Abstract
Anaplastic large cell lymphomas (ALCLs) are CD30-positive T-cell non-Hodgkin lymphomas that bear chromosomal rearrangements of the TP53 homologue, TP63, in a subset of cases that demonstrate aggressive clinical behavior. In the present study, we examined the relationship between p63 protein expression by immunohistochemistry and the results of fluorescence in situ hybridization (FISH) using TP63 probes in 116 ALCLs. We also determined the relative expression of full-length TAp63 and truncated ΔNp63 isoforms (e.g. p40) in ALCL cell lines and a subset of clinical cases. Overall, 35.3% of ALCLs were positive for p63 protein. Primary cutaneous and ALK-negative ALCLs were positive more frequently than ALK-positive ALCLs (p=0.0034). As previously reported, cases with TP63 gene rearrangements expressed p63 uniformly. p63 expression in non-rearranged cases was associated with extra copies of TP63 on 3q28 (p<0.0001). Extra copies of TP63 correlated with extra copies of the DUSP22 locus on 6p25.3 (p<0.0001). Results of immunohistochemistry, Western blotting, and RNA sequencing indicated that p63 expression in non-rearranged cases was entirely attributable to TAp63 isoforms. Taken together, these findings indicate that ALCLs without TP63 rearrangements may express TAp63 isoforms of p63 and that this expression is associated with extra copies of TP63, probably due to widespread genomic copy number abnormalities rather than focal gains. Immunohistochemistry for p63 in ALCL is not specific for TP63 rearrangements, but is useful clinically as a screening test to select cases for further testing by FISH. Immunohistochemistry for ΔNp63 (p40) is not informative in the evaluation of ALCL.
Keywords: Anaplastic large cell lymphoma, p63, p40, TP63, copy number abnormality, fluorescence in situ hybridization
1. Introduction
Anaplastic large cell lymphomas (ALCLs) represent a heterogeneous group of T-cell non-Hodgkin lymphomas that share morphologic and phenotypic features, including CD30 expression [1]. About half of systemic ALCLs have rearrangements of the ALK locus and express ALK fusion proteins, the detection of which has diagnostic, prognostic, and therapeutic significance [2–5]. Detection of ALK fusion proteins by immunohistochemistry (IHC) has obviated the need for genetic studies of the ALK locus in most cases. However, about half of systemic ALCLs and nearly all primary cutaneous (pc) ALCLs lack ALK rearrangements, and no surrogate test analogous to ALK IHC currently exists to aid in their diagnosis and subclassification.
Recently, we reported that systemic ALK-negative ALCLs were genetically heterogeneous, with rearrangements involving the DUSP22 locus on 6p25.3 in 30% of cases and rearrangements of the TP63 locus on 3q28 in 8% [6–9]. While ALK-negative ALCLs with DUSP22 rearrangements had a favorable overall survival (OS) rate similar to ALK-positive ALCLs, ALK-negative ALCLs with TP63 rearrangements had clinically aggressive disease and a 5-year OS rate of only 17%. ALCLs lacking rearrangements of ALK, DUSP22, and TP63 (“triple-negative” ALCLs) had an intermediate prognosis [9]. Of note, DUSP22 and TP63 rearrangements also can occur in pcALCL [6–8, 10, 11].
The TP63 gene encodes p63, a member of the p53 family of transcription factors with multiple isoforms, some of which share tumor suppressor function with p53 [12, 13]. p63 isoforms comprise combinations of various C-termini with one of two N-termini, which differ based on whether a transactivation (TA) domain or an N-truncated (ΔN) domain is located directly upstream of a common DNA-binding domain (DBD). Thus, these groups of isoforms are referred to as TAp63 and ΔNp63 isoforms, respectively. TP63 rearrangements in ALCL cause gene fusions, most commonly with TBL1XR1 on 3q26, that lead to expression of p63 fusion proteins [8]. These fusion proteins contain elements encoded by the partner gene directly upstream of the p63 DBD, and lack both TA and ΔN domains. Antibodies generated from epitopes in or near the p63 DBD, such as the 4A4 clone, react with all known p63 isoforms as well as p63 fusion proteins (“pan-p63” antibodies); in contrast, antibodies directed at the ΔN domain react only with ΔNp63 isoforms (including so-called “p40”) [8, 14].
TP63 rearrangements can be detected by genetic studies, such as fluorescence in situ hybridization (FISH) [8, 9, 15–17]. Although p63 fusion proteins can be detected by IHC in ALCL [8, 9, 15], the role of p63 IHC in the evaluation of ALCLs remains unclear. Gualco et al reported that 44% of ALCLs showed p63 nuclear positivity [18]. Although this study preceded the discovery of TP63 rearrangements, this expression frequency is unlikely to be solely attributable to TP63 rearrangements based on our reported rearrangement frequency of 8%. We have confirmed that a subset of ALCLs lacking TP63 rearrangements but positive for p63 by IHC [9]. We undertook the present study in an extended series of cases to characterize the spectrum of p63 protein expression in the context of TP63 FISH results; to investigate the relative expression of TAp63 and ΔNp63 isoforms; and to clarify the clinical role of p63 IHC in the evaluation of ALCL.
