Abstract
Objective
Myeloproliferative disorders (MPD) are clonal hematopoietic diseases that include polycythemia vera (PV), essential thrombocytosis (ET) and primary myelofibrosis (PMF). Mutations in JAK2 are present in many MPD patients. Additional genomic abnormalities are not fully examined in MPD.
Materials and Methods
We used single-nucleotide polymorphism DNA microarray (SNP-chip) to analyze 43 patients with MPD (10 PV, 17 ET, and 16 PMF) for genomic aberrations.
Results
Genomic abnormalities were rare in ET. The region containing either RB (13q14) or NF1 (17q11) was deleted in four of the 16 PMF, especially PMF with no JAK2 mutations. All five cases of PV having homozygous JAK2V617F had loss of heterozygosity with normal copy number [uniparental disomy (UPD)] involving the gene. A subpopulation with 9p UPD was detected in 11 MPD (3 PV, 1 ET, 7 PMF). UPD at 1p was found in one PV and three PMF. A novel mutation of MPL (Y591D) which was involved in this UPD was found in 1 PV with JAK2 mutation. The other three cases of PMF with 1p UPD had point mutations of the MPL gene (a novel mutation, S240F, as well as, W515L).
Conclusion
Genomic abnormalities including 9p UPD/JAK2 point mutations, 1p UPD/MPL point mutations, deletions of RB1 are common alteration in MPD, especially in PMF.
Keywords: SNP-chip, uniparental disomy, MPL, NF1, RB1
Introduction
Myeloproliferative disorders (MPD) are clonal hematopoietic diseases that are classified, based on their clinical features, into subtypes that include polycythemia vera (PV), essential thrombocytosis (ET), and primary myelofibrosis (PMF) [1–3]. Recently, activating point mutations of JAK2 kinase in exon 14 (V617F) [4–8] and exon 12 [9] have been detected in MPDs. These alterations transform JAK2 into a constitutively active kinase, leading to dysregulated proliferation of hematopoietic cells. JAK2V617F is detected in greater than 90% of cases of PV and ~50% of cases of ET and PMF [4–8]. Further, a point mutation of the MPL gene (W515L) has been detected in PMF [10,11]. This point mutation also activates Jak2/Stat signal transduction and induces proliferation of hematopoietic cells and an MPD phenotype in murine models of disease [10].
Nevertheless, clonality analysis suggests that at least in some cases, the JAK2V617F allele is acquired as a secondary event [12]. Thus, further genetic analysis is required to understand the pathophysiology of JAK2V617F negative cases of ET and PMF, and to identify other acquired somatic mutations that contribute to disease pathogenesis in collaboration with JAK2V617F.
Recently, genomic microarray has been developed in which oligonucleotide probes corresponding to 250,000 – 1,000,000 chromosomal locations over the whole genome are placed on small glass plates. DNA extracted from cancer cells and normal cells are fragmented and labeled with fluorescence. The fluorescence labeled DNA fragments are hybridized on the microarray and amount of DNA hybridized to each probe is measure on the scanner, leading to quantification of alleleic dosage at each genomic site. This technique allows detection of subcytogenetic deletions and amplification. Further, new screening platforms using arrayed oligonucleotide probes that contain single nucleotide polymorphisms (SNP), so-called “SNP-chip”, have been developed and employed for analyzing the genetic abnormalities in cancers [13–15]. SNP-chip is able to distinguish each parental alleles at heterozygous site and sensitively detect alleleic imbalance at the sites. This technique allows detection of loss of heterozygosity with neutral allelic dosage [uniparental disomy (UPD)] that cannot be identified by conventional methods [14, 15]. We employed this technique for analysis of a set of JAK2V617F positive (24 cases) or negative (19 cases) MPDs to identify novel genetic abnormalities.
Materials and Methods
Clinical samples and DNA preparation
Forty-three patients with MPD, including 10 PV (all JAK2V617F positive), 17 ET including 7 JAK2V617F positive and 10 JAK2V617F negative cases, and 16 PMF including 10 JAK2V617F negative and 6 JAK2V617F positive cases were enrolled in this study after informed consents were obtained. DNA was extracted from neutrophils in the peripheral blood, using standard proteinase K-phenol-chloroform extraction method.
