Abstract
Insulin receptor (InsR) and the type I insulin-like growth factor (IGF1R) are homologous receptors necessary for signal transduction by their cognate ligands insulin, insulin-like growth factor-I and –II (IGF-I and IGF-II). IGF1R monoclonal antibodies, intended to inhibit malignant phenotypic signaling, failed to show benefit in patients with endocrine-resistant tumors in phase III clinical trials. Our previous work showed that in tamoxifen-resistant cells IGF1R expression was lacking but InsR inhibition effectively blocked growth. In endocrine-sensitive breast cancer cells, insulin was not growth stimulatory, likely due to the presence of hybrid InsR/IGF1R, which has high affinity for IGF-I, but not insulin. Combination inhibition of InsR and IGF1R showed complete suppression of the system in endocrine-sensitive breast cancer cells. To develop InsR-binding agents, we employed a small protein scaffold, T7 phage Gene 2 Protein (Gp2) with the long-term goal of creating effective InsR inhibitors and diagnostics. Using yeast display and directed evolution, we identified three Gp2 variants (Gp2 #1, #5 and #10) with low nanomolar affinity and specific binding to cell-surface InsR. These Gp2 variants inhibited insulin-mediated monolayer proliferation in both endocrine-sensitive and -resistant breast cancer, but did not downregulate InsR expression. Gp2 #5 and Gp2#10 disrupted InsR function by inhibiting ligand-induced receptor activation. In contrast, Gp2 #1 did not block InsR phosphorylation. Notably, Gp2 #1 binding was enhanced by pre-treatment of cells with insulin suggesting a unique receptor-ligand binding mode. These Gp2 variants are the first non-immunoglobulin protein scaffolds to target insulin receptor and present compelling opportunity for modulation of InsR signaling.
Keywords: Insulin receptor inhibition, endocrine-resistant breast cancer, yeast surface display, directed evolution, engineered Gp2 domain
Introduction
Numerous studies have shown the insulin-like growth factor (IGF) system plays a major role in enhancement of malignant phenotype and in drug resistance, including endocrine resistant breast cancer [1–3]. This system is composed of multiple receptors and ligands including insulin receptor (InsR), type I IGF receptor (IGF1R) and the ligands IGF-I, IGF-II and insulin. Phase III clinical trials testing IGF1R monoclonal antibodies failed to show benefits in breast cancer patients with endocrine-resistant tumors. Our data suggest that tamoxifen-resistant (TamR) cells exhibited a lack of IGF1R expression and an ability to block cell growth via InsR inhibition [4, 5]. Tissues obtained from tamoxifen-treated patients also show a decrease in IGF1R in resistant tumors [6]. Thus, IGF1R may not be the most appropriate target in endocrine resistant breast cancers.
InsR functions in breast cancer biology have been well-documented [7]. InsR content was approximately 80% higher in breast cancer compared to normal breast tissue [8]. Suppression of IGF1R sensitizes cells to insulin in model systems [5, 9]. Elevated insulin levels were identified in breast cancer patients compared to control. Further, elevated levels of insulin in non-diabetic patients were a negative predictor of progression free survival in breast cancer patients [10]. Breast cancer patients who suffer from obesity and type 2 diabetes, insulin resistance and hyperinsulinemia are more likely to develop metastatic disease and have high mortality compared to non-diabetic patients [11–13]. IGF ligands and IGF1R have mostly been the targets within this family for therapeutic intervention in cancer in the past, while InsR has been purposely avoided as a cancer therapy due to unwanted metabolic side effects [14]. However, preclinical and clinical evidence suggests insulin is a key regulator of metabolic homeostasis, and is also a cellular growth factor. This collection of evidence suggests that controlled modulation of InsR signaling presents an intriguing opportunity in breast cancer therapy.
While systemic inhibition of InsR may disrupt metabolism in an unselected patient group, certain subpopulations of cancer patients including non-diabetic endocrine-resistant breast cancer patients with increased InsR expression or insulin sensitivity may benefit from InsR inhibition. Yet, there are no reliable tools available for InsR quantification or specific InsR inhibition. The only drugs that inhibit InsR – linsitinib (OSI-906) and BMS-754807 – exist as dual IGF1R/InsR tyrosine kinase inhibitors Evidence suggested OSI-906 was well tolerated and antitumor activity was observed in phase I clinical trial [15]. A phase II trial testing BMS-754807 in ER+ breast cancer patients resistant to aromatase inhibitors has completed (NCT01225172) but the results have not been disclosed.
Antibodies, the most commonly studied target specific binding molecules have long been used as therapeutic and diagnostic tools in medicine as well as in basic and applied research. Numerous antibodies have been approved against many diseases from cancer to viral and immunologic diseases [16–18]. Despite their valuable and widespread applicability, antibodies suffer from several limitations including limited tissue penetration [19–21], complex bispecific generation, suboptimal pharmacokinetic clearance for diagnostic imaging [20, 22], reduced ease to target select epitopes [23], modest instability and high production cost [24]. Non-immunoglobulin protein scaffolds have arisen as an alternative strategy including single domains that are relatively small (<15 kDa), structurally stable, tolerable to mutation, and do not require disulfide bonds and post-translational modification for activity [25]. A number of non-immunoglobulin scaffolds have been developed and tested in clinical trials [25]. Adnectin CT-322, an engineered 10th type III domain of human fibronectin targeted against VEGFR-2, showed tolerable side effects and promising antitumor activity in patients with variety of solid tumors in phase I [26] although phase II trial of CT-322 did not show efficacy in recurrent glioblastoma [27]. More studies are underway with other adnectins (NCT02515669). ABY-025, an engineered affibody targeting HER2 receptor was able to discriminate HER2 status in metastatic breast cancer in patients under positron emission tomography (PET) imaging [28]. These and other examples have shown that small protein scaffolds can be engineered and selected against a wide array of targets and their applications can range from basic science to diagnostic and therapeutic functions.
In this study, we used yeast surface display [29, 30], a platform for library screening and binder isolation to develop binders to InsR based on the T7 gene 2 protein (Gp2) scaffold [31]. Gp2 is a 45-amino-acid domain with two solvent-exposed loops (Supplementary Figure 1) capable of mutation to generate new binding function while retaining structural and thermal stability with midpoints of thermal denaturation of 65–80°C [31]. A Gp2 domain engineered to bind epidermal growth factor receptor was effectively used as a PET probe for in vivo imaging in murine xenograft tumor models [32]. Although Gp2 domain has been engineered against various targets, no Gp2 binder has so far been shown to exhibit biological activity on cancer cells. Here, we evolved three Gp2 variants exhibiting strong binding affinity against cell-surface InsR. The domains inhibit insulin-regulated growth in endocrine-resistant and –sensitive breast cancer models and exhibit differential inhibition of receptor and downstream signaling. Notably this inhibition is observed even though one of the variants exhibits enhanced binding upon insulin co-treatment. These agents are the first non-immunoglobulin proteins that inhibit InsR function in breast cancer.