2. Materials and methods
2.1. Patients
Patients included 116 ALCLs comprising 22 systemic ALK-positive ALCLs, 62 systemic ALK-negative ALCLs, and 32 pcALCLs. There were 75 males and 41 females, with an overall mean age of 56 years (range, 6–93 years). Additional demographic data and the distribution of DUSP22 and TP63 rearrangements are shown in Table 1. Studies were approved by the respective Institutional Review Boards at the participating institutions.
Table 1.
Demographic and genetic characteristics of ALCLs in the present study
| WHO subtype
|
All cases | |||
|---|---|---|---|---|
| ALK-positive | ALK-negative* | Primary cutaneous** | ||
| N | 22 | 62 | 32 | 116 |
| Age (years) | ||||
| Mean | 29 | 62 | 62 | 56 |
| Range | 6–74 | 24–87 | 13–93 | 6–93 |
| Sex | ||||
| Male | 18 | 40 | 17 | 75 |
| Female | 4 | 22 | 15 | 41 |
| Genetics | ||||
| DUSP22 rearrangement | ||||
| Present | 0 | 19 | 13 | 32 |
| Absent | 20 | 43 | 19 | 82 |
| Not done | 2 | 0 | 0 | 2 |
| TP63 rearrangement | ||||
| Present | 0 | 8 | 2 | 10 |
| Absent | 22 | 54 | 30 | 106 |
| Not done | 0 | 0 | 0 | 0 |
Abbreviations: ALCL: anaplastic large cell lymphoma; ALK: anaplastic lymphoma kinase; WHO: World Health Organization.
In 1 case of systemic ALK-negative ALCL the specimen available for analysis was a skin biopsy, though the patient had documented systemic disease.
In 3 cases of primary cutaneous ALCL the specimens available for analysis were locoregional lymph nodes, though the patients had documented primary cutaneous disease.
2.2. IHC and FISH
IHC for pan-p63 was performed in all cases using the 4A4 clone (Biocare, Concord, CA), which recognizes all known native p63 isoforms as well as p63 fusion proteins, as previously described [8]. IHC for ΔNp63 (“p40”) was performed in a subset of cases with available tissue sections using a rabbit polyclonal antibody (Biocare) as previously described [16]. p63 expression was scored by decile for percentage of tumor cell nuclei with positive staining. IHC also was performed on paraffin tissue sections of cell blocks prepared from ALCL cell lines (see below). Interphase FISH on paraffin tissue sections using a breakapart probes to the TP63 and DUSP22 loci and/or a dual-fusion probe for TBL1XR1/TP63 either was performed as previously described, or had been performed for clinical purposes or in prior studies [6, 8–10, 15] One hundred interphase nuclei were evaluated for each case. Upper limits of normal for extra copies were based on 95% confidence intervals derived from FISH results in 25 normal samples and were: DUSP22 breakapart probe, 8%; TP63 breakapart probe, 9%; and TP63 dual-fusion probe, 5%. Copy numbers for TP63 and DUSP22 loci were recorded when these loci were not involved in rearrangements. When a range of copy numbers was present in different cells, the highest value in the range was used in comparative analyses.
2.3. Western Blotting
The ALCL cell lines Karpas 299 (ATCC, Gaithersburg, MD), SU-DHL-1 (DSMZ, Braunschweig, Germany), MAC1 (established by M.E.K. [19]), and FE-PD (generously provided by Dr. K. Pulford, Oxford, U.K., with kind permission from Dr. A., Del Mistro, Padova, Italy) were maintained in DMEM medium containing 10% fetal bovine serum. HEK-293 cells (ATCC) were transfected with pcDNA3 (Invitrogen) either as an empty vector or containing FLAG-tagged ΔNp63α (a gift from David Sidransky; Addgene plasmid #26979) [20] using Lipofectamine 2000 (Invitrogen) per the manufacturer’s instructions. Western blotting was performed on cell lysates using antibodies to p63 (clone 4A4; Biocare), ΔNp63 (goat polyclonal; Santa Cruz Biotechnology, Santa Cruz, CA), and β-actin (clone AC-15; Novus Biologicals, Littleton, CO).
2.3. RNA Sequencing
RNA sequencing was performed on total RNA extracted from ALCL cell lines, and normalized read-count data and bridging reads that spanned exons were used to evaluate TP63 splicing patterns and assess the relative presence of isoforms containing the TA domain and ΔN leader sequence, respectively, as previously described [21, 22].
2.4. Statistics
Relationships between groups were evaluated using the Wilcoxon test, Pearson χ2 test, or Cochran-Mantel-Haenszel non-parametric trend test (modified to use Ridit scores), as appropriate, and p values less than 0.05 were considered statistically significant.
