+ mice), indicative of chimeras containing both genotypes, were obtained (Supplementary information, Table S2). Sequencing of FACS-enriched tail mT+ and mG+ cells from newborn pups showed that 24 mice carried clear one or two mutant sequences (Figure 1G, Supplementary information, Figure S2C and Table S2). To examine the whole-body chimeric rate in mT+/mG+ mice, cells from brain, thymus, liver and testis of seven mT+/mG+ mice were FACS sorted, and all found to contain both mT+ and mG+ cells (Figure 1H and Supplementary information, Figure S2D). Importantly, TA cloning and sequencing showed that all tested organs in four mice exhibited the same genotype as the tail, demonstrating successful induction of mutagenesis by CRISPR-Cas9 at an early developmental stage (Figure 1H, Supplementary information, Figure S2D and S2E). In an adult mT+/mG+ mouse (5 months) carrying Tet3 mutation, mT+ and mG+ cells were still present in multiple organs, and mG+cells carried the same mutation in all organs examined (Figure 1H and Supplementary information, Figure S2F). To test germline transmission of the mutated genes, sperm from a Tet1-mutant chimeric mouse was used for intracytoplasmic injection, leading to generation of blastocysts carrying mutant allele with the same sequence or WT allele (Supplementary information, Figure S2G). Moreover, when a Tet1-mutant mouse was crossed with a WT mouse to obtain heterozygous F1 offspring, the intercrossing of F1 offspring generated viable F2 Tet1 whole-body mutant mice (Supplementary information, Figure S2H). In contrast, F2 or F3 Tet3 homozygous mutant offspring from one mT+/mG+ mouse carrying Tet3 mutations were neonatally lethal (Supplementary information, Figure S2I). This result is consistent with the previously reported phenotype of the Tet3-knockout mice21. Together, our results demonstrate that blastomere injection of CRISPR-Cas9 and Cre mRNA into two-cell stage embryos, together with the use of reporter lines such as mT/mG, can be used to generate chimeric mice, in which mT+ cells are WT and mG+ cells are mutant. Moreover, genotyping results from tail can reflect the mutant state of the chimeric mice, allowing us to choose the mice carrying one or two mutant peaks in mG+ cells for further analysis.
Multiple sgRNAs efficiently generate chimeras carrying large deletions or insertions through 2CC method
Recent studies have shown that zygotic injection of multiple sgRNAs targeting the same gene can efficiently induce large deletion in the gene of interest27. This strategy can not only improve the deletion rate in founder animals, but also simplify genotype analysis to PCR-based approaches. We thus tested whether it can be employed in our system to produce large deletion in the targeted gene. We first designed four sgRNAs targeting exon 6 and 4 sgRNAs targeting exon 9 of Tet3 (termed IJKL and MNOP, respectively; Figure 2A). Using our two-step injection method, mT+/mG+ blastocysts were efficiently derived (Figure 2B). PCR analyses showed that the IJKL and MNP sgRNAs induced large deletions more efficiently than other sgRNA combinations (Figure 2C). Embryos injected with these sgRNAs were then transplanted into recipients' oviducts, and the resulting mT+/mG+ mice were genotyped. Analysis showed that most mT+/mG+ mice (9 of 10 from IJKL and 6 of 8 from MNP) carried large deletions or insertions in mG+ cells, indicating that 2CC using multiple sgRNAs could efficiently generate Tet3-mutant chimeric mice (Figure 2D and 2E; Supplementary information, Table S3A). We further validated multiple sgRNAs-mediated gene targeting by successful mutagenesis of the coat color gene Tyr, encoding tyrosinase, to generate mT+/mG+ mice with chimeric agouti-white coat color (Supplementary information, Figure S3A-S3D, Table S3A). Similarly, mutagenesis of the transcription factor Gata4, which is essential for embryonic development28,29, resulted in mT+/mG+ mice, in which mG+ cells carried mutated Gata4 gene with large deletions (Supplementary information, Figure S3E-S3L, Table S3A).
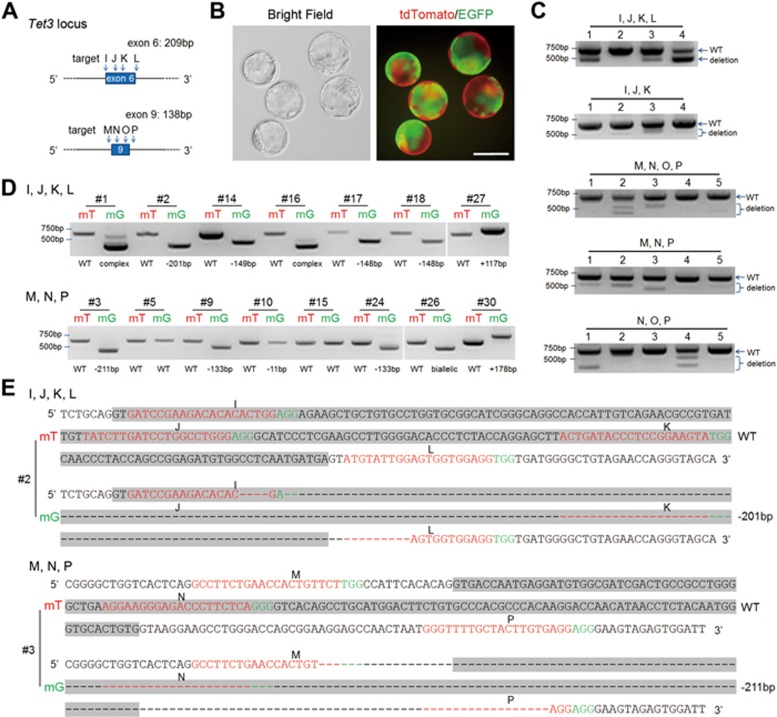
Figure 2.
Gene editing using multiple sgRNAs efficiently produced chimeras carrying large deletions or insertions. (A) Schematic of the Cas9/sgRNA-targeting sites in exons 6 and 9 of Tet3. (B) Representative mT+/mG+ blastocysts generated by combinatorial MNP sgRNAs targeting Tet3 in one blastomere of two-cell stage embryos through 2CC. Scale bar, 100 μm. (C) Single blastocyst PCR genotyping of blastocysts derived from different combinations of sgRNAs via 2CC method. (D) Representative PCR results of mT+/mG+ chimeric mice generated using the IJKL or MNP sgRNA combinations. Most mT+/mG+ mice carried large deletions or insertions in mG+ cells. “Complex” denotes multiple mutations (three or more). “Biallelic” here denotes one apparent PCR band from mouse #26, which upon sequencing showed two distinct mutations. (E) Representative sequencing results of mT+/mG+ chimeric mice generated using the IJKL or MNP sgRNA combinations. The sgRNA-targeting sequence is labeled in red, and the protospacer-adjacent motif (PAM) sequence is labeled in green. Sequences of exons 6 and 9 are highlighted in gray.
