Dear Editor,
Lysine crotonylation is a posttranslational modification (PTM) of histone proteins originally identified by Tan et al.1. This novel evolutionarily conserved histone modification was identified on 28 lysine sites on various histones1. Lysine crotonylation occurs primarily on the ε-amino group of lysine, but its planar orientation and four-carbon length distinguish it from lysine acetylation. Histone crotonylation specifically labels the enhancers and transcription starting sites of active genes in both the human somatic cell genome and the murine male germ cell genome. Histone crotonylation, like acetylation, affects chromatin structure and facilitates histone replacement in elongating spermatids1. Recently YEATS domain-containing protein YEATS2 was found to be a selective histone crotonylation reader2.
PTMs such as acetylation, phosphorylation and methylation have been found on both histone and non-histone proteins3,4,5. However, it is unknown whether non-histone proteins can also be modified by crotonylation.
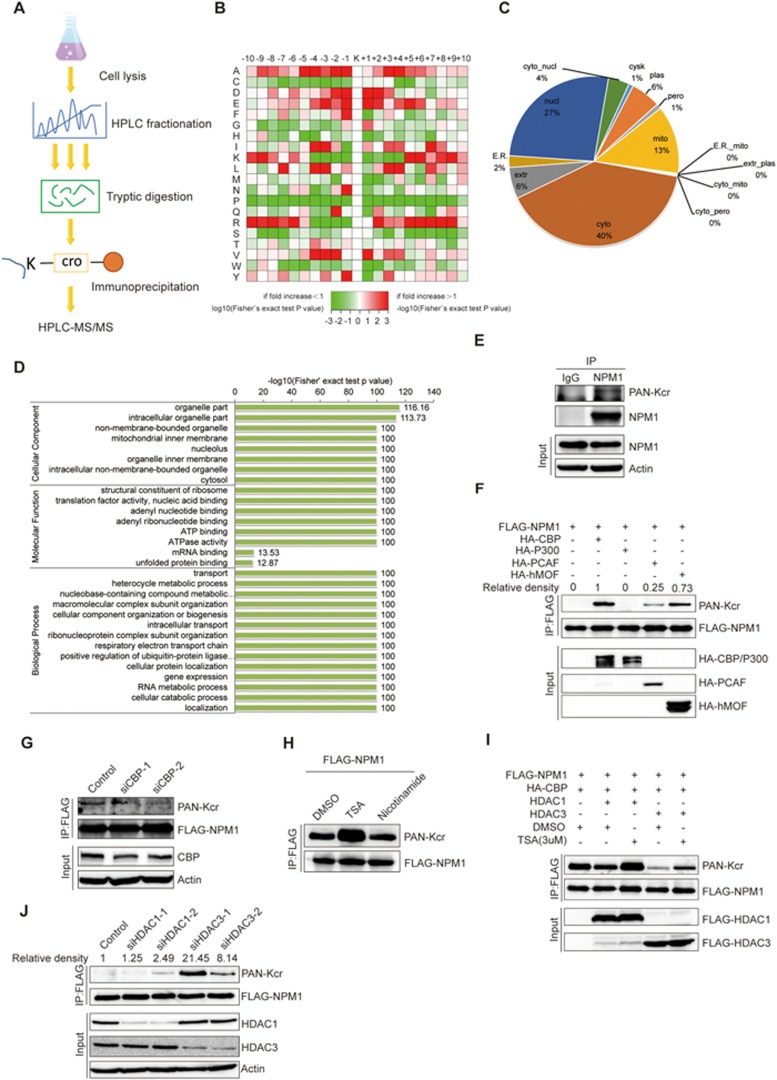
To determine whether non-histone proteins are modified by crotonylation, lysine-crotonylated peptides from trypsin-digested whole-cell lysates of the human lung adenocarcinoma cell line H1299 were enriched with an antibody against crotonylated lysine. This was followed by high-resolution liquid chromatography-tandem MS (LC-MS/MS) (Figure 1A). In order to validate the MS data, we first checked the mass error of the identified peptides. The distribution of mass errors was near zero, and most errors were < 0.02 Da, indicating accuracy of the MS data (Supplementary information, Figure S1A). The length of most of the peptides distributed between 8 and 20, which is consistent with the length of tryptic peptides, indicating that sample preparation achieved a reasonable standard (Supplementary information, Figure S1B). To assess the distribution of crotonylation sites, the number of crotonylation sites identified per protein was calculated (Supplementary information, Figure S1C). Altogether, 2 696 lysine crotonylation sites were identified on 1 024 proteins (Supplementary information, Table S1). To determine whether there are any specific amino acid biases adjacent to crotonylation sites, we analyzed the flanking sequences of these sites. This analysis showed that glutamate (E) residues were overrepresented at the −1 and +1 positions surrounding the lysine crotonylation sites (Figure 1B). Further characterization of the motifs surrounding lysine crotonylation sites showed a series of different patterns of crotonylation motifs (Supplementary information, Figure S1D). These data showed for the first time that a large number of non-histone proteins are modified by crotonylation.