Materials and Methods
Binder Selection and Directed Evolution
The naïve Gp2 library contains 4×108 unique variants in which the sequences of two adjacent loops are diversified to six to eight amino acids with composition biased to mimic natural antibody repertoires (17% tyrosine, 13% serine, 11% aspartic acid, 9% asparagine, 6% alanine and histidine, 5% cysteine and threonine, 4% glycine and proline, 3% phenylalanine, arginine, valine,2% leucine, isoleucine, glutamic acid, lysine, 1% glutamine, tryptophan and <1% methionine) [31]. Binder selection and directed evolution were generally adopted from [31, 33]. Recombinant insulin receptor ectodomain (rInsR) (R&D Systems, Minneapolis MN), which served as a target antigen for screening, was biotinylated with EZ-link® NHS-PEG4-Biotin (Thermo Scientific, Rockford IL) resulting in nine biotin moieties per receptor. Yeast displaying the naïve Gp2 library underwent magnetic sorting, which consisted of two negative depletions (first with avidin-coated beads, then beads with immobilized biotinylated transferrin) to remove any non-specific binders, followed by a positive selection to enrich binders to immobilized biotinylated rInsR. Magnetic sorts on the naïve library were performed at 4°C with two washes during rInsR selections. Bound yeast were grown, re-induced and sorted more stringently with another round of magnetic sorting (two depletions and positive selection, as above) at room temperature with three washes. The resultant population was sorted by flow cytometry to isolate full length Gp2 mutants. Induced yeast were labeled with 10 μg/mL mouse anti-c-MYC antibody (9E10, BioLegend, San Diego CA) followed by 10 μg/mL fluorescein-conjugated goat anti-mouse antibody (Sigma Aldrich, St. Louis, MO). All fluorescein-positive yeast were collected via BD FACS Aria II. Sorted yeast were grown and plasmid DNA was extracted using Zymoprep yeast plasmid miniprep kit II (Zymo Research Corp., Irvine CA). The enriched Gp2 population was diversified by random mutagenesis to the full gene and the loops of Gp2 in parallel via error-prone PCR using nucleotide analogs 2′-deoxy-P-nucleoside-5′-triphosphate and 8-oxo-2′-deoxyguanosine-5′-triphosphate (Trilink Biotechnologies, San Diego CA) [9]. Mutant genes and gene fragments were retransformed into yeasts via electroporation with homologous recombination with linearized pCT vector. The process results in shuffling of mutated Gp2 loops.
The mutagenized Gp2 population underwent two rounds of magnetic sorting (both with three washes at room temperature), one flow cytometry sort against biotinylated rInsR and one flow cytometry sort against mammalian cell lysates expressing GFP-conjugated InsR. For first flow cytometry sort, the yeast library was labeled with anti-c-MYC antibody 9E10 and 50 nM biotinylated rInsR, followed by fluorescein-conjugated goat anti-mouse antibody and Alexa Fluor 647 (AF647)-conjugated streptavidin (Life Technologies, Eugene, OR). Yeast clones that showed double positive signals for fluorescein and AF647, indicating full length Gp2 expression and rInsR binding, were collected. To ensure selected Gp2 clones showed consistent binding capacity to cell surface InsR, the yeast population underwent another round of flow cytometry against mammalian cell lysates [34]. HEK293T cells were plated, transiently transfected with InsR-GFP plasmid (Addgene #22286) using Lipofectamine 2000 (Life Technologies) according to manufacturer’s protocol and lysed (1% Triton-X, 2 mM EDTA, 1x protease inhibitor in PBS). Plasmid expressing GFP alone (Addgene #19319) was used to create a GFP control. The induced yeast population was incubated in the cell lysate expressing either InsR-GFP or GFP-control for one hour, washed twice with cold 1% PBSA (1% bovine serum albumin in PBS) and labeled with 9E10 and AF647-conjugated anti-mouse antibodies. All yeast clones that showed GFP and AF647 signals were collected.
This enriched Gp2 population underwent mutagenesis and two rounds of flow cytometry sorts against cell lysates (two fold diluted relative to the first cell lysate sort) for affinity maturation. For each sort, yeast exhibiting the top 2% of GFP:AF647 ratio (InsR binding per displayed Gp2) were collected. These cells were grown and zymoprepped to isolate plasmid DNA. Clonal plasmid was obtained by transforming extracted DNA into E. coli One Shot® TOP10 E. coli (Invitrogen, Carlsbad, CA). Single colonies were grown in LB medium and miniprepped (Invitrogen, Carlsbad, CA). Purifed DNA was sequenced by Eurofins Genomics (Huntsville, AL).
Gp2 production and purification
NheI and BamHI restriction enzymes were used to transfer the Gp2 gene from pCT yeast surface display vector into a pET vector with a C-terminal His6 tag (Novagen, EMD Milipore, Billerica, MA). The resultant plasmids were transformed into T7 express competent E. coli and grown in LB medium containing 100 μg/mL kanamycin. One liter of LB medium was inoculated with 5 mL of overnight culture, grown at 37°C to an optical density at 600nm of 0.6–1.0 units, and induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside overnight at 20°C. Cells were pelleted, resuspended in 20 mL of lysis buffer (50 mM sodium phosphate, pH 8.0, 0.5 M sodium chloride, 5 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, 25 mM imidazole) and underwent five freeze-thaw cycles. The soluble fraction was isolated by centrifugation at 10,000 g for 40 min at 4°C and filtered through a 0.45 μm syringer filter. Gp2 was purified by nickel-nitrilotriacetic acid affinity chromatography (Qiagen, Hilden, Germany) and by size exclusion/gel filtration chromatography using PD-10 column packed with Sephadex G-25 (GE Healthcare Bio-Sciences, Pittsburgh, PA)
Cell lines and culture
MCF-7L and T47D are human estrogen receptor positive breast cancer cell lines. MCF-7L (parental cell line) was kindly provided by C. Kent Osborne (Baylor College of Medicine, Houston, TX) and maintained in improved MEM Richter’s modification medium (zinc option) supplemented with 5% FBS, and 11.25 nM insulin. They have been maintained in the Yee laboratory since 1989. MCF-7L karyotyping and gene expression profiling have shown that these cells are consistent with the originally described cell line. T47D (parental cell line) was obtained from ATCC in 1999 and maintained in MEM supplemented with 5% FBS, 1X nonessential amino acids and 1 nM insulin. MCF-7L TamR and T47D TamR cells were generated as described [4]. MCF-7L TamR cells were maintained in phenol-red free IMEM (zinc option) supplemented with 11.25 nM insulin, 5% charcoal/dextran-treated FBS and 100 nM 4-OH tamoxifen; while T47D TamR cells were maintained in phenol-red free IMEM supplemented with 6 ng/mL insulin, 1X nonessential amino acids, 5% charcoal/dextran-treated FBS and 100 nM 4-OH tamoxifen. Growth media above were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin and purchased from Gibco®. HEK293T were maintained in HyCloneTM Dulbecco’s Modified Eagles Medium supplemented with 10% FBS. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2.
