Abstract
Ankylosing spondylitis (AS) is a type of autoimmune disease that predominantly affects the spine and sacroiliac joints. However, the pathogenesis of AS remains unclear. Some evidence indicates that infection with bacteria, especially Gram-negative bacteria, may have an important role in the onset and progression of AS. Recently, many studies have demonstrated that mesenchymal stem cells (MSCs) dysfunction may contribute to the pathogenesis of many rheumatic diseases. We previously demonstrated that MSCs from AS patients exhibited markedly enhanced osteogenic differentiation capacity in vitro under non-inflammatory conditions. However, the properties of MSCs from AS patients in an inflammatory environment have never been explored. Lipopolysaccharide (LPS), a proinflammatory substance derived from the outer membrane of Gram-negative bacteria, can alter the status and function of MSCs. However, whether MSCs from AS patients exhibit abnormal responses to LPS stimulation has not been reported. Autophagy is a lysosome-mediated catabolic process that participates in many physiological and pathological processes. The link between autophagy and AS remains largely unknown. The level of autophagy in ASMSCs after LPS stimulation remains to be addressed. In this study, we demonstrated that although the basal level of autophagy did not differ between MSCs from healthy donors (HDMSCs) and ASMSCs, LPS-induced autophagy was weaker in ASMSCs than in HDMSCs. Specifically, increased TRAF4 expression in ASMSCs impaired LPS-induced autophagy, potentially by inhibiting the phosphorylation of Beclin-1. These data may provide further insight into ASMSC dysfunction and the precise mechanism underlying the pathogenesis of AS.
Introduction
Ankylosing spondylitis (AS) is a type of autoimmune disease that predominantly affects the spine and sacroiliac joints.1 The central pathological characteristics of AS are chronic inflammation and ectopic ossification.1 Several hypotheses for the mechanism underlying AS, including genetic susceptibility related to HLA-B27 or ERAP1 as well as environmental factors have been proposed.2, 3, 4, 5, 6 However, these hypotheses cannot fully explain the entire process of AS onset and progression. Recent evidence indicates that infection with bacteria, especially Gram-negative bacteria, may have an important role in the onset and progression of AS.7, 8, 9 However, the precise relationship between Gram-negative bacterial infection and AS must be explored further.
Mesenchymal stem cells (MSCs) are a cell type with strong potential for immunosuppression and tri-lineage differentiation.10, 11 Recently, many studies have demonstrated that MSCs dysfunction may contribute to the pathogenesis of many rheumatic diseases.12, 13, 14 Our previous study demonstrated that MSCs from AS patients exhibited markedly enhanced osteogenic differentiation capacity in vitro under non-inflammatory conditions.15 However, the properties of MSCs from patients with inflammatory diseases such as AS in an inflammatory environment have never been explored. Lipopolysaccharide (LPS), a proinflammatory substance derived from the outer membrane of Gram-negative bacteria,16 can alter the status and function of MSCs.17, 18 However, whether MSCs from AS patients exhibit aberrant responses to LPS stimulation has never been reported.
Autophagy is a lysosome-mediated catabolic process that participates in many physiological and pathological processes.19, 20 Recently, many studies have demonstrated that the level of autophagy, which can be affected by various factors in vivo,21, 22 including LPS,23 reflects the status of MSCs. Moreover, several studies have demonstrated that abnormal autophagy contributes to the onset and progression of multiple autoimmune diseases,24, 25, 26 including inflammation in the gut of AS patients.27 However, the link between autophagy and AS remains largely unknown. The level of autophagy in ASMSCs after LPS stimulation remains to be addressed.
In this study, we first measured LPS-induced autophagy in both ASMSCs and healthy donors (HDMSCs), and then investigated the mechanisms of impaired LPS-induced autophagy in ASMSCs. These data may provide further insight into ASMSCs dysfunction and clues concerning the precise mechanism underlying the pathogenesis of AS.
Materials and methods
Isolation and culture of MSCs
This study was approved by the ethics committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, China. Written informed consent was obtained from all subjects included in the study (30 healthy donors and 30 patients with AS (diagnosed according to the modified New York criteria28)). The characteristics of the subjects of this study are presented in Table 1. MSCs were isolated and cultured as previously described.15 Briefly, MSCs were isolated and purified from bone marrow using density gradient centrifugation at 12 000 r.p.m. for 30 min. The MSCs were then resuspended in Dulbecco’s modified Eagle’s medium (DMEM, GIBCO, Waltham, MA, USA) containing 10% fetal bovine serum (FBS, Hangzhou Sijiqing Biological Engineering Material Company, Hangzhou, China), seeded in flasks and cultured in incubators at 37 °C and 5% CO2. After 48 h, the culture medium was replaced to remove the nonadherent cells. The medium was replaced every 3 days thereafter. When the cells reached 80–90% confluence, the MSCs were digested using 0.25% trypsin containing 0.53 mM ethylene diamine tetraacetic acid (EDTA) and reseeded in new flasks as passage 1. The MSCs were expanded and used for subsequent experiments at passages 3–5.
Table 1. Characteristics of the study subjects.
| Healthy donors | AS patients | |
|---|---|---|
| Number | 30 | 30 |
| Age, years | 30.9±9.9 | 33.5±10.3 |
| Male, n (%) | 17 (56.7%) | 19 (63.3%) |
| HLA-B27 positive, n (%) | 0 | 28 (93.3%)a |
| Disease duration, years | NA | 7.3±6.48a |
| CRP level, mg l−1 | 3±0.9 | 19.8±9.7a |
| ESR, mm h−1 | 10.4±4.2 | 29.7±14.2a |
| BASDAI | 1.05±0.93 | 4.33±0.99a |
| BASMI | 1.1±0.82 | 3.45±0.87a |
| BASFI | 0.91±0.58 | 3.46±0.63a |
Abbreviations: AS, ankylosing spondylitis; BASDAI, Bath Ankylosing Spondylitis Disease Activity Index; BASMI, Bath Ankylosing Spondylitis Metrology Index; BASFI, Bath Ankylosing Spondylitis Functional Index; CRP, C-reactive protein; ESR, erythrocyte sedimentation rate; HLA-B27, human leukocyte antigen B27.
Indicates P<0.05 compared to the healthy donor group.
Mean±s.d.
Stimulation of MSCs with LPS
MSCs were separately seeded in 6-well plates at a density of 1.5 × 104 cells cm−2 in culture medium as described above. After 12 h, when cells were adherent to the wells, the culture medium was replaced with culture medium containing LPS (Sigma-Aldrich, St Louis, MO, USA) at final concentration of 1-20 μg ml−1 and stimulated from 0–24 h.
Flow cytometry
MSCs were trypsinized for identification of several surface markers using human anti-CD29-PE, anti-CD44-V450A, anti-CD105-FITC, anti-CD45-FITC, anti-CD14-APC and anti-HLA-DR-PE antibodies (all purchased from BD Pharmingen, Franklin Lakes, NJ, USA). Flow cytometry was performed using a BD Influx cell sorter (BD Biosciences, Franklin Lakes, NJ, USA).
Cell proliferation assay
The proliferation rate of MSCs was determined via the Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) assay according to the manufacturer’s protocol. Medium lacking MSCs was used as a negative control.