Figure 1.
Identification of crotonylation on non-histone proteins. (A) Flowchart illustrating the experimental procedure for identification of crotonylated proteins by proteomics. (B) Motif analysis of all identified crotonylated sites. (C) Subcellular localization of the proteins with lysine crotonylation. (D) GO-based enrichment analysis of crotonylated proteins. (E) Validation of endogenous crotonylated NPM1. H1299 cells were treated with 3 μM TSA and 5 mM nicotinamide for 12 h, and cell lysates were immunoprecipitated with an anti-NPM1 antibody, followed by immunoblotting with a pan-Kcr antibody. (F) FLAG-tagged NPM1 was cotransfected into H1299 cells with a variety of acetyltransferases. Forty-eight hours post transfection, cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by western blot analysis with a pan-Kcr antibody. (G) H1299 cells were transfected with FLAG-NPM1, and endogenous CBP was knocked down with siRNA. Cells were treated with 3 μM TSA for 12 h, and cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by western blot analysis with a pan-Kcr antibody. (H) The HDAC inhibitor TSA increased the crotonylation level of NPM1. H1299 cells were transfected with FLAG-NPM1 and HA-CBP, and then treated with 3 μM TSA or 5 mM nicotinamide for 12 h. Cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by western blot analysis with a pan-Kcr antibody. (I) H1299 cells were co-transfected with FLAG-NPM1, HA-CBP, and FLAG-HDAC1 or FLAG-HDAC3, and then treated with 3 μM TSA for 12 h. Western blot analysis was performed with a pan-Kcr antibody. (J) H1299 cells were co-transfected with FLAG-NPM1 and HA-CBP. Endogenous HDAC1 or HDAC3 was knocked down with siRNAs. Cell lysates were immunoprecipitated with an anti-FLAG antibody, followed by western blot analysis with a pan-Kcr antibody.
To further evaluate crotonylated proteins, these proteins were classified according to their subcellular location, cellular component, molecular function and associated biological processes. Forty percent of crotonylated proteins were located in the cytoplasm, 27% were located in the nucleus and 13% were in the mitochondria (Figure 1C). The cellular component of crotonylated proteins was classified according to the gene ontology (GO) annotation. Crotonylated proteins were distributed in the cell (30%), the organelles (27%), the macromolecular complexes (15%), membranes (12%) and membrane-enclosed compartments (12%) (Supplementary information, Figure S1E). The molecular function of these crotonylated proteins was analyzed based on the GO annotation. Crotonylated proteins were assigned to several groups. Forty-nine percent of these proteins were involved in binding, 28% were associated with catalytic activity and 8% were related to structural molecule activity (Supplementary information, Figure S1F). Biological process analysis of crotonylated proteins showed that 18% of crotonylated proteins were involved in cellular processes, 14% in metabolism, 13% were single-organism proteins and 11% were biological regulation proteins (Supplementary information, Figure S1G). These results indicate the relatively broad distributions of crotonylated proteins in subcellular location, cellular component, molecular function and biological processes.
