Abstract
Circular RNAs (circRNAs) were only recently discovered as a new class of noncoding RNAs, functionally still largely uncharacterized. Three publications that appeared concurrently in Cell Research and Molecular Cell provide initial evidence for certain endogenous circRNAs coding for proteins.
CircRNA was established only a few years ago as a new and intriguing class of noncoding RNA1. Over the last decades some isolated cases of circRNAs were identified in diverse biological systems, but only systematic deep-sequencing and bioinformatic approaches uncovered large classes of circRNAs in animal and plant systems, each comprising hundreds to thousands of individual examples1.
Despite this rapid initial progress in setting up circRNA catalogs in many different species, little is known about circRNA biogenesis and function. Regarding RNA processing, most circRNAs are generated from canonical mRNA precursors, using the standard splice signals, two-step mechanism, and spliceosome machinery to join the ends of a single or several adjacent exons in a covalently closed and circular configuration (also called back or circular splicing2). Thereby, instead of the “normal”, linear protein-coding mRNA, a circular isoform is generated. Beyond this basic mechanism, many open questions remain: What determines linear versus circular processing of the same pre-mRNA; what signals in RNA sequence and structure are involved, what protein factors, and what makes this decision regulated by cell type, tissue, and development?
In terms of the biological relevance of circRNAs, as well as the significance in human disease mechanisms, even less is known. This is partly due to experimental difficulties related to the low copy number of many circRNAs in the cell. At least for two specific cases, however, a miRNA sponge role has been convincingly demonstrated, based on multiple repeated copies of binding sites for the same miRNA3,4. Other than that, there are for the most part only hypotheses and speculations on the functional spectrum of circRNAs, ranging from protein sponges, antisense vectors, to biochemical effector molecules. In addition, circRNAs, although generally considered “noncoding” so far, may in fact serve as templates for protein translation. The idea that translation of a circular RNA template in vitro is possible had been shown already in 1995 by Chen and Sarnow5, using constructs with a so-called IRES element inserted (internal ribosome entry site) that promotes cap-independent translation.
This situation changed with three publications that appeared concurrently in Cell Research6 and Molecular Cell7,8, applying different approaches, but coming to the same conclusion that, at least in some instances, endogenous circRNAs are in fact protein-coding.
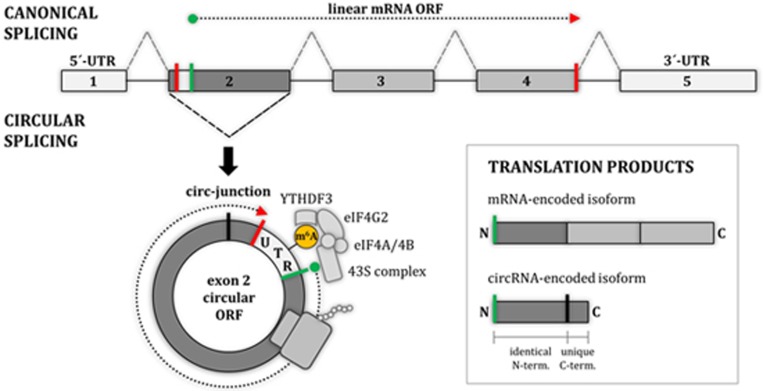
Yang et al.6 report in Cell Research that RNA base modification by N6-methyladenosine (m6A) provides an important signal mediating cap-independent translation initiation on circRNAs. Their work is based on recent reports from several other groups that m6A modification is an important determinant for translation of canonical linear mRNAs and can even promote their cap-independent translation9. As a logical extension, they show, by using circRNA-expressing minigene reporters in conjunction with mutational and knockdown analyses, that the m6A modification indeed affects translation also from circular RNA templates. Moreover, the initiation mechanism on circRNAs relies on a known m6A recognition factor, YTHDF3, and, similarly to cap-independent mRNA translation, an IRES-specialized translation initiation factor, eIF4G2 (Figure 1). In addition, Yang et al. were able to show that the m6A methyltransferase METTL3/14 and the demethylase FTO can modulate m6A-dependent translation efficiency, and that at least 13% of all identified circRNAs are m6A-modified.
Figure 1.
Cap-independent translation initiation on circular RNAs. Canonical splicing of a pre-mRNA produces the canonical linear RNA, with an open reading frame (ORF) encoding the standard protein. Alternatively, circular splicing can generate a covalently closed, circular RNA molecule (circRNA) with a continuous ORF spanning the circ-junction. This circular ORF encodes a shortened protein isoform with an N-terminus as in the standard protein and a unique C-terminus encoded downstream of the circ-junction. As shown by Yang et al.6, cap-independent translation of m6A-modified circRNAs can be initiated by m6 A recognition by the YTHDF3 reader protein and involves initiation factor eIF4G2.
