Abstract
Emerging evidence has emphasized the importance of cancer therapies targeting an abnormal metabolic state of tumor-initiating cells (TICs) in which they retain stem cell-like phenotypes and nicotinamide adenine dinucleotide (NAD+) metabolism. However, the functional role of NAD+ metabolism in regulating the characteristics of TICs is not known. In this study, we provide evidence that the mitochondrial NAD+ levels affect the characteristics of glioma-driven SSEA1+ TICs, including clonogenic growth potential. An increase in the mitochondrial NAD+ levels by the overexpression of the mitochondrial enzyme nicotinamide nucleotide transhydrogenase (NNT) significantly suppressed the sphere-forming ability and induced differentiation of TICs, suggesting a loss of the characteristics of TICs. In addition, increased SIRT3 activity and reduced lactate production, which are mainly observed in healthy and young cells, appeared following NNT-overexpressed TICs. Moreover, in vivo tumorigenic potential was substantially abolished by NNT overexpression. Conversely, the short interfering RNA-mediated knockdown of NNT facilitated the maintenance of TIC characteristics, as evidenced by the increased numbers of large tumor spheres and in vivo tumorigenic potential. Our results demonstrated that targeting the maintenance of healthy mitochondria with increased mitochondrial NAD+ levels and SIRT3 activity could be a promising strategy for abolishing the development of TICs as a new therapeutic approach to treating aging-associated tumors.
Introduction
Nicotinamide adenine dinucleotide (NAD+) is a crucial electron acceptor during glycolysis and has an essential role in redox and non-redox reactions that regulate diverse biological functions, including energy metabolism, DNA damage response, transcriptional control, cell proliferation/differentiation/death control and mitochondrial functions.1, 2, 3 Depletion or disturbance of NAD+ homeostasis leads to a failure of key processes in normal physiology and results in various dysfunctions and pathologies, including cancer and aging.3, 4 NAD+ is reduced to NADH during cytosolic glycolysis and the mitochondrial tricarboxylic acid cycle; then, NADH is utilized by the mitochondrial electron transport chain for ATP generation.2 Thus, a deficient amount of cytosolic NAD+ for glycolysis impairs glucose utilization even when a sufficient supply of glucose is available, resulting in cell death.5 Maintenance of NAD+/NADH ratios and optimal NAD+ levels in each subcellular compartment (nucleus, cytoplasm and mitochondria) is critical for basic cellular processes.1, 6 Moreover, major metabolic pathways in mitochondria highly depend on NAD+ availability.6 The mitochondrial NAD+ content in cardiac myocytes, which have dense mitochondria, accounts for up to 70% of the total cellular NAD+.7,8,9
Sirtuins (SIRTs) are major NAD+-consuming enzymes and play fundamental roles in metabolic regulation and are mainly involved in protective functions.10 Deacetylation targets for nuclear SIRT1 are related to genomic stability11, 12 and mitochondrial metabolism.13, 14 Mitochondrial SIRT3 activity is closely linked to SIRT1, which senses NAD+ and induces mitochondrial biogenesis, anti-oxidant defense mechanisms and life extension.15, 16 In contrast, the loss of function of SIRT1 or SIRT3 induces metabolic and age-related complications.17, 18, 19 Age-dependent decreases in NAD+ content followed by a reduction in SIRT1/3 activities is reported to contribute to stem cell decline and dysfunction, and their reverse by calorie restriction, exercise, calorie restriction mimetics (metformin and so on) and NAD+ precursors (nicotinamide riboside, nicotinamide mononucleotide and so on) improve stem cell function and lifespan.20, 21, 22, 23 Accumulated studies have provided evidence that maintenance of healthy mitochondrial fitness in response to metabolic energy stress is essential to recover declined stem cell function associated with aging and to contribute to enhanced health span and lifespan.22, 23, 24 We have previously demonstrated that the restoration of enzymes that can increase mitochondrial NAD+ levels delays stem cell senescence and facilitates reprogramming of aged somatic cells.25 Nicotinamide nucleotide transhydrogenase (NNT) produces NADPH with NAD+ in the mitochondrial matrix26 and nicotinamide mononucleotide adenylyltransferase 3 (NMNAT3) is an enzyme that is responsible for mitochondrial NAD+ synthesis,27 which appears to be susceptible to aging. Restoration of this enzyme was sufficient to have an effect on overcoming age-associated barriers.25
Recent studies strongly suggest that understating the metabolic state of tumor-initiating cells (TICs) is crucial to improve therapeutic and diagnostic opportunity in tumor research and to potentially open novel avenues for anti-tumor therapies.28, 29, 30 In normal developmental process, proliferating neural progenitors mainly utilize glycolysis for their energy production upon activation of quiescent adult neural stem cells and then switch the metabolic state into mitochondrial oxidative phosphorylation upon differentiation.31, 32 However, unlike controlled developmental process, unprogrammed and heterogeneous tumor cells exhibit a survival-prone metabolic plasticity, which is able to adapt the fluctuating metabolic microenvironment of the tumor.33 The TIC-specific energy metabolism, which distinguishes them from the non-TICs, is still controversial, accompanied by the issue of their quiescence/frequency.30, 34, 35, 36 Thus, the dual blockade of the bioenergetics of TICs, as the study shows with glycolysis and oxidative phosphorylation inhibitors, could be more effective in the treatment of tumor initiation and progression.37 More importantly, although the maintenance of mitochondrial function and cellular NAD+ levels is critical in redox homeostasis and metabolic integrity in TICs,38 the coupling between NAD+ status and tumor-initiating capacity of TICs have not yet been elucidated. In the present study, we sought to determine whether the mitochondrial NAD+ levels could influence the fundamental features of TICs. We used a neural stem/progenitor cell marker, stage-specific embryonic antigen 139 (SSEA1/CD15)+, which we previously identified as an enrichment marker for TICs in human glioblastoma, cells as a TIC population.40 The frequencies of SSEA1+ cells in freshly isolated patient samples are quite variable between 2.4 and 70.5% that 8 out of 12 samples had over 10% for SSEA1 expression, indicating SSEA1+ cells not as a rare subpopulation and a general enrichment marker for glioma TICs.40 SSEA1+ cells are highly clonogenic and proliferative compared to their SSEA1− counterpart.40 In this context, we found that the NNT overexpression-mediated increase of mitochondrial NAD+ levels negatively supported SSEA1+ TIC maintenance, which was evidenced by the decreased in vitro sphere formation and in vivo tumorigenic potential of glioma TICs. The knockdown of NNT facilitated TIC maintenance, which was supported by improved in vitro sphere formation and in vivo tumorigenic potential of glioma TICs. Our data suggest that mitochondrial NAD+ metabolism is crucial in the regulation of TIC characteristics, and maintenance of mitochondrial fitness as a youthful metabolic state can impair the clonogenicity of SSEA1+ glioma TICs.
Materials and methods
Cell culture and reagents
U373MG glioma cells were obtained from the Korean Collection for Type Culture (Jeollabuk-do, Korea), cultured in Neurobasal media (Invitrogen, Grand Island, NY, USA) and supplemented with N2 and B27 supplements (0.5 × each; Invitrogen), human recombinant basic fibroblast growth factor and epidermal growth factor (25 ng ml−1 each; R&D Systems, Minneapolis, MN, USA), and 1% penicillin/streptomycin (Invitrogen). Nicotinamide and FK866 were purchased from Sigma-Aldrich (Saint Louis, MO, USA) and Cayman Chemical (Ann Arbor, MI, USA), respectively. Short interfering RNA and Dharmafect I transfection reagent used for NNT or NMNAT3 knockdown were purchased from Dharmacon (Lafayette, CO, USA).