Affinity titration
Lentiviral pLenti-InsR-GFP containing full length human InsR cDNA and control vector were purchased from Applied Biological Materials Inc. (British Columbia, Canada). They were referred to InsR-GFP and GFP-ctrl, respectively and were used to generate stable InsR overexpressing cell lines. Lentivirus production and transduction were carried out in HEK293T cells with 10 μg/mL polybrene to increase transduction efficiency. Cells underwent selection and were maintained in 2 μg/mL puromycin (Sigma-Adrich, St. Louis, MO). Stable HEK293T cell lines with either GFP-tagged InsR overexpression or GFP-control were labeled with soluble Gp2 at increasing concentrations for one hour at 4°C, washed with PBSA and labeled with AF647-conjugated anti-His5 antibody (Qiagen, Valencia, CA) for one hour at 4°C. Fluorescence was analyzed on a BD Accuri C6 flow cytometer and quantified using FlowJo software. The equilibrium dissociation constant, KD, was determined by minimizing the sum of squared errors assuming a 1:1 binding interaction.
Cell surface binding assay
Cells were detached using trypsin, washed with PBS, labeled with soluble Gp2 at varying concentrations for one hour at 4°C, washed with cold PBSA and labeled with AF647-conjugated anti-His5 antibody for one hour at 4°C. An anti-InsR antibody, clone 83-7 (hybridoma was kindly provided by Ken Siddle from University of Cambridge, UK) was used as a positive control for cell surface receptor binding. Fluorescence was analyzed on a BD Accuri C6 flow cytometer and quantified using FlowJo software.
Immunoblotting analysis
Cells were plated at a density of 8 × 105 cells in 60 mm diameter dishes and incubated overnight in complete medium. Cells were washed and incubated in serum-free medium for 24 hours. Cells were pre-treated with Gp2 overnight, treated with native ligands for 10 minutes, washed twice with ice-cold PBS and lysed with lysis buffer of 50 mM Tris-Cl (pH 7.4), 1% Nonidet P-40, 2 mM EDTA (pH 8.0), 100 mM NaCl, 10 mM sodium orthovanadate, and complete protease inhibitor cocktail (Roche Diagnostics). Lysates were centrifuged at 12,000 g for 30 minutes at 4°C. Protein concentrations were measured using bicinchoninic acid protein assay reagent kit (Pierce). Whole cell lysates (60 μg) were boiled in 5X Laemmli loading buffer, separated by 8% SDS-PAGE, transferred to PVDF membrane and immunoblotted for primary antibodies overnight at 4°C and secondary antibody for one hour in room temperature. The primary antibodies against total IGF1R (#3027), phosphorylated AKT S473 (#9271), phosphorylated IGF1R Y1135 (#3918), p44/42 MAPK (#9102), phosphorylated p44/42 MAPK T202/Y204 (#4376), and phosphorylated p70 S6 kinase T389 (#9205) were purchased from Cell Signaling Technology (Beverly, MA). InsR antibody (sc-711) was purchased from Santa Cruz Biotechnology (Dallas, Texas). Horseradish peroxidase-conjugated anti-phosphotyrosine (pY-20) (#610012) was purchased from BD transduction Lab (San Jose, CA) and was used to identify phosphorylation of insulin receptor substrate [35]. Anti-rabbit horseradish peroxidase-conjugated secondary antibody (#NA934V) was purchased from GE Health Life Science.
Monolayer growth assay
Cells were plated at a density of 15,000 cells per well in 24-well plates and allowed to attach overnight. Complete medium was then replaced by serum-free medium for 24 hours. Cells were treated with insulin (Eli Lilly, Indianapolis IN), IGF-I (Gemini, West Sacramento CA), IGF-II (Gemini), and/or Gp2 as indicated. After 5 or 6 days of treatment, growth was assessed via MTT assay. Sixty μL of 5 mg/mL thiazolyl blue tetrazolium bromide solution (MTT) (Sigma Adrich) was added to each well in serum-free medium for 4 hours at 37°C in dark. Media were aspirated and purple formazan crystals were dissolved with 500 μL solubilization solution (95% dimethylsulfoxide and 5% improved MEM). Absorbance was measured with a plate reader at 570 nm using a 650 nm differential filter.
Competitive binding assay
HEK293T cells transduced with pLenti-InsR were trypsinized, washed with PBS and pre-treated with or without insulin or IGF-II in 1% PBSA at 4°C for one hour. Cells were then labeled with Gp2 followed by AF-647 conjugated anti-His5 antibody at 4°C for one hour each. Fluorescence signal was detected using BD Accuri C6 flow cytometer and analyzed using FlowJo software.
Please see Supplementary Materials and Methods for additional information.
Results
Discovery and evolution of InsR-specific Gp2 binders through yeast surface display
InsR-specific Gp2 domains were discovered and evolved through yeast surface display and various enrichment and depletion technologies including magnetic bead selection and fluorescence activated cell sorting (FACS) with recombinant InsR ectodomain (rInsR) and detergent solubilized lysate of cells expressing InsR-GFP fusion (Figure 1A). Yeast displaying the naïve Gp2 library underwent two sorts of rInsR binding enrichment via magnetic beads and one FACS for full-length Gp2 before parallel error-prone PCR targeting the loops and the genes of enriched Gp2 variants. The resultant Gp2 population was further enriched with two sorts on rInsR-coated magnetic beads, one FACS with rInsR and one FACS with InsR-expressing cell lysate before undergoing a second round of mutations. During the final evolutionary round, the mutagenized InsR-enriched Gp2 population was sorted against cell lysate containing InsR-GFP and the top 2% of InsR binders were isolated via FACS (Figure 1B). Appreciable binding was not observed for the enriched Gp2 population to GFP-only cell lysates or yeast displaying non-enriched Gp2 control incubated with InsR-GFP lysates, indicating that the population evolved a Gp2—InsR interaction rather than binding between other elements of the selection process (Figure 1B).
Figure 1. Identification of InsR-specific Gp2 through yeast surface display.
(A) Schematic diagram of Gp2 scaffold engineering.
 indicates negative depletion;
indicates negative depletion;
 indicates InsR positive selection;
indicates InsR positive selection;
 indicates gene and loop mutations of Gp2, followed by electroporation via homologous recombination with linearized yeast surface display pCT vector. (B) Yeast surface display of Gp2 libraries against mammalian cell lysates containing GFP-conjugated InsR. HEK293T cells were plated, transfected with either InsR-GFP or GFP-control plasmids and collected as whole cell lysates. Yeast library displaying evolved population (−Gp2αInsR) was pre-incubated in either InsR-GFP or GFP-control contained lysates for 1 hour before labeling for a mouse c-MYC antibody, followed by AF647-conjugated anti-mouse antibody (Gp2 expression). Binding of Gp2 to InsR-GFP was detected and isolated by flow cytometry. Gp2 against rabbit IgG was used as a Gp2-control and was tested against InsR-GFP contained lysates under similar conditions.
indicates gene and loop mutations of Gp2, followed by electroporation via homologous recombination with linearized yeast surface display pCT vector. (B) Yeast surface display of Gp2 libraries against mammalian cell lysates containing GFP-conjugated InsR. HEK293T cells were plated, transfected with either InsR-GFP or GFP-control plasmids and collected as whole cell lysates. Yeast library displaying evolved population (−Gp2αInsR) was pre-incubated in either InsR-GFP or GFP-control contained lysates for 1 hour before labeling for a mouse c-MYC antibody, followed by AF647-conjugated anti-mouse antibody (Gp2 expression). Binding of Gp2 to InsR-GFP was detected and isolated by flow cytometry. Gp2 against rabbit IgG was used as a Gp2-control and was tested against InsR-GFP contained lysates under similar conditions.