Tet3 deficiency does not affect the differentiation of major cell types in the cerebral cortex
Having demonstrated that our 2CC method efficiently generated mT+/mG+ mice, in which mT+ cells are WT and mG+ cells are mutant, we attempted to address our question of interest, what is the function of Tet3 in the postnatal brain. Multiple lines of evidence suggest that Tet3 plays important roles in neuronal differentiation and function, including maintenance of neural progenitor cells, development of dendritic arbors in cerebellar granule cells and regulation of homeostatic plasticity in cultured hippocampal neurons26,30,31,32. However, as Tet3-knockout mice are neonatally lethal10,21, previous studies were mostly carried out in culture or using shRNA-mediated knockdown, leaving its in vivo postnatal function relatively unexplored. Here, we switched to the Ai9 reporter mouse line8, which expresses cytoplasmic tdTomato upon Cre induction, and thus is more suitable for visualizing the cell bodies of different cell types, especially neurons33. Embryos sequentially injected with Cas9 mRNA, followed by Cre mRNA and IJKL sgRNA cocktail (Supplementary information, Figure S4A), efficiently developed into blastocysts containing both non-fluorescent cells and tdTomato+ (Ai9+) cells (Supplementary information, Figure S4B). Deep sequencing of six late-stage blastocysts showed that the IJKL sgRNA cocktail led to large deletions (> 120 bp) in all tested embryos. Four embryos carried one or two main mutation sequences, whereas the remaining sequences were the mixture of various mutations, each of which accounted for < 5% of the total sequence, likely due to PCR and/or sequencing error (Supplementary information, Figure S4C)34. Importantly, over 99% of mutations in three embryos were frameshift deletions (3n + 1 or 2; Supplementary information, Figure S4C). These results indicate that 2CC method with multiple sgRNAs can efficiently induce gene mutation at early stage of Ai9 embryos. We next transferred the injected embryos into pseudopregnant females to obtain chimeric mice containing tdTomato+ cells (Supplementary information, Table S3B). Sanger sequencing analysis of multiple organs from five chimeric mice showed that tdTomato+ cells all exhibited the same genotype as that of the tail (Figure 3A), suggesting homogeneity of Tet3 genomic editing in tdTomato+ cells. We then performed deep sequencing of three of them and found that over 96% of the target sequences in all tested organs carried the deletions/insertions that were detected in tails (Figure 3B). To further analyze the gene mutation at the single-cell resolution, we isolated single tdTomato− and tdTomato+ cells from different tissues of another two mice and performed single-cell PCR, followed by sequencing analysis. The results showed that one mouse had the same Tet3 mutation in all tdTomato+ cells and the other had two types of tdTomato+ cells carrying different mutations in all tested tissues (Figure 3C). As expected, all tdTomato− cells were WT (Figure 3C). Off-target analysis of two chimeric mice showed no obvious mutations at a total of 40 potential “off-target” sites of IJKL sgRNAs (Supplementary information, Table S4). Taken together, these data indicate that successful induction of mutagenesis by multiple sgRNAs occurs at an early developmental stage, resulting in chimeric mice with tdTomato+ cells carrying homozygous mutant Tet3 gene. Moreover, these data demonstrate that genotype analysis of tail genome by PCR-based sequencing can reflect the genotype of the mutant cells from other organs in the 2CC system.
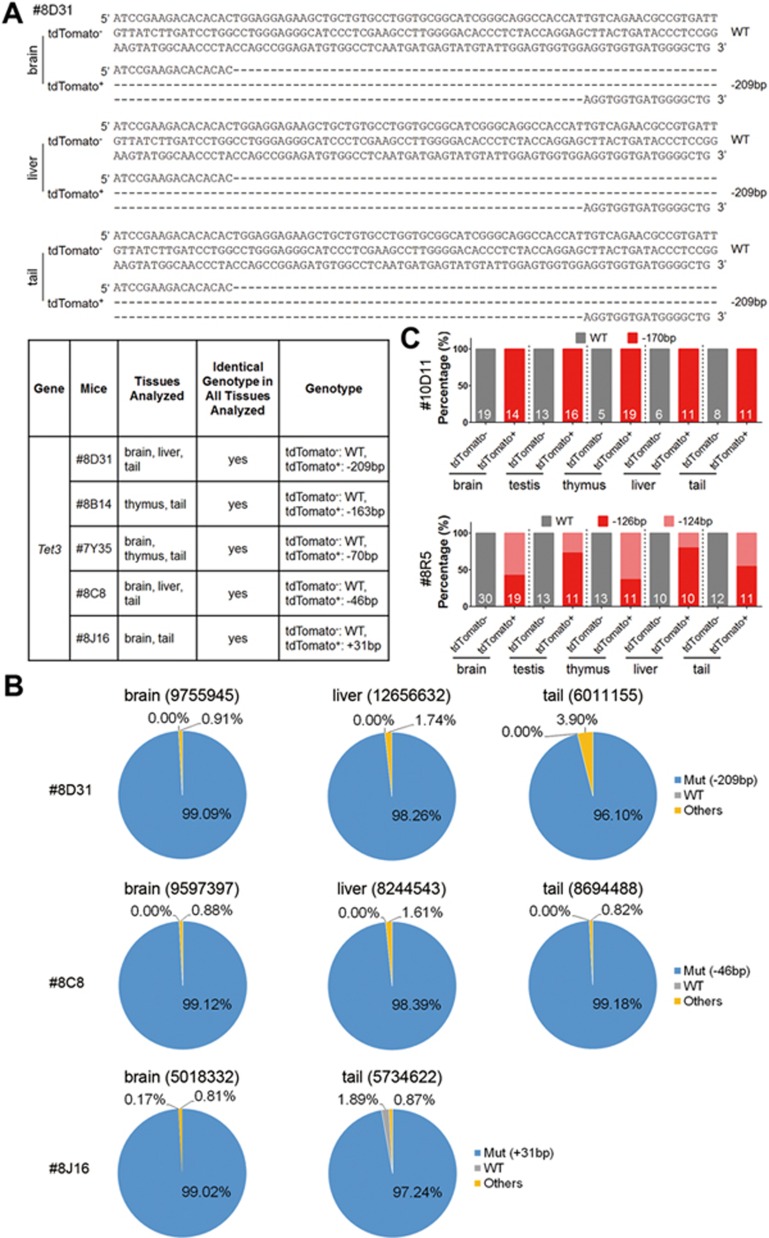
Figure 3.