Quantitative real-time PCR
High-throughput microfluidic fluorescence quantitative real-time PCR (qRT-PCR) was performed as previously described.15 Briefly, total RNA was extracted from MSCs using TRIzol (TaKaRa, Kusatsu, Japan) and then converted into cDNA using a PrimeScriptTM RT reagent kit (Takara). The sample pre-mix and assay pre-mix were prepared and added to the respective inlets of a 48.48 Dynamic Array integrated fluidic circuit (IFC, Fludigm, South San Francisco, CA, USA) that was primed and loaded in an IFC Controller MX (Fluidigm). qRT-PCR was performed in a BioMark HD System (Fluidigm), and the resulting data were analyzed using analysis software (Fluidigm). Real-time PCR was performed using a LightCycler 480 Real-Time PCR Detection System (Roche, Basel, Switzerland) using SYBR Green I Master Mix (Roche). The specific primers used for each gene are shown in Table 2. Data were first normalized by glyceraldehyde-3-hosphate dehydrogenase (GAPDH) expression and then by the values in HDMSCs before LPS stimulation. The relative expression levels of each gene were determined using the 2−ΔΔCt method.
Table 2. Primers used for qRT-PCR.
| Gene | Accession No. | Forward primer (5′–3′) | Reverse primer (5′–3′) | Product size (bp) |
|---|---|---|---|---|
| A20 | NM_006290 | TCCTCAGGCTTTGTATTTGAGC | TGTGTATCGGTGCATGGTTTTA | 124 |
| AP1 | NM_002228 | TCCAAGTGCCGAAAAAGGAAG | CGAGTTCTGAGCTTTCAAGGT | 78 |
| ATG5 | NM_004849 | AAAGATGTGCTTCGAGATGTGT | CACTTTGTCAGTTACCAACGTCA | 144 |
| ATG7 | NM_001144912 | ATGATCCCTGTAACTTAGCCCA | CACGGAAGCAAACAACTTCAAC | 114 |
| ATG12 | NM_004707 | CTGCTGGCGACACCAAGAAA | CGTGTTCGCTCTACTGCCC | 97 |
| ATG16L1 | NM_198890 | ATGCGCGGATTGTCTCAGG | GTCCACTCATTACACATTGCTCT | 138 |
| BCL2 | NM_000657 | GGTGGGGTCATGTGTGTGG | CGGTTCAGGTACTCAGTCATCC | 89 |
| BECN1 | NM_003766 | GGTGTCTCTCGCAGATTCATC | TCAGTCTTCGGCTGAGGTTCT | 121 |
| GAPDH | NM_001256799 | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG | 197 |
| IKKα | NM_001278 | GGCTTCGGGAACGTCTGTC | TTTGGTACTTAGCTCTAGGCGA | 87 |
| IKKβ | NM_001556 | GGAAGTACCTGAACCAGTTTGAG | GCAGGACGATGTTTTCTGGCT | 147 |
| IKKε | NM_001193322 | TGCCTGAGGATGAGTTCCTG | CGATGCACAATGCCGTTCT | 82 |
| IKKγ | NM_003639 | AAGAGCCAACTGTGTGAGATG | TTCGCCCAGTACGTCCTGA | 69 |
| IRAK4 | NM_001145258 | CCTGACTCCTCAAGTCCAGAA | ACAGAAATGGGTCGTTCATCAAA | 119 |
| IRAKM | NM_007199 | CTGCGGGATCTCCTTAGAGAA | GCAGAGAAATTCCGAGGGCA | 101 |
| IRF3 | NM_001197128 | AGAGGCTCGTGATGGTCAAG | AGGTCCACAGTATTCTCCAG | 100 |
| IRF4 | NM_002460 | GCTGATCGACCAGATCGACAG | CGGTTGTAGTCCTGCTTGC | 111 |
| IRF5 | NM_001098630 | GGGCTTCAATGGGTCAACG | GCCTTCGGTGTATTTCCCTG | 138 |
| IRF7 | NM_004029 | GCTGGACGTGACCATCATGTA | GGGCCGTATAGGAACGTGC | 84 |
| IRF8 | NM_002163 | ATGTGTGACCGGAATGGTGG | AGTCCTGGATACATGCTACTGTC | 77 |
| IRF9 | NM_006084 | GCCCTACAAGGTGTATCATTG | TGCTGTCGCTTTGATGGTACT | 84 |
| MyD88 | NM_001172569 | GGCTGCTCTCAACATGCGA | CTGTGTCCGCACGTTCAAGA | 61 |
| SOCS1 | NM_003745 | CACGCACTTCCGCACATTC | TAAGGGCGAAAAAGCAGTTCC | 191 |
| RIP1 | NM_003804 | GGGAAGGTGTCTCTGTGTTTC | CCTCGTTGTGCTCAATGCAG | 91 |
| TAK1 | NM_003298 | CCTCTGGCCCATTGAGTGTTT | GTCCACTGCGGTGACTATCTG | 98 |
| TBK1 | NM_013254 | TGGGTGGAATGAATCATCTACGA | GCTGCACCAAAATCTGTGAGT | 124 |
| TIRAP | NM_148910 | GACCCCTGGTGCAAGTACC | CGACGTAGTACATGAATCGGAG | 133 |
| TRAF1 | NM_001190945 | TCCTGTGGAAGATCACCAATGT | GCAGGCACAACTTGTAGCC | 117 |
| TRAF3 | NM_003300 | GCGTGTCAAGAGAGCATCGTT | GCAGATGTCCCAGCATTAACT | 133 |
| TRAF4 | NM_004295 | TATTGGGCCTGCCTATCCG | CAAAACTCGCACTTGAGGGG | 205 |
| TRAF6 | NM_145803 | ATGCGGCCATAGGTTCTGC | TCCTCAAGATGTCTCAGTTCCATD | 198 |
Abbreviations: A20, tumor necrosis factor-alpha induced protein 3; AP1, activator protein 1; ATG5, autophagy-related 5; ATG7, autophagy-related 7; ATG12, autophagy-related 12; ATG16L1, autophagy-related 16 like 1; BECN1, beclin-1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IKKα, inhibitor of kappaB kinase alpha; IKKβ, inhibitor of kappaB kinase beta; IKKε, inhibitor of kappaB kinase epsilon; IKKγ, inhibitor of kappaB kinase gamma; IRAK4, interleukin-1 receptor-associated kinase 4; IRAKM, interleukin-1 receptor-associated kinase M; IRF3, interferon regulatory factor 3; IRF4, interferon regulatory factor 4; IRF5, interferon regulatory factor 5; IRF7, interferon regulatory factor 7; IRF8, interferon regulatory factor 8; IRF9, interferon regulatory factor 9; MyD88, myeloid differentiation primary response 88; SOCS1, suppressor of cytokine signaling 1; RIP1, rieske iron–sulfur polypeptide 1; TAK1, transforming growth factor-beta activated kinase 1; TBK1, TANK binding kinase 1; TIRAP, toll/interleukin-1 receptor-like protein domain-containing adaptor protein; TRAF1, tumor necrosis factor receptor-associated factor 1; TRAF3, tumor necrosis factor receptor-associated factor 3; TRAF4, tumor necrosis factor receptor-associated factor 4; TRAF6, tumor necrosis factor receptor-associated factor 6.
Western blot
Cells were washed three times with ice-cold PBS. Then, the cells were collected and lysed in RIPA buffer (Sigma) containing a cocktail of protease inhibitors (Roche) for 30 min on ice. Lysates were obtained via centrifugation at 12 000 r.p.m. at 4 °C for 30 min. The protein concentration in the supernatant was determined using a BCA Protein Assay Kit (Sigma). Equal amounts of each sample diluted in 5 × SDS loading buffer were subjected to SDS-polyacrylamide gel electrophoresis and were subsequently transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). The membranes were blocked in 5% nonfat dry milk dissolved in TBST (150 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 0.05% Tween-20) at room temperature for 1 h and then incubated with primary antibodies against GAPDH (1:1000, Cell Signaling Technology, Danvers, MA, USA), LC3B (1:1000, Sigma), P62 (1:1000, Cell Signaling Technology), TRAF4 (2.5 μg ml−1, R&D systems, Minneapolis, MN, USA), Beclin-1 (1:1000, Cell Signaling Technology) phospho-Beclin-1 (1:1000, Cell Signaling Technology) or specific K63-linked polyubiquitin (1:1000, Cell Signaling Technology) overnight at 4 °C. After washing three times in TBST, the membranes were incubated with the appropriate HRP-conjugated secondary antibodies (1:3000, Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h at room temperature, followed by an additional three washes with TBST. The immunoreactive bands on the membranes were detected using chemiluminescent reagents (Millipore) according to the instructions provided by the manufacturer.