Sorting of TICs
Spheres were dissociated into single-cell suspensions, incubated with anti-SSEA1 (CD15) microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) for 15 min at 4 °C, and then immunomagnetic sorted by using the magnetic-activated cell sorting (MACS) separation system (Miltenyi Biotec) according to the manufacturer's instructions. Two separation columns were consecutively used for ensuring high purity of sorting, and the purity of sorted populations was confirmed by a BD Accuri C6 flow cytometer (BD, Franklin Lakes, NJ, USA).
Soft-agar colony-forming assay
The bottom agar layer (1%) was made by mixing boiled low-melting point agar (Lonza, Rockland, ME, USA; 4% in phosphate-buffered saline (Invitrogen)) with culture media; 2 ml of the mixture were poured in each well of a six-well plate and allowed to solidify at room temperature (RT) for 20 min. The top agar layer (0.4%) was prepared by mixing 0.8% agar and 1 × 105 cells in culture media at a 1:1 ratio; this mixture was placed on top of the bottom agar layer. Plates were incubated at RT for 10 min to solidify the agar and then incubated at 37 °C for 3–4 weeks until colonies were formed. Media with growth factors (10 ×, 200 μl) were added twice per week, and the total number of colonies in the six-well plates were counted at the time of quantification.
Limiting dilution assay
Single-cell suspensions of each group were seeded into 96-well plates with various densities (5, 10, 20 and 50 cells per well). More than 60 wells were prepared for each condition and incubated at 37 °C for 3–4 weeks until colonies were formed. Images of colonies were obtained using an inverted microscope (Olympus, Tokyo, Japan) and the number of wells with/without spheres was counted. Sphere size was measured by diameter of spheres in wells (initial seeding number=50 cells) at the end point of the limiting dilution analysis (4-week incubation). Serial secondary sphere-forming assay was performed. The primary spheres were passaged and then re-treated each condition.
Cell fractionation
Mitochondria, nuclear and cytosolic fractions of cells were separated using a cell fractionation kit following the manufacturer’s protocols (Abcam, Cambridge, UK). Protein concentrations of total or each fraction were determined using a BCA protein assay kit (Bio-Rad, Hercules, CA, USA).
NAD assay
NAD levels were determined using the NAD/NADH assay kit according to the manufacturer’s instructions (Abcam). Briefly, 10 μg of lysate was used for each enzyme cycling reaction and incubated at RT for 1–2 h. The color was measured at 450 nm every hour using a SpectraMax microplate reader (Molecular Devices, Sunnyvale, CA, USA). NAD contents were calculated by subtracting NADH contents from the total NAD and NADH.
Western blot analysis
Sorted each populations were lysed with RIPA buffer (Thermo Fisher Scientific, Waltham, MA, USA)/protease inhibitor cocktail (Roche, Basel, Switzerland) by vortexing shortly and then incubated for 5 min on ice as previously described.41 Lysates were centrifuged at 20 000 g for 15 min at 4 °C. Protein concentrations of cell lysates were determined using a BCA protein assay kit (Bio-Rad). Lysates (20 μg) were electroseparated using Mini-PROTEAN TGX Gels (Bio-Rad) and transferred by Trans-Blot Turbo Mini PVDF Transfer Packs (Bio-Rad), using a Trans-Blot Turbo Transfer System (Bio-Rad). The membranes were blocked with 5% skim milk (BD) at RT for 2 h and blotted with specific primary antibodies at 4 °C for overnight and then incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology, Dallas, Texas, USA). The membranes were washed using 0.1% Tween-20 (Sigma, St Louis, MO, USA) in phosphate-buffered saline during each step. The signals were detected by LAS-3000 (Fujifilm, Minato, Tokyo, Japan) after incubation of a SuperSignal West Femto Chemiluminescent Substrate (Thermo Fisher Scientific) solution. The antibodies used in this experiments are listed in Supplementary Table S1.
SIRT3 activity assay
SIRT3 activity was determined using a SIRT3 fluorometric drug discovery kit (Enzo Life Sciences, Farmingdale, NY, USA) according to the manufacturer’s protocol. Mitochondrial fractions (0.2 μg) was reacted with Fluor de Lys-SIRT2 substrate at 37 °C for 10 min and then stopped by incubation with developer/nicotinamide at 37 °C for 45 min. Fluorescence was measured at 37 °C, 460 nm at 5-min intervals using a SpectraMax microplate reader (Molecular Devices) for 1 h.
Lactate assays
Lactate production was quantified using the Lactate Assay Kit (BioVision, Milpitas, CA, USA) following the manufacturer’s protocol. Ten micrograms of protein were incubated with the lactate enzyme mix at RT for 30 min in the dark, and then the color was measured at 450 nm using a SpectraMax microplate reader (Molecular Devices).
RNA isolation and real-time PCR
Total RNA was isolated using an RNeasy Mini kit (Qiagen, Valencia, CA, USA) following the manufacturer’s protocol. Reverse transcription was performed with a SuperScript First-strand Synthesis System kit (Invitrogen). Quantitative real-time PCR was performed by a 7500 Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) using Fast SYBR Green Master Mix (Applied Biosystems) as previously described.42 The primer sequences used in this study are presented in Supplementary Table S2.
Virus production
GP2-293 cells (Clontech, Mountain View, CA, USA) were transfected with pMX vectors containing NNT or NMNAT3 (OriGene, Rockville, MD, USA) and the VSV-G envelope gene using Lipofectamine 2000 transfection reagent (Invitrogen). The supernatants collected at 2–5 days after transfection were filtered using a 0.45-μm pore size filter (Millipore, Billerica, MA, USA) and concentrated using ultracentrifugation (Beckman Coulter, Brea, CA, USA) at 25 000 r.p.m. (rotor: SW32Ti) for 90 min. Titration was measured by quantifying green fluorescent protein-transduced cells using an IncuCyte live-cell analysis system (EssenBioscience, Ann Arbor, MI, USA).
Overexpression or knockdown experiments
For overexpression experiments, U373MG SSEA1+ cells were sorted and pre-incubated for 1 h with 8 μg ml−1 of Polybrene (Sigma) and then transduced with concentrated NNT or NMNAT3 retroviruses at a multiplicity of infection of 5 in the presence of Polybrene (8 μg ml−1). For knockdown experiments, sorted U373MG SSEA1+ cells were transfected with short interfering RNA for NNT (siNNT) or NMNAT3 (siNMNAT3) using Dharmafect I transfection reagent (Dharmacon) according to the manufacturer’s instructions.