Detailed genotype analysis of Tet3-mutant chimeras. (A) Representative sequencing results of organs/tissues from mouse #8D31 and the summary of genotyping results of five Tet3-targeted chimeric mice using the Ai9 reporter line. Five mice with one mutant peak of Tet3 in tail tdTomato+ cells were used for genotyping analysis. (B) Deep sequencing of tdTomato+ cells from three independent Tet3-mutant mice from A. Numbers beside the organs/tissues indicate deep sequencing reads. “Others” denotes the undefined reads that may be caused by PCR and/or sequence error. (C) Single-cell genotyping of the isolated tdTomato− and tdTomato+ cells from brain, testis, thymus, liver and tail of two Tet3-mutant chimeras, respectively. Single cell from each tissue was PCR amplified and sequenced. The number indicated inside bar graphs represents the number of successfully genotyped cells. In total, 122 and 140 cells from the two Tet3-mutant chimeras were successfully genotyped. A Tet3-mutant chimera with homozygous tail (#10D11) showed 170-bp deletions in all the tdTomato+ cells, whereas another Tet3-mutant chimera (#8R5) carrying two types of mutations showed 126- and 124-bp deletions, respectively.
We next tested the function of Tet3 in mutant cells. First, qPCR analysis of Tet3 in FACS-sorted tdTomato+ cells of the cerebral cortex showed that Tet3 mRNA level, but not that of related Tet1 or Tet2 was significantly lowered in mutant cells (Figure 4A). Second, mass spectrometry analysis indicated that 5hmC levels were significantly decreased in chimeric mutant mice, as compared to WT controls (Figure 4B). Taken together, our results indicate that the function of Tet3 protein has been attenuated in Tet3 loss-of-function cells.
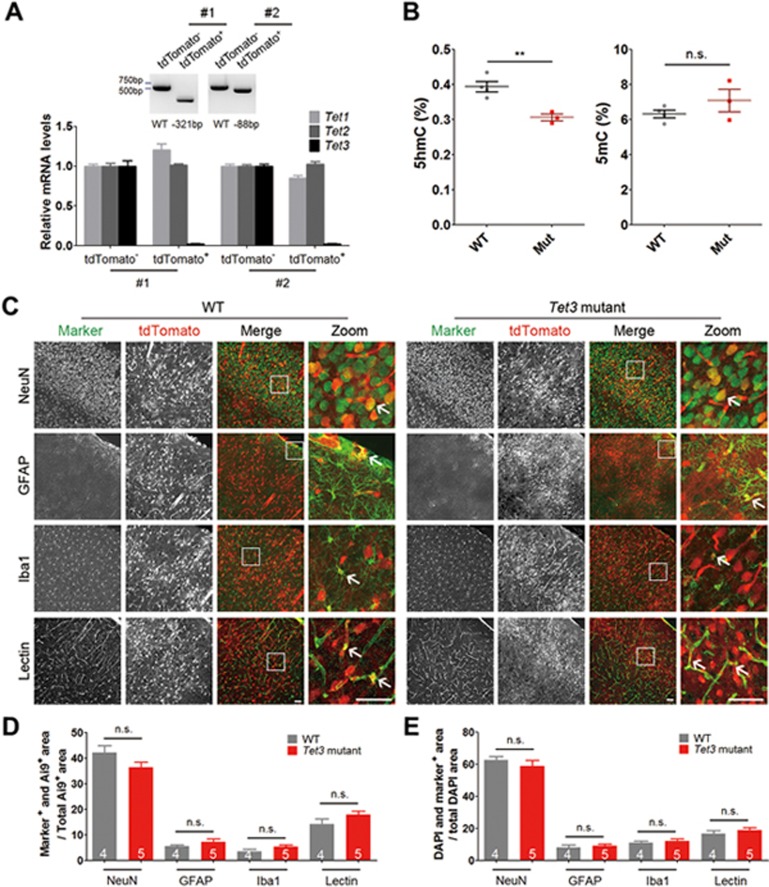
Figure 4.
Loss of Tet3 function does not affect the development of major cell types in the cerebral cortex. (A) qPCR analysis of Tet3 in the cerebral cortex of chimeras. Top, representative genotyping results of two Tet3-mutant mice. Sequencing result of the PCR bands showed that tdTomato− cells were WT, whereas tdTomato+ cells in mouse #1 and #2 carried 321- and 88-bp deletions, respectively. Bottom, relative mRNA levels (normalized to GAPDH) of Tet1, Tet2 and Tet3 in tdTomato− and tdTomato+ cells from the cerebral cortex of Tet3-mutant mice shown in Top. Tet3 mRNA expression was hardly detectable in tdTomato+ cells sorted from the cerebral cortex of Tet3-mutant mouse #1 and #2, whereas Tet1 and Tet2mRNA levels in tdTomato+ cells remained comparable to those in tdTomato− WT cells. Error bars, SD of three reactions. (B) Global genomic levels of 5hmC and 5mC in WT or Tet3-mutant chimeric cortices. Four WT and three Tet3-mutant mice (chimeric rate > 50%) were analyzed by mass spectrometry. **P< 0.01; n.s., not significant, P > 0.05 by unpaired t-test. (C) Representative images of colocalization between tdTomato+ cells and various cell type markers (NeuN, GFAP, Iba1 and Lectin) in brain slices from the somatosensory cortex of WT and Tet3-mutant mice. Arrows indicate colocalization between tdTomato and cell markers. Scale bar, 50 μm. (D) Quantification of the percentage colocalization between tdTomato+ cells and various cell type markers as indicated. (E) Quantification of the percentage colocalization between the nuclear marker DAPI and various cell type markers as indicated. n (4 or 5) as indicated inside bar graphs represents the number of mice, error bars denote SEM; n.s., not significant, P> 0.05 by unpaired t-test.
