Abstract
5-methylcytosine was shown before to be an epitranscriptomic mark. Yang et al. now explored the unique topology of this mRNA modification, identified its writer and demonstrated its involvement in nuclear-cytoplasmic shuttling mediated by a specific reader.
More than one hundred modified nucleotides, in addition to the four canonical A, C, G and U letters, are found in RNA molecules, mainly in highly abundant RNA species such as rRNA and tRNA. The last decade witnessed the mapping and characterization of a growing number of mRNA-decorating modified nucleotides collectively constituting the nascent field known as epitranscriptomics1. Starting with the deciphering of inosine (I) and N6-methyladenosine (m6A) and followed by the identification of 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), pseudouridine (ψ), N1-methyadenosine (m1A), 2′-O-methylnucleotides (Nm) and N6-2′-O-dimethyladenosine (m6Am), the epitranscriptome alphabet is rapidly expanding1.
m5C is the time-honored prototypic DNA modification, a major player in gene expression control and epigenetic regulation. It turned out that RNA is also decorated by m5C. rRNA marking by m5C was demonstrated in bacteria, archaea and eukaryotes whereas tRNA marking was found only in the latter two2. Archaeal and viral mRNA m5C decoration was clearly demonstrated, and some indications for eukaryotic mRNA marking also surfaced2. In recent years, based on the advance of bisulfite deep RNA sequencing and additional high throughput methodologies, more than 10 000 m5C sites were identified in the human transcriptome, and were reported to be enriched in the untranslated regions (UTRs) of mRNA transcripts3. A prominently augmented m5C marking was reported in the vicinity of Argonaute protein binding sites, suggesting a role in gene expression regulation. m5C decoration of mRNA was found to be dynamic, with some indications for a role in protein translation. Yet, much remained to be studied regarding cellular machineries involved in regulation of m5C RNA decoration and in mediation of m5C-regulated activities.
The extent of information available regarding the biological significance of the various epitranscriptomic marks differs widely. Some criteria enable us to evaluate, based on current knowledge, the relevance of each type of modification to cellular and organismal functions in the context of RNA epigenetics. These criteria include characteristic topology, evolutionary conservation, dynamic and reversible marking, identification of specific writers, readers and erasers, and documented links to biochemical and cellular outcomes. m6A marking currently provides the gold standard for a key player in the epitranscriptomic network. This modification is non-randomly distributed along mRNA landmarks, with enhanced decoration of stop codon and the proximal 3′ UTR regions as well as of long exons4. m6A is dynamic in response to environmental signals and is highly conserved4. Both writers and erasers of m6A are known, further alluding to the dynamic nature of this new type of epigenetic regulation1,4. A major key to deciphering cellular functions mediated by m6A was the identification of specific readers, members of the YTH and the HNRNP protein families, that are recruited to the modified nucleotide embedded in a typical motif and mediate its downstream activities4. Much like DNA methyl-CpG-binding proteins at the time, it was the discovery of m6A-binding proteins that proved instrumental in uncovering its functions and mechanisms of action. After revealing how cells read m6A, the first molecular mechanisms soon followed. m6A recognized by specific readers was shown to be involved in the regulation of RNA splicing, mRNA recruitment to P bodies and degradation, translation, 3′ UTR processing, microRNA biogenesis and activity, and X-chromosome inactivation1. m6A was shown to be essential for cell fate decisions in early stages of embryogenesis5, was linked to circadian control6, and was found to be relevant to diseases such as cancer, neurodegeneration, infertility and obesity1.
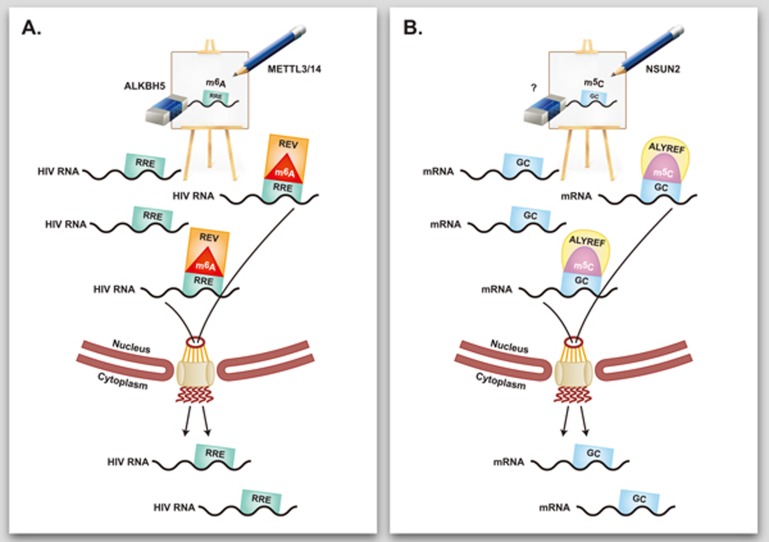
The extensive study by Yang et al. in May issue of Cell Research significantly upgrades the standing of m5C in the epitranscriptome field7. The detailed unique m5C topology, as explored in this study, differs from that reported before. Enhanced m5C deposition was found not only in the neighborhood of Argonaute protein binding sites, but also in regions located immediately after translation initiation sites. No specific conserved methylation-directing motif was identified, yet the m5C-decorated sequences are characterized by high GC content. The location of m5C in GC-rich sequences in the vicinity of translation start sites resembles the recently unraveled topology of m1A8, suggesting that m5C marking, similar to m1A marking, may affect translation efficiency. Another major finding of the study is that NSUN2 is the main RNA methyltransferase that mediates m5C installation on mRNA. The discovery of this m5C writer opens the way to experiments exploring the role of this modification by enhancing or silencing its activity. An additional important finding is the identification of the mRNA export adaptor ALYREF as a bona fide mRNA m5C reader. The authors convincingly show that ALYREF-mediated nuclear-cytoplasmic shuttling of mRNA is dependent on m5C marking by NSUN2 and precisely map K171 as the lysine residue that is essential for recruitment of the methylated transcripts. There are some indications that m6A also affects mRNA nuclear-cytoplasmic shuttling6, however no clear mechanistic insight is available that connects a specific endogenous cellular component of the export machinery in this case. Interestingly, the HIV-related Rev protein was shown to mediate viral RNA nuclear export, which is needed for viral replication, in an m6A-dependent manner9. The clear implication of ALYREF in m5C-regulated nuclear export provides an excellent mechanistic illustration for modification-dependent mRNA localization mediated by an endogenous export protein (Figure 1). Having in mind the location of both m5C and m1A in GC-rich sequences in the vicinity of translation start sites, it is tempting to explore the possibility that m1A decoration may also affect transcript export.