GFP-LC3B assay
The assay was performed as previously described.29, 30, 31, 32 MSCs were separately seeded in 6-well plates at a density of 1.5 × 104 cells cm−2 in culture medium for 24 h. The cells were then transfected with lentiviral vector containing green fluorescent protein-light chain 3B (GFP LC3B, GenePharma, Suzhou, China) at a multiplicity of infection of 50 with polyprene at a concentration of 5 μg ml−1. After 24 h, the culture medium was replaced according to the protocol provided by the manufacturer. At 72 h after transfection, when GFP-LC3B was sufficiently expressed in the MSCs, GFP-LC3B was observed in MSCs before and after LPS stimulation via fluorescence microscopy (Zeiss, Oberkochen, Germany). GFP-LC3B dots in autophagic cells were much more numerous and larger, referred to as GFP-LC3B punctate. A minimum of 100 cells per sample was observed, and for quantification, the number of GFP-LC3B dots per cell was counted in 100 cells per sample using Image Pro Software (Roper Technologies, Sarasota, FL, USA).
Lentivirus construction and infection
A lentivirus encoding short hairpin RNA (shRNA, GenePharma) targeting TRAF4 with the sequence 5′-GCACTAAGGAGTTCGTCTTTG-3′ (Lv-TRAF4) was constructed. The negative control shRNA sequence was 5′-TTCTCCGAACGTGTCACGTTTC-3′ (Lv-NC). Lentivirus (109 TU ml−1) and polybrene (5 μg ml−1, Sigma) were added to the medium and incubated with MSCs for 24 h at a multiplicity of infection of 50.
Immunoprecipitation
MSCs were quickly harvested and homogenized on ice in modified RIPA buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% (vol/vol) Triton X-100, 0.5% (wt/vol) sodium deoxycholate, 0.1% (wt/vol) SDS, 1 mM EDTA, 50 mM N-ethylmaleimide, 1 mM NaF, 1 mM Na3VO4, 1 mM PMSF and 1 μg ml−1 each of aprotinin, leupeptin and pepstatin. The cell extracts (~200 μg of total protein) were incubated with an antibody against Beclin-1 (2.5 μl, Cell Signaling Technology) at 4 °C overnight. Then, protein-G agarose beads (40 μl, Beyotime Biotechnology, Shanghai, China) were added, and the mixture was incubated at 4 °C for 3 h. The agarose beads were collected, washed, and resuspended in 60 μl of sample buffer containing 50 mM Tris-HCl, pH 7.6, 2% (wt/vol) SDS, 10% (vol/vol) glycerol, 10 mM DTT and 0.2% bromophenol blue. Afterwards, the samples were boiled for 5 min and analyzed via western blot.
Statistical analysis
Experiments and subsequent analysis were performed separately by different authors in this study. All data are expressed as the means±s.d. Statistical analysis was performed using SPSS 18.0 software (SPSS, Chicago, IL, USA). For statistical analysis, Student’s t-test was performed to compare the means between two groups, or one-way analysis of variance followed by Bonferroni correction was performed for multiple comparisons among groups. P<0.05 was considered statistically significant.
Results
ASMSCs and HDMSCs showed similar morphologies, phenotypes and proliferative capacities
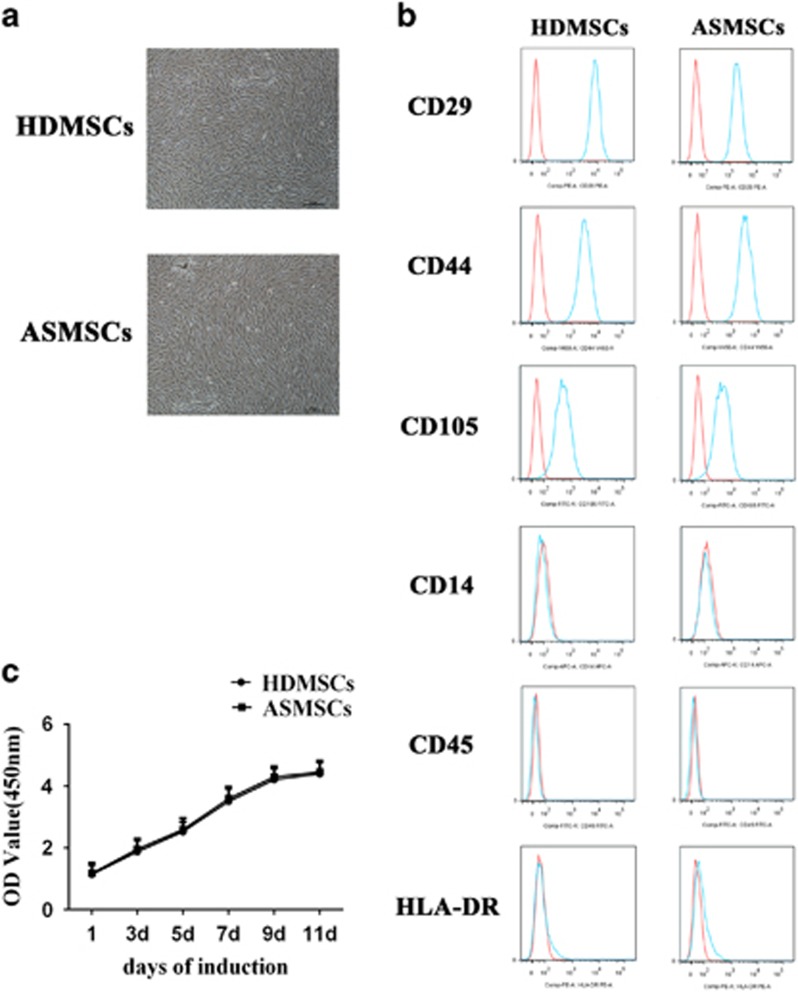
The morphologies of both HDMSCs and ASMSCs were plastic-adherent and spindle-shaped (Figure 1a). Regarding surface marker expression, both HDMSCs and ASMSCs were positive for CD29, CD44 and CD105 but negative for CD45, CD14 and HLA-DR; this expression pattern is characteristic of MSCs (Figure 1b). The growth curve of ASMSCs showed the same trend as that of HDMSCs (Figure 1c). In conclusion, there were no differences in morphology, phenotype or proliferative capacity between HDMSCs and ASMSCs.
Figure 1.
ASMSCs and HDMSCs showed comparable morphologies, proliferative activities and phenotypes. (a) The morphologies of MSCs were observed via microscopy (× 40, scale bar=50 μm); ASMSCs (n=30) and HDMSCs (n=30) shared the same morphologies. (b) Both ASMSCs (n=30) and HDMSCs (n=30) were positive for CD29, CD44 and CD105 but negative for CD14, CD45 and HLA-DR; this surface marker expression pattern is characteristic of MSCs. (c) The growth curve of ASMSCs (n=30) was similar to that of HDMSCs (n=30) cultured in medium from 0 to 11 days. Values in c are presented as the means±s.d. ASMSCs, ankylosing spondylitis mesenchymal stem cells; HDMSCs, healthy donors mesenchymal stem cells; MSCs, mesenchymal stem cells.