Cells from the early embryos can differentiate into all cell types with similar likelihoods. We first examined whether loss of Tet3 function affected the proportion of tdTomato+ cells corresponding to different cell types in the brain. Brain slices from 4 control mice and 5 Tet3-mutant mice with homozygous genomic deletions were immunostained against NeuN, GFAP, Iba1 and Lectin to label neurons, astrocytes, microglia and endothelial cells, respectively (Figure 4C). No significant differences in the proportion of tdTomato+ cells corresponding to neurons, astrocytes, microglia and endothelial cells were observed between control and Tet3-mutant mice (Figure 4D and 4E; P > 0.05 between WT and Tet3-mutant mice for all markers), suggesting that loss of Tet3 function very early in development did not significantly affect the differentiation and/or survival of major cell types in the cerebral cortex.
Changes in excitatory and inhibitory synaptic transmission in Tet3-mutant chimeric mice
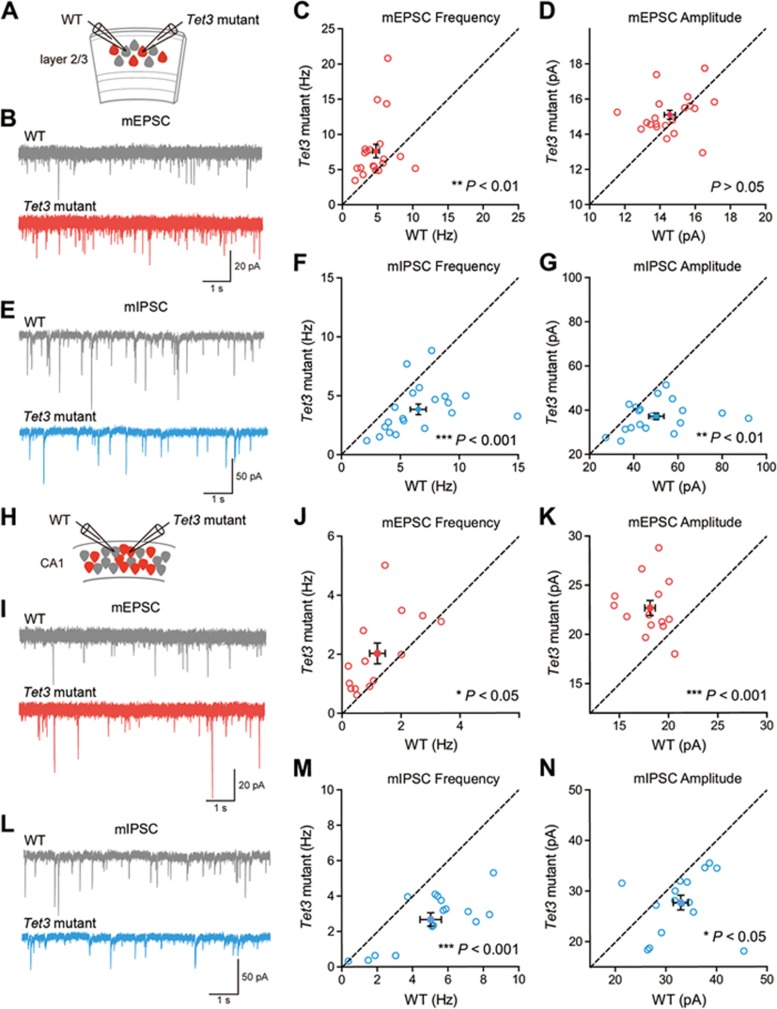
We next used whole-cell patch clamp recordings to assay the effect of loss of Tet3 function on neural circuit formation during the period of rapid synapse formation in the cerebral cortex (Figure 5A)35. For this purpose, we chose chimeric mice with tdTomato+ cells that carried only one type of mutant Tet3 gene identified by sequencing analysis. Recording AMPAR-mediated miniature excitatory post-synaptic currents (mEPSCs) from layer 2/3 pyramidal neurons of the primary somatosensory cortex of P14 chimeric mice, we found that tdTomato+ Tet3-mutant neurons had a significantly higher mEPSC frequency and a similar mEPSC amplitude, as compared to neighboring non-fluorescent WT neurons (Figure 5B-5D), suggesting an important role of endogenous Tet3 in negatively regulating excitatory synaptic transmission in young mice. As control, tdTomato+ cells from mice injected with Cre mRNA had similar mEPSC frequency and amplitude to neighboring non-fluorescent cells (Supplementary information, Figure S5A-S5C). Loss of Tet3 function (tdTomato+ cells) in layer 2/3 pyramidal neurons also significantly reduced GABAAR-mediated inhibitory synaptic transmission, as measured by miniature inhibitory post-synaptic currents (mIPSCs). Both the frequency and amplitude of Tet3-mutant cells were significantly reduced as compared to neighboring WT cells (Figure 5E-5G), suggesting a promoting role of endogenous Tet3 in regulating inhibitory synaptic transmission. No significant differences in mIPSC amplitude or frequency were detected between tdTomato+ cells injected with Cre mRNA and neighboring WT cells (Supplementary information, Figure S5D-S5F). To investigate whether the above effects applied more generally to the developing neural circuits, we recorded from hippocampal CA1 neurons, and found that mEPSC frequency and amplitude were significantly increased, whereas those of mIPSCs were significantly reduced (Figure 5H-5N). These results are similar to those of the primary somatosensory cortex and are consistent with a previous report recording from cultured hippocampal neurons with Tet3 knockdown by RNAi30. We also performed qPCR analysis of candidate target genes of Tet3 described in previous reports30,31. As shown in Supplementary information, Figure S6A, some genes changed in the same direction as previously reported, for example, the mRNA level of Mapk6 in chimeric Tet3-mutant samples was significantly increased, supporting previous observations in hippocampal neuronal cultures with Tet3 knockdown30. Finally, bisulfite sequencing of several candidate genes showed slightly increased CpG methylation at the Bdnf promoter IV, IX and Wfdc2 promoter regions, but not the Npas4 promoter-exon 1 junction or the Fgf1G and Ndst1 promoter regions (Supplementary information, Figure S6A and S6B), consistent with previous reports30,31. Together, these results demonstrate a critical role of endogenous Tet3 in regulating both excitatory and inhibitory synaptic transmission in the developing cerebral cortex.