3. Results
3.1. p63 Protein Expression Varies by ALCL Subtype
Representative ALCLs with and without p63 protein expression are shown in Figures 1–3. Among all 116 ALCLs the mean percentage (± standard deviation) of tumor cells with positive nuclear staining for p63 was 27.3% ± 38.1%. When stratified by WHO subtype, p63 protein expression was significantly lower in ALK-positive ALCL (5.0% ± 17.4%) than either ALK-negative ALCL (33.4% ± 40.6%) or pcALCL (30.9% ± 38.6%; p=0.0065, Wilcoxon test; Figure 4a). Even after excluding cases with TP63 rearrangements, p63 expression in the 3 ALCL subtypes was 5.0% ± 17.4%, 24.3% ± 35.0%, and 27.0% ± 36.6%, respectively (p=0.026). Therefore, the lower expression of p63 in ALK-positive ALCLs could not be attributed only to the lack of TP63 rearrangements in this subtype (Table 1 and references [8, 9]). Similar results were obtained when p63 protein expression was assessed as a dichotomous variable using a cutoff for p63 positivity of ≥30% staining as previously used [15]. Using this criterion, 35.3% of ALCLs were positive for p63. Stratified by ALCL subtype, percentages of positive cases were 4.6%, 43.6%, and 40.6%, for ALK-positive, ALK-negative, and pcALCL, respectively (p=0.0034, Pearson χ2 test; Supplemental Figure 1). Excluding cases with TP63 rearrangements, these values were 4.6%, 35.2%, and 36.7%, respectively (p=0.017). DUSP22 rearrangements were present in 30.6% of ALK-negative ALCLs and 40.6% of pcALCLs (Table 1). No significant association was observed between DUSP22 rearrangement status and either percentage of p63-positive cells or percentage of p63-positive cases (data not shown).
Figure 1.
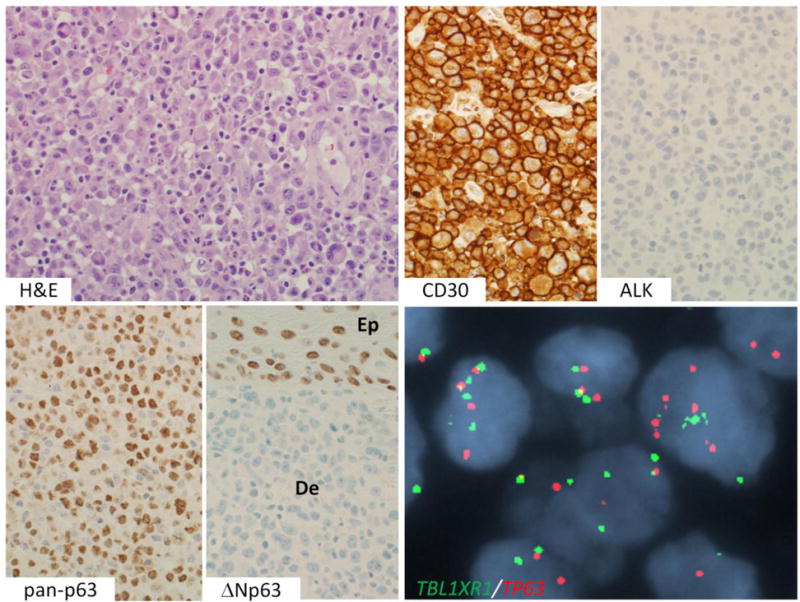
p63 protein-positive primary cutaneous ALCL without TP63 rearrangement. An H&E stain shows sheets of large tumor cells, including “hallmark” cells. By IHC, the cells are positive for CD30 and negative for ALK. They show uniform nuclear staining using a pan-p63 antibody (4A4 clone). The tumor cells in the dermis (De) are negative for ΔNp63 (“p40”); epithelial cells in the basal layer of the epidermis (Ep) are positive. Interphase FISH using a dual-fusion probe for TBL1XR1 (green) and TP63 (red) shows extra copies of both genes without evidence of TBL1XR1-TP63 fusion.
Figure 3.