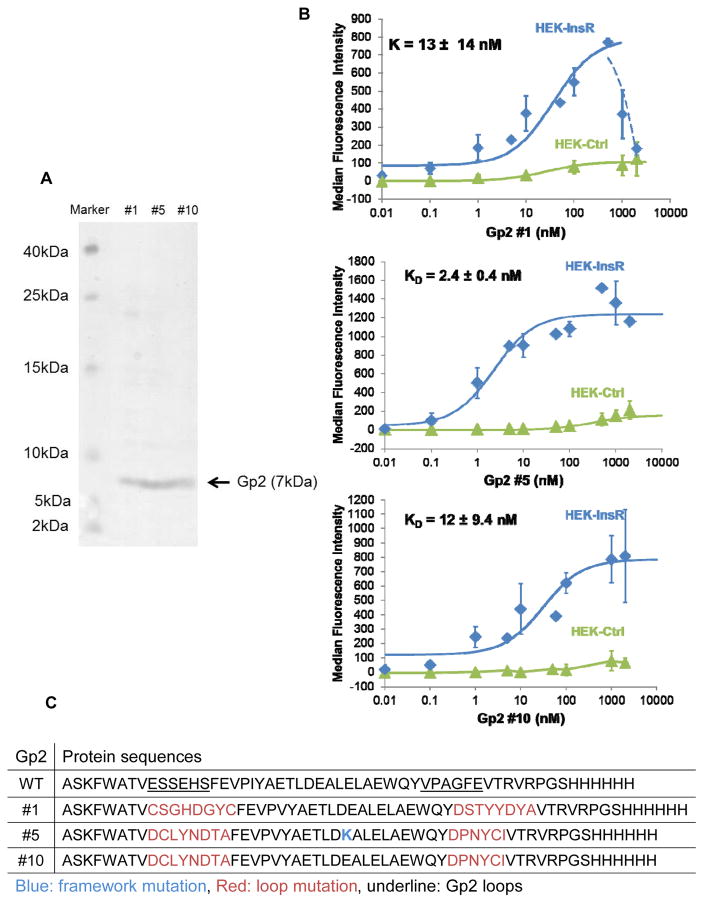
Among twenty selected top InsR binders from FACS, eight clones had unique protein sequences and proceeded for protein production in E. coli. Six variants had suitable protein yield in the soluble fraction of E. coli and were evaluated for binding to InsR overexpressing HEK293T cells. Two variants showed weak binding against cell-surface InsR; while a third variant showed no InsR specificity as it was not able to differentiate binding between InsRhigh and InsRlow cell lines. Three variants, name Gp2 #1, #5 and #10 exhibited strong binding to cell-surface InsR (Figure 2B). Gp2 #1 has unique loop sequences whereas Gp2s #5 and #10 have equivalent loops but a single amino acid difference at framework position 24 (Figure 2C). The purity and molecular weights (7 kDa with C-terminal His6 tag) of purified proteins were verified using SDS-PAGE (Figure 2A).
Figure 2. Affinity titration of Gp2 variants.
(A) Coomassie blue staining of soluble Gp2 clones. Identified Gp2 variants were purified using Ni-NTA resin and size exclusion filter, separated by 15% SDS-PAGE. The gel was stained with Coomassie blue. (B) HEK293T lentivirus transduced with either pLenti-InsR-GFP or pLent-GFP-ctrl were labeled with increasing concentrations of indicated soluble Gp2. Binding was detected by AF647-conjugated anti-His antibody via flow cytometry. Fluorescence signal was subtracted from the basal signal. KD values represented median ± standard deviation of three independent experiments. Some data points were not repeated and thus no standard deviation indicated. Note that HEK-Ctrl cells express modest levels of InsR (Supplementary Figure 2A), which accounts for the non-zero signal at higher concentrations. (C) Protein sequences of Gp2 variants.
 indicates a loop mutation;
indicates a loop mutation;
 indicates a framework mutation relative to initial library.
indicates a framework mutation relative to initial library.
Affinity titration of Gp2 variants
To measure the affinity of soluble Gp2 variants against cell-surface InsR and further evaluate specificity, two cell types (Supplementary Figure 2A) — InsR-GFP overexpression of HEK293T (InsRhigh) and GFP-control transduced HEK293T (InsRlow) – were labeled with ranging concentrations of soluble Gp2 proteins. Titration curves indicate strong binding of soluble Gp2 variants against cell-surface InsR with low nanomolar affinity. The KD values for Gp2 variants #1, #5 and #10 are 13 ± 14 nM, 2.4 ± 0.4 nM and 12 ± 9.0 nM, respectively (Figure 2B). Unevolved Gp2 control does not bind InsR overexpressing cells (Supplementary Figure 2B). Interestingly, Gp2 #1 reduced binding capacity after reaching a saturating point at very high concentration (1 μM), perhaps because of a high-dose “hook effect” limiting antibody-based detection [36, 37] or a reversible conformational change at high concentration. Notably, other functional assays in the study were performed at nanomolar concentrations where this phenomenon is not observed. Moreover, this scenario was not observed with Gp2 #5 and #10.
Minimal binding of Gp2 variants to IGF1R
Since InsR is closely related to IGF1R, sharing 45–65% domain dependent sequence and activating almost identical downstream signaling pathways [3, 38, 39], it is important to examine whether the Gp2 variants are able to distinguish IGF1R from InsR. The tamoxifen resistant MCF-7L TamR and T47D TamR cell lines were shown to have mild to moderate increased levels of InsR expression compared to their parental cell lines, but lack IGF1R expression [4], making them a good model for this study. Cells were labeled with either anti-InsR antibody, anti-IGF1R antibody or 100 nM soluble Gp2, followed by AF647-conjugated anti-His antibody to better understand the binding capacity of these Gp2 relative to the commercially available antibodies. Gp2#5 and Gp2#10 bound greater to the TamR cells compared to their parental cells (Figure 3B and 3D), which is consistent to that of anti-InsR antibody (Figure 3A and 3C). While antibody labelling demonstrated relatively similar IGF1R and InsR expressions in both MCF-7Ls and T47Ds, Gp2#1 also showed comparable binding to the parental and TamR in both cell lines, suggesting that Gp2#1 may have some weak cross-reaction with IGF1R.
Figure 3. Minimal binding of Gp2 variants to IGF1R.
MCF-7Ls (A) and T47Ds (C) were labeled with a mouse PE-conjugated anti-IGF1R antibody or a mouse anti-InsR antibody followed by an AF647-conjugated anti-mouse antibody for an hour each at 4°C. MCF-7Ls (B) and T47Ds (D) were labeled with 100 nM of indicated Gp2 variants and AF647-conjugated anti-His antibody for an hour each at 4°C. Bindings were measured via flow cytometry. Values were normalized to their respected controls and represented as geometry mean ± standard deviation in duplicates.