To investigate the role(s) of the crotonylation pathway, enrichment analysis was used to classify all the crotonylated protein groups based on the GO, KEGG and Pfam databases. GO enrichment analysis of the cellular component showed that crotonylated proteins were significantly enriched in the organelles, intracellular organelles and non-membrane-bound organelles (Figure 1D, upper). GO-based functional enrichment analysis of molecular function showed that crotonylated proteins were significantly enriched in the structural constituents of ribosomes, translation factor activity and adenyl nucleotide binding (Figure 1D, middle). Enrichment analysis of biological processes indicated these modified proteins were enriched in a variety of biological processes including transport, heterocycle metabolic process and nucleobase-containing compound metabolic process (Figure 1D, lower). KEGG enrichment analysis showed that crotonylated proteins were related to multiple metabolic pathways, including ribosome, spliceosome, proteasome and Parkinson's disease pathways (Supplementary information, Figure S1H). Pfam domain analysis indicated that crotonylated proteins were enriched in RNA recognition motif, nucleotide-binding and p-loop containing nucleoside triphosphate hydrolase domains (Supplementary information, Figure S1I). The STRING database was used to search for protein-protein interaction among some of the crotonylated proteins associated with special complexes. The crotonylated proteins that were identified belonged to multiple protein complexes, including the nop56p-associated pre-rRNA complex, ribosome, the C complex spliceosome, and PA700-20S-PA28, H2AX, and TRBP-containing complexes (Supplementary information, Figure S1J). The interaction network was visualized in Cytoscape (Supplementary information, Figure S1K). GO annotation, the KEGG pathway and Pfam domain analyses suggest that crotonylated proteins participate in a variety of important cellular pathways and perform different functions in H1299 cells.
In order to confirm that living cells harbor crotonylation modifications, we performed immunofluorescence and immunohistochemical staining with a pan anti-Kcr antibody. Immunofluorescent staining showed that crotonylated proteins were widely localized in the cytoplasm and nuclei of H1299 and HeLa cells (Supplementary information, Figure S2A). We also found that crotonylated proteins were widely detected in multiple mouse tissues, including lung, kidney, liver, colon, uterus and ovary (Supplementary information, Figure S2B).
To validate the crotonylated proteins identified by our MS data, we randomly selected 12 non-histone proteins including NPM1, FHL1, ACTN1, Vinculin, VASP, Integrin β1, GAPDH, OTUB1, SIRT2, Cofilin-1, ERK2 and CDK1, and examined their crotonylation status. Out of these 12 proteins, endogenous NPM1, FHL1, ACTN1, Integrin β1, Vinculin, ERK2 and CDK1 were found to be crotonylated, with histone H3 as a positive control (Figure 1E and Supplementary information, Figure S2C-S2H). In addition, we confirmed that GAPDH and OTUB1 were also crotonylated in living cells using lysates from transfected cells (Supplementary information, Figure S2I). According to the previous report, P300 acts as a histone crotonyltransferase6, and we thus hypothesized that members of the acetyltransferase superfamily may catalyze the crotonylation of non-histone proteins. To this end, we selected two non-histone proteins NPM1 and DDX5 that were proved to be crotonylated in our study, and used them to identify candidate crotonyltransferases for non-histone proteins. Expression vectors encoding these two proteins were separately co-transfected with a panel of acetyltransferases including CBP, P300, PCAF and hMOF. We found NPM1 is strongly crotonylated by CBP and hMOF, and moderately crotonylated by PCAF (Figure 1F). However, DDX5 crotonylation can be catalyzed by CBP but not the other acetyltransferases that were tested (Supplementary information, Figure S2J). Moreover, when endogenous CBP was knocked down, the crotonylation level of FLAG-NPM1 decreased, indicating that CBP is indeed responsible for the crotonylation of NPM1 (Figure 1G).