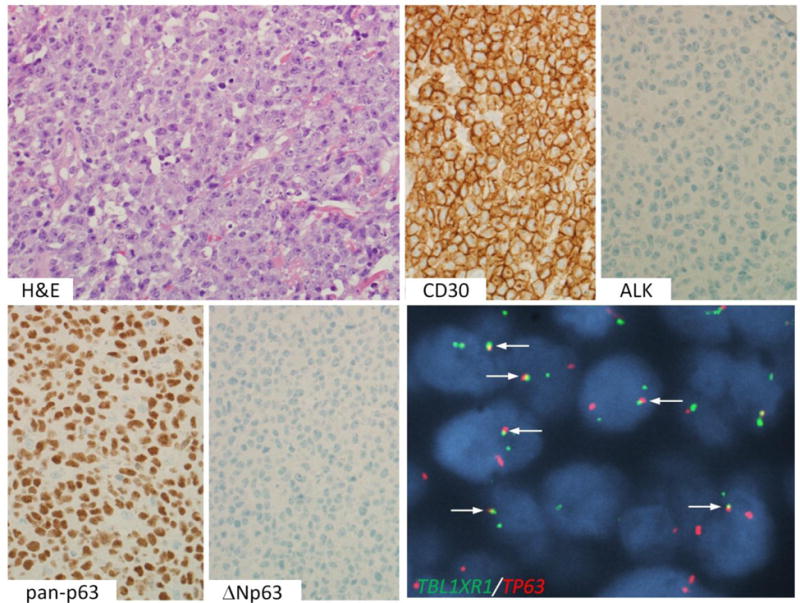
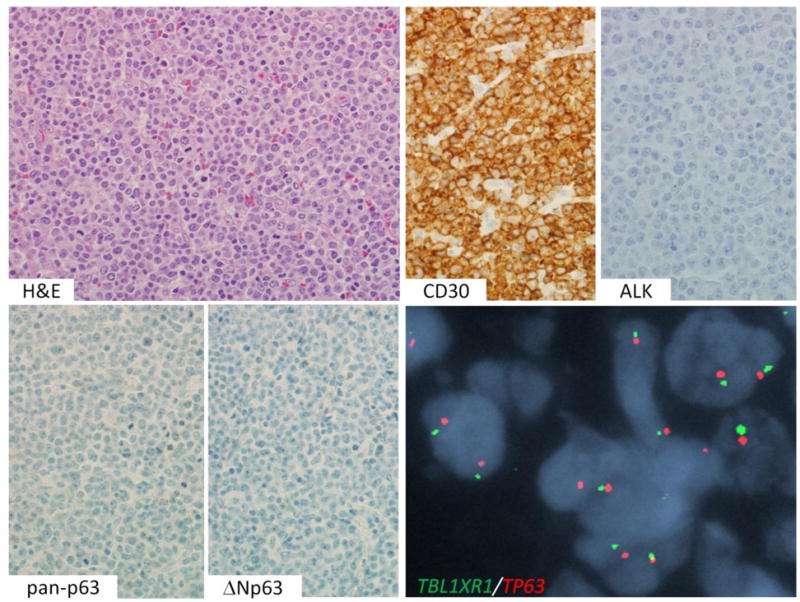
ALK-negative ALCL with TP63 rearrangement. An H&E stain shows sheets of tumor cells. By IHC, the cells are positive for CD30 and pan-p63, and negative for ALK and ΔNp63. Dual-fusion FISH shows abnormal fusion signals (arrows), indicative of TBL1XR1-TP63 fusion.
Figure 4.
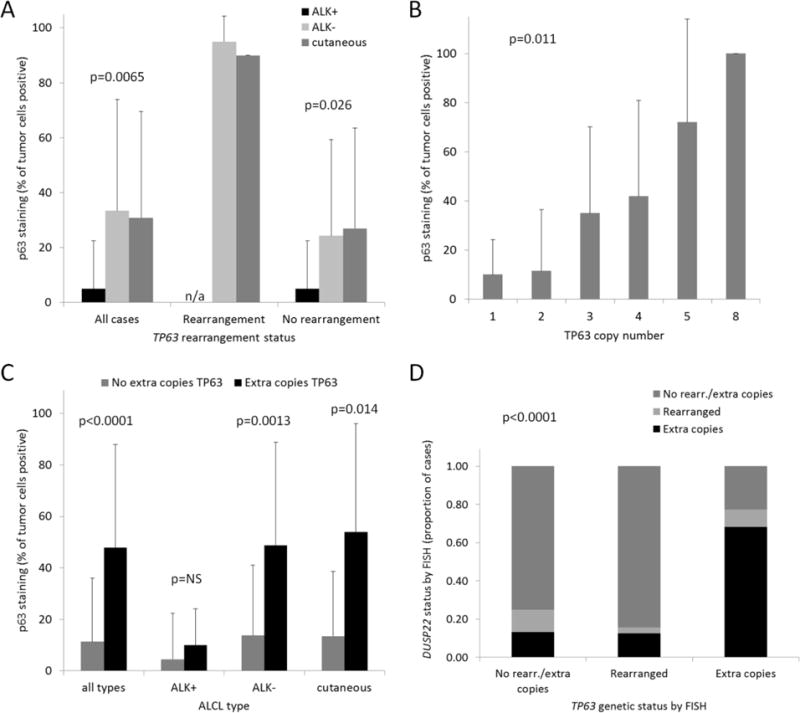
p63 protein expression in ALCL. (A) Percentages of tumor cell nuclei staining for pan-p63 in ALCL, stratified by WHO subtype and TP63 rearrangement status, and expressed as mean ± standard deviation. n/a, not applicable (TP63 rearrangements were not observed in ALK-positive ALCLs). (B) Percentages of tumor cell nuclei staining for p63 in ALCL, stratified by number of copies of the TP63 locus (n=106; ALCLs with TP63 rearrangements were excluded). (C) Percentages of tumor cell nuclei staining for p63 in ALCL subtypes with and without extra copies of TP63. NS, not significant. (D) Proportions of cases with extra copies or rearrangements of DUSP22, stratified by the presence of extra copies or rearrangements of TP63.
