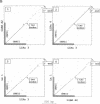
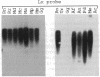
Abstract
Each mammalian genus examined so far contains 50,000-100,000 members of an L1 (LINE 1) family of long interspersed repeated DNA elements. Current knowledge on the evolution of L1 families presents a paradox because, although L1 families have been in mammalian genomes since before the mammalian radiation approximately 80 million years ago, most members of the L1 families are only a few million years old. Accordingly it has been suggested either that the extensive amplification that characterizes present-day L1 families did not occur in the past or that old members were removed as new ones were generated. However, we show here that an ancestral rodent L1 family was extensively amplified approximately 10 million years ago and that the relics (approximately 60,000 copies) of this amplification have persisted in modern murine genomes (Old World rats and mice). This amplification occurred just before the divergence of modern murine genera from their common ancestor and identifies the murine node in the lineage of modern muroid rodents. Our results suggest that repeated amplification of L1 elements is a feature of the evolution of mammalian genomes and that ancestral amplification events could provide a useful tool for determining mammalian lineages.
Full text
PDF
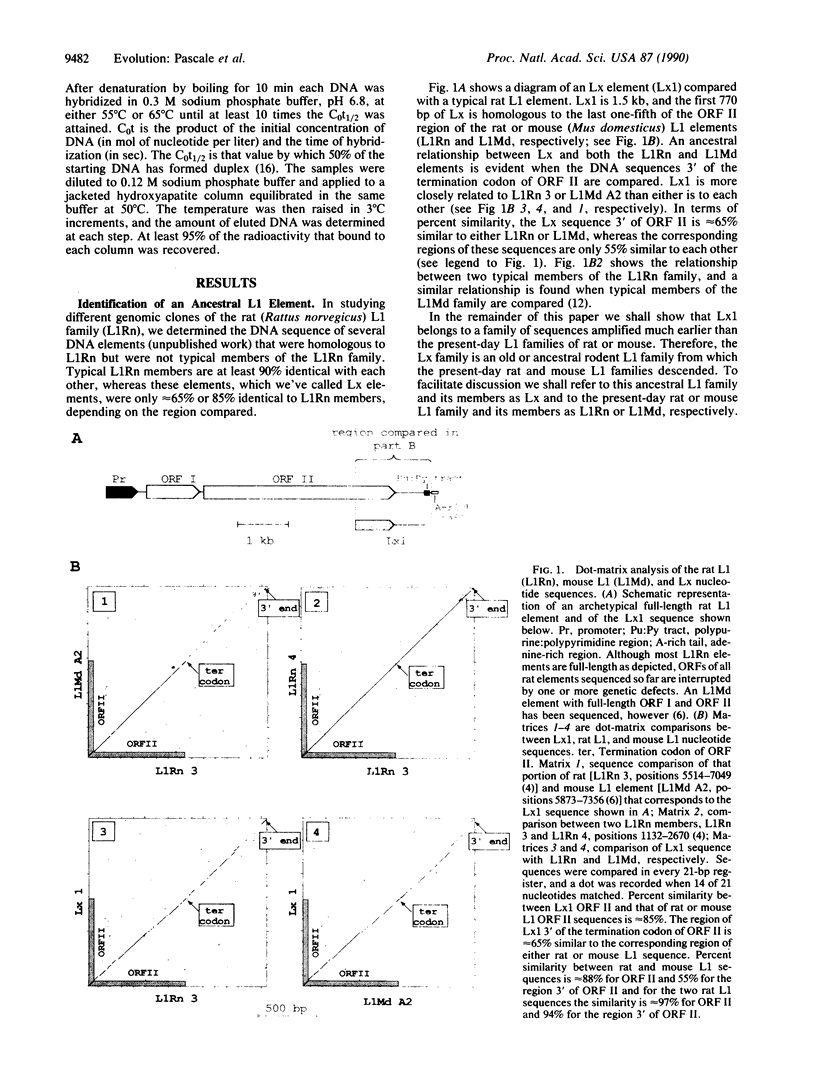


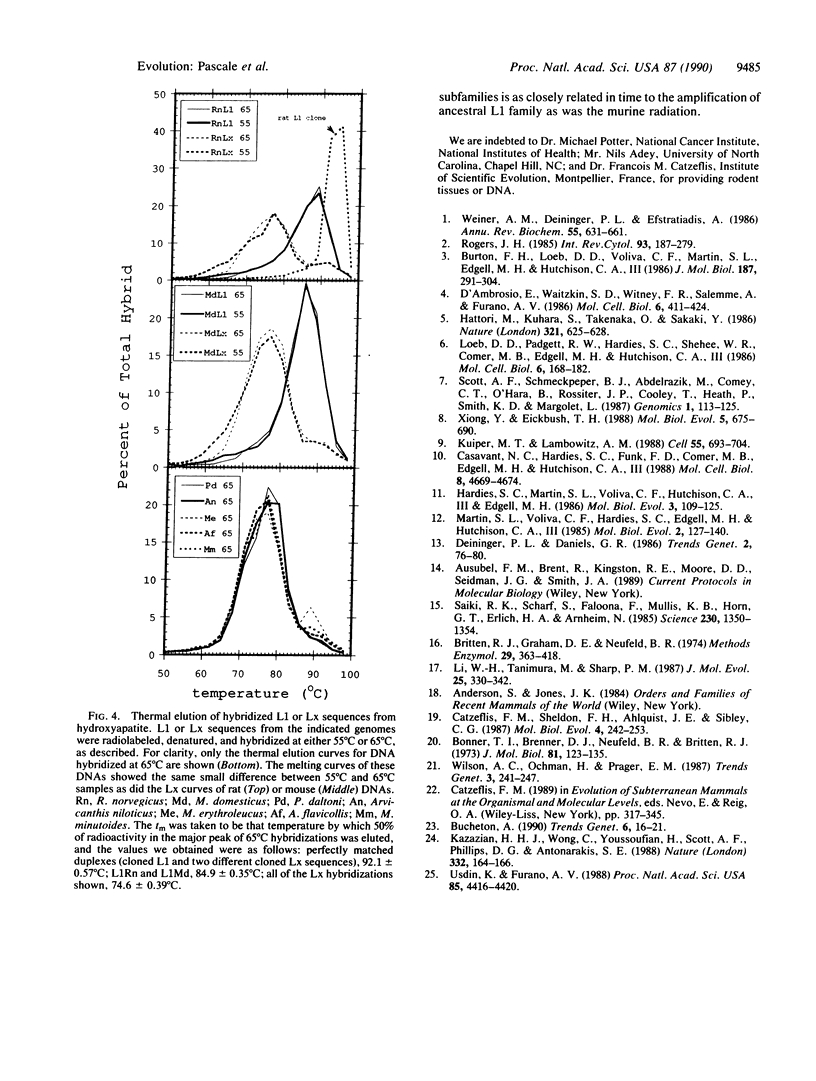
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonner T. I., Brenner D. J., Neufeld B. R., Britten R. J. Reduction in the rate of DNA reassociation by sequence divergence. J Mol Biol. 1973 Dec 5;81(2):123–135. doi: 10.1016/0022-2836(73)90184-8. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Bucheton A. I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet. 1990 Jan;6(1):16–21. doi: 10.1016/0168-9525(90)90044-7. [DOI] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., Voliva C. F., Martin S. L., Edgell M. H., Hutchison C. A., 3rd Conservation throughout mammalia and extensive protein-encoding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol. 1986 Jan 20;187(2):291–304. doi: 10.1016/0022-2836(86)90235-4. [DOI] [PubMed] [Google Scholar]
- Casavant N. C., Hardies S. C., Funk F. D., Comer M. B., Edgell M. H., Hutchison C. A., 3rd Extensive movement of LINES ONE sequences in beta-globin loci of Mus caroli and Mus domesticus. Mol Cell Biol. 1988 Nov;8(11):4669–4674. doi: 10.1128/mcb.8.11.4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catzeflis F. M., Sheldon F. H., Ahlquist J. E., Sibley C. G. DNA-DNA hybridization evidence of the rapid rate of muroid rodent DNA evolution. Mol Biol Evol. 1987 May;4(3):242–253. doi: 10.1093/oxfordjournals.molbev.a040444. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio E., Waitzkin S. D., Witney F. R., Salemme A., Furano A. V. Structure of the highly repeated, long interspersed DNA family (LINE or L1Rn) of the rat. Mol Cell Biol. 1986 Feb;6(2):411–424. doi: 10.1128/mcb.6.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardies S. C., Martin S. L., Voliva C. F., Hutchison C. A., 3rd, Edgell M. H. An analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol Biol Evol. 1986 Mar;3(2):109–125. doi: 10.1093/oxfordjournals.molbev.a040386. [DOI] [PubMed] [Google Scholar]