Figure 5.
Regulation of excitatory and inhibitory synaptic transmission by Tet3 as revealed by 2CC targeting Tet3 in one blastomere of two-cell stage embryos. (A) Schematic of whole-cell recordings of cortex layer 2/3 tdTomato− (WT) and tdTomato+ (Tet3 mutant) pyramidal cells in acute slices of Tet3-mutant chimeric mice. (B-D) Representative mEPSC traces and X-Y plots of mEPSC frequency and amplitude of WT and Tet3-mutant cells in Tet3 chimeric mice. mEPSC frequency: WT, 4.77 ± 0.47 Hz; Tet3 mutant, 7.64 ± 0.96 Hz; P < 0.01. mEPSC amplitude: WT, 14.56 ± 0.31 pA; Tet3 mutant, 15.10 ± 0.26 pA; P > 0.05. n = 20 cell pairs from four mice. (E-G) Same as B-D for mIPSC. mIPSC frequency: WT, 6.52 ± 0.67 Hz; Tet3 mutant, 3.85 ± 0.45 Hz; P < 0.001. mIPSC amplitude: WT, 50.18 ± 3.47 pA; Tet3 mutant, 37.19 ± 1.49 pA; P < 0.01. n = 20 cell pairs from five mice. (H) Schematic of whole-cell recordings of hippocampal CA1 tdTomato− (WT) and tdTomato+ (Tet3mutant) cells in acute slices of Tet3-mutant chimeric mice. (I-K) Representative mEPSC traces and X-Y plots of mEPSC frequency and amplitude of WT and Tet3-mutant hippocampal CA1 neurons in Tet3 chimeric mice. mEPSC frequency: WT, 1.21 ± 0.26 Hz; Tet3 mutant, 2.03 ± 0.35 Hz; P < 0.05. mEPSC amplitude: WT, 18.13 ± 0.53 pA; Tet3 mutant, 22.68 ± 0.76 pA; P < 0.001. n = 14 cell pairs from three mice. (L-N) Same as I-K for mIPSC. mIPSC frequency: WT, 5.03 ± 0.60 Hz; Tet3 mutant, 2.67 ± 0.37 Hz; P < 0.001. mIPSC amplitude: WT, 32.97 ± 1.49 pA; Tet3 mutant, 27.69 ± 1.45 pA; P < 0.05. n = 16 cell pairs from three mice. Data are presented as mean ± SEM, paired t-test.
Discussion
In summary, using CRISPR-Cas9-mediated genome editing in one blastomere of two-cell stage mouse embryos, a method termed 2CC, we identify a critical role of essential gene Tet3 in regulating synaptic transmission during early neural circuit wiring (Figure 5). To our knowledge, this is the first report of Tet3 function in regulating inhibitory synaptic transmission. In terms of excitatory synapse function, a previous study reporting the effect of Tet3 knockdown in cultured hippocampal neurons30 identified an important role of Tet3 in mediating homeostatic plasticity. Here, through in vivo Tet3 mutagenesis, we demonstrate that Tet3 is critical for regulating the development of excitatory synaptic transmission during normal neural circuit wiring. The effects of Tet3 in downregulating excitatory synaptic transmission and upregulating inhibitory synaptic transmission, as deduced from its loss-of-function effects, suggest a critical role of endogenous Tet3 in regulating excitation-inhibition balance and in maintaining neural circuit stability. In our assays, tdTomato+ Tet3-mutant neurons were compared with neighboring tdTomato− cells, and the effects were similar regardless of whether tdTomato+ cells were clustered together or sparsely labeled, suggesting that the observed effects are most likely cell autonomous. Finally, as the proportion of tdTomato+ neurons, astrocytes, microglia and endothelial cells in Tet3-mutant mice were similar to those of Cre mRNA-injected control mice (Figure 4C-4E), Tet3 expression during early embryonic development31 likely does not affect the differentiation and/or survival of major cells types in the brain.
In more general terms, we believe that 2CC, our two-step injection method of generating chimeric two-cell stage embryos using CRISPR-Cas9-mediated mutagenesis complements existing genome editing methods1,2,3,8 for the following reasons: (1) for essential genes with unknown functions, our method can rapidly screen through multiple tissues and cell types, saving the time required for at least two generations of crosses with multiple drivers using the Cre/LoxP system6,7,8; (2) by measuring the proportion of different cell types, in combination with lineage tracing methods, this method can determine, in a well-controlled system, whether the gene of interest affects the differentiation and survival of different cell lineages starting from early developmental stages; (3) by assaying the phenotype of mutant cells in sparsely and densely labeled regions, as compared to neighboring cells, cell-autonomous and non-cell-autonomous effects can be distinguished; (4) as compared to existing genome editing methods in mature neurons36,37,38,39,40, this method offers homogeneous mutations across cell types early in development, simplifying the interpretation of the results; (5) by mating founder chimeric mice with WT mice, this method can rapidly generate knockout progeny of non-essential genes. However, application of this approach for analyzing gene function in vivo needs to exclude the possibility of confounding off-target effects induced by CRISPR-Cas9, especially when multiple sgRNAs are used. Besides specificity and efficiency improvement of CRISPR-Cas9 technology41 and analysis of in silico predicted off-targets, one important improvement is to obtain similar phenotypes from different mice with mutations generated by different combinations of sgRNAs. Meanwhile, the variability in the degree of mosaicism within the embryos should be taken into consideration when analyzing the results of 2CC experiments. To overcome this problem, the chimeric mice with only one type of mutant gene by genotyping analysis of tail DNA should be selected for further functional analysis; and the same phenotype should be observed in multiple mice. Moreover, the overall efficiency of generating homogenous mutants through 2CC method needs to be further improved. Finally, the two-step injection protocol may be a significant hurdle for many groups. Therefore, a future task is to optimize and simplify the protocol, which would facilitate the potential applications of our method. One potential strategy is to adopt Cas9 ribonucleoprotein (RNP) complex by defining appropriate injection conditions that would enable one-step injection into the blastomere with Cre mRNA25, which will not only simplify the 2CC protocol, but may also reduce off-target effects.
In summary, we have demonstrated that our system is an important addition to the current battery of genetic tools, particularly for rapidly assaying the effects of essential genes with unknown functions in multiple cell types.