3.2. p63 Protein Expression in ALCL is Associated with Extra Copies of the TP63 Locus
We previously have demonstrated that p63 protein expression is associated with extra copies of the TP63 locus in lung adenocarcinomas [16]. Thus, we investigated whether this association was observed in ALCLs lacking TP63 rearrangements. Indeed, cases with increasing copies of the TP63 locus demonstrated increasing percentages of p63-positive tumor cells (p=0.011, Cochran-Mantel-Haenszel trend test; Figure 4b). Overall, ALCLs with extra (≥3) copies of TP63 had 47.9% ± 40% p63-positive cells, compared to 11.4% ± 24.6% for cases without extra copies (p<0.0001, Wilcoxon test; Figure 4c). Stratified by WHO subtype, significant differences were observed both for ALK-negative ALCL (48.8% ± 40% vs. 13.9% ± 27.2%, respectively; p-0.0013) and pcALCL (54.0% ± 42.0% vs. 13.5% ± 25.2, respectively; p=0.014); a trend in the same direction was observed for ALK-positive ALCL (10.0% ± 14.1% vs. 4.5% ± 17.9%, respectively), but this was not statistically significant. To explore whether extra copies of TP63 were more likely due to focal gains or more widespread genomic complexity and/or aneuploidy, we then examined the correlation with the number of copies of the DUSP22 locus. We found that 15/27 ALCLs (55.6%) with extra copies of TP63 had extra copies of DUSP22. In contrast, 2/10 cases (20.0%) with TP63 rearrangements and only 5/77 cases (6.5%) without rearrangements or extra copies had extra copies of DUSP22 (p<0.0001, Pearson χ2 test; Figure 4d and Supplemental Table 1). These findings suggest that extra copies of TP63 are more likely due to aneuploidy or other structural complexity than to isolated, focal gains in the majority of cases.
3.3. p63 Protein Expression in ALCLs without TP63 Rearrangements Represents TAp63 Isoforms
Slides were available to perform immunohistochemistry for ΔNp63 in 59 ALCLs (Supplemental Table 2). All cases were negative for ΔNp63, regardless of p63 protein expression status or TP63 rearrangement status (Figures 1–3). We also examined cell block sections derived from 4 ALCL cells, including 2 ALK-positive lines (Karpas 299 and SU-DHL-1) and 2 ALK-negative lines (MAC1 and FE-PD). Karpas 299 and MAC1 were positive for p63 (stronger in Karpas 299) and SU-DHL-1 and FE-PD were negative (Figure 5a). Sections from all 4 cell lines were negative for ΔNp63. To further confirm these results, we performed Western blotting on cell line lysates. Bands detectable with the 4A4 pan-p63 antibody were seen for Karpas 299 (stronger) and MAC1 (weaker), but not for SU-DHL-1 or FE-PD (Figure 5b). No band corresponding to ΔNp63 was seen in any of the cell lines. Finally, we examined exonic and junctional reads mapping to the TP63 locus in RNAseq data from all 4 cell lines. The results corroborated those from IHC and Western blot (Figure 5c). Notably, no exonic reads were detected that mapped to the ΔNp63-specific exon 3′, nor were junctional reads spanning exons 3′ and 4 identified. Taken together with the lack of ΔNp63 staining in any clinical ALCL sample, these findings indicate that ΔNp63 is not expressed in ALCL, and strongly suggest that positivity for p63 by IHC in ALCLs lacking TP63 rearrangements is due to expression of TAp63 isoforms.
Figure 5.
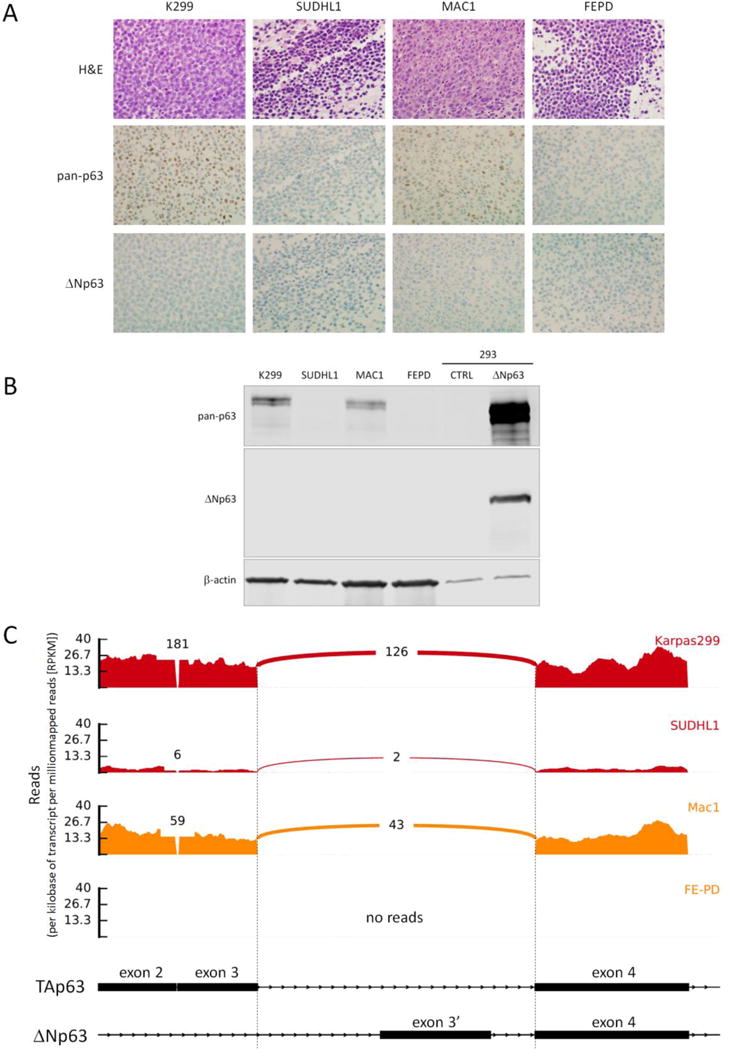
p63 isoform distribution in ALCL. (A) Results of pan-p63 and ΔNp63 immunohistochemistry in sections of cell blocks prepared from ALCL cell lines. (B) Western blots of protein lysates from ALCL cell lines using antibodies to pan-p63, ΔNp63, and β-actin (loading control). The positive control for ΔNp63 is a lysate from 293 cells transfected with a ΔNp63 expression plasmid. (C) Representation of exons 2–4 of the TP63 locus on 3q28 showing RNA sequencing data from ALCL cell lines. ALK-positive cell lines are shown in red and ALK-negative lines in orange. Both exon-level read counts and exon-exon junctional read counts are shown. The schematic diagram is not to scale (exons are exaggerated with respect to introns to facilitate visualization).