Oxygen consumption measurement and ATP assays
U373MG SSEA1+ cells were transduced with NNT retroviruses or transfected with siNNT as described above. After 72 h, cells were dissociated and seeded at 4.5 × 105 cells per well in black wells of 96-well plates (Thermo Scientific). One micromole of MitoXpress Xtra HS (Luxcel Biosciences, Cork, Ireland) was added and then overlaid by mineral oil at 30 °C. Fluorescence was measured using a SpectraMax microplate reader (Molecular Devices) at 30 °C. ATP was measured using an ATP assay mix solution (Sigma) according to the manufacturer’s instructions. One mirogram of total protein was used for each reaction in white well and luminescence intensity was measured by a SpectraMax microplate reader (Molecular Devices).
Orthotopic and subcutaneous xenograft models
All animal experimental protocols were approved by the bioethics committee of KRIBB. For intracranial orthotopic xenograft model, single-cell suspension of control or siNNT-transfected U373MG SSEA1+ cells were prepared and injected stereotactically into the striatum of 10-week-old Balb/c-Nude mice (Dae han BioLink, Chungbuk, Korea). One million cells were injected by using Hamilton syringe (Sigma) with stereotactic device (coordinates, 2 mm anterior, 2 mm lateral, 2.5 mm depth from the dura). Six weeks after injection, the brains were perfused with 4% paraformaldehyde by cardiac perfusion and further fixed at 4 °C overnight. For subcutaneous xenograft model, single-cell suspension of 5 × 106 control, retrovial NNT-transduced or siNNT-transfected U373MG SSEA1+ cells were mixed with Matrigel (BD) and injected into the flank of 7-week-old Balb/c-Nude mice (Dae han BioLink). Tumor volume was measured using caliper daily and the tumors were excised at 6 weeks after injection and then fixed with 4% paraformaldehyde.
Immunohisto/cytochemistry
For immunohistochemistry, fixed brains were immersed in 30% sucrose and then embed in OCT compound (Sakura Finetek USA Inc, Torrance, CA, USA). Frozen sections were stained with hematoxylin and eosin or with respective antibodies. For immunocytochemistry, cells were fixed with 4% paraformaldehyde, permeabilized 0.1% Triton X-100 (Sigma) in phosphate-buffered saline and blocked with 4% bovine serum albumin (Sigma) for 2 h at RT. The samples were stained with respective primary antibodies diluted in blocking buffer at 4 °C and washed with 0.05% Tween-20 (Sigma) in phosphate-buffered saline and then incubated with Alexa Fluor-conjugated secondary antibodies (Thermo Fisher Scientific) for 1 h at RT. Florescence images were captured with an Olympus microscope or Axiovert 200M microscope (Carl Zeiss, Oberkochen, Germany). The antibodies used are listed in Supplementary Table S1.
Statistical analysis
The data are each representative of at least three independent biological replicates. The graphs present the means±s.e.’s of quadruplicate samples for the soft-agar colony-forming assay, triplicate samples for the limiting dilution assay and PCR analysis, and duplicate samples for the NAD, the SIRT3, and the lactate assay from one representative experiment (technical replicates). Student’s t-test was used to evaluate inter-group comparisons, and a value of P<0.05 was considered statistically significant.
Results
TICs represent enhanced SIRT3 activity and lactate production
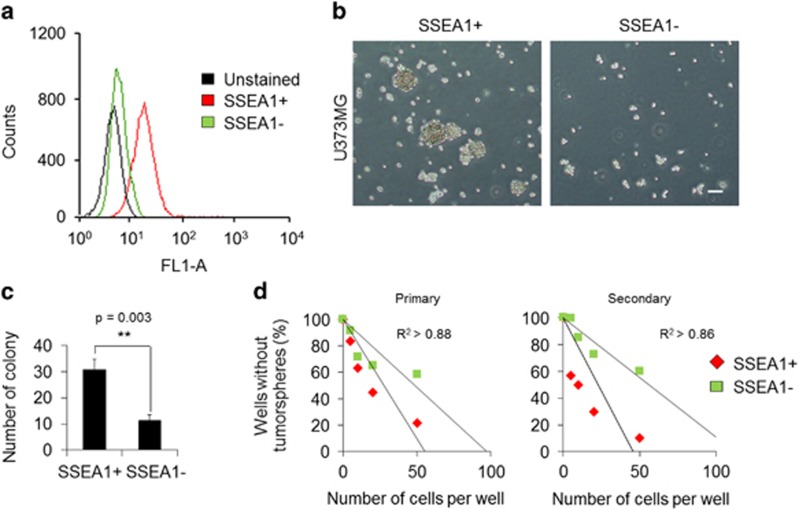
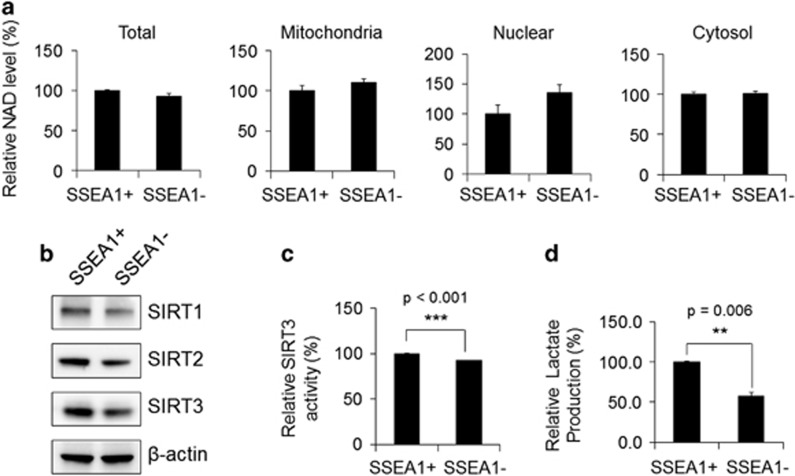
To understand the role of NAD+ metabolism in TICs, SSEA1+ TIC populations40 were sorted from U373MG glioma cells. The purity rate of the SSEA1+ cell population was >96%, which was confirmed by flow cytometry (Figure 1a). As important features of TICs, the SSEA1+ cell population, but not SSEA1− cells, displayed sphere-forming capacity 3 days after cell sorting (Figure 1b) and exhibited higher clonogenic potential, which was confirmed by a standard soft-agar colony-forming assay (Figure 1c) and a limiting dilution assay (Figure 1d). After successive culture, the secondary sphere-forming ability was clearly distinguished between the two populations (Figure 1d). We then primarily determined the NAD+ availability in SSEA1+ and SSEA1− populations and found no significant differences in total and subcellular (mitochondrial, nuclear and cytosol) NAD+ levels between the two populations (Figure 2a). As NAD+ acts as a rate-limiting co-substrate for SIRTs, we further confirmed the involvement of SIRTs in TICs. SIRT1 (nuclear), SIRT2 (cytosolic) and SIRT3 (mitochondria) expression was clearly decreased in SSEA1− population (Figure 2b; Supplementary Figure S1a). Notably, the activity of SIRT3 (a major mitochondrial NAD+-dependent deacetylase) was higher in the SSEA1+ TICs than in the SSEA1− non-TICs (Figure 2c). In addition, the production of lactate (end product of glycolysis) was significantly higher in the SSEA1+ TICs than in SSEA1− non-TICs, suggesting higher glycolytic activity in SSEA1+ TICs (Figure 2d).
Figure 1.