- Hattori M., Kuhara S., Takenaka O., Sakaki Y. L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature. 1986 Jun 5;321(6070):625–628. doi: 10.1038/321625a0. [DOI] [PubMed] [Google Scholar]
- Kazazian H. H., Jr, Wong C., Youssoufian H., Scott A. F., Phillips D. G., Antonarakis S. E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988 Mar 10;332(6160):164–166. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- Kuiper M. T., Lambowitz A. M. A novel reverse transcriptase activity associated with mitochondrial plasmids of Neurospora. Cell. 1988 Nov 18;55(4):693–704. doi: 10.1016/0092-8674(88)90228-0. [DOI] [PubMed] [Google Scholar]
- Li W. H., Tanimura M., Sharp P. M. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J Mol Evol. 1987;25(4):330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- Loeb D. D., Padgett R. W., Hardies S. C., Shehee W. R., Comer M. B., Edgell M. H., Hutchison C. A., 3rd The sequence of a large L1Md element reveals a tandemly repeated 5' end and several features found in retrotransposons. Mol Cell Biol. 1986 Jan;6(1):168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. L., Voliva C. F., Hardies S. C., Edgell M. H., Hutchison C. A., 3rd Tempo and mode of concerted evolution in the L1 repeat family of mice. Mol Biol Evol. 1985 Mar;2(2):127–140. doi: 10.1093/oxfordjournals.molbev.a040340. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Scott A. F., Schmeckpeper B. J., Abdelrazik M., Comey C. T., O'Hara B., Rossiter J. P., Cooley T., Heath P., Smith K. D., Margolet L. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987 Oct;1(2):113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usdin K., Furano A. V. Rat L (long interspersed repeated DNA) elements contain guanine-rich homopurine sequences that induce unpairing of contiguous duplex DNA. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4416–4420. doi: 10.1073/pnas.85.12.4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. M., Deininger P. L., Efstratiadis A. Nonviral retroposons: genes, pseudogenes, and transposable elements generated by the reverse flow of genetic information. Annu Rev Biochem. 1986;55:631–661. doi: 10.1146/annurev.bi.55.070186.003215. [DOI] [PubMed] [Google Scholar]
- Xiong Y., Eickbush T. H. Similarity of reverse transcriptase-like sequences of viruses, transposable elements, and mitochondrial introns. Mol Biol Evol. 1988 Nov;5(6):675–690. doi: 10.1093/oxfordjournals.molbev.a040521. [DOI] [PubMed] [Google Scholar]