4. Discussion
This study is the first to address in a comprehensive fashion the distribution and origin of p63 protein expression in ALCL in the context of the recent discovery of TP63 rearrangements in a subset of these cases. The prognostic role of TP63 and DUSP22 rearrangements in ALCL has become recognized [9, 23], and has led to clinical FISH testing for these alterations in some centers. ALCLs with TP63 rearrangements produce p63 fusion proteins that can be detected by routine p63 IHC, have been exclusively ALK-negative, and have poor outcomes. Here, we show unequivocally that a subset of ALCLs without TP63 rearrangements express p63 protein and that this expression occurs predominantly in ALK-negative ALCL and pcALCL, is associated with extra copies of the TP63 gene, and represents TAp63 isoforms. Overall, these findings support the use of p63 IHC as a screening test in the evaluation of ALCL, but also emphasize the need for TP63 FISH in IHC-positive cases for prognostic purposes.
Gualco et al reported in 2008 that p63 was detected by IHC in a subset of ALCLs, but not cases of classical Hodgkin lymphoma [18]. However, that study preceded our group’s discovery of TP63 rearrangements in ALCL and therefore the proportions of p63-positive cases attributable to rearranged and non-rearranged cases in the series reported by Gualco et al cannot be ascertained. Of note and in contrast to our study, Gualco et al did not identify a significant association between p63 positivity and ALK negativity in systemic ALCLs. This may be due in part to the lower threshold for p63 positivity in the earlier study (nuclear staining in >5% of the neoplastic cells, in contrast to ≥30% in the current study); when Gualco et al used a cutoff of >50% positive cells the proportions of positive cases were 4.1% and 14.8% for systemic ALK-positive and -negative cases, respectively. In addition to systemic ALCL and pcALCL, breast implant-associated ALCL represents a new provisional entity in the WHO classification [23]. The present study did not include any examples of this entity and further study of the p63 status of these neoplasms is warranted.
In a previous study, we reported a p63 protein expression in a TP63-rearranged lung adenocarcinoma and in non-rearranged cases with extra copies of the intact TP63 locus [16]. Therefore, we examined p63 expression and TP63 copy number in ALCL and found these to be correlated significantly. p63 expression also has been reported in B-cell lymphomas with or without TP63 rearrangements, but an association with TP63 copy number has not been reported [8, 17, 24–29]. Though our FISH approach did not allow precise mapping of the regions of chromosomal gain in ALCLs with extra copies of TP63 (on 3q28), this finding was associated significantly with extra copies of the DUSP22 locus on 6p25.3, suggesting aneuploidy rather than focal gains as the cause of these extra copies. Interestingly, Gualco et al reported that 3 of 5 cases of T-cell ALCL with aberrant expression of the B-cell transcription factor PAX5 also were p63 positive, a finding likely explained by structural complexity in p63-positive cases given our previous finding that PAX5 expression in ALCL is associated with extra copies of the PAX5 gene on 9p13.2 [18, 30].
Immunohistochemical staining for ΔNp63 (p40) is used widely in surgical pathology practice but its expression in ALCL has not been reported. We found that p63 expression in ALCLs lacking TP63 rearrangements is attributable solely to the expression of TAp63 isoforms. Importantly, this result was not assumed based only on the lack of immunohistochemical staining for ΔNp63, but also was validated by analysis of TP63 transcripts in RNA sequencing in ALCL cell lines. In fact, no exonic or junctional reads corresponding to a ΔNp63 isoform were identified. TAp63 isoforms have tumor suppressor properties in some experimental models, and their expression has been associated with favorable outcomes in diffuse large B-cell lymphoma [27, 29]. However, while TP63 rearrangements are associated with poor prognosis in ALCL, we previously reported a lack of significant differences in outcomes between p63 protein-positive and -negative cases [9]. Similarly, ALCLs included in the present study, excluding those with TP63 rearrangements, did not demonstrate a significant association between p63 IHC result and overall survival (data not shown). The functional roles of p63 isoforms in ALCL cells, as well as the possibility that any tumor suppressor effect conferred by TAp63 expression is negated by prognostic effects relating to aneuploidy and/or underlying genomic instability, merit further investigation.