The SSEA1+ population from U373MG glioma cells was more clonogenic than the SSEA1− population. (a) The purity of the SSEA1+ population sorted by immunomagnetic selection was confirmed by flow cytometer. (b) Representative cell morphology of each population three days after cell sorting is shown. (c) Total numbers of colonies were determined in SSEA1+ versus SSEA1− populations using the soft-agar colony-forming assay. (d) Numbers of wells without tumor spheres were quantified in SSEA1+ versus SSEA1− populations using the limiting dilution sphere-forming assay (primary). Serial secondary sphere forming assay was performed (secondary). **P<0.01 (Student’s t-test). Scale bar=50 μm.
Figure 2.
Nicotinamide adenine dinucleotide (NAD+) levels, SIRT3 expression and activity, and lactate production in SSEA1+ and SSEA1− populations from U373MG glioma cells are shown. (a) SSEA1+ and SSEA1− populations were sorted from U373MG glioma cells (total, unfractionated) and then fractionated into mitochondrial, nuclear and cytosolic fractions. The NAD+ levels in each lysate were quantified. (b) Western blot analysis of SIRT1, 2 and 3 in total protein of each subpopulations. β-Actin was used as an internal control. (c) SIRT3 enzyme activity and (d) lactate contents in each population were determined. **P<0.01; ***P<0.001 (Student’s t-test).
NAD+ depletion is fatal in maintaining the clonogenicity
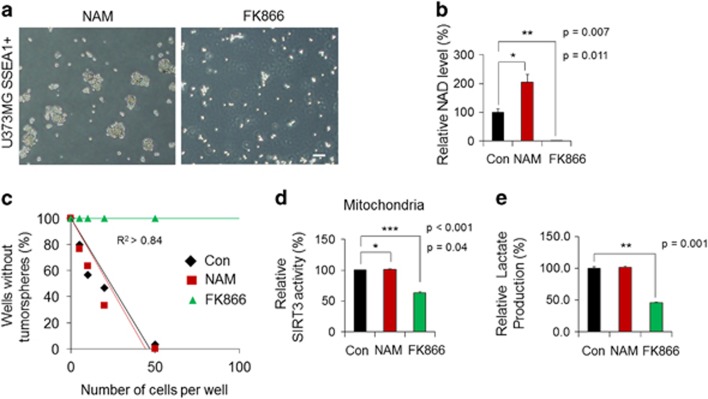
To further investigate the contribution of NAD+ content to TIC identity or properties, we examined the effects of nicotinamide (NAM, a precursor of NAD+: 1 mM) and FK866 (a pharmacological inhibitor of nicotinamide phosphoribosyltransferase, the rate-limiting enzyme in NAD+ salvage biosynthesis: 0.1 nM) in SSEA1+ TICs (Figure 3a). Only 48 h of treatment with FK866 was able to completely deplete intracellular NAD+ levels in SSEA1+ TICs (Figure 3b). The sphere-forming ability of SSEA1+ TICs was completely abolished by FK866-mediated NAD+ depletion; however, increasing NAD+ levels with NAM did not notably affect the sphere-forming ability of SSEA1+ TICs in limiting dilution analysis (Figure 3c) and the secondary sphere-forming ability also was not changed by addition of NAM (Supplementary Figure S1b). As expected, SIRT3 activity (Figure 3d) and lactate production (Figure 3e) significantly decreased as a result of FK866-mediated NAD+ depletion. (Figure 3b). In the case of SSEA1− population, a substantial NAD+ decrease by FK866 treatment (Supplementary Figure S1c and d) and followed abolition of sphere formation (Supplementary Figure S1e), reduction of SIRT3 activity (Supplementary Figure S1f) and lactate production (Supplementary Figure S1g) was also detected. However, increased NAD+ levels (Supplementary Figure S1d) and SIRT3 activity (Supplementary Figure S1f) with the addition of NAM in SSEA1− non-TICs exhibited rather a decreased sphere-forming ability (Supplementary Figure S1e). These results suggest that maintaining a proper level of intracellular NAD+ content is essential for the identity and clonogenicity of TICs.
Figure 3.
Adequate intracellular nicotinamide adenine dinucleotide (NAD+) levels are required to maintain the identity of tumor-initiating cells. (a) U373MG SSEA1+ cells were sorted and then incubated with 1 mM of nicotinamide (NAM; NAD+ precursor) or 0.1 nM of FK866 (inhibitor of NAD+ biosynthesis). Representative cell images at 48 h after treatment are shown. (b) The NAD+ levels in each lysate were quantified. (c) Clonogenicity was determined in each condition using limiting dilution analysis. (d) SIRT3 enzyme activity in the mitochondrial fraction and (e) lactate production in each condition were determined. *P<0.05; **P<0.01; ***P<0.001 (Student’s t-test). Scale bar=50 μm.
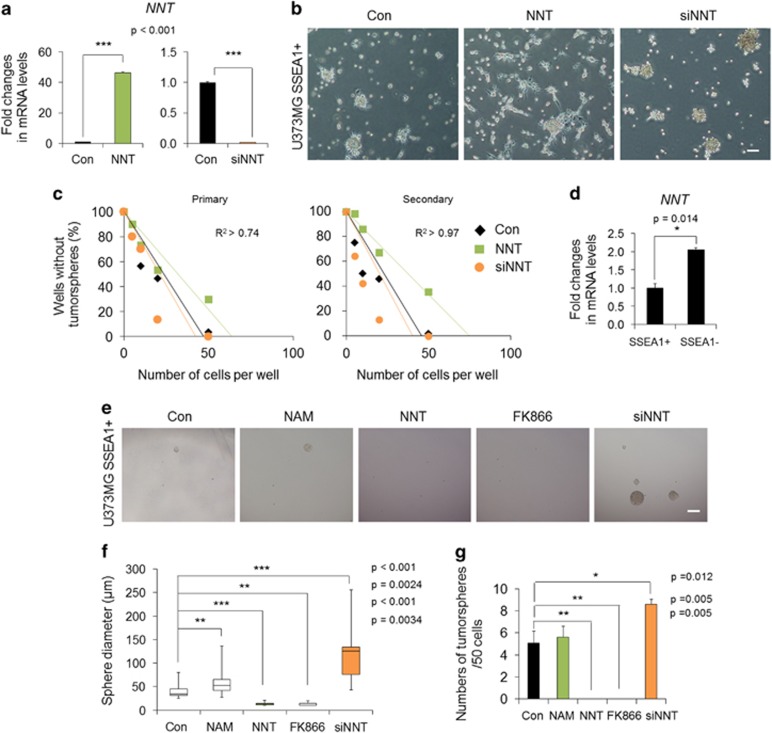
NNT or NMNAT3 overexpression impair the sphere-forming ability of TICs
Previously, we found that mitochondrial enzymes, namely, NNT and NMNAT3, can delay the senescence of stem cells via a restoration of mitochondrial NAD+ levels.25 Thus, we further questioned whether regulation of mitochondrial NAD+ levels mediated by NNT and NMNAT3 can influence the identity or properties of TICs. To examine the influences of NNT in TICs, we overexpressed and knocked down NNT in SSEA1+ TICs (Figure 4a). Importantly, NNT-overexpressed cells underwent a morphological change to an attached and differentiated phenotype (Figure 4b) and displayed decreased sphere-forming ability in limiting dilution analysis (Figure 4c), suggesting loss of TIC characteristics. After successive culture, the secondary sphere-forming ability was much less in NNT-transduced cells (Figure 4c). We confirmed that these effects were not due to the retroviral transduction, as demonstrated by transduction with retroviral green fluorescent protein as a control (data not shown). Interestingly, the basal expression of NNT was about twofold higher in the SSEA1− non-TICs compared to SSEA1+ TICs (Figure 4d), and NNT-overexpressed SSEA1− cells hardly formed tumor spheres (Supplementary Figure S2a–c). Meanwhile, NNT knockdown significantly increased the sphere-forming ability of SSEA1+ and SSEA1− cells in limiting dilution analysis (Figure 4c; Supplementary Figure S2c). More importantly, sphere size was markedly larger in NNT short interfering RNA-treated cells than in controls at the end point of limiting dilution analysis (4-week incubation; Figure 4e and f; Supplementary Figure S2d and e) as well as sphere number increase (Figure 4g; Supplementary Figure S2f). Similar to NNT-overexpressed cells, NMNAT3-overexpressed cells displayed an attached and differentiated phenotype (Supplementary Figure S3a), whereas NMNAT3 knockdown strongly increased the sphere-forming ability and sphere size (Supplementary Figure S3b and c).
