Abstract
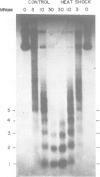
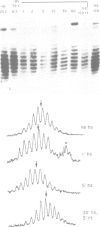
Coding regions of transcribed and non-transcribed genes typically differ in chromatin structure. However, it is not known what kind of alterations in nucleosome or chromatin structure these differences reflect. To determine whether changes in nucleosome topology accompany transcription, we introduced into yeast a multicopy plasmid bearing the gene coding for the heat shock protein HSP26. The plasmid-borne gene is assembled into chromatin, and is induced by heat shock in the same manner as the endogenous HSP26 gene. A small change in linking number in the HSP26 plasmid accompanies heat shock. This change is consistent with that previously reported for thermal untwisting of DNA in yeast chromatin, and is equivalent in magnitude to that observed in control plasmids which lack heat-shock response elements. These data indicate that no stable alteration in nucleosome topology accompanies transcription of the heat shock gene. Moreover, the kinetics of the observed changes in linking number indicate that topoisomerase relaxes the thermally induced torsional stress in 1-5 min. We conclude that if alterations in nucleosome topology accompany polymerase passage, recovery must take place within this time period.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambrose C., McLaughlin R., Bina M. The flexibility and topology of simian virus 40 DNA in minichromosomes. Nucleic Acids Res. 1987 May 11;15(9):3703–3721. doi: 10.1093/nar/15.9.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazett-Jones D. P., Brown M. L. Electron microscopy reveals that transcription factor TFIIIA bends 5S DNA. Mol Cell Biol. 1989 Jan;9(1):336–341. doi: 10.1128/mcb.9.1.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benezra R., Cantor C. R., Axel R. Nucleosomes are phased along the mouse beta-major globin gene in erythroid and nonerythroid cells. Cell. 1986 Mar 14;44(5):697–704. doi: 10.1016/0092-8674(86)90835-4. [DOI] [PubMed] [Google Scholar]
- Bloom K. S., Anderson J. N. Hormonal regulation of the conformation of the ovalbumin gene in chick oviduct chromatin. J Biol Chem. 1982 Nov 10;257(21):13018–13027. [PubMed] [Google Scholar]
- Bloom K. S., Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982 Jun;29(2):305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- Bossier P., Fitch I. T., Boucherie H., Tuite M. F. Structure and expression of a yeast gene encoding the small heat-shock protein Hsp26. Gene. 1989 May 30;78(2):323–330. doi: 10.1016/0378-1119(89)90234-5. [DOI] [PubMed] [Google Scholar]
- Brill S. J., Sternglanz R. Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell. 1988 Jul 29;54(3):403–411. doi: 10.1016/0092-8674(88)90203-6. [DOI] [PubMed] [Google Scholar]
- Chen T. A., Allfrey V. G. Rapid and reversible changes in nucleosome structure accompany the activation, repression, and superinduction of murine fibroblast protooncogenes c-fos and c-myc. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5252–5256. doi: 10.1073/pnas.84.15.5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi A., Widmer R. M., Koller T., Sogo J. M. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989 Jun 2;57(5):753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- De Bernardin W., Koller T., Sogo J. M. Structure of in-vivo transcribing chromatin as studied in simian virus 40 minichromosomes. J Mol Biol. 1986 Oct 5;191(3):469–482. doi: 10.1016/0022-2836(86)90142-7. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germond J. E., Hirt B., Oudet P., Gross-Bellark M., Chambon P. Folding of the DNA double helix in chromatin-like structures from simian virus 40. Proc Natl Acad Sci U S A. 1975 May;72(5):1843–1847. doi: 10.1073/pnas.72.5.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G. N., Wang J. C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988 Dec 2;55(5):849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- Gilmour D. S., Dietz T. J., Elgin S. C. TATA box-dependent protein-DNA interactions are detected on heat shock and histone gene promoters in nuclear extracts derived from Drosophila melanogaster embryos. Mol Cell Biol. 1988 Aug;8(8):3204–3214. doi: 10.1128/mcb.8.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri C. P., Gorovsky M. A. DNase I sensitivity of ribosomal genes in isolated nucleosome core particles. Nucleic Acids Res. 1980 Jan 11;8(1):197–214. doi: 10.1093/nar/8.1.197-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C., Meeks-Wagner D. W., Fangman W. L., Botstein D. A rapid, efficient method for isolating DNA from yeast. Gene. 1986;42(2):169–173. doi: 10.1016/0378-1119(86)90293-3. [DOI] [PubMed] [Google Scholar]