LPS-induced autophagy is impaired in ASMSCs compared with that in HDMSCs
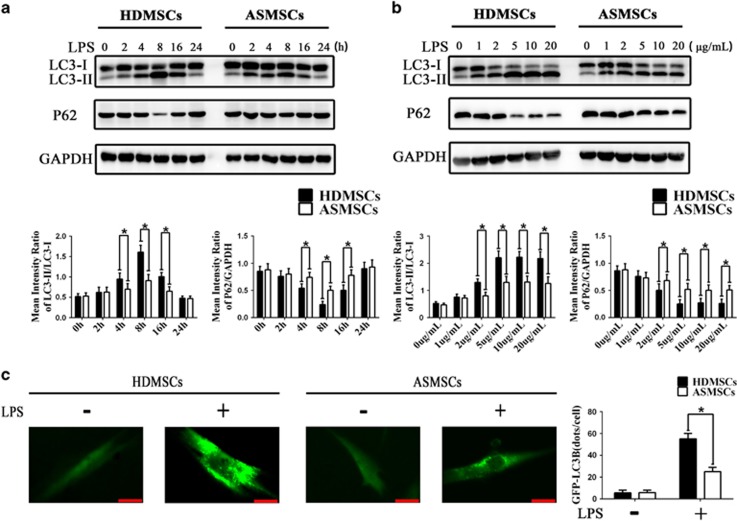
Because LC3-II accumulation in the cytoplasm is a specific hallmark of autophagy induction,33 we examined the effects of LPS on LC3-II accumulation in ASMSCs and HDMSCs. We first evaluated the expression of LC3-II in ASMSCs and HDMSCs after stimulation with 5 μg ml−1 LPS for 0–24 h. Western blot analysis revealed that the endogenous LC3-II levels peaked after LPS stimulation for 8 h and declined thereafter in both ASMSCs and HDMSCs (Figure 2a). However, the level of LC3-II expression was higher in HDMSCs than in ASMSCs after LPS stimulation for 4, 8 or 16 h (Figure 2a). Next, we measured the expression of LC3-II in ASMSCs and HDMSCs after LPS stimulation for 8 h at different concentrations ranging from 1 to 20 μg ml−1. LPS-induced autophagy in both ASMSCs and HDMSCs in a dose-dependent manner at concentrations of 1–5 μg ml−1, but the autophagy upon stimulation with LPS was similar between concentrations ranging from 5 to 20 μg ml−1 (Figure 2b). The expression of LC3-II was weaker in ASMSCs than in HDMSCs at LPS concentrations of 2–20 μg ml−1, and the difference in autophagy between ASMSCs and HDMSCs peaked at 5 μg ml−1 LPS (Figure 2b). The P62 protein, a specific autophagic substrate, is also used to monitor autophagy, and a lower level of P62 indicates stronger autophagy induction.34 In our study, the trend of P62 expression was opposite to that of LC3-II expression (Figures 2a and b). According to the above-mentioned results, subsequent experiments were conducted under conditions of LPS stimulation for 8 h at 5 μg ml−1. Following LPS stimulation, fluorescence microscopy revealed a significant strong and punctate pattern of GFP-LC3 expression in HDMSCs but much weaker and more diffuse GFP-LC3 expression in ASMSCs (Figure 2c). In summary, these data demonstrated that LPS-induced autophagy was impaired in ASMSCs compared with that in HDMSCs.
Figure 2.
LPS-induced autophagy is decreased in ASMSCs compared with that in HDMSCs. (a) MSCs were treated with LPS (5 μg ml−1) for 0–24 h. The expression levels of LC3-II were lower in ASMSCs (n=30) than in HDMSCs (n=30) after LPS stimulation for 4, 8 and 16 h. The expression pattern of P62 was opposite to that of LC3-II: P62 expression was higher in ASMSCs (n=30) than in HDMSCs (n=30) after LPS stimulation for 4, 8 or 16 h. (b) MSCs were treated with different concentrations (1–20 μg ml−1) of LPS for 8 h. The expression of LC3-II was lower in ASMSCs (n=30) than in HDMSCs (n=30) in the presence of LPS at concentrations from 2 to 20 μg ml−1, and the expression of P62 was higher in ASMSCs (n=30) than in HDMSCs (n=30) at LPS concentrations from 2 to 20 μg ml−1. (c) MSCs were transfected with GFP-LC3B and treated with LPS (5 μg ml−1) for 8 h. Autophagosome formation was indicated by GFP-labeled punctate. Fluorescence microscopy (× 400, scale bar=20 μm) revealed a markedly strong and punctate pattern of GFP-LC3B expression in HDMSCs (n=30) but much weaker and more diffuse GFP-LC3B expression in ASMSCs (n=30). The quantitative results are presented at right. The figures in a, b are typical examples of MSCs from one healthy donor and one AS subject. The values in a, b are presented as the means±s.d.; *P<0.05. AS, ankylosing spondylitis; ASMSCs, ankylosing spondylitis mesenchymal stem cells; HDMSCs, healthy donors mesenchymal stem cells; LPS, lipopolysaccharide; MSCs, mesenchymal stem cells.
Gene and protein expression patterns of HDMSCs and ASMSCs after LPS stimulation
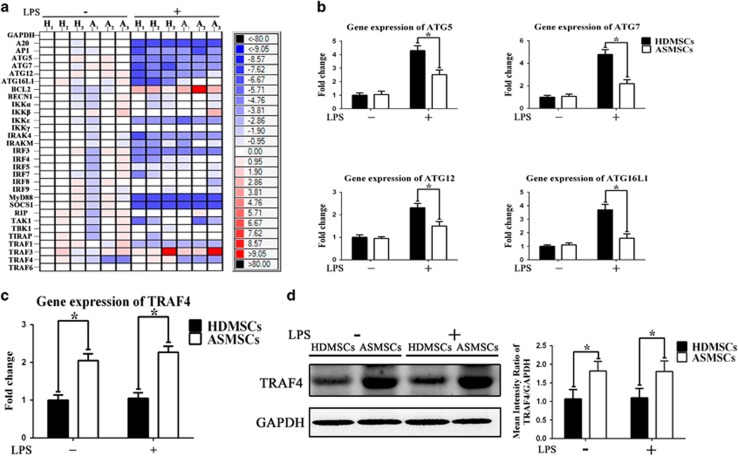
To confirm the levels of autophagy induced by LPS simulation and to explore the mechanism underlying the above-mentioned difference between ASMSCs and HDMSCs, we analyzed the gene expression of HDMSCs and ASMSCs after LPS stimulation using high-throughput qRT-PCR and confirmed the results via standard qRT-PCR. Our results demonstrated that the expression levels of autophagy-related genes including ATG5, ATG7, ATG12 and ATG16L1 were lower in ASMSCs after LPS stimulation (Figures 3a and b), confirming that LPS-induced autophagy was impaired in ASMSCs. In addition, we found that the mRNA expression of TRAF4, an important suppressor of signaling pathways,35, 36 was much higher in ASMSCs than in HDMSCs both before and after LPS stimulation (Figures 3a and c). We then examined the protein levels of TRAF4 in ASMSCs and HDMSCs. Consistent with pattern of TRAF4 mRNA expression, TRAF4 protein expression was much higher in ASMSCs than in HDMSCs both before and after LPS stimulation (Figure 3d).
Figure 3.