Inhibition of IGF-II/insulin-stimulated cell proliferation in breast cancer cells
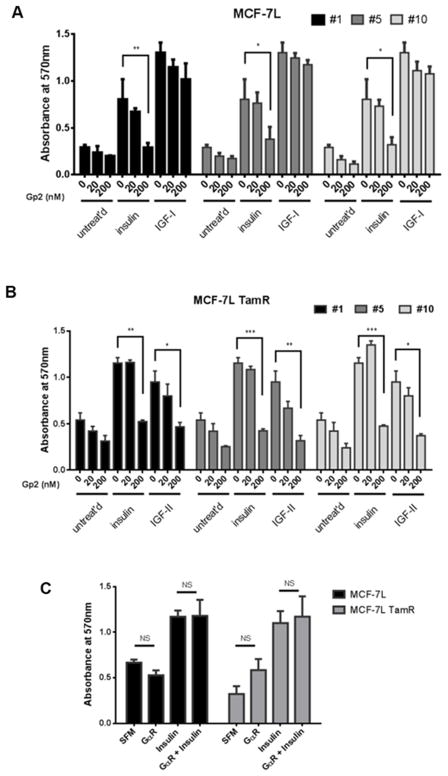
TamR cells derived from estrogen receptor positive cell lines not only lack IGF1R expression but also depend on InsR signaling for cell growth compared to their parental cells [5]. To elucidate how these Gp2 variants affect biological function in these breast cancer cells, an MTT assay was used to access cell monolayer growth in MCF-7L (Figure 4A) and MCF-7L TamR cells (Figure 4B). All three Gp2 variants inhibited insulin-stimulated growth in both MCF-7L and MCF-7L TamR cells in a dose-dependent manner, whereas non-binding Gp2 control elicited no effect (Figure 4C). All three Gp2 variants inhibited IGF-II-stimulated monolayer growth in MCF-7L TamR cells but insignificant inhibition – even at 200 nM – was observed on IGF-I regulated growth in MCF-7L cells. In the absence of IGF1R in MCF-7L TamR cells, IGF-II signals through InsR, whereas IGF-I signals via IGF1R in MCF-7L [40]. These results suggest that the inhibitory effects of Gp2 variants are specific to InsR-regulated growth. Gp2 variants yield insignificant reduction on the basal growth in MCF-7L and MCF-7L TamR cells.
Figure 4. Inhibition of IGF-II/insulin-stimulated cell proliferation in breast cancer cells by Gp2 variants.
Cell monolayer growth of (A) MCF-7L, MCF-7L TamR (B) T47D and T47D TamR were measured using MTT proliferation assay. Cells were plated, starved for 24 hours and then treated with indicated concentrations of soluble Gp2 in combination with either 10 nM insulin, 10 nM IGF-1 or 10 nM IGF-II. (C) Gp2 against rabbit IgG was used as a Gp2 control in affecting cell growth in MCF-7L and MCF-7L TamR. Readings were taken 5 days later and the values were represented as mean ± standard deviation in triplicates. Two-way ANOVA with Bonferroni comparison was performed to identify significance among untreated versus treated groups. *, p<0.05; **, p<0.01; ***, p<0.001.
The analogous experiment was repeated with T47D (Supplementary Figure 3A) and T47D TamR cells (Supplementary Figure 3B). Overall, T47D TamR cells have a higher basal growth compared to T47D parental cells and yield results consistent to those of MCF-7L TamR, where the Gp2 variants completely blocked insulin-regulated cell growth. Since IGF-II-stimulated growth was not strong in these cells, the inhibitory effects of Gp2 #5 and #10 were not significant compared to the Gp2 untreated groups. In contrast, toxicity was observed in T47D TamR cells treated with 200 nM of Gp2 #1 as the growth dropped below the basal level of the comparable untreated group. The T47D parental cell line exhibited some analogous behavior to MCF-7L in that Gp2 #1 inhibited insulin-driven growth and Gp2 #5 and #10 did not significant inhibit IGF-I-driven growth. However, T47D also showed several divergent outcomes relative to MCF-7L. Gp2 #1 had a strong inhibitory effect on IGF-I-regulated growth and Gp2 # 5 and #10 were not appreciable inhibitors of insulin-driven growth. Consistent to MCF-7L’s growth result, Gp2 variants blocked insulin-driven, but not IGF-I-driven, cell cycle progression in MDA-MB-231 cells and kept the cells arrested at G0/G1 phases (Supplementary Figure 4). To further examine if these variants inhibited growth by inducing cell death, cleaved PARP was measured by flow cytometry. Results showed that Gp2 variants did not significantly induce apoptosis in MCF-7L nor MDA-MB-231, suggesting that Gp2 itself is not cytotoxic (Supplementary Figure 5).
Differential effects of Gp2 variants on insulin/IGF signaling in breast cancer cells
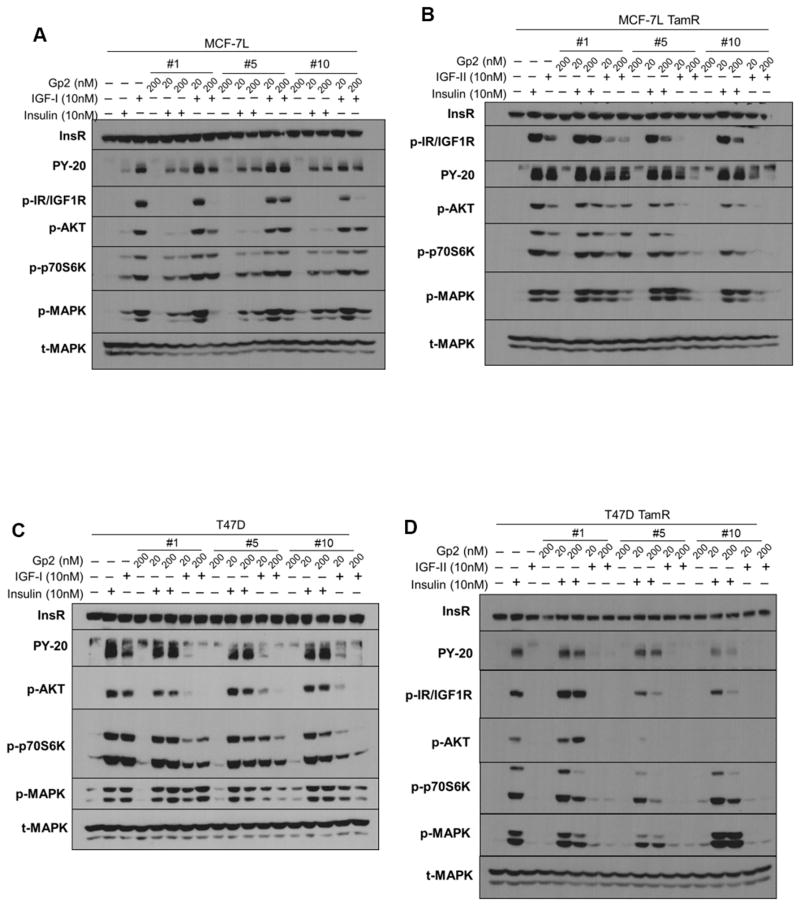
To better understand the underlying mechanism of Gp2-mediated growth inhibition on breast cancer cells, receptor and effector phosphorylation analysis was carried out after various treatments with growth factors and Gp2 variants in MCF-7L, T47D and their TamR derivatives (Figure 5). None of the Gp2 variants exhibited receptor activation in any of the cell lines. Overall, despite inhibiting growth, Gp2 #1 did not block insulin-mediated signaling in MCF-7L and T47D parental cells, as measured by phosphorylation of InsR/IGF1R, IRS (pY-20), AKT, p70S6K and MAPK. Similarly, in TamR cells, Gp2 #1 inhibits insulin-driven growth but is predominantly passive towards the tested signaling pathways with the exception of p70S6K inhibition in T47D TamR. Conversely, inhibition of InsR/IGF1R and MAPK phosphorylation are in accord with growth inhibition in MCF-7L TamR with IGF-II treatment. IGF-II driven signaling in T47D TamR was below the detection limit.