- Ip Y. T., Granner D. K., Chalkley R. Hormonal regulation of phosphoenolpyruvate carboxykinase gene expression is mediated through modulation of an already disrupted chromatin structure. Mol Cell Biol. 1989 Mar;9(3):1289–1297. doi: 10.1128/mcb.9.3.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen B. K., Pelham H. R. Constitutive binding of yeast heat shock factor to DNA in vivo. Mol Cell Biol. 1988 Nov;8(11):5040–5042. doi: 10.1128/mcb.8.11.5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpov V. L., Preobrazhenskaya O. V., Mirzabekov A. D. Chromatin structure of hsp 70 genes, activated by heat shock: selective removal of histones from the coding region and their absence from the 5' region. Cell. 1984 Feb;36(2):423–431. doi: 10.1016/0092-8674(84)90235-6. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Elgin S. C. Patterns of DNA structural polymorphism and their evolutionary implications. Cell. 1984 Jan;36(1):121–129. doi: 10.1016/0092-8674(84)90080-1. [DOI] [PubMed] [Google Scholar]
- Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W., Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- Lacy E., Axel R. Analysis of DNA of isolated chromatin subunits. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3978–3982. doi: 10.1073/pnas.72.10.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Torchia T. Structure of the chromosomal copy of yeast ARS1. Biochemistry. 1988 May 31;27(11):3961–3965. doi: 10.1021/bi00411a011. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987 Apr 24;49(2):203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- Lorch Y., LaPointe J. W., Kornberg R. D. On the displacement of histones from DNA by transcription. Cell. 1988 Dec 2;55(5):743–744. doi: 10.1016/0092-8674(88)90128-6. [DOI] [PubMed] [Google Scholar]
- Losa R., Brown D. D. A bacteriophage RNA polymerase transcribes in vitro through a nucleosome core without displacing it. Cell. 1987 Aug 28;50(5):801–808. doi: 10.1016/0092-8674(87)90338-2. [DOI] [PubMed] [Google Scholar]
- Morse R. H., Cantor C. R. Nucleosome core particles suppress the thermal untwisting of core DNA and adjacent linker DNA. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4653–4657. doi: 10.1073/pnas.82.14.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Pederson D. S., Dean A., Simpson R. T. Yeast nucleosomes allow thermal untwisting of DNA. Nucleic Acids Res. 1987 Dec 23;15(24):10311–10330. doi: 10.1093/nar/15.24.10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse R. H., Simpson R. T. DNA in the nucleosome. Cell. 1988 Jul 29;54(3):285–287. doi: 10.1016/0092-8674(88)90190-0. [DOI] [PubMed] [Google Scholar]
- Nacheva G. A., Guschin D. Y., Preobrazhenskaya O. V., Karpov V. L., Ebralidse K. K., Mirzabekov A. D. Change in the pattern of histone binding to DNA upon transcriptional activation. Cell. 1989 Jul 14;58(1):27–36. doi: 10.1016/0092-8674(89)90399-1. [DOI] [PubMed] [Google Scholar]
- Nedospasov S. A., Georgiev G. P. Non-random cleavage of SV40 DNA in the compact minichromosome and free in solution by micrococcal nuclease. Biochem Biophys Res Commun. 1980 Jan 29;92(2):532–539. doi: 10.1016/0006-291x(80)90366-6. [DOI] [PubMed] [Google Scholar]
- Osborne B. I., Guarente L. Transcription by RNA polymerase II induces changes of DNA topology in yeast. Genes Dev. 1988 Jun;2(6):766–772. doi: 10.1101/gad.2.6.766. [DOI] [PubMed] [Google Scholar]
- Pavlovic J., Banz E., Parish R. W. The effects of transcription on the nucleosome structure of four Dictyostelium genes. Nucleic Acids Res. 1989 Mar 25;17(6):2315–2332. doi: 10.1093/nar/17.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. S., Shupe K., Gorovsky M. A. Changes in chromatin structure accompany modulation of the rate of transcription of 5S ribosomal genes in Tetrahymena. Nucleic Acids Res. 1984 Nov 26;12(22):8489–8507. doi: 10.1093/nar/12.22.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson D. S., Thoma F., Simpson R. T. Core particle, fiber, and transcriptionally active chromatin structure. Annu Rev Cell Biol. 1986;2:117–147. doi: 10.1146/annurev.cb.02.110186.001001. [DOI] [PubMed] [Google Scholar]
- Pederson D. S., Venkatesan M., Thoma F., Simpson R. T. Isolation of an episomal yeast gene and replication origin as chromatin. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7206–7210. doi: 10.1073/pnas.83.19.7206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone F., Ballario P. Stimulation of yeast RNA polymerase II transcription by critical values of supercoiling. Biochemistry. 1984 Jan 3;23(1):69–73. doi: 10.1021/bi00296a011. [DOI] [PubMed] [Google Scholar]