Figure 4.
The effect of nicotinamide nucleotide transhydrogenase (NNT) overexpression or knockdown on clonogenicity of SSEA1+ tumor-initiating cells (TICs) is shown. (a) U373MG SSEA1+ cells were sorted and then transduced with retroviral NNT or transfected with short interfering RNA for NNT (siNNT). Overexpression or knockdown of NNT was confirmed by real-time PCR analysis. (b) Representative morphology of each population at 3 days after treatment is shown. Scale bar=50 μm. (c) Clonogenicity of each condition was compared using primary and secondary limiting dilution analysis. (d) Basal expression of NNT in each subpopulation was examined by real-time PCR analysis. (e) Representative sphere images, (f) the diameter of spheres, (g) sphere numbers per well (initial seeding number=50 cells) in each condition were quantitated at the end point of the limiting dilution analysis (4-week incubation). Box plots indicate the median and 25th and 75th percentiles. Whiskers indicate the minimum and maximum values. Nicotinamide (NAM, 1 mM) and FK866 (0.1 nM) were used as the comparison groups. *P<0.05; **P<0.01; ***P<0.001 (Student’s t-test). Scale bar=200 μm.
NNT overexpression abolishes tumorigenic potential of SSEA1+ TICs
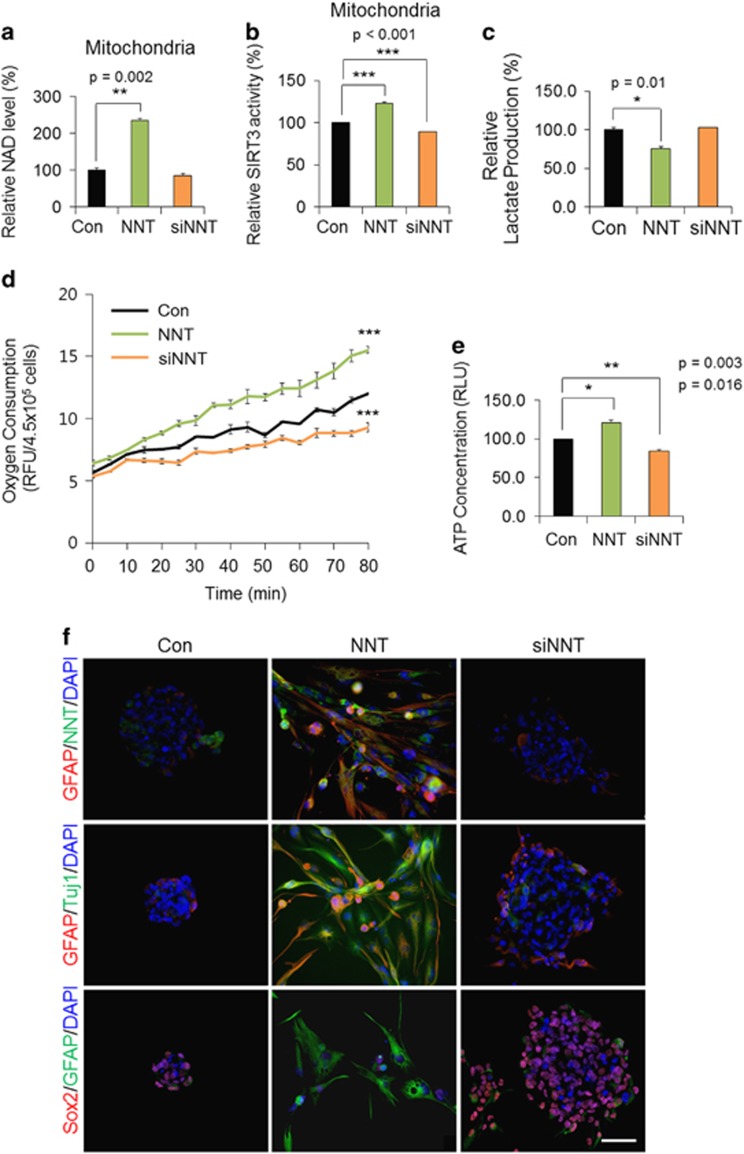
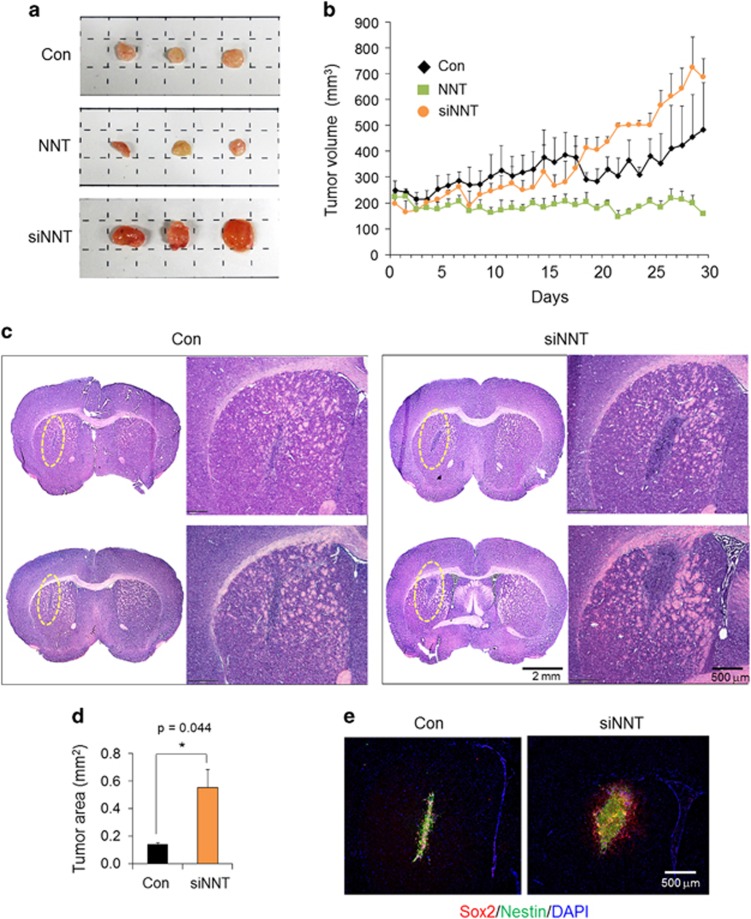
Significantly, NNT overexpression in SSEA1+ TICs induced prominent increases in mitochondrial NAD+ levels (Figure 5a) and SIRT3 activity (Figure 5b), while lactate production was clearly decreased (Figure 5c). In consistent with this reduced glycolytic capacity, oxygen consumption (Figure 5d) and ATP production (Figure 5e) were increased by NNT overexpression, suggesting the mitochondria-dependent metabolic change upon NNT overexpression. Moreover, NNT-overexpressed SSEA1+ TICs showed robust NNT expression in mitochondria and preferentially displayed differentiation, resulting in an attached and spread-out morphology and increased expressions of differentiation markers such as GFAP, a glial marker and Tuj1, a neuronal marker (Figure 5f). Expressions of neural stem cell markers, namely, Sox2, Nestin, and SSEA1, were concurrently decreased by NNT overexpression in TICs (Figure 5f and Supplementary Figure S4). Finally, in vivo tumorigenic potential of SSEA1+ TICs was prominently inhibited by NNT overexpression (Figure 6). In a subcutaneous xenograft model, NNT-overexpressed TIC-injected mice showed substantial delay of tumor growth, whereas control cell-injected mice showed continuous tumor growth (Figure 6a and b; Supplementary Figure S5). Meanwhile, NNT-knockdowned TICs showed a remarkable increase in tumor growth both in subcutaneous (Figure 6a and b) and orthotopic xenograft model (Figure 6c and d). Human cell-specific Nestin- and Sox2-stained tumor cells were clearly discrete in injected mouse brain (Figure 6e). Collectively, these data demonstrate that NNT knockdown enriches the tumorigenic potential of SSEA1+ TICs and boosted mitochondrial NAD+ levels and SIRT3 activity over the level observed in SSEA1+ TICs by NNT overexpression is capable of preventing the tumorigenic potential of TICs via the induction of differentiation (Figure 7; Supplementary Figure S6).