Materials and Methods
Animal use and care
All animal procedures were performed under the ethical guidelines of Shanghai Institute of Biochemistry and Cell Biology and Institute of Neuroscience. Mice were kept under a 12 h-12 h light-dark cycle with food and water provided at libitum from the cage lid. Both males and females were used, unless otherwise stated. Animal ages are stated in the text and figure legends.
Production of mRNA and sgRNAs
Cre and Cas9 with T7 promoter were amplified by PCR from pZ4f-Cre and pX260 (Addgene) using KOD-Plus-Neo (TOYOBO, Cat# KOD-401), respectively. PCR products were purified with Universal DNA Purification Kit (TIANGEN, Cat# DP214) and in vitro transcribed using mMESSAGE mMACHINE T7 ULTRA kit (Life Technologies, Cat# AM1345). sgRNAs with T7 promoter were amplified by PCR from pX330 (Addgene) using KOD-Plus-Neo, purified with Universal DNA Purification Kit and in vitro transcribed using MEGAshortscript T7 kit (Life Technologies, Cat# AM1354). After transcription, Cre mRNA, Cas9 mRNA and sgRNAs were purified with MEGAclear kit (Life Technologies, Cat# AM1908) according to the manufacturer's instructions. Purified Cre mRNA, Cas9 mRNA and sgRNAs were eluted in elution buffer supplied with the MEGAclear kit and then diluted with RNase-free water to desired concentrations for injection. The primers used for preparation of Cre and Cas9 template were (5′-3′):
CAS9-F TAATACGACTCACTATAGGGAGATTTCAGG-TTGGACCGGTG
CAS9-R GACGTCAGCGTTCGAATTGC
Cre-F TAATACGACTCACTATAGGGAGACACACCA-TTGTCACCACACTCG
Cre-R CTCCCCCTGAACCTGAAAC
Zygote and two-cell embryo injection
Two-month-old B6D2F1 (C57BL/6 × DBA2) female mice were superovulated by injecting 5-7.5 IU of pregnant mare serum gonadotropin (PMSG), followed by 5-7.5 IU of hCG 48 h later. Superovulated female mice were mated to mT/mG (full name B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J; JAX strain 007676)22 or Ai9 (full name B6.Cg-Gt(ROSA)26Sortm9(CAG-td-Tomato)Hze/J; JAX strain 007909)8 male mice, and zygotes (one-cell embryos) were collected from oviducts 22.5-23.5 h post hCG injection using hyaluronidase (Sigma, Cat# H3884). Heterozygous mT/mG or Ai9 male mice were used for mating. For Cre expression, Cre mRNA was microinjected into the cytoplasm of one blastomere 46-48 h post hCG injection as previously reported, to ensure that the two blastomeres were completely separated24. For the two-step injection protocol, 25 or 50 ng/μl Cas9 mRNA was microinjected into the zygotic cytoplasm 23-26 h post hCG injection, followed by Cre mRNA/sgRNA mixture microinjection into one blastomere of two-cell embryos 46-48 h or 47.5-49.5 h post hCG injection for mT/mG or Ai9 stain, respectively. The zygote and two-cell embryo injection procedures took about 1 h for 150 embryos, respectively. In the two-step injection protocol, 2.5 ng/μl Cre mRNA was used together with 25 or 50 ng/μl sgRNAs as a mixture for injection into one blastomere of the two-cell stage embryos.
ES cell derivation
Dually injected embryos were cultured in KSOM+AA with 𝒟-glucose (Millipore, Cat# MR-106-D) at 37 °C under 5% CO2 until 3.5-day blastocysts formed. Zona pellucida was removed using Acidic Tyrode's solution (Sigma, Cat# C6762) and each blastocyst was transferred into one well of a 96-well plate to culture in ESC medium containing 2i (Selleck, Cat# S2924, S1036)42 for ES cell derivation. One week later, the colonies were dissociated using 0.05% Trypsin-EDTA (Gibco, Cat# 25300054) and expanded. For routine culture, the established ESC lines were cultured in 2i medium and passaged every 2-3 days. ESC lines were free of mycoplasma as determined by PCR-based detection method.
Embryo transfer
Injected two-cell embryos were cultured in KSOM+AA with 𝒟-glucose for 1 h and then transferred into oviducts of pseudopregnant ICR females at 0.5 day postcoitum (dpc; related to Figures 2,3,4,5). Alternatively, injected two-cell embryos were cultured to blastocyst stage and transferred into uteruses of 2.5 dpc ICR females (related to Figure 1).
Deep-sequencing analysis
PCR products were amplified from mouse tissues or blastocysts and purified with Universal DNA Purification Kit (TIANGEN, Cat# DP214). Illumina library was constructed using the NEBNext Ultra DNA Library Prep Kit for Illumina (New England Biolabs) according to the manufacturer's instructions. Qualified libraries were applied to 2× 150 bp paired-end sequencing on Illumina HiSeq X-ten platform (Illumina) at Shanghai Biotechnology Corporation. Primers used were (5′-3′):
Tet3 deep F2 TACAGGTCTGTGGGATAGTGAGG
Tet3 deep R2 AAGTTGAGCCAGGGCCAGA
Tet3 deep F4 GTGGCTCAGTCCCATGTCT
Tet3 deep R4 TGCCTGCTACCCTGGTTCTA
Tet3 deep F5 TGCAGGTGATCCGAAGACA
Tet3 deep R5 CTACAGCCCCATCACCACC
Tet3-exon6-OF (1st) CTCTGCCCAAGTGTAAAACG
Tet3-exon6-OR (1st) GCTAGTTCCAGGACAGCAAGAC
Tet3 deep6 F (2nd) TCTGCAGGTGATCCGAAGA
Tet3 deep6 R (2nd) TGCCTGCTACCCTGGTTCTA
Mass spectrometry analysis
We mainly followed the published protocol to analyze global 5hmC and 5mC levels using mass spectrometry43. Briefly, extracted genomic DNA (600-1 200 ng) was heat-denatured and then hydrolyzed with Nuclease P1 (Sigma, Cat# N8630) for > 4 h, followed by CIAP (Takara, Cat# 2250A ) treatment for another 4 h or overnight. The amount of 5hmC, 5mC and C in each sample was measured through a multiple reaction monitoring (MRM) method using an Agilent 1290 UHPLC system coupled with a G6490 triple quadrupole mass spectrometer (Agilent Technologies, Palo Alto, CA, USA). A Zorbax SB-C18 column (100 mm × 2.1 mm I.D., 1.8 μm particle size, Agilent Technologies) was used for UHPLC separation. The mobile phase (at a flow rate of 0.25 ml/min) consisted of 5% methanol in 0.1% formic acid solution. The mass spectrometer was operated at positive ion mode. The nebulization gas was set at 20 psi, the flow rate of desolvation gas was 12 l/min, and the source temperature was set at 200 °C. And the temperature and flow rate of sheath gas were set at 400 °C and 12 l/min, respectively. Capillary voltage was set at 3 500 V. High-purity nitrogen (99.999%) was used as collision gas. Transitions used for detection were m/z 258→142 for 5hmC (collision energy, 3 eV); m/z 242→126 for 5mC (3 eV); m/z 228→112 for C (5 eV).