- Petko L., Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986 Jun 20;45(6):885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- Prior C. P., Cantor C. R., Johnson E. M., Littau V. C., Allfrey V. G. Reversible changes in nucleosome structure and histone H3 accessibility in transcriptionally active and inactive states of rDNA chromatin. Cell. 1983 Oct;34(3):1033–1042. doi: 10.1016/0092-8674(83)90561-5. [DOI] [PubMed] [Google Scholar]
- Pruss G. J., Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989 Feb 24;56(4):521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- Pérez-Ortin J. E., Matallana E., Franco L. Chromatin structure of yeast genes. Yeast. 1989 Jul-Aug;5(4):219–238. doi: 10.1002/yea.320050404. [DOI] [PubMed] [Google Scholar]
- Pérez-Ortín J. E., Estruch F., Matallana E., Franco L. Fine analysis of the chromatin structure of the yeast SUC2 gene and of its changes upon derepression. Comparison between the chromosomal and plasmid-inserted genes. Nucleic Acids Res. 1987 Sep 11;15(17):6937–6956. doi: 10.1093/nar/15.17.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds W. F., Gottesfeld J. M. 5S rRNA gene transcription factor IIIA alters the helical configuration of DNA. Proc Natl Acad Sci U S A. 1983 Apr;80(7):1862–1866. doi: 10.1073/pnas.80.7.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard-Foy H., Hager G. L. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987 Aug;6(8):2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose S. M., Garrard W. T. Differentiation-dependent chromatin alterations precede and accompany transcription of immunoglobulin light chain genes. J Biol Chem. 1984 Jul 10;259(13):8534–8544. [PubMed] [Google Scholar]
- Ryoji M., Worcel A. Chromatin assembly in Xenopus oocytes: in vivo studies. Cell. 1984 May;37(1):21–32. doi: 10.1016/0092-8674(84)90297-6. [DOI] [PubMed] [Google Scholar]
- Saavedra R. A., Huberman J. A. Both DNA topoisomerases I and II relax 2 micron plasmid DNA in living yeast cells. Cell. 1986 Apr 11;45(1):65–70. doi: 10.1016/0092-8674(86)90538-6. [DOI] [PubMed] [Google Scholar]
- Schroth G. P., Cook G. R., Bradbury E. M., Gottesfeld J. M. Transcription factor IIIA induced bending of the Xenopus somatic 5S gene promoter. Nature. 1989 Aug 10;340(6233):487–488. doi: 10.1038/340487a0. [DOI] [PubMed] [Google Scholar]
- Sigurdson D. C., Gaarder M. E., Livingston D. M. Characterization of the transmission during cytoductant formation of the 2 micrometers DNA plasmid from Saccharomyces. Mol Gen Genet. 1981;183(1):59–65. doi: 10.1007/BF00270139. [DOI] [PubMed] [Google Scholar]
- Simpson R. T., Thoma F., Brubaker J. M. Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell. 1985 Oct;42(3):799–808. doi: 10.1016/0092-8674(85)90276-4. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Lewis M. J., Pelham H. R. Heat shock factor is regulated differently in yeast and HeLa cells. Nature. 1987 Sep 3;329(6134):81–84. doi: 10.1038/329081a0. [DOI] [PubMed] [Google Scholar]
- Stein A., Holley K., Zeliff J., Townsend T. Interactions between core histones and chromatin at physiological ionic strength. Biochemistry. 1985 Mar 26;24(7):1783–1790. doi: 10.1021/bi00328a032. [DOI] [PubMed] [Google Scholar]
- Studitsky V. M., Belyavsky A. V., Melnikova A. F., Mirzabekov A. D. The structure of nucleosomal core particles within transcribed and repressed gene regions. Nucleic Acids Res. 1988 Dec 9;16(23):11187–11205. doi: 10.1093/nar/16.23.11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Bergman L. W., Simpson R. T. Nuclease digestion of circular TRP1ARS1 chromatin reveals positioned nucleosomes separated by nuclease-sensitive regions. J Mol Biol. 1984 Aug 25;177(4):715–733. doi: 10.1016/0022-2836(84)90046-9. [DOI] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Villeponteau B., Lundell M., Martinson H. Torsional stress promotes the DNAase I sensitivity of active genes. Cell. 1984 Dec;39(3 Pt 2):469–478. doi: 10.1016/0092-8674(84)90454-9. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Worcel A., Alberts B. A model for chromatin based upon two symmetrically paired half-nucleosomes. Cell. 1976 Nov;9(3):409–417. doi: 10.1016/0092-8674(76)90085-4. [DOI] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- Yaniv M., Cereghini S. Structure of transcriptionally active chromatin. CRC Crit Rev Biochem. 1986;21(1):1–26. doi: 10.3109/10409238609113607. [DOI] [PubMed] [Google Scholar]