Figure 1.
Epitranscriptomic marks regulate nuclear export. (A) Decoration of the HIV RNA REV Response Element (RRE) by m6A recruits the viral REV reader protein, which accelerates nuclear export of viral transcripts9. m6A writing is mediated by METTL3/14 and its erasure is mediated by ALKBH5. (B) Decoration with m5C of GC-rich regions of mRNA molecules by the NSUN2 writer enables recruitment of the ALYREF m5C reader, which mediates nuclear export.
The precise localization of m5C marks to a unique region brings to mind the preferred targeting of m1A and m6A around the start and the end of coding regions, respectively. Recent studies indicate that m1A is located just upstream of the first exonic junction8 whereas m6A is found immediately downstream of the last exonic junction10, suggesting that components of the exonic junction complex (EJC) are involved in the exact positioning of these modifications. It can be speculated that the EJC is also involved in the precise deposition of m5C in the vicinity of the translation initiation site. The assembly of nuclear export-competent messenger ribonucleoprotein complexes comprises the recruitment of export-facilitating factors to the mature mRNA. It was shown lately that a core set of export proteins containing UAP56, DDX and importantly, ALYREF, nucleate and associate with spliced transcripts in an EJC- and cap-dependent manner11. Further studies are needed to explore the hypothesis that EJC and ALYREF play a major role in both m5C deposition and nuclear-cytoplasmic shuttling. Additional immediate questions that are raised by this study include the role of m5C in gene expression control in general and translation in particular, a possible interaction or overlap with other modified nucleotides such as m1A and m6A in a kind of “epitranscriptomic code”, whether there is a specific eraser that can demethylate m5C (TET protein?) and what the consequences of removal of this mark are.
The data obtained by Yang et al. and previous researchers concerning the unique telltale topology of m5C, its evolutionary conservation, dynamic nature and tissue specificity as well as the identification of specific writer and reader proteins and the unequivocal mechanistic role in nuclear export, now turn m5C into a very respected citizen of the epitranscriptome world, similar to the well-established position of m6A. The findings of this paper will enable to study the role of m5C in physiological and pathological states. Similar to the translation of DNA and histone epigenetics into established therapies, the clarification of the players and mechanisms involved in m5C decoration and reading may open the way for novel future therapeutic interventions.
References
- Zhao BS, Roundtree IA, He C. Nat Rev Mol Cell Biol 2017; 18:31–42. [DOI] [PMC free article] [PubMed]
- Hoernes TP, Erlacher MD. Wiley Interdiscip Rev RNA 2017; 8. 10.1002/wrna.1375. [DOI] [PMC free article] [PubMed]
- Squires JE, Patel HR, Nousch M, et al. Nucleic Acids Res 2012; 40:5023–5033. [DOI] [PMC free article] [PubMed]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Nature 2012; 485:201–206. [DOI] [PubMed]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, et al. Science 2015; 347:1002–1006. [DOI] [PubMed]
- Fustin JM, Doi M, Yamaguchi Y, et al. Cell 2013; 155:793–806. [DOI] [PubMed]
- Yang X, Yang Y, Sun BF, et al. Cell Res 2017; 27:606–625. [DOI] [PMC free article] [PubMed]
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, et al. Nature 2016; 530:441–446. [DOI] [PMC free article] [PubMed]
- Lichinchi G, Gao S, Saletore Y, et al. Nat Microbiol 2016; 1:16011. [DOI] [PMC free article] [PubMed]
- Ke S, Alemu EA, Mertens C, et al. Genes Dev 2015; 29:2037–2053. [DOI] [PMC free article] [PubMed]
- Gromadzka AM, Steckelberg AL, Singh KK, et al. Nucleic Acids Res 2016; 44:2348–2361. [DOI] [PMC free article] [PubMed]