Figure 5. Differential signaling effects of Gp2 variants on insulin/IGF signaling in breast cancer cells.
Cells were plated, serum-starved overnight, pre-treated with indicated concentrations of Gp2 overnight for (A) MCF-7L, (B) MCF-7L TamR and 4 hours Gp2 pretreatment for (C) T47D, (D) T47D TamR before treating with either 10 nM IGF-I, 10 nM IGF-II or 10 nM insulin for 15 minutes. Whole cell lysates were separated by SDS-PAGE and immunoblotted with indicated antibodies.
In T47D expressing IGF1R, Gp2 #1’s inhibition of IGF-I-driven growth is in accord with AKT and p70S6K inhibition. Despite not impacting growth, Gp2 #1 strongly blocked IGF-I-mediated InsR/IGF1R and MAPK activation, and weakly inhibited AKT activation in MCF-7L parental cells. Gp2 #5 and #10 inhibited insulin- and IGF-II-driven growth in both TamR cell types in accord with blocking phosphorylation of most tested molecules and exhibited minimal inhibition of insulin-mediated MAPK activation in MCF-7L TamR. IGF-II-driven phosphorylation in T47D TamR cells was below the detection limit. In parental cells, Gp2 #5 and #10 did not impact insulin-driven signaling, which coordinated with sustained growth in T47D but countered the observed growth inhibition in MCF-7L. Consistent with growth experiments, both Gp2 #5 and #10 had little effect on IGF-I signaling in MCF-7L parental cells except that Gp2 #10 blocked receptor phosphorylation at higher concentration and both Gp2s modestly inhibited AKT. Both variants inhibited IGF-I-mediated signaling to MAPK, AKT and p70S6K activation in T47D parental cells.
As the signaling results of MCF-7L and T47D were varying, two other triple negative cell lines, MDA-MB-231 and MDA-MB-435 were tested. The Gp2 variants blocked insulin-, but not IGF-I-stimulated AKT activation in MDA-MB-435 cells. Surprisingly, an opposite result was observed in MDA-MB-231. The Gp2 variants strongly blocked IGF-I-mediated signaling, but exhibited little to no inhibition on insulin-mediated signaling (Supplementary Figure 6A). These diverse signaling outcomes in different cell lines are consistent with different abundance of IGF1R/InsR hybrid receptors measured by immunoprecipitation (Supplementary Figure 6B). When the cells express predominantly holo-InsR and minimal hybrid receptors – as in MDA-MB-435 – the inhibition of InsR signaling is straightforward. In contrast, in MCF-7L and MDA-MB-231 cells that express an abundance of hybrid receptors, Gp2 may have minimal inhibitory effects on insulin but overlapping impacts on IGF-I signaling, as IGF-I is the preferred ligand for hybrid receptors. Therefore, in term of signal transduction, it is not surprise that InsR-evolved Gp2 variants failed to block InsR existed in a hybrid context.
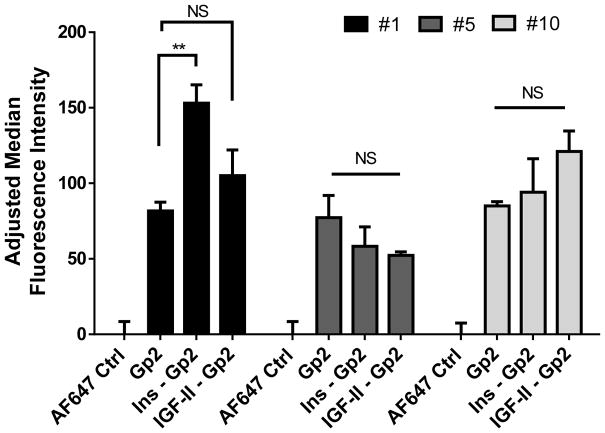
Positive cooperativity of insulin on binding of Gp2 #1, but not Gp2 #5 and #10
A competitive binding assay was carried out to assess the impact of natural ligands on Gp2 binding. HEK293T cells overexpressing InsR were treated with either insulin or IGF-II for 1 hour prior to Gp2 treatments. Gp2 binding was evaluated by flow cytometry. Results indicated that insulin, but not IGF-II enhanced Gp2#1 binding (Figure 6). Neither insulin nor IGF-II impacted binding by Gp2 #5 and #10. These results are additional evidence that Gp2 #1 has a different mode of action than Gp2 #5 and #10.
Figure 6. Non-competitive binding of Gp2 variants against InsR.
HEK293T pLenti-InsR-GFP cells were trypsinized and pre-incubated with either 1 μM of insulin or IGF-II for 1 hour before labeling with 100 nM Gp2 variants, followed by AF647-conjugated anti-His antibody. Binding was measured using flow cytometry. Results represented adjusted median ± standard deviation in triplicates, where the basal values were subtracted from all samples. Paired two-tailed t tests were used to compare Gp2-labeled versus ligands-Gp2-labeled samples. NS, not significant; **, p<0.01.
Discussion
Gp2 is a new protein scaffold that is a promising candidate for ligand engineering as it offers a smaller size in a robust, stable structural framework with a large available binding surface amenable to mutation [31]. Directed evolution yielded three Gp2 variants directed against InsR. Notably, all three carry a pair of cysteines, located at sites adjacent to each other in the wild-type structure, suggesting a disulfide bond in the proteins (Supplementary Figure 1B and 1C). The Gp2 variants were able to be produced in T7 express E. coli (Figure 2A) and their monomers showed InsR-specific binding (Figure 2B) and InsR inhibition in breast cancer cells (Figure 4).