SNP-chip analysis
SNP-chip of GeneChip Human mapping 50k array XbaI 240 was used for this study (Affymetrix, Tokyo, Japan). Fragmentation and labeling of DNAs were performed using GeneChip resequencing kit (Affymetrix) according to the manufacturer’s protocols. Hybridzation, washing and signal detection were performed on a GeneChip Fluidics Station 400 and GeneChip scanner 3000 according to the manufacturer’s protocols (Affymetrix). The data were analyzed by the CNAG program as previously described [16].
In addition, we developed a new algorithm to compare the parental allele-specific gene dosages of the samples with non-matched control DNA as previously described [17], allowing us to measure gene dosage levels of each parental allele.
Allele specific PCR for JAK2 mutation
Allele specific PCR was performed according to the method previously reported to detect JAK2 V617F mutations [4]. All 43 MPD cases were examined using mutation specific primers and wild type specific primers for the JAK2 gene as previously reported [4]. All PCR products were electrophoresed in 2% agarose gels containing ethidium bromide. PCR bands were visualized under UV lights and photographed.
Quantitative genomic PCR
Quantitative genomic PCR was performed using real-time PCR technique as previously described. Gene dosage of the NF1 and RB was measured [14]. All primers used for the quantitative genomic PCR are listed in Supplement Table.
PCR and sequencing
All coding exons of FGR1, TIE1, and MPL genes were amplified from selected samples using the PCR technique. The primers used for amplification of the MPL gene are listed in the Supplement Table. The PCR conditions were as follows: 40 cycles of 94 °C, 30 sec for denaturing, 55 °C, 30 sec for annealing, and 73 °C, 30 sec for extension. All PCR products were electrophoresed in a 2% agarose gel; PCR bands were excised, and purified using Qiagen Gel extraction kit (Qiagen, Valencia, CA) according to the manufacturer’s protocols. Purified PCR bands were directly sequenced using the Big-dye sequence reaction (Applied Biosystems, Foster City, CA) and analyzed on an Autosequencer 3100 (Applied Biosystems). PCR products which had mutations of S204F-MPL (MPL exon 4), W515L (MPL exon 10), Y591D (MPL exon 12) as detected by direct sequencing were cloned into pGEM-T vectors (Promega) and transformed into E.coli competent cells. Plasmids were extracted from eight independent clones of each PCR product and their nucleotide sequences were determined.
Screening of normal DNA
DNA from one hundred normal volunteers and 42 MPD patients were screened for S204F and Y591D mutations of the MPL gene. PCR products of exon 4 of MPL were digested with AlwNI for the S204F mutation (wild-type is digested and mutant is not digested); PCR products of exon 12 of MPL were digested with BsiEI for the Y591D mutation (wild-type is not digested and mutant is digested). The treated PCR products were run in 2% agarose gel.
Expression of MPL in BaF/3 cells
Human full-length MPL cDNA was cloned into pMSCV vector (Clontech, Mountain View, CA) from normal human bone marrow cells using PCR. EGFP cDNA (Clontech) was ligated into the pMSCV vector as a marker protein driven by the pGK promoter. S204F, W515L, and Y591D mutations were introduced into pMSCV-EGFP-hMPL vector by the PCR mutagenesis method. Expression of wild-type and mutant MPL was driven by LTR. The constructs were electroporated into the murine IL3-dependent B-cell line, BaF/3, and cultured in media containing IL3 for 2 days. GFP positive cells were selected by FACS and cultured in media without IL3. Cell numbers were counted on days 2, 4, and 6.