Figure 5.
The effect of nicotinamide nucleotide transhydrogenase (NNT) overexpression or knockdown in SSEA1+ tumor-initiating cells is shown. Mitochondria were fractionated from retroviral NNT-overexpressed or siNNT-transfected U373MG SSEA1+ cells, and (a) nicotinamide adenine dinucleotide (NAD+) levels and (b) SIRT3 enzyme activity were determined at 3 days after treatment. (c) Lactate production was examined in the total lysate in each condition. (d) Each condition at 3 days after culture, the oxygen consumption was measured in over 80 min using an oxygen probe and (e) the ATP concentration in cells of each group was determined. (f) Immunocytochemistry of each condition was fixed at 7 days after treatment. Antibodies used in each condition are shown on the left. 4,6-Diamidino-2-phenylindole (DAPI) staining (blue) was used to identify nuclei.
Figure 6.
The tumorigenic potential of nicotinamide nucleotide transhydrogenase (NNT) overexpression or knockdown in SSEA1+ tumor-initiating cells is shown. (a) Control (Con), retroviral NNT-transduced or short interfering RNA for NNT (siNNT)-transfected U373MG SSEA1+ cells were injected subcutaneously and tumors were excised at 6 weeks after injection. Representative images of excised tumor mass in each group were shown. (b) Tumor volume was measured daily after tumor mass detected. (c) Intracranial orthotopic injection of control or siNNT-transfected U373MG SSEA1+ cells. Representative hematoxylin and eosin images of each two mice brain (upper and lower) and higher magnification of each section (each right in group). (d) Tumor area was quantified. (e) Immunohistochemistry of Sox2 and Nestin in each brain. 4,6-Diamidino-2-phenylindole (DAPI) staining (blue) was used to identify nuclei. *P<0.05; **P<0.01; ***P<0.001 (Student’s t-test).
Figure 7.
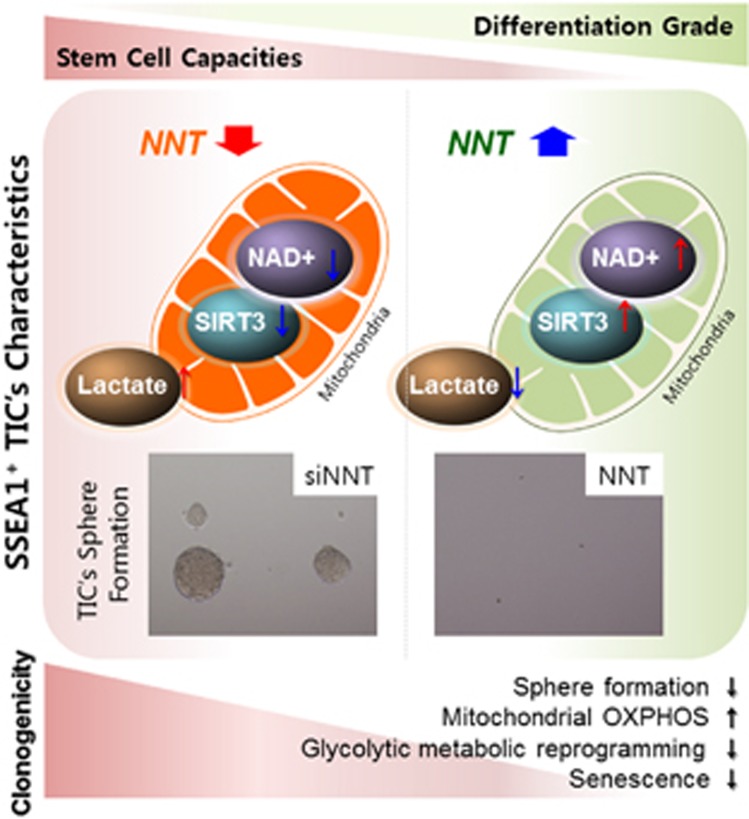
A proposed model for the importance of the maintenance of mitochondrial fitness in the prevention of tumor initiation is shown. An increase of mitochondrial NAD+ level with NNT overexpression impairs SSEA1+ tumor-initiating cell’s (TIC’s) characteristics such as sphere-forming ability and glycolytic metabolic reprogramming to produce lactate.
Discussion
Maintenance of mitochondrial fitness as a youthful metabolic state confers advantages for the functional continuation of the healthy condition22, 24 by maintaining the number, self-renewal/differentiation capacity and quality of stem cells, which can possibly alleviate disease conditions and even aging.20, 43, 44 In the present study, when we upregulated mitochondrial NAD+ levels and SIRT3 activity of U373MG glioma-driven SSEA1+ TICs via NNT overexpression, differentiation occurred, and the clonogenicity of TICs was distinctly prevented, representing a loss of TIC characteristics. Recently, a ‘geroncogenesis’ hypothesis has been proposed that certain conditions that are bioenergetically cancer-prone, such as metabolic reprogramming toward glycolysis, have been developed during aging, contributing to tumor initiation.45 Previously, we have shown that the knockdown of NNT in human mesenchymal stem cells induced cellular senescence, whereas restoration of mitochondrial NAD+ levels through methods such as NNT overexpression delayed the replicative senescence of stem cells.25 Our results do not directly support ‘geroncogenesis’ however, it is obvious, at least in part, that maintaining mitochondria in a similar state with healthy cells by replenishing NAD+ decreases the tumorigenicity of TICs as well as delaying stem cell senescence (Figure 7; Supplementary Figure S6). Therefore, the mitochondrial NAD+ level increase may be a safe and beneficial method for maintaining a healthy condition in normal cells, with delayed senescence and no risk of tumor initiation.