Gene and protein expression patterns of HDMSCs and ASMSCs after LPS stimulation. (a) High-throughput qRT-PCR was performed to determine the gene expression of important molecules related to the LPS-induced autophagy process in ASMSCs (n=30) and HDMSCs (n=30). H1-H3 and A1-A3 represent three samples randomly selected from the 30 healthy donors and the 30 AS patients, respectively. The heat maps show the ΔΔCT values. Each column represents one sample, and each row represents one gene. The results indicated that ASMSCs (A) had lower expression of ATG5, ATG7, ATG12 and ATG16L1 than HDMSCs (H) after LPS stimulation but higher expression of TRAF4 both before and after LPS stimulation. (b, c) Standard qRT-PCR was performed to confirm the results of high-throughput qRT-PCR. The standard qRT-PCR results were consistent with those of high-throughput qRT-PCR (n=30). (d) The protein level of TRAF4 was evaluated in ASMSCs (n=30) and HDMSCs (n=30) before and after LPS stimulation. The TRAF4 protein expression levels were consistent with the TRAF4 mRNA expression levels: TRAF4 protein expression was much higher in ASMSCs than in HDMSCs both before and after the LPS stimulation. The figures in d are typical examples of MSCs from one healthy donor and one AS subject. The values in b–d are presented as the means±s.d.; *P<0.05. The high-throughput and standard qRT-PCR data were first normalized to GAPDH expression and then normalized to the values of H1 before LPS stimulation, and the relative expression levels of each gene were analyzed using the 2−ΔΔCt method. AS, ankylosing spondylitis; ASMSCs, ankylosing spondylitis mesenchymal stem cells; HDMSCs, healthy donors mesenchymal stem cells; LPS, lipopolysaccharide; MSCs, mesenchymal stem cells; qRT-PCR, quantitative real-time PCR.
Silencing TRAF4 expression using Lv-TRAF4 abolished the discrepancy in LPS-induced autophagy between HDMSCs and ASMSCs
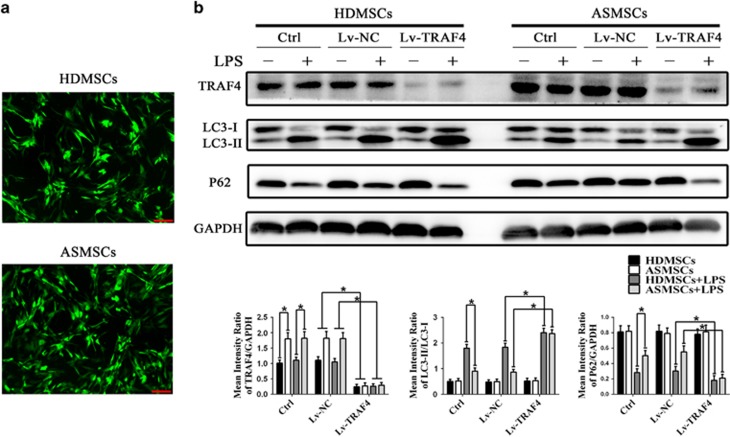
To explore the role of TRAF4 in the deficiency in LPS-induced autophagy in ASMSCs, we used Lv-TRAF4 to silence the expression of TRAF4 in ASMSCs and HDMSCs. As shown in Figure 4a, this lentivirus effectively transduced both HDMSCs and ASMSCs, and western blot confirmed that Lv-TRAF4 transduction markedly decreased the protein expression of TRAF4 to comparable levels between ASMSCs and HDMSCs (Figure 4b). Silencing TRAF4 increased the level of LC3-II but decreased the level of P62 in both ASMSCs and HDMSCs after LPS stimulation, and the difference in the expression of these markers between ASMSCs and HDMSCs was abolished by Lv-TRAF4 transduction (Figure 4b). These data indicated that elevated expression of TRAF4 in ASMSCs was the primary cause of the difference in LPS-induced autophagy between ASMSCs and HDMSCs.
Figure 4.
Inhibiting TRAF4 expression using Lv-TRAF4 abolished the difference in LPS-induced autophagy between HDMSCs and ASMSCs. (a) GFP-positive cells were observed under a confocal microscope (× 50, scale bar=200 μm), and the transduction efficiencies were equivalent between HDMSCs (n=30) and ASMSCs (n=30). (b) The expression levels of TRAF4, LC3-II and P62 in ASMSCs (n=30) and HDMSCs (n=30) were determined. Lv-TRAF4 effectively decreased the expression of TRAF4 in both ASMSCs and HDMSCs compared with that in the control and Lv-NC treatments before and after LPS stimulation. Lv-TRAF4 transduction led to increased LC3-II expression but decreased P62 expression in both ASMSCs and HDMSCs after LPS stimulation. In addition, silencing TRAF4 abolished the differences in LC3-II and P62 expression between HDMSCs and ASMSCs after LPS stimulation. The figures in b are typical examples of MSCs from one healthy donor and one AS subject. (b) are presented as the means±s.d.; *P<0.05. The control group consisted of MSCs not transduced with lentivirus. The Lv-NC group consisted of MSCs transduced with lentivirus encoding a negative control shRNA sequence. The Lv-TRAF4 group consisted of MSCs transduced with lentivirus encoding shRNA targeting TRAF4. AS, ankylosing spondylitis; ASMSCs, ankylosing spondylitis mesenchymal stem cells; HDMSCs, healthy donors mesenchymal stem cells; LPS, lipopolysaccharide; MSCs, mesenchymal stem cells.
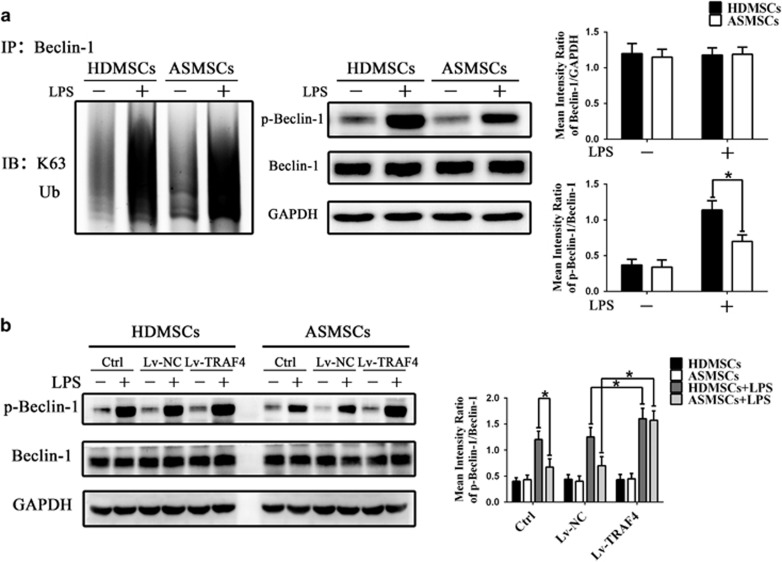
ASMSCs showed decreased phosphorylation of Beclin-1 after LPS stimulation due to elevated TRAF4 expression
To our knowledge, few studies have explored the relationship between TRAF4 and autophagy. Previous reports have demonstrated that TRAFs have a role in mediating the function of Beclin-1,37 which is an essential molecule for autophagy initiation.38 Therefore, we examined whether Beclin-1 is involved in the deficiency in LPS-induced autophagy in ASMSCs. The expression and K63-linked ubiquitination of Beclin-1 were comparable between ASMSCs and HDMSCs after LPS stimulation (Figure 5a). Next, we assessed the level of Beclin-1 phosphorylation in ASMSCs and HDMSCs. As shown in Figure 5a, after LPS stimulation, the level of phosphorylated Beclin-1 was much lower in ASMSCs than in HDMSCs. In addition, Lv-TRAF4 transduction increased the levels of phosphorylated Beclin-1 in ASMSCs to that in HDMSCs after LPS stimulation (Figure 5b). Because phosphorylation of Beclin-1 is important for its autophagic activity,39, 40 these results indicated that ASMSCs exhibit impaired LPS-induced autophagy due to their increased expression of TRAF4, which alters the phosphorylation of Beclin-1.
Figure 5.