As Sirt1, Sirt2 and Sirt3 are involved in the decrotonylation of histones7,8, and HDAC3 has decrotonylase activity9, it was of particular interest to identify candidate decrotonylases for non-histone proteins. To this end, NPM1 expression vector was transfected into H1299 cells with CBP and cells were treated with TSA, an inhibitor of HDACs, or nicotinamide, an inhibitor of SIRT family deacetylases. We found that the crotonylation level of NPM1 increased after TSA treatment, indicating that HDACs may influence NPM1 crotonylation (Figure 1H). NPM1 and CBP were then co-transfected with HDAC family members including HDAC1, HDAC2 and HDAC3. We found that HDAC1 and HDAC3, but not HDAC2, decrotonylate NPM1, and this effect can be reversed upon TSA treatment (Supplementary information, Figure S2K and Figure 1I). Similarly, another two HDAC inhibitors SAHA and LBH589 could enhance the crotonylation of NPM1 (Supplementary information, Figure S2L). In addition, crotonylation of FLAG-NPM1 was strengthened when endogenous HDAC3 or HDAC1 was knocked down (Figure 1J). These results suggest that the acetyltransferases CBP, PCAF and hMOF act as crotonyltransferases for non-histone proteins, and deacetylases HDAC1 and HDAC3 also act as decrotonylases for non-histone proteins. According to the previous report, the cellular concentration of crotonyl-CoA can regulate the crotonylation of histone6. We thus hypothesized that the balance of crotonylation and acetylation of a non-histone protein depends on the relative concentration of crotonyl-CoA and acetyl-CoA in a cell. HDAC1/3 possess both deacetylase and decrotonylase activities and these two activities may be coordinated by the levels of crotonylation and/or acetylation of the substrates. In addition, these two acylations may take place at the same time on a protein. The ability of HDAC1/3 to remove both types of acylation suggests that decrotonylation and deacetylation may occur at the same time on the same protein. In this way, the functional shift of the substrate can be simplified and accelerated.
In this report, we describe heretofore unidentified crotonylation of non-histone proteins. We show that some enzymes that catalyze acetylation or deacetylation also mediate crotonylation or decrotonylation of non-histone proteins. These non-histone proteins are involved in diverse cellular functions and signaling pathways. Identification of crotonylation on non-histone proteins expands our understanding of this PTM at a proteomics scale, and may facilitate the elucidation of the precise modulation of protein functions.
Acknowledgments
This study was supported by the Ministry of Science and Technology of China (2016YFC1302103, 2015CB553906, and 2013CB910501), and the National Natural Science Foundation of China (81230051, 81472734, 31170711, 81321003, 81301802 and 30830048), the Beijing Natural Science Foundation (7171005), the 111 Project of the Ministry of Education, Peking University (BMU20120314 and BMU20130364) and a Leading Academic Discipline Project of Beijing Education Bureau to HZ.
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
(A) The mass error of the whole identified peptides.
(A)The crotonylation proteins were detected in H1299 and Hela cells by immunofluorescence using an anti-Kcrantibody (green), and the nuclei was stained with DAPI (blue), followed by visualization with confocal microscopy. Scale bars: 10 μm.
Lysine crotonylation sites on non-histone proteins
Materials and Methods
References
- Tan M, Luo H, Lee S, et al. Cell 2011; 146:1016–1028. [DOI] [PMC free article] [PubMed]
- Zhao D, Guan H, Zhao S, et al. Cell Res 2016; 26:629–632. [DOI] [PMC free article] [PubMed]
- Glozak MA, Sengupta N, Zhang X, Seto E. Gene 2005; 363:15–23. [DOI] [PubMed]
- Latham JA, Dent SY. Nat Struct Mol Biol 2007; 14:1017–1024. [DOI] [PubMed]
- Yang XJ, Seto E. Mol Cell 2008; 31:449–461. [DOI] [PMC free article] [PubMed]
- Sabari BR, Tang Z, Huang H, et al. Mol Cell 2015; 58:203–215. [DOI] [PMC free article] [PubMed]
- Feldman JL, Baeza J, Denu JM. J Biol Chem 2013; 288:31350–31356. [DOI] [PMC free article] [PubMed]
- Bao X, Wang Y, Li X, et al. eLife 2014; 3:e02999.
- Madsen AS, Olsen CA. Angew Chem Int Ed Engl 2012; 51:9083–9087. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) The mass error of the whole identified peptides.
(A)The crotonylation proteins were detected in H1299 and Hela cells by immunofluorescence using an anti-Kcrantibody (green), and the nuclei was stained with DAPI (blue), followed by visualization with confocal microscopy. Scale bars: 10 μm.
Lysine crotonylation sites on non-histone proteins
Materials and Methods