Boosting mitochondrial NAD+ improves the adaptive cellular response under stress associated with energy status, and increases the oxidative phosphorylation-dependent ATP synthesis (Figure 5d and e), fatty acid metabolism and mitochondria biogenesis by PGC1α induction.22, 24, 46 In addition, these changes toward mitochondria-dependent metabolism with the concurrent reduction of lactate are closely correlated with the transition of the cell from a proliferative to a differentiated state.47 There are controversial reports that quiescent or slow cycling-glioma stem cells36 and -leukemia stem cells48 primarily depended on mitochondrial oxidative phosphorylation.30 However, highly proliferative cells such as SSEA1+ TICs and pluripotent stem cells prefer to undergo glycolysis for their energy production, which provides favorable conditions for the production of the biomass needed for rapid cell division and avoidance of mitochondrial oxidative stress.49 Consistent high rates of glycolysis reduce cellular NAD+ contents,43 resulting in a loss of cellular metabolic integrity by the disturbance of redox homeostasis and further contribution to tumor progression.50, 51 More importantly, intramitochondrial NAD+ levels can reach mM concentrations, whereas nuclear NAD+ levels are measured in μM concentrations.1 In this respect, NAD+ concentrations could limit activities of mitochondrial SIRTs,1, 52 whereby mitochondrial NAD+-dependent signaling most prominently involved in SIRT3 activity is devastated by the depletion of total cellular NAD+. SIRT3 has demonstrated its role as a context-dependent tumor suppressive or promotive factor,53, 54 whereas in brain tumors such as glioblastomas55 and medulloblastomas,56 SIRT3 has been reported to act as a tumor suppressor. SIRT3 could prevent metabolic reprogramming of cancer cells by hypoxia-inducible factor 1α destabilization, which works through reducing the overproduction of reactive oxygen species56 and mice that lack SIRT3 are cancer-prone.55 Furthermore, SIRT3 has been observed to be associated with myoblast differentiation57 and brown adipocyte differentiation.58 SIRT1 suppressed the self-renewal of adult hippocampal neural stem cells59 and promoted astrogenesis.60 In our present data, a differentiated phenotype was exhibited following an increase of mitochondrial NAD+ levels and SIRT3 activity in NNT-overexpressed TICs of glioma cells (Figures 4 and 5). Thus, it is possible that neuronal lineage differentiation might be regulated by SIRT3.
Although there is a limitation when using SSEA1+ TICs isolated from a cell line rather than using cells sorted from freshly isolated patient tumor specimens, clonogenicity control upon overexpression or knockdown of NNT was similarly ascertained in CD133+, a well-known glioma stem cell marker,61 the populations of which were sorted from U373MG cells (data not shown). In addition, the effects of NNT expression on the clonogenic capacity of TICs were equivalently established by NMNAT3, another mitochondrial NAD+-synthesizing enzyme (Supplementary Figure S3). These results strongly suggest the inverse correlation between mitochondrial NAD+ levels and the clonogenicity of TICs, which may not be an SSEA1- or NNT-specific effect. Even in SSEA1− cells, the knockdown of NNT by short interfering RNA transfection facilitated the transition of non-TIC properties to tumor-initiating state, whereas NNT overexpression still counteracted to clonogenicity of SSEA1− cells (Supplementary Figure S2). Therefore, the maintenance of an appropriate level of mitochondrial NAD+ content may be critical to the prevention of tumor initiation.
Overall, it has been suggested that the metabolic control of TICs can be beneficial for the treatment of tumors in humans;62 however, the coupling between the NAD+ status and tumor-initiating capacity of TICs has currently not been well described. In the present study, we uncovered a specific role of mitochondrial NAD+ in SSEA1+ glioma TICs and targeting mitochondrial NAD+ levels was effective in treating tumor-initiating populations; especially, in the case of TICs with glycolytic and proliferative phenotype such as SSEA1+ subpopulation. Further precise and in-depth studies of the correlations among the perturbation and distribution of particular (cytosolic, nuclear or mitochondrial) pools of NAD+, aging and tumor initiation would provide mechanistic insight into tumorigenesis and finally contribute to the development of patient specific personalized therapeutic options deliberating metabolic state for treating aging-related diseases, including tumors.
Acknowledgments
This work was supported by the NRF grant (2012M3A9C7050224) and the NST grant (CRC-15-02-KRIBB) from the Korean government (MSIP); the KRIBB Initiative of the Korea Research Council of Fundamental Science and Technology; the Korea Health Technology R&D Project through the KHIDI, funded by the Ministry of Health & Welfare (HI16C0312); and the Korea Basic Science Institute research program (D36402).
Footnotes
Supplementary Information accompanies the paper on Experimental & Molecular Medicine website (http://www.nature.com/emm)
The authors declare no conflict of interest.