To identify translated circRNAs more directly, they provide two independent lines of evidence: First, they designed a computational pipeline to select circRNA translation candidates, based on m6A modification, pre-mapped translation initiation sites, appropriate open reading frames, as well as polysome association, which yielded 25 circRNA candidates. The second approach relied directly on polysome fractionation and RNA-seq, which identified 250 circRNA candidates.
Finally, convincing evidence on circRNA-derived proteins was sought by another experimental strategy: identifying endogenous peptide sequences that are derived from and conform to circRNA junction sequences. Therefore they link two databases with each other: a customized database of peptides encoded by circ-junction spanning RNA sequences of all known circRNAs and the UniProt database of all human protein sequences for HEK293 cells. They succeed to identify by this approach 19 peptides that are unique to circular junctions, and validate two of them by targeted mass spectrometry.
In parallel to this highlighted study, two publications by Legnini et al.7 and Pamudurti et al.8 in Molecular Cell provide independent evidence for translation of circRNAs, also relying on multiple lines of evidence.
Legnini et al. identified cap-independent translation of a circRNA derived from the ZNF609 pre-mRNA, which is expressed in murine and human myoblasts. Besides that, they can link the expression of ZNF609 circRNA to modulation of myoblast proliferation; however, whether this is due to the circRNA-encoded protein, remains open. Notably, their evidence also includes polysome fractionation of the ZNF circRNA, as well as mass spectrometry, based on expression of a genome-engineered circ-ZNF609.
As circRNAs are highly expressed in brain, Pamudurti et al. employed Drosophila heads to analyze ribosome-footprinting data, which resulted in a subset of 122 circRNAs possibly associated with actively translating ribosomes. They extensively analyzed this subset of circRNAs for their characteristic features. Concentrating on a specific circRNA isoform of the muscleblind gene, they demonstrate IRES activity of this circRNA flanking region in an in vitro translation system.
Altogether a large amount of data has been accumulated by Yang et al.6, and the two other studies7,8, coming from different and strategically independent lines of evidence, all in favor of a positive answer to the central circRNA-translation question and adding important new information on the coding-potential of circRNAs. Are we definitively convinced? It should be noted that this issue was recently also addressed in other studies, based on small subsets of abundant human circRNAs and polysome gradients10,11, on global analysis of ribosome-footprinting data from human U2OS cells12, and on polysome fractionation combined with RNA-seq in mouse brain13. These studies came to a negative answer and leave us with a controversial impression.
Clearly, important questions are left open after all: What is the overall biological significance of circRNA translation as shown in the three new publications; are there many examples where protein or peptide isoforms are generated from circRNAs in biologically significant amounts and with novel functions, and where is the borderline to some kind of pervasive translational background? Not unexpected for such a fundamental question, even the three papers combined do not answer all of that, but highlight important advances; therefore more work is required, which these initial studies will hopefully stimulate. The underlying questions of circRNA functions, not only in translation, will certainly remain an experimental and intellectual challenge for the next few years.
References
- Chen LL. Nat Rev Mol Cell Biol 2016; 17:205–211. [DOI] [PubMed]
- Starke S, Jost I, Rossbach O, et al. Cell Rep 2014; 10:103–111. [DOI] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Nature 2013; 495:384–388. [DOI] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Nature 2013; 495:333–338. [DOI] [PubMed]
- Chen CY, Sarnow P. Science 1995; 268:415–417. [DOI] [PubMed]
- Yang Y, Fan X, Mao M, et al. Cell Res 2017; 27: 626–641. [DOI] [PMC free article] [PubMed]
- Legnini I, Di Timoteo G, Rossi F, et al. Mol Cell 2017; 66:22–37. [DOI] [PMC free article] [PubMed]
- Pamudurti NR, Bartok O, Jens M, et al. Mol Cell 2017; 66:9–21. [DOI] [PMC free article] [PubMed]
- Zhao BS, Roundtree IA, He C. Nat Rev Mol Cell Biol 2017; 18:31–42. [DOI] [PMC free article] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. RNA 2013; 19:141–157. [DOI] [PMC free article] [PubMed]
- Schneider T, Hung LH, Schreiner S, et al. Sci Rep 2016; 6:31313. [DOI] [PMC free article] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Genome Biol 2014; 15:409–414. [DOI] [PMC free article] [PubMed]
- You X, Vlatkovic I, Babic A, et al. Nat Neurosci 2015; 18:603–610. [DOI] [PMC free article] [PubMed]