We conclude that p63 protein expression in ALCLs without TP63 rearrangements is a phenomenon associated with extra copies of TP63 and other structural chromosomal abnormalities, is due to expression of TAp63 isoforms, and is of uncertain clinical and biologic significance. Immunohistochemistry for ΔNp63 (p40) is not informative in the evaluation of ALCL. IHC for p63 using pan-p63 antibodies is a useful screening test to select cases of systemic ALK-negative ALCL for TP63 FISH, which is more costly and less readily available. In our experience, a threshold for positivity of ≥30% has 100% sensitivity for ALCLs with TP63 rearrangements (Supplemental Figure 2). A possible algorithm for the use of p63 IHC and TP63 FISH in the evaluation of ALCL is shown in Supplemental Figure 3. The lack of specificity of IHC for TP63 rearrangements should be emphasized, as should the fact that no prognostic significance of p63 IHC has yet been demonstrated in ALCL.
Supplementary Material
Figure 2.
p63 protein-negative ALCL without TP63 rearrangement. An H&E stain shows sheets of tumor cells. By IHC, the cells are positive for CD30 and negative for ALK, pan-p63, and ΔNp63. Dual-fusion FISH shows two copies each of TBL1XR1 and TP63 (normal signal pattern).
Highlights.
IHC for p63 is highly sensitive but not specific for TP63 rearrangements in ALCL.
p63-positive ALCLs lacking TP63 rearrangements often have extra copies of TP63.
IHC for p63 can be used to select cases for FISH testing for TP63 rearrangements.
IHC for ΔNp63 (p40) is not informative in the evaluation of ALCL.
Acknowledgments
Funding/Support: This study was supported by Award Numbers R01 CA177734 (ALF), P30 CA15083 (Mayo Clinic Cancer Center), and P50 CA97274 (University of Iowa/Mayo Clinic Lymphoma SPORE) from the National Cancer Institute; CTSA Grant Number UL1 TR000135 from the National Center for Advancing Translational Science (NCATS); Award Number CI-48-09 from the Damon Runyon Cancer Research Foundation (ALF); and the Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN. XW was supported by a scholarship award from the China Scholarship Council.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests: The authors have no conflicts of interest to disclose.
References
- 1.Xing X, Feldman AL. Anaplastic large cell lymphomas: ALK positive, ALK negative, and primary cutaneous. Adv Anat Pathol. 2015;22:29–49. doi: 10.1097/PAP.0000000000000047. [DOI] [PubMed] [Google Scholar]
- 2.Gascoyne RD, Aoun P, Wu D, et al. Prognostic significance of anaplastic lymphoma kinase (ALK) protein expression in adults with anaplastic large cell lymphoma. Blood. 1999;93:3913–21. [PubMed] [Google Scholar]
- 3.Jaffe ES. Anaplastic large cell lymphoma: the shifting sands of diagnostic hematopathology. Mod Pathol. 2001;14:219–28. doi: 10.1038/modpathol.3880289. [DOI] [PubMed] [Google Scholar]
- 4.Delsol G, Falini B, Muller-Hermelink HK, et al. Anaplastic large cell lymphoma, ALK-positive. In: Swerdlow S, editor. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer; Lyon: 2008. pp. 312–6. [Google Scholar]
- 5.Bennani-Baiti N, Ansell S, Feldman AL. Adult systemic anaplastic large-cell lymphoma: recommendations for diagnosis and management. Expert Rev Hematol. 2016;9:137–50. doi: 10.1586/17474086.2016.1122514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldman AL, Law M, Remstein ED, et al. Recurrent translocations involving the IRF4 oncogene locus in peripheral T-cell lymphomas. Leukemia. 2009;23:574–80. doi: 10.1038/leu.2008.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman AL, Dogan A, Smith DI, et al. Discovery of recurrent t(6;7)(p25.3;q32.3) translocations in ALK-negative anaplastic large cell lymphomas by massively-parallel genomic sequencing. Blood. 2011;117:915–9. doi: 10.1182/blood-2010-08-303305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasmatzis G, Johnson SH, Knudson RA, et al. Genome-wide analysis reveals recurrent structural abnormalities of TP63 and other p53-related genes in peripheral T-cell lymphomas. Blood. 2012;120:2280–9. doi: 10.1182/blood-2012-03-419937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parrilla Castellar ER, Jaffe ES, Said JW, et al. ALK-negative anaplastic large cell lymphoma is a genetically heterogeneous disease with widely disparate clinical outcomes. Blood. 2014;124:1473–80. doi: 10.1182/blood-2014-04-571091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wada DA, Law ME, Hsi ED, et al. Specificity of IRF4 translocations for primary cutaneous anaplastic large cell lymphoma: a multicenter study of 204 skin biopsies. Mod Pathol. 2011;24:596–605. doi: 10.1038/modpathol.2010.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pham-Ledard A, Prochazkova-Carlotti M, Laharanne E, et al. IRF4 Gene Rearrangements Define a Subgroup of CD30-Positive Cutaneous T-Cell Lymphoma: A Study of 54 Cases. J Invest Dermatol. 2010;130:816–825. doi: 10.1038/jid.2009.314. [DOI] [PubMed] [Google Scholar]