Our previous study showed that InsR inhibition alone (using either shRNA, small molecule inhibitory insulin analog or anti-InsR antibody) was only effective in blocking insulin-stimulated signaling and growth in MCF-7L TamR and T47D TamR, but not their parental cells [5]. In this study, Gp2 variants inhibited growth in TamR cells and cell cycle progression in MDA-MB-231. The Gp2 variants were also able to efficiently block insulin-stimulated cell growth in MCF-7L parental with some variation in T47D parental cells. However, the signaling data in Figure 5A and 5C suggest that growth effects were not due to short term inhibition of InsR biochemical signaling or apoptosis induction. In parental cells, single chain of the InsR can exist as holoreceptors or InsR/IGF1R hybrid receptors. If InsR was predominantly contained in a hybrid form, then a Gp2 InsR specific binder may not inhibit the hybrid. Since the hybrid receptors have high affinity for IGF-I, but not insulin, this could explain why the Gp2 binders do not blocked IGF action. Additional studies need to be performed to determine if the Gp2 binders affect signaling in the TamR cells after long term exposure to explain the differences between the signaling and growth results we have shown here.
Gp2 #5 and #10 exhibit similar functional impact on growth, which is consistent with their almost identical sequences (an E24K framework mutation in #5 is the only difference). Both demonstrated moderate signaling inhibition and strong cell growth inhibition. Gp2 #1 appears to operate via a different mechanism of action than Gp2 #5 and #10. Unlike Gp2 #5 and #10, Gp2 #1 had three significant differences. Gp2 #1 completely blocked IGF-I and insulin-mediated growth in T47D parental cells, inhibited signaling in a different fashion and exhibited positive cooperative binding with insulin, which suggests a unique insulin/InsR binding mechanism. An insignificant reduction on the basal growth in the MCF-7L and MCF-7L TamR cells could possibly be due to indirect effects of InsR inhibition on glucose metabolism (Figure 4).
InsR exists in two isoforms: isoform A and B. InsR-A, which lacks 12 amino acid via alternative splicing of exon 11 is predominantly expressed in fetal and cancer cells with high affinity to insulin and IGF-II; whereas, InsR-B is mainly expressed in metabolic organs such as adult muscle, liver and fat and binds only insulin at physiological concentration [41, 42]. In the cells we used, the predominant isoform is InsR-A [43] and further study will be needed to determine if the molecules we have described also have high affinity for InsR-B. Since Gp2 is one of the smaller protein scaffolds, it may be at advantage in the design strategy for targeting against a challenging epitope to develop isoform-specific binders.
In summary, targeting InsR is needed to completely disrupt the IGF1R/InsR network, which is especially evident in tamoxifen resistant breast cancers. The continued function of InsR may explain why targeting IGF1R was unsuccessful in phase III clinical trials. The emergence of precision medicine increasingly recognizes the development of new cancer interventions that may one day improve clinical outcomes. We demonstrate small InsR-evolved Gp2 variants not only exhibit strong binding affinity and specificity to InsR, but also promising biological outcomes in breast cancer cells. Before clinical translation, additional work including in vivo study is needed to understand the mechanism, potential side effects and efficacy of these Ins-targeted inhibitory Gp2 domains.
Supplementary Material
Acknowledgments
We acknowledge the assistance of the Flow Cytometry Shared Resource of the Masonic Cancer Center, University of Minnesota.
Financial support:
DY and JYC supported by NIH/NCI P50 CA116201, P30 CA 077598, and Komen for the Cure SAC110039. BJH supported by the University of Minnesota.
Footnotes
Disclosure of any potential conflicts of interest:
The authors declare no conflict of interest.
References
- 1.Vigneri PG, Tirro E, Pennisi MS, Massimino M, Stella S, Romano C, et al. The Insulin/IGF System in Colorectal Cancer Development and Resistance to Therapy. Frontiers in oncology. 2015;5:230. doi: 10.3389/fonc.2015.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bieghs L, Johnsen HE, Maes K, Menu E, Van Valckenborgh E, Overgaard MT, et al. The insulin-like growth factor system in Multiple Myeloma: diagnostic and therapeutic potential. Oncotarget. 2016 doi: 10.18632/oncotarget.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Y, Yee D. Targeting insulin and insulin-like growth factor signaling in breast cancer. Journal of mammary gland biology and neoplasia. 2012;17(3–4):251–261. doi: 10.1007/s10911-012-9268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fagan DH, Uselman RR, Sachdev D, Yee D. Acquired resistance to tamoxifen is associated with loss of the type I insulin-like growth factor receptor: implications for breast cancer treatment. Cancer research. 2012;72(13):3372–3380. doi: 10.1158/0008-5472.CAN-12-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan JY, LaPara K, Yee D. Disruption of insulin receptor function inhibits proliferation in endocrine-resistant breast cancer cells. Oncogene. 2016;35(32):4235–4243. doi: 10.1038/onc.2015.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drury SC, Detre S, Leary A, Salter J, Reis-Filho J, Barbashina V, et al. Changes in breast cancer biomarkers in the IGF1R/PI3K pathway in recurrent breast cancer after tamoxifen treatment. Endocrine-related cancer. 2011;18(5):565–577. doi: 10.1530/ERC-10-0046. [DOI] [PubMed] [Google Scholar]
- 7.Vigneri R, Goldfine ID, Frittitta L. Insulin, insulin receptors, and cancer. Journal of endocrinological investigation. 2016 doi: 10.1007/s40618-016-0508-7. [DOI] [PubMed] [Google Scholar]
- 8.Papa V, Pezzino V, Costantino A, Belfiore A, Giuffrida D, Frittitta L, et al. Elevated insulin receptor content in human breast cancer. The Journal of clinical investigation. 1990;86(5):1503–1510. doi: 10.1172/JCI114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zaccolo M, Williams DM, Brown DM, Gherardi E. An approach to random mutagenesis of DNA using mixtures of triphosphate derivatives of nucleoside analogues. Journal of molecular biology. 1996;255(4):589–603. doi: 10.1006/jmbi.1996.0049. [DOI] [PubMed] [Google Scholar]
- 10.Ferroni P, Riondino S, Laudisi A, Portarena I, Formica V, Alessandroni J, et al. Pretreatment Insulin Levels as a Prognostic Factor for Breast Cancer Progression. The oncologist. 2016;21(9):1041–1049. doi: 10.1634/theoncologist.2015-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, et al. Diabetes and cancer: a consensus report. CA: a cancer journal for clinicians. 2010;60(4):207–221. doi: 10.3322/caac.20078. [DOI] [PubMed] [Google Scholar]
- 12.Rose DP, Haffner SM, Baillargeon J. Adiposity, the metabolic syndrome, and breast cancer in African-American and white American women. Endocrine reviews. 2007;28(7):763–777. doi: 10.1210/er.2006-0019. [DOI] [PubMed] [Google Scholar]
- 13.Josefson D. High insulin levels linked to deaths from breast cancer. BMJ : British Medical Journal. 2000;320(7248):1496–1496. [PMC free article] [PubMed] [Google Scholar]