TRAF4 overexpression decreased Beclin-1 phosphorylation in ASMSCs after LPS stimulation. (a) K63-linked ubiquitination, expression and phosphorylation of Beclin-1 were detected via western blot. K63-linked ubiquitination and expression of Beclin-1 were comparable between ASMSCs (n=30) and HDMSCs (n=30). LPS-induced phosphorylation of Beclin-1 was reduced in ASMSCs (n=30) compared with that in HDMSCs (n=30). (b) Lv-TRAF4 transduction increased LPS-induced phosphorylation of Beclin-1 to identical levels in ASMSCs (n=30) and HDMSCs (n=30). The figures in a, b are typical examples of MSCs from one healthy donor and one AS subject. The values in a, b are presented as the means±s.d.; *P<0.05. AS, ankylosing spondylitis; ASMSCs, ankylosing spondylitis mesenchymal stem cells; HDMSCs, healthy donors mesenchymal stem cells; LPS, lipopolysaccharide; MSCs, mesenchymal stem cells.
Discussion
In this study, we demonstrated that even though there was no difference in the basal level of autophagy between HDMSCs and ASMSCs, LPS-induced autophagy was weaker in ASMSCs than in HDMSCs. Specifically, increased TRAF4 expression in ASMSCs impaired LPS-induced autophagy, potentially by inhibiting the phosphorylation of Beclin-1.
Recently, some authors have postulated that autoimmune diseases are stem cell-related disorders.12, 41 Consistent with this hypothesis, our previous work demonstrated that ASMSCs have enhanced osteogenic differentiation potential in the absence inflammatory stimulation, which may be a mechanism of pathological osteogenesis in AS.15 These data indicated that ASMSCs were abnormal. However, the status and functions of MSCs are plastic in response to multiple factors, such as inflammation and hypoxia.42, 43 Considering that AS is a type of inflammatory disease, studying the abnormalities in ASMSCs in an inflammatory microenvironment will have clinical and scientific research value.
Infection with bacteria, especially Gram-negative bacteria, is thought to actively participate in the pathogenesis of AS.7, 8, 9 LPS is an important proinflammatory substance derived from the outer membrane of Gram-negative bacteria.16 It has been reported that AS patients had significantly elevated serum levels of antibodies against LPS compared to those of healthy controls and that LPS antibody levels strongly correlated with the levels of the acute-phase reactant C-reactive protein.44 Therefore, LPS should be considered an important component of the inflammatory microenvironment in AS. In addition, other studies have demonstrated that stimulation with LPS modulates the status or the function of MSCs.17, 18 Therefore, we explored the status and function of ASMSCs under conditions of LPS stimulation and assessed whether ASMSCs exhibit any abnormalities in response to LPS stimulation compared with HDMSCs.
Autophagy is a lysosome-mediated catabolic process that maintains cellular homeostasis by degrading and recycling cytoplasmic components or organelles.45 The biological implications of insufficient or excessive autophagic activity are a rapidly growing area of study.46 Previous hypotheses that autophagy is strictly a catabolic recycling process are now being expanded. Although the association between autophagy and rheumatic diseases has been widely studied in past years, the relationship between autophagy and AS remains largely unknown. The two central features in AS that require explanation are inflammation and new bone formation.1 However, the pathogenesis of these two features remains largely unknown. The potential role of autophagy in the pathogenesis of these two features requires further exploration. As in peripheral blood mononuclear cells (PBMCs), defective autophagy can contribute to increased inflammation through a failure to regulate the release of several proinflammatory cytokines, including IL-1β, IL-17 and IL-23.47, 48, 49 Autophagy can regulate multiple functions of MSCs.50, 51, 52 One of the characteristics of MSCs is their immunoregulation ability, and autophagy has an important role in modulating this ability.52 Defects of MSCs in immune regulation may contribute to the pathogenesis of inflammation.53, 54 Our data demonstrated that LPS-induced autophagic activity was much weaker in ASMSCs than in HDMSCs. Thus, the impairment of LPS-induced autophagy in ASMSCs may suppress their immunoregulation ability in an inflammatory microenvironment and, in turn, contribute to the pathogenesis of inflammation in AS. As mentioned above, another central feature of AS is new bone formation, and there are few studies about the pathogenesis of pathological osteogenesis in AS. MSCs are precursor cells of osteoblasts,55 and we have previously demonstrated that ASMSCs have enhanced osteogenic differentiation potential in the absence of inflammatory stimulation, which may be a mechanism of pathological osteogenesis in AS. Autophagy also modulates the differentiative capacity of MSCs.51, 56 Thus, the impairment of LPS-induced autophagy in ASMSCs may influence their osteogenesis in an inflammatory microenvironment and, in turn, contribute to the pathogenesis of pathological osteogenesis in AS. Conversely, other studies have shown that the level of autophagic activity is elevated in the subclinically inflamed gut of AS patients,27 indicating the opposite association between autophagy and AS. Similarly, both insufficient and excessive autophagy have been reported in rheumatoid arthritis (RA); for example, one study found that reduced autophagy in T cells resulted in immune system abnormalities,57 whereas another study found that enhanced autophagy contributed to hyperplasia in synovium from RA patients.25 Therefore, autophagy could have a dual role in the pathogenesis of RA, as both enhanced and reduced autophagy contribute to malfunctions observed in RA. Considering the complex role of autophagy in regulating different physiological and pathological processes, this ‘discrepancy’ in the function of autophagy in rheumatic diseases including AS is very likely caused by differences in cell types or tissues examined between studies. To identify the role of autophagy in AS, the level of autophagic activity in different types of cells and tissues related to AS should be studied. Elucidating the precise role of the impairment of LPS-induced autophagy in AS will be a focus of our future studies.
In addition, our study raises an intriguing question. Decreased protein levels of P62 are often used as a typical marker of autophagy.58 However, some studies have demonstrated that LPS-induced autophagy is P62-dependent; upon LPS stimulation, the protein level of P62 increases, resulting in induction of autophagy.59, 60 However, in our study, LPS-induced autophagy in MSCs was also accompanied by degradation of P62, which appears to contradict this previous observation. Indeed, the expression pattern of P62 during LPS-induced autophagy appears to be controversial. Some studies have demonstrated that the expression of P62 is increased after LPS stimulation as mentioned above, whereas others have demonstrated that the protein level of P62 is decreased after LPS stimulation following the typical expression pattern during autophagy induction.23, 61 However, we have identified several differences among these studies. First, the types of cells used in these studies are very different. Most studies that demonstrated that P62 is increased during LPS-induced autophagy used macrophages in their experiments, whereas studies demonstrating that P62 is decreased after LPS stimulation, consistent with our results, used epithelial or endothelial cells. Second, although the LPS stimulation times in these studies were similar, the concentrations of LPS differed. The concentration of LPS used in the studies showing an increase in P62 ranged from 0.1 to 1 μg ml−1, whereas the concentrations in the studies in which P62 decreased were often >1 μg ml−1. Thus, differences in cell type and LPS concentration may be responsible for the discrepancies in P62 levels.
To elucidate the mechanism underlying this abnormal LPS-induced autophagy, we measured the expression of several critical molecules related to this process. Our results demonstrated that TRAF4 expression was elevated in ASMSCs. After silencing TRAF4 using a lentivirus, the difference in LPS-induced autophagy between ASMSCs and HDMSCs was abolished. This result suggested that elevated expression of TRAF4 was the main cause of the abnormality in LPS-induced autophagy in ASMSCs. TRAF4 belongs to the TRAF family, which consists of signal transducers for multiple signaling pathways.62 In contrast to other TRAF members, TRAF4 acts as a suppressor of many signaling pathways and thus prevents excessive activation of these pathways.35, 36 Similarly, our results indicated that TRAF4 negatively regulated LPS-induced autophagy. Therefore, TRAF4 acts as a suppressor of LPS-induced autophagy and thus prevents overreactions to LPS stimulation under normal conditions. However, upon pathological upregulation of TRAF4 in ASMSCs, TRAF4 impairs LPS-induced autophagy in ASMSCs compared with HDMSCs. In addition, we observed an interesting phenomenon: TRAF4 was also upregulated in ASMSCs under basal conditions, but there was no difference in the basal status of autophagy between HDMSCs and ASMSCs. It is possible that the basal status of autophagy even in HDMSCs is very weak that the elevated expression of TRAF4 in ASMSCs can hardly further suppress the basal level of autophagy. Thus, the basal status of autophagy is comparable between ASMSCs and HDMSCs. However, upon LPS stimulation, autophagy is strongly induced in HDMSCs, but this process is suppressed by upregulated TRAF4 in ASMSCs, in turn causing impairment in autophagic activity in ASMSCs in an environment of LPS stimulation.