Acute slice preparation and whole-cell recordings
After mice were deeply anesthetized with 0.7% sodium pentobarbital (0.14 g/kg body weight), brains were quickly transferred into ice-cold (0-4 °C) choline-based cutting solutions containing choline-Cl (110 mM), KCl (2.5 mM), NaH2PO4 (1.3 mM), MgCl2 (7 mM), CaCl2 (0.5 mM), NaHCO3 (25 mM) and 𝒟-glucose (20 mM), bubbled with 95% O2/5% CO2. Coronal slices of 300 μm thick were cut on a Leica VT 1200s vibratome (Leica Microsystems, Nussloch, Germany). Brain slices were recovered in a submerged chamber containing artificial cerebrospinal fluid (aCSF) at 37 °C for 30 min and then at room temperature (25-28 °C) for 1 h. aCSF contained NaCl (125 mM), KCl (2.5 mM), NaH2PO4 (1.3 mM), MgCl2 (1.3 mM), CaCl2 (2 mM), NaHCO3 (25 mM) and 𝒟-glucose (20 mM), bubbled with 95% O2/5% CO2. All slices were used within 6 h of the first recording.
Whole-cell recordings of layer 2/3 pyramidal neurons in cortex S1BF and hippocampus CA1 neurons were made with a MultiClamp 700B amplifier (Molecular Devices, Sunnyvale, CA, USA) and 3-4 MΩ pipettes, on a Nikon FN1 microscope (Nikon, Tokyo, Japan) with a 40× water immersion objective under infrared differential interference contrast optics, in aCSF at 28.5-30.0 °C as previously described44. Signals were low-pass filtered at 2 kHz and digitized at 10 kHz using Axon Digidata 1332A (Molecular Devices). For mEPSC recordings, a Cs+-based internal solution containing CsMeSO4 (130 mM), CsCl (5 mM), HEPES (10 mM), EGTA (0.5 mM), Na2ATP (15 mM), MgATP (4 mM), Na3GTP (0.3 mM) and Na2-phosphocreatine (10 mM) was used. For mIPSC recordings, high chloride Cs+-based internal solution containing CsCl (110 mM), NaCl (10 mM), MgCl2 (5 mM), EGTA (0.5 mM), MgATP (2 mM), Na3GTP (0.3 mM) and HEPES (40 mM) was used. Internal solutions were adjusted to pH 7.4, 270-280 mOsm. For recording AMPAR-mediated mEPSC, cells were held at −70 mV in voltage-clamp mode, in aCSF containing TTX (0.5 μM), picrotoxin (50 μM) and 𝒟-APV (50 μM) to block Na+ channels, GABAARs and NMDARs, respectively. For recording GABAAR-mediated mIPSC, cells were held at −60 mV with aCSF containing TTX (0.5 μM), NBQX (10 μM) and 𝒟-APV (50 μM) to block Na+ channels, AMPARs and NMDARs, respectively. Series resistance and input resistance were monitored throughout whole-cell recordings by giving a brief hyperpolarizing pulse (−10 mV, 100 ms) every 10 s. Only cells with series resistance < 15 MΩ and with changes of input resistance < 20% were analyzed. Series resistances were not significantly different between experimental groups and control groups. Liquid junction potential and series resistance were uncompensated.
tdTomato− cells and tdTomato+ cells were recorded in a random order. Data were analyzed blinded to the genotype in pClamp 10.3 (Molecular Devices) and MiniAnalysis (Synaptosoft) with detection thresholds of 5 and 6 pA for mEPSC and mIPSC, respectively.
Statistics
All the morphological data and electrophysiological data analyses were performed blindly. Statistical tests were performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA). Two-tailed paired Student's t-test was used for paired recording data and two-tailed unpaired Student's t-test was used for all other experiments. In the quantitative reverse transcription PCR experiment, the error bars represent SD of triplicate reactions in Figure 4A or SEM of several mice in Supplementary information, Figure S6A. In the morphological and electrophysiological experiments, data are represented as mean ± SEM, where n refers to the number of mice (for immunohistochemistry) or the number of cell pairs (for electrophysiological experiments). All conditions statistically different from control are indicated as *P < 0.05; **P < 0.01; ***P< 0.001.
Other methods are described in detail in Supplementary information, Data S1.
Author Contributions
JL and LW conceived the project. LW, ML, CQ, WM, Q Yin, JYL, HC, MH, KW, EZ, GP, SZ, GC, QL, KT, Q Yu and ZL performed the experiments. JL, LW, ML, CQ, WM, XY, NJ, GX and CW analyzed and interpreted data. JL, LW and XY wrote the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Acknowledgments
We thank Yi Zeng (Shanghai Institute of Biochemistry and Cell Biology) for kindly providing mT/mG mice. This study was supported by the Ministry of Science and Technology of China (2014CB964803 and 2015AA020307 to JL), the National Natural Science Foundation of China (31530048 and 81672117 to JL, 31530030 to XY and 31601163 to KW), the Chinese Academy of Sciences (XDB19010204 to JL and XDB02010000 to XY), Shanghai Municipal Commission for Science and Technology (16JC1420500 to JL) and China Postdoctoral Science Foundation (2016M601661 to KW).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
In vitro and in vivo Development of Two-cell Embryos after Injection of Cre mRNA into One Blastomere*.
CRISPR-Cas9-Mediated Tet1 or Tet3 Targeting in One Blastomere of Two-cell Embryos*.
Multiple sgRNAs Mediated Tet3, Tyr or Gata4 Targeting in One Blastomere of Two-cell Stage Embryos*.