- 12.Yang A, Kaghad M, Wang Y, et al. p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2:305–16. doi: 10.1016/s1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 13.Guo X, Keyes WM, Papazoglu C, et al. TAp63 induces senescence and suppresses tumorigenesis in vivo. Nat Cell Biol. 2009;11:1451–7. doi: 10.1038/ncb1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petitjean A, Ruptier C, Tribollet V, et al. Properties of the six isoforms of p63: p53-like regulation in response to genotoxic stress and cross talk with DeltaNp73. Carcinogenesis. 2008;29:273–81. doi: 10.1093/carcin/bgm258. [DOI] [PubMed] [Google Scholar]
- 15.Chavan RN, Bridges AG, Knudson RA, et al. Somatic rearrangement of the TP63 gene preceding development of mycosis fungoides with aggressive clinical course. Blood Cancer J. 2014;4:e253. doi: 10.1038/bcj.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aubry MC, Roden A, Murphy SJ, et al. Chromosomal rearrangements and copy number abnormalities of TP63 correlate with p63 protein expression in lung adenocarcinoma. Mod Pathol. 2015;28:359–66. doi: 10.1038/modpathol.2014.118. [DOI] [PubMed] [Google Scholar]
- 17.Scott DW, Mungall KL, Ben-Neriah S, et al. TBL1XR1/TP63: a novel recurrent gene fusion in B-cell non-Hodgkin lymphoma. Blood. 2012;119:4949–52. doi: 10.1182/blood-2012-02-414441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gualco G, Weiss LM, Bacchi CE. Expression of p63 in anaplastic large cell lymphoma but not in classical Hodgkin’s lymphoma. Hum Pathol. 2008;39:1505–10. doi: 10.1016/j.humpath.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davis TH, Morton CC, Miller-Cassman R, et al. Hodgkin’s disease, lymphomatoid papulosis, and cutaneous T-cell lymphoma derived from a common T-cell clone. N Engl J Med. 1992;326:1115–22. doi: 10.1056/NEJM199204233261704. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee A, Upadhyay S, Chang X, et al. U-box-type ubiquitin E4 ligase, UFD2a attenuates cisplatin mediated degradation of DeltaNp63alpha. Cell Cycle. 2008;7:1231–7. doi: 10.4161/cc.7.9.5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feldman AL, Vasmatzis G, Asmann YW, et al. Novel TRAF1-ALK fusion identified by deep RNA sequencing of anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2013;52:1097–102. doi: 10.1002/gcc.22104. [DOI] [PubMed] [Google Scholar]
- 22.Boddicker RL, Razidlo GL, Dasari S, et al. Integrated mate-pair and RNA sequencing identifies novel, targetable gene fusions in peripheral T-cell lymphoma. Blood. 2016;128:1234–45. doi: 10.1182/blood-2016-03-707141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–90. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Como CJ, Urist MJ, Babayan I, et al. p63 expression profiles in human normal and tumor tissues. Clin Cancer Res. 2002;8:494–501. [PubMed] [Google Scholar]
- 25.Pruneri G, Fabris S, Dell’Orto P, et al. The transactivating isoforms of p63 are overexpressed in high-grade follicular lymphomas independent of the occurrence of p63 gene amplification. J Pathol. 2005;206:337–45. doi: 10.1002/path.1787. [DOI] [PubMed] [Google Scholar]
- 26.Fukushima N, Satoh T, Sueoka N, et al. Clinico-pathological characteristics of p63 expression in B-cell lymphoma. Cancer Sci. 2006;97:1050–5. doi: 10.1111/j.1349-7006.2006.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hallack Neto AE, Siqueira SA, Dulley FL, et al. p63 protein expression in high risk diffuse large B-cell lymphoma. J Clin Pathol. 2009;62:77–9. doi: 10.1136/jcp.2008.059519. [DOI] [PubMed] [Google Scholar]
- 28.Robson A, Shukur Z, Ally M, et al. Immunocytochemical p63 expression discriminates between primary cutaneous follicle centre cell and diffuse large B cell lymphoma-leg type, and is of the TAp63 isoform. Histopathology. 2016;69:11–9. doi: 10.1111/his.12855. [DOI] [PubMed] [Google Scholar]
- 29.Xu-Monette ZY, Zhang S, Li X, et al. p63 expression confers significantly better survival outcomes in high-risk diffuse large B-cell lymphoma and demonstrates p53-like and p53-independent tumor suppressor function. Aging (Albany NY) 2016;8:345–65. doi: 10.18632/aging.100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feldman AL, Law ME, Inwards DJ, et al. PAX5-positive T-cell anaplastic large cell lymphomas associated with extra copies of the PAX5 gene locus. Mod Pathol. 2010;23:593–602. doi: 10.1038/modpathol.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