To our knowledge, few studies have explored the relationship between TRAF4 and autophagy. Previous studies have demonstrated that other TRAFs modulate the function of Beclin-1.37 Beclin-1 is the key molecule for induction of autophagy that initiates the formation of autophagosomes by helping localize other autophagy-related proteins to the pre-autophagosomal membrane.38 Thus, we examined whether Beclin-1 is involved in the impairment in LPS-induced autophagy in ASMSCs. In this study, we evaluated the expression and K63-linked ubiquitination and phosphorylation of Beclin-1 after LPS stimulation in ASMSCs and HDMSCs. We did not observe any difference in the expression or K63-linked ubiquitination of Beclin-1 between ASMSCs and HDMSCs. However, after LPS stimulation, the induction of Beclin-1 phosphorylation was much weaker in ASMSCs than in HDMSCs. Considering that Beclin-1 acts as an initiator of autophagy, K63-linked ubiquitination and phosphorylation of Beclin-1 are essential for its induction of autophagic activity,37, 40 and decreasing either K63-linked ubiquitination or phosphorylation of Beclin-1 leads to impairment of autophagy induction.63, 64 In our study, LPS-induced K63-linked ubiquitination of Beclin-1 was comparable between ASMSCs and HDMSCs, and this finding indicated that this ubiquitination process was not involved in the reduction in LPS-induced autophagy in ASMSCs. However, LPS-induced phosphorylation of Beclin-1 was much weaker in ASMSCs than in HDMSCs. Moreover, silencing TRAF4 in ASMSCs significantly enhanced the level of phosphorylated Beclin-1 in ASMSCs after LPS stimulation to a level comparable to that in HDMSCs. This observation was consistent with the above results that silencing TRAF4 in ASMSCs increased LPS-induced autophagic activity to the same level as that measured in HDMSCs. Therefore, our results indicated that phosphorylation of Beclin-1 might be involved in the impairment in LPS-induced autophagy in ASMSCs caused by increased TRAF4 expression.
In conclusion, our results have confirmed that LPS-induced autophagy was weaker in ASMSCs than in HDMSCs because of elevated expression of TRAF4, potentially by altering the phosphorylation of Beclin-1. These findings imply a possible role of MSC abnormalities in AS. However, there are many limitations to the present study. First, the mechanism by which TRAF4 suppresses LPS-induced phosphorylation of Beclin-1 remains unclear. Second, we have confirmed only that LPS-induced autophagy was reduced in ASMSCs, but the association of this reduction with ASMSC dysfunction in an inflammatory environment has yet to be concretely established. Our future research aims to address these limitations.
Acknowledgments
This study was financially supported by the National Natural Science Foundation of China (81672128, 81672097) and the Science and Technology Project of Guangdong Province (2015B020228001, 2015B090903059).
Footnotes
The authors declare no conflict of interest.
References
- Braun J, Sieper J. Ankylosing spondylitis. Lancet 2007; 369: 1379–1390. [DOI] [PubMed] [Google Scholar]
- Caffrey MF, James DC. Human lymphocyte antigen association in ankylosing spondylitis. Nature 1973; 242: 121. [DOI] [PubMed] [Google Scholar]
- Davidson SI, Wu X, Liu Y, Wei M, Danoy PA, Thomas G et al. Association of ERAP1, but not IL23R, with ankylosing spondylitis in a Han Chinese population. Arthritis Rheum 2009; 60: 3263–3268. [DOI] [PubMed] [Google Scholar]
- Evans DM, Spencer CC, Pointon JJ, Su Z, Harvey D, Kochan G et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet 2011; 43: 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MA, Kennedy LG, Macgregor AJ, Darke C, Duncan E, Shatford JL et al. Susceptibility to ankylosing spondylitis in twins the role of genes, HLA, and the environment. Arthritis Rheum 1997; 40: 1823–1828. [DOI] [PubMed] [Google Scholar]
- Calin A, Elswood J. Relative role of genetic and environmental factors in disease expression: sib pair analysis in ankylosing spondylitis. Arthritis Rheum 1989; 32: 77–81. [DOI] [PubMed] [Google Scholar]
- Martínez A, Pacheco-Tena C, Vázquez-Mellado J, Burgos-Vargas R. Relationship between disease activity and infection in patients with spondyloarthropathies. Ann Rheum Dis 2004; 63: 1338–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zochling J, Bohl-Bühler MH, Baraliakos X, Feldtkeller E, Braun J. The high prevalence of infections and allergic symptoms in patients with ankylosing spondylitis is associated with clinical symptoms. Clin Rheumatol 2006; 25: 648–658. [DOI] [PubMed] [Google Scholar]
- Ebringer RW, Cawdell DR, Cowling P, Ebringer A. Sequential studies in ankylosing spondylitis. Association of Klebsiella pneumoniae with active disease. Ann Rheum Dis 1978; 37: 146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal S, Pittenger MF. Human mesenchymal stem cells modulate allogeneic immune cell responses. Blood 2005; 105: 1815–1822. [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD et al. Multilineage potential of adult human mesenchymal stem cells. Science 1999; 284: 143–147. [DOI] [PubMed] [Google Scholar]
- Sun LY, Zhang HY, Feng XB, Hou YY, Lu LW, Fan LM. Abnormality of bone marrow-derived mesenchymal stem cells in patients with systemic lupus erythematosus. Lupus 2007; 16: 121–128. [DOI] [PubMed] [Google Scholar]
- Sun Y, Deng W, Geng L, Zhang L, Liu R, Chen W et al. Mesenchymal stem cells from patients with rheumatoid arthritis display impaired function in inhibiting Th17 cells. J Immunol Res 2015; 2015: 284215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzanti B, Aldinucci A, Biagioli T, Barilaro A, Urbani S, Dal Pozzo S et al. Differences in mesenchymal stem cell cytokine profiles between MS patients and healthy donors: implication for assessment of disease activity and treatment. J Neuroimmunol 2008; 199: 142–150. [DOI] [PubMed] [Google Scholar]
- Xie ZY, Wang P, Li YX, Deng W, Zhang X, Su HJ et al. Imbalance between bone morphogenetic protein 2 and noggin induces abnormal osteogenic differentiation of mesenchymal stem cells in ankylosing spondylitis. Arthritis Rheum 2016; 68: 430–440. [DOI] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem 2002; 71: 635–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SH, Cho HK, Park SH, Lee W, Lee HJ, Lee DC et al. Toll like receptor 3 & 4 responses of human turbinate derived mesenchymal stem cells: stimulation by double stranded RNA and lipopolysaccharide. PLoS ONE 2014; 9: e101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wu T, Xiong J, Su Y, Zhang C, Wang S et al. Porphyromonas gingivalis lipopolysaccharides regulate functions of bone marrow mesenchymal stem cells. Cell Prolif 2015; 48: 239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6: 463–477. [DOI] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science 2004; 306: 990–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouysségur J et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009; 29: 2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang ZF, Klionsky DJ. Endoplasmic reticulum stress triggers autophagy. J Biol Chem 2006; 281: 30299–30304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng N, Wu LL, Gao JG, Zhao J, Su L, Su H et al. Lipopolysaccharide induces autophagy through BIRC2 in human umbilical vein endothelial cells. J Cell Physiol 2010; 225: 174–179. [DOI] [PubMed] [Google Scholar]
- Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M et al. Autophagy is activated in systemic lupus erythematosus and required for plasmablast development. Ann Rheum Dis 2015; 74: 912–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Xu P, Yao JF, Zhang YG, Hou WK, Lu SM. Reduced apoptosis correlates with enhanced autophagy in synovial tissues of rheumatoid arthritis. Inflamm Res 2013; 62: 229–237. [DOI] [PubMed] [Google Scholar]
- Zhou XJ, Zhang H. Autophagy in immunity: implications in etiology of autoimmune/autoinflammatory diseases. Autophagy 2012; 8: 1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia F, Accardo-Palumbo A, Rizzo A, Guggino G, Raimondo S, Giardina A et al. Evidence that autophagy, but not the unfolded protein response, regulates the expression of IL-23 in the gut of patients with ankylosing spondylitis and subclinical gut inflammation. Ann Rheum Dis 2014; 73: 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum 1984; 27: 361–368. [DOI] [PubMed] [Google Scholar]
- Inokuchi-Shimizu S, Park EJ, Roh YS, Yang L, Zhang B, Song JY et al. TAK1-mediated autophagy and fatty acid oxidation prevent hepatosteatosis and tumorigenesis. J Clin Invest 2014; 124: 3566–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. MyD88 and Trif target Beclin 1 to trigger autophagy in macrophages. J Biol Chem 2008; 283: 33175–33182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N, Sasaki T, Iemura S, Natsume T, Hara T, Mizushima N. Atg101, A novel mammalian autophagy protein interacting with Atg13. Autophagy 2009; 5: 973–979. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Ueno T, Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol 2004; 36: 2503–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Ichimura Y. Physiological significance of selective degradation of p62 by autophagy. FEBS Lett 2010; 584: 1374–1378. [DOI] [PubMed] [Google Scholar]
- Takeshita F, Ishii KJ, Kobiyama K, Kojima Y, Coban C, Sasaki S et al. TRAF4 acts as a silencer in TLR-mediated signaling through the association with TRAF6 and TRIF. Eur J Immunol 2005; 35: 2477–2485. [DOI] [PubMed] [Google Scholar]
- Marinis JM, Hutti JE, Homer CR, Cobb BA, Cantley LC, McDonald C et al. I kappa B kinase alpha phosphorylation of TRAF4 downregulates innate immune signaling. Mol Cell Biol 2012; 32: 2479–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 2010; 3: ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ 2011; 18: 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 2009; 10: 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell RC, Tian Y, Yuan HX, Park HW, Chang YY, Kim J et al. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15: 741–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehara S, Kawamura M, Takao F, Inaba M, Yasumizu R, Than S et al. Organ-specific and systemic autoimmune diseases originate from defects in hematopoietic stem cells. Proc Natl Acad Sci USA 1990; 87: 8341–8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Han ZP, Zhang SS, Jing YY, Bu XX, Wang CY et al. Effects of inflammatory factors on mesenchymal stem cells and their role in the promotion of tumor angiogenesis in colon cancer. J Biol Chem 2011; 286: 25007–25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grayson WL, Zhao F, Bunnell B, Ma T. Hypoxia enhances proliferation and tissue formation of human mesenchymal stem cells. Biochem Biophys Res Commun 2007; 358: 948–953. [DOI] [PubMed] [Google Scholar]
- Ahmadi K, Wilson C, Tiwana H, Binder A, Ebringer A. Antibodies to Klebsiella pneumoniae lipopolysaccharide in patients with ankylosing spondylitis. Br J Rheumatol 1998; 37: 1330–1333. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290: 1717–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA et al. Activation of autophagy by inflammatory signals limits IL-1 beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13: 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peral de Castro C, Jones SA, Ní Cheallaigh C, Hearnden CA, Williams L, Winter J et al. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J Immunol 2012; 189: 4144–4153. [DOI] [PubMed] [Google Scholar]
- Brereton CF, Sutton CE, Ross PJ, Iwakura Y, Pizza M, Rappuoli R et al. Escherichia coli heat-labile enterotoxin promotes protective Th17 responses against infection by driving innate IL-1 and IL-23 production. J Immunol 2011; 186: 5896–5906. [DOI] [PubMed] [Google Scholar]
- Hou J, Han ZP, Jing YY, Yang X, Zhang SS, Sun K et al. Autophagy prevents irradiation injury and maintains stemness through decreasing ROS generation in mesenchymal stem cells. Cell Death Dis 2013; 4: e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver L, Hue E, Priault M, Vallette FM. Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev 2012; 21: 2779–2788. [DOI] [PubMed] [Google Scholar]
- Dang SP, Xu HB, Xu CF, Cai W, Li Q, Cheng YJ et al. Autophagy regulates the therapeutic potential of mesenchymal stem cells in experimental autoimmune encephalomyelitis. Autophagy 2014; 10: 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prockop DJ, Oh JY. Mesenchymal stem/stromal cells (MSCs): role as guardians of inflammation. Mol Ther 2012; 20: 14–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo ME, Fibbe WE. Mesenchymal stromal cells: sensors and switchers of inflammation. Cell Stem Cell 2013; 13: 392–402. [DOI] [PubMed] [Google Scholar]
- Heino TJ, Hentunen TA. Differentiation of osteoblasts and osteocytes from mesenchymal stem cells. Curr Stem Cell Res Ther 2008; 3: 131–145. [DOI] [PubMed] [Google Scholar]
- Tao J, Wang HN, Zhai Y, Park H, Wang J, Ji F et al. Downregulation of Nrf2 promotes autophagy-dependent osteoblastic differentiation of adipose-derived mesenchymal stem cells. Exp Cell Res 2016; 349: 221–229. [DOI] [PubMed] [Google Scholar]
- Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med 2013; 210: 2119–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282: 24131–24145. [DOI] [PubMed] [Google Scholar]
- Fujita KI, Srinivasula SM. TLR4-mediated autophagy in macrophages is a p62-dependent type of selective autophagy of aggresome-like induced structures (ALIS). Autophagy 2011; 7: 552–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Yuan JX, Yao SQ, Jin YL, Chen G, Tian W et al. Lipopolysaccharides may aggravate apoptosis through accumulation of autophagosomes in alveolar macrophages of human silicosis. Autophagy 2015; 11: 2346–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SY, Guo L, Qian P, Zhao YF, Liu A, Ji FY et al. Lipopolysaccharide induces autophagic cell death through the PERK-Dependent branch of the unfolded protein response in human alveolar epithelial A549 cells. Cell Physiol Biochem 2015; 36: 2403–2417. [DOI] [PubMed] [Google Scholar]
- Inoue JI, Ishida T, Tsukamoto N, Kobayashi N, Naito A, Azuma S et al. Tumor necrosis factor receptor-associated factor (TRAF) family: adapter proteins that mediate cytokine signaling. Exp Cell Res 2000; 254: 14–24. [DOI] [PubMed] [Google Scholar]
- Xia PY, Wang S, Du Y, Zhao ZN, Shi L, Sun L et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J 2013; 32: 2685–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara N, Usui T, Ohama T, Sato K. Regulation of Beclin 1 protein phosphorylation and autophagy by protein phosphatase 2A (PP2A) and death-associated protein kinase 3 (DAPK3). J Biol Chem 2016; 291: 10858–10866. [DOI] [PMC free article] [PubMed] [Google Scholar]