Supplementary Material
References
- Canto C, Menzies KJ, Auwerx J. NAD(+) metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab 2015; 22: 31–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Sauve AA. NAD+ metabolism: bioenergetics, signaling and manipulation for therapy. Biochim Biophys Acta 2016; 1864: 1787–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdin E. The many faces of sirtuins: coupling of NAD metabolism, sirtuins and lifespan. Nat Med 2014; 20: 25–27. [DOI] [PubMed] [Google Scholar]
- Schultz MB, Sinclair DA. Why NAD(+) declines during aging: it’s destroyed. Cell Metab 2016; 23: 965–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying W, Alano CC, Garnier P, Swanson RA. NAD+ as a metabolic link between DNA damage and cell death. J Neurosci Res 2005; 79: 216–223. [DOI] [PubMed] [Google Scholar]
- VanLinden MR, Dolle C, Pettersen IK, Kulikova VA, Niere M, Agrimi G et al. Subcellular distribution of NAD+ between cytosol and mitochondria determines the metabolic profile of human cells. J Biol Chem 2015; 290: 27644–27659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LR, Imai S. The dynamic regulation of NAD metabolism in mitochondria. Trends Endocrinol Metab 2012; 23: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lisa F, Menabo R, Canton M, Barile M, Bernardi P. Opening of the mitochondrial permeability transition pore causes depletion of mitochondrial and cytosolic NAD+ and is a causative event in the death of myocytes in postischemic reperfusion of the heart. J Biol Chem 2001; 276: 2571–2575. [DOI] [PubMed] [Google Scholar]
- Alano CC, Tran A, Tao R, Ying W, Karliner JS, Swanson RA. Differences among cell types in NAD(+) compartmentalization: a comparison of neurons, astrocytes, and cardiac myocytes. J Neurosci Res 2007; 85: 3378–3385. [DOI] [PubMed] [Google Scholar]
- Guarente L. The many faces of sirtuins: sirtuins and the Warburg effect. Nat Med 2014; 20: 24–25. [DOI] [PubMed] [Google Scholar]
- Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell 2008; 14: 312–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Luo J. SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell 2010; 39: 247–258. [DOI] [PubMed] [Google Scholar]
- Price NL, Gomes AP, Ling AJ, Duarte FV, Martin-Montalvo A, North BJ et al. SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metab 2012; 15: 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature 2005; 434: 113–118. [DOI] [PubMed] [Google Scholar]
- Satoh A, Brace CS, Rensing N, Cliften P, Wozniak DF, Herzog ED et al. Sirt1 extends life span and delays aging in mice through the regulation of Nk2 homeobox 1 in the DMH and LH. Cell Metab 2013; 18: 416–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R et al. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem 2009; 284: 20408–20417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhang F, Yan M, Wu D, Yu Q, Zhang Y et al. WldS enhances insulin transcription and secretion via a SIRT1-dependent pathway and improves glucose homeostasis. Diabetes 2011; 60: 3197–3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutant M, Canto C. SIRT1 metabolic actions: Integrating recent advances from mouse models. Mol Metab 2014; 3: 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E, Grueter CA, Collins AM, Aouizerat B et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol Cell 2011; 44: 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ryu D, Wu Y, Gariani K, Wang X, Luan P et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 2016; 352: 1436–1443. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Mitchell SJ, Fang EF, Iyama T, Ward T, Wang J et al. A high-fat diet and NAD(+) activate Sirt1 to rescue premature aging in cockayne syndrome. Cell Metab 2014; 20: 840–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchiroud L, Houtkooper RH, Moullan N, Katsyuba E, Ryu D, Canto C et al. The NAD(+)/sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell 2013; 154: 430–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes AP, Price NL, Ling AJ, Moslehi JJ, Montgomery MK, Rajman L et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell 2013; 155: 1624–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirinen E, Canto C, Jo YS, Morato L, Zhang H, Menzies KJ et al. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell Metab 2014; 19: 1034–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MJ, Kwon Y, Son T, Cho YS. Restoration of mitochondrial NAD+ levels delays stem cell senescence and facilitates reprogramming of aged somatic cells. Stem Cells 2016; 34: 2840–2851. [DOI] [PubMed] [Google Scholar]
- Lopert P, Patel M. Nicotinamide nucleotide transhydrogenase (Nnt) links the substrate requirement in brain mitochondria for hydrogen peroxide removal to the thioredoxin/peroxiredoxin (Trx/Prx) system. J Biol Chem 2014; 289: 15611–15620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka K, Ikutani M, Shito M, Kazuma K, Gulshan M, Nagai Y et al. Deficiency of nicotinamide mononucleotide adenylyltransferase 3 (nmnat3) causes hemolytic anemia by altering the glycolytic flow in mature erythrocytes. J Biol Chem 2014; 289: 14796–14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer 2016; 114: 1305–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res 2016; 18: 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E, Pajonk F. The metabolic state of cancer stem cells-a valid target for cancer therapy? Free Radic Biol Med 2015; 79: 264–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development 2013; 140: 2535–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, Martin-Villalba A. Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 2015; 17: 329–340. [DOI] [PubMed] [Google Scholar]
- Jang H, Yang J, Lee E, Cheong JH. Metabolism in embryonic and cancer stemness. Arch Pharm Res 2015; 38: 381–388. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhou Y, Shingu T, Feng L, Chen Z, Ogasawara M et al. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J Biol Chem 2011; 286: 32843–32853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Wu Q, Hitomi M, Rahim N, Kim Y, Sloan AE et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat Neurosci 2013; 16: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA 2011; 108: 16062–16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JH, Park ES, Liang J, Dennison JB, Tsavachidou D, Nguyen-Charles C et al. Dual inhibition of tumor energy pathway by 2-deoxyglucose and metformin is effective against a broad spectrum of preclinical cancer models. Mol Cancer Ther 2011; 10: 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab 2016; 23: 27–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capela A, Temple S. LeX is expressed by principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev Biol 2006; 291: 300–313. [DOI] [PubMed] [Google Scholar]
- Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell 2009; 4: 440–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MJ, Son MY, Seol B, Kim MJ, Yoo CH, Han MK et al. Nicotinamide overcomes pluripotency deficits and reprogramming barriers. Stem Cells 2013; 31: 1121–1135. [DOI] [PubMed] [Google Scholar]
- Son MJ, Jeong BR, Kwon Y, Cho YS. Interference with the mitochondrial bioenergetics fuels reprogramming to pluripotency via facilitation of the glycolytic transition. Int J Biochem Cell Biol 2013; 45: 2512–2518. [DOI] [PubMed] [Google Scholar]
- Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol 2014; 15: 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryall JG, Dell'Orso S, Derfoul A, Juan A, Zare H, Feng X et al. The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 2015; 16: 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LE, Gomes AP, Sinclair DA. Geroncogenesis: metabolic changes during aging as a driver of tumorigenesis. Cancer Cell 2014; 25: 12–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez RM, Fernandez AF, Fraga MF. Role of sirtuins in stem cell differentiation. Genes Cancer 2013; 4: 105–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 2013; 12: 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son MJ, Kwon Y, Son MY, Seol B, Choi HS, Ryu SW et al. Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency. Cell Death Differ 2015; 22: 1957–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikal AA. Intracellular coenzymes as natural biomarkers for metabolic activities and mitochondrial anomalies. Biomark Med 2010; 4: 241–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008; 7: 11–20. [DOI] [PubMed] [Google Scholar]
- Harvey AJ, Rathjen J, Gardner DK. Metaboloepigenetic regulation of pluripotent stem cells. Stem Cells Int 2016; 2016: 1816525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhazzazi TY, Kamarajan P, Verdin E, Kapila YL. SIRT3 and cancer: tumor promoter or suppressor? Biochim Biophys Acta 2011; 1816: 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Fu LL, Wen X, Wang XY, Liu J, Cheng Y et al. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis 2014; 5: e1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD et al. SIRT3 is a mitochondria-localized tumor suppressor required for maintenance of mitochondrial integrity and metabolism during stress. Cancer Cell 2010; 17: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1alpha destabilization. Cancer Cell 2011; 19: 416–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel Khalek W, Cortade F, Ollendorff V, Lapasset L, Tintignac L, Chabi B et al. SIRT3, a mitochondrial NAD(+)-dependent deacetylase, is involved in the regulation of myoblast differentiation. PLoS ONE 2014; 9: e114388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giralt A, Hondares E, Villena JA, Ribas F, Diaz-Delfin J, Giralt M et al. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J Biol Chem 2011; 286: 16958–16966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CY, Yao MJ, Zhai QW, Jiao JW, Yuan XB, Poo MM. SIRT1 suppresses self-renewal of adult hippocampal neural stem cells. Development 2014; 141: 4697–4709. [DOI] [PubMed] [Google Scholar]
- Wang W, Esbensen Y, Kunke D, Suganthan R, Rachek L, Bjoras M et al. Mitochondrial DNA damage level determines neural stem cell differentiation fate. J Neurosci 2011; 31: 9746–9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T et al. Identification of human brain tumour initiating cells. Nature 2004; 432: 396–401. [DOI] [PubMed] [Google Scholar]
- Pacini N, Borziani F. Cancer stem cell theory and the warburg effect, two sides of the same coin? Int J Mol Sci 2014; 15: 8893–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.