- 14.Yee D. Insulin-like growth factor receptor inhibitors: baby or the bathwater? Journal of the National Cancer Institute. 2012;104(13):975–981. doi: 10.1093/jnci/djs258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macaulay VM, Middleton MR, Eckhardt SG, Rudin CM, Juergens RA, Gedrich R, et al. Phase I Dose-Escalation Study of Linsitinib (OSI-906) and Erlotinib in Patients with Advanced Solid Tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(12):2897–2907. doi: 10.1158/1078-0432.CCR-15-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan AC, Carter PJ. Therapeutic antibodies for autoimmunity and inflammation. Nat Rev Immunol. 2010;10(5):301–316. doi: 10.1038/nri2761. [DOI] [PubMed] [Google Scholar]
- 17.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12(4):278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 18.Sanna PP, Burton DR. Role of Antibodies in Controlling Viral Disease: Lessons from Experiments of Nature and Gene Knockouts. Journal of Virology. 2000;74(21):9813–9817. doi: 10.1128/jvi.74.21.9813-9817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thurber GM, Schmidt MM, Wittrup KD. Antibody tumor penetration: transport opposed by systemic and antigen-mediated clearance. Advanced drug delivery reviews. 2008;60(12):1421–1434. doi: 10.1016/j.addr.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmidt MM, Wittrup KD. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Molecular cancer therapeutics. 2009;8(10):2861–2871. doi: 10.1158/1535-7163.MCT-09-0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minchinton AI, Tannock IF. Drug penetration in solid tumours. Nat Rev Cancer. 2006;6(8):583–592. doi: 10.1038/nrc1893. [DOI] [PubMed] [Google Scholar]
- 22.Stern LA, Case BA, Hackel BJ. Alternative Non-Antibody Protein Scaffolds for Molecular Imaging of Cancer. Current opinion in chemical engineering. 2013;2(4) doi: 10.1016/j.coche.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz KR, Bagchi A, Roovers RC, van Bergen en Henegouwen PM, Ferguson KM. Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure. 2013;21(7):1214–1224. doi: 10.1016/j.str.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies: successes, limitations and hopes for the future. British journal of pharmacology. 2009;157(2):220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skrlec K, Strukelj B, Berlec A. Non-immunoglobulin scaffolds: a focus on their targets. Trends in biotechnology. 2015;33(7):408–418. doi: 10.1016/j.tibtech.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 26.Tolcher AW, Sweeney CJ, Papadopoulos K, Patnaik A, Chiorean EG, Mita AC, et al. Phase I and Pharmacokinetic Study of CT-322 (BMS-844203), a Targeted Adnectin Inhibitor of VEGFR-2 Based on a Domain of Human Fibronectin. Clinical Cancer Research. 2011;17(2):363. doi: 10.1158/1078-0432.CCR-10-1411. [DOI] [PubMed] [Google Scholar]
- 27.Schiff D, Kesari S, de Groot J, Mikkelsen T, Drappatz J, Coyle T, et al. Phase 2 study of CT-322, a targeted biologic inhibitor of VEGFR-2 based on a domain of human fibronectin, in recurrent glioblastoma. Investigational new drugs. 2015;33(1):247–253. doi: 10.1007/s10637-014-0186-2. [DOI] [PubMed] [Google Scholar]
- 28.Sorensen J, Sandberg D, Sandstrom M, Wennborg A, Feldwisch J, Tolmachev V, et al. First-in-human molecular imaging of HER2 expression in breast cancer metastases using the 111In-ABY-025 affibody molecule. Journal of nuclear medicine : official publication, Society of Nuclear Medicine. 2014;55(5):730–735. doi: 10.2967/jnumed.113.131243. [DOI] [PubMed] [Google Scholar]
- 29.Chao G, Lau WL, Hackel BJ, Sazinsky SL, Lippow SM, Wittrup KD. Isolating and engineering human antibodies using yeast surface display. Nat Protocols. 2006;1(2):755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 30.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypeptide libraries. Nature biotechnology. 1997;15(6):553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 31.Kruziki MA, Bhatnagar S, Woldring DR, Duong VT, Hackel BJ. A 45-Amino-Acid Scaffold Mined from the PDB for High-Affinity Ligand Engineering. Chemistry & biology. 2015;22(7):946–956. doi: 10.1016/j.chembiol.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kruziki MA, Case BA, Chan JY, Zudock EJ, Woldring DR, Yee D, et al. A 64Cu-labeled Gp2 Domain for PET Imaging of Epidermal Growth Factor Receptor. Molecular Pharmaceutics. 2016 doi: 10.1021/acs.molpharmaceut.6b00538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackel BJ, Wittrup KD. The full amino acid repertoire is superior to serine/tyrosine for selection of high affinity immunoglobulin G binders from the fibronectin scaffold. Protein engineering, design & selection : PEDS. 2010;23(4):211–219. doi: 10.1093/protein/gzp083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho YK, Shusta EV. Antibody library screens using detergent-solubilized mammalian cell lysates as antigen sources. Protein engineering, design & selection : PEDS. 2010;23(7):567–577. doi: 10.1093/protein/gzq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byron SA, Horwitz KB, Richer JK, Lange CA, Zhang X, Yee D. Insulin receptor substrates mediate distinct biological responses to insulin-like growth factor receptor activation in breast cancer cells. Br J Cancer. 2006;95(9):1220–1228. doi: 10.1038/sj.bjc.6603354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tate J, Ward G. Interferences in Immunoassay. The Clinical Biochemist Reviews. 2004;25(2):105–120. [PMC free article] [PubMed] [Google Scholar]
- 37.Schiettecatte J, Anckaert E, Smitz J. In Tech, editor. Interferences in Immunoassays. In: Chiu NHL, Christopoulos TK, editors. Advances in Immunoassay Technology. 2012. pp. 45–62. [Google Scholar]
- 38.Lou M, Garrett TP, McKern NM, Hoyne PA, Epa VC, Bentley JD, et al. The first three domains of the insulin receptor differ structurally from the insulin-like growth factor 1 receptor in the regions governing ligand specificity. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12429–12434. doi: 10.1073/pnas.0605395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddle K, Urso B, Niesler CA, Cope DL, Molina L, Surinya KH, et al. Specificity in ligand binding and intracellular signalling by insulin and insulin-like growth factor receptors. Biochemical Society transactions. 2001;29(Pt 4):513–525. doi: 10.1042/bst0290513. [DOI] [PubMed] [Google Scholar]
- 40.Morrione A, Valentinis B, Xu S-q, Yumet G, Louvi A, Efstratiadis A, et al. Insulin-like growth factor II stimulates cell proliferation through the insulin-receptor. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(8):3777–3782. doi: 10.1073/pnas.94.8.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moller DE, Yokota A, Caro JF, Flier JS. Tissue-specific expression of two alternatively spliced insulin receptor mRNAs in man. Molecular endocrinology (Baltimore, Md) 1989;3(8):1263–1269. doi: 10.1210/mend-3-8-1263. [DOI] [PubMed] [Google Scholar]
- 42.Frasca F, Pandini G, Scalia P, Sciacca L, Mineo R, Costantino A, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Molecular and cellular biology. 1999;19(5):3278–3288. doi: 10.1128/mcb.19.5.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Fagan DH, Zeng X, Freeman KT, Sachdev D, Yee D. Inhibition of cancer cell proliferation and metastasis by insulin receptor downregulation. Oncogene. 2010;29(17):2517–2527. doi: 10.1038/onc.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.