Off-target Analysis in Chimeric Mice Carrying Tet3 Mutation Generated Using Multiple sgRNAs.
Chimerism Analysis of Embryos and Mice Derived after Cre mRNA Injection into One Blastomere of Two-cell Stage Embryos.
Analysis of Tet1 or Tet3 Targeted mT+/mG+ ESC Lines and Mice.
Multiple sgRNAs Mediated Tyr or Gata4 Targeting.
CRISPR-Cas9-mediated Tet3 Targeting Using the Ai9 Reporter Line.
Whole-cell Recordings from the Somatosensory Cortex of Chimeric Ai9 mice.
Gene Expression And Local Methylation Analysis of Candidate Target Genes in Tet3 Mutant Chimeric Mice.
Supplemental experimental procedures
References
- Griffiths AJF. An introduction to genetic analysis. 7th Edition. New York: WH Freeman, 2000.
- Capecchi MR. Altering the genome by homologous recombination. Science 1989; 244:1288–1292. [DOI] [PubMed] [Google Scholar]
- Nagy A. Manipulating the mouse embryo: a laboratory manual. 3rd Edition. Cold Spring Harbor: Cold Spring Harbor Laboratory Press, 2003.
- Luo H, Lin Y, Gao F, Zhang CT, Zhang R. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res 2014; 42:D574–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picciotto MR, Wickman K. Using knockout and transgenic mice to study neurophysiology and behavior. Physiol Rev 1998; 78:1131–1163. [DOI] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger BJr, et al. Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA 1992; 89:6232–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryding AD, Sharp MG, Mullins JJ. Conditional transgenic technologies. J Endocrinol 2001; 171:1–14. [DOI] [PubMed] [Google Scholar]
- Nagy A, Mar L, Watts G. Creation and use of a cre recombinase transgenic database. Methods Mol Biol 2009; 530:365–378. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013; 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiacovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods 2013; 10:741–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Qiu Z, Shao Y, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol 2013; 31:681–683. [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014; 156:836–843. [DOI] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014; 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014; 157:1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol 2013; 31:684–686. [DOI] [PubMed] [Google Scholar]
- Edwards RG, Beard HK. Oocyte polarity and cell determination in early mammalian embryos. Mol Hum Reprod 1997; 3:863–905. [DOI] [PubMed] [Google Scholar]
- Tan L, Shi YG. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 2012; 139:1895–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago M, Antunes C, Guedes M, Sousa N, Marques CJ. TET enzymes and DNA hydroxymethylation in neural development and function — how critical are they? Genomics 2014; 104:334–340. [DOI] [PubMed] [Google Scholar]
- Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011; 477:606–610. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis 2007; 45:593–605. [DOI] [PubMed] [Google Scholar]
- Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development 2003; 130:5113–5122. [DOI] [PubMed] [Google Scholar]
- Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development 2001; 128:3739–3748. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Yamashita Y, Takemoto T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev Biol 2016; 418:1–9. [DOI] [PubMed] [Google Scholar]
- Li T, Yang D, Li J, Tang Y, Yang J, Le W. Critical role of Tet3 in neural progenitor cell maintenance and terminal differentiation. Mol Neurobiol 2015; 51:142–154. [DOI] [PubMed] [Google Scholar]
- Yen ST, Zhang M, Deng JM, et al. Somatic mosaicism and allele complexity induced by CRISPR/Cas9 RNA injections in mouse zygotes. Dev Biol 2014; 393:3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 1997; 11:1061–1072. [DOI] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev 1997; 11:1048–1060. [DOI] [PubMed] [Google Scholar]
- Yu H, Su Y, Shin J, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat Neurosci 2015; 18:836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Girardo D, Govek EE, et al. Role of Tet1/3 genes and chromatin remodeling genes in cerebellar circuit formation. Neuron 2016; 89:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen TJ, Ji F, Jiao JW. Epigenetics: major regulators of embryonic neurogenesis. Sci Bull 2015; 60:1734–1743. [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010; 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Yamashita Y, Takemoto T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev Biol 2016; 418:1–9. [DOI] [PubMed] [Google Scholar]
- Micheva KD, Beaulieu C. Quantitative aspects of synaptogenesis in the rat barrel field cortex with special reference to GABA circuitry. J Comp Neurol 1996; 373:340–354. [DOI] [PubMed] [Google Scholar]
- Swiech L, Heidenreich M, Banerjee A, et al. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat Biotechnol 2015; 33:102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, et al. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell 2014; 159:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron 2014; 83:1051–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub C, Granger AJ, Saulnier JL, Sabatini BL. CRISPR/Cas9-mediated gene knock-down in post-mitotic neurons. PLoS One 2014; 9:e105584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikuni T, Nishiyama J, Sun Y, Kamasawa N, Yasuda R. High-throughput, high-resolution mapping of protein localization in mammalian brain by in vivo genome editing. Cell 2016; 165:1803–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Joung JK. Defining and improving the genome-wide specificities of CRISPR-Cas9 nucleases. Nat Rev Genet 2016; 17:300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature 2008; 453:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He YF, Li BZ, Li Z, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333:1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JJ, Li SJ, Zhang XD, et al. Oxytocin mediates early experience-dependent cross-modal plasticity in the sensory cortices. Nat Neurosci 2014; 17:391–399. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vitro and in vivo Development of Two-cell Embryos after Injection of Cre mRNA into One Blastomere*.
CRISPR-Cas9-Mediated Tet1 or Tet3 Targeting in One Blastomere of Two-cell Embryos*.
Multiple sgRNAs Mediated Tet3, Tyr or Gata4 Targeting in One Blastomere of Two-cell Stage Embryos*.
Off-target Analysis in Chimeric Mice Carrying Tet3 Mutation Generated Using Multiple sgRNAs.
Chimerism Analysis of Embryos and Mice Derived after Cre mRNA Injection into One Blastomere of Two-cell Stage Embryos.
Analysis of Tet1 or Tet3 Targeted mT+/mG+ ESC Lines and Mice.
Multiple sgRNAs Mediated Tyr or Gata4 Targeting.
CRISPR-Cas9-mediated Tet3 Targeting Using the Ai9 Reporter Line.
Whole-cell Recordings from the Somatosensory Cortex of Chimeric Ai9 mice.
Gene Expression And Local Methylation Analysis of Candidate Target Genes in Tet3 Mutant Chimeric Mice.
Supplemental experimental procedures