Results
Chromosomal duplications and deletions in MPDs
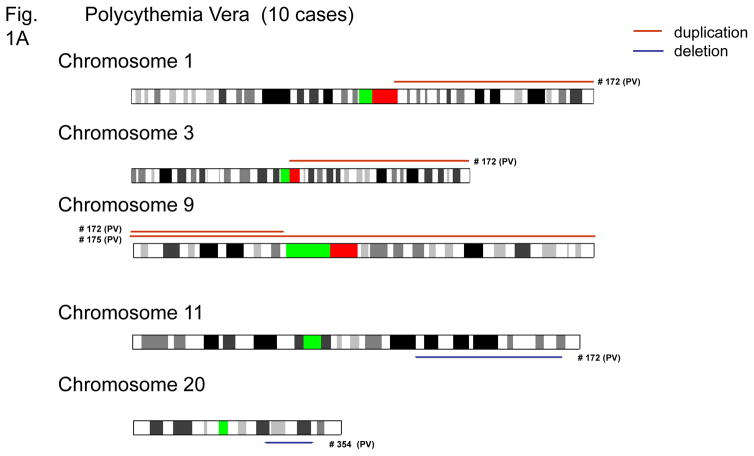
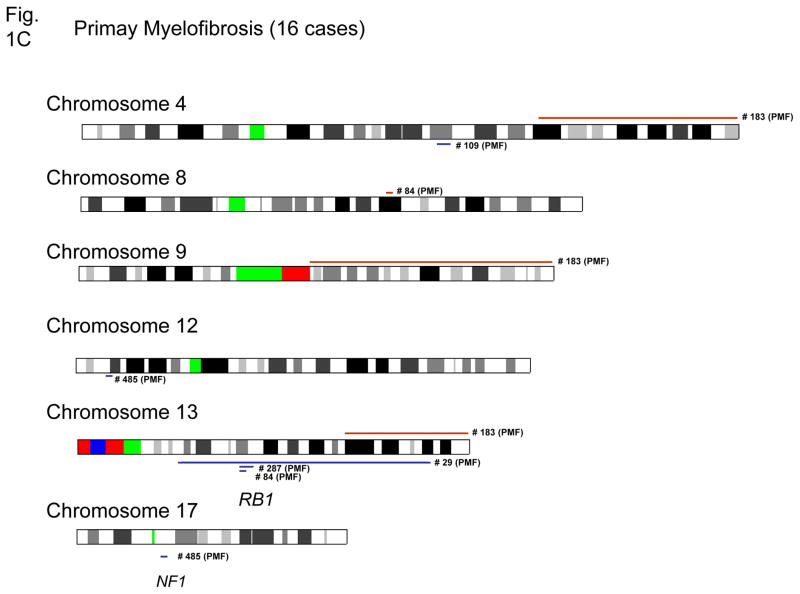
Genetic abnormalities, including deletions and duplications are shown in Figures 1A–C and summarized in Table 1. Gain of genomic materials was detected in five of 43 MPD; loss of genomic materials was detected in nine of 43 MPD cases. Trisomy 9 was detected in one PV and one ET; duplication of 9p was detected in one PV; duplication of 9q21.11-qter was detected in one PMF.
Figure 1. Chromosomal duplications and deletions in MPD samples as detected by SNP-chip.
Duplications are indicated as brown lines above the chromosomes; deletions are displayed as blue lines under the chromosomes. Case numbers are shown to the right of the chromosomes. Results of polycythemia vera (A), essential thrombocytosis (B) and myelofibrosis with myeloid metaplasia (C) are shown.
Table 1.
Genomic abnormalities detected by SNP-Chip in MPDs
| Disease | case # | JAK2 | UPD | UPD* | Recurrent | Other abnormalities | MPL** |
|---|---|---|---|---|---|---|---|
|
| |||||||
| PV | 78 | 1 | 9p | dup 9p, dup 3q, Trisomy 4, del 11q | |||
|
|
|||||||
| 172 | 1 | Trisomy 9 | |||||
|
|
|||||||
| 175 | 1 | ||||||
|
|
|||||||
| 298 | 1 | 1p/9p | Y591D | ||||
|
|
|||||||
| 375 | 1 | 9p | |||||
|
| |||||||
| 86 | 2 | 9p | |||||
|
|
|||||||
| 249 | 2 | 9p | |||||
|
|
|||||||
| 307 | 2 | 9p | |||||
|
|
|||||||
| 327 | 2 | 9p | |||||
|
|
|||||||
| 354 | 2 | 9p | Del 20q | ||||
|
| |||||||
| ET | 114 | 0 | |||||
|
|
|||||||
| 115 | 0 | ||||||
|
|
|||||||
| 146 | 0 | ||||||
|
|
|||||||
| 201 | 0 | ||||||
|
|
|||||||
| 240 | 0 | ||||||
|
| |||||||
| 281 | 0 | ||||||
|
|
|||||||
| 285 | 0 | ||||||
|
|
|||||||
| 314 | 0 | ||||||
|
|
|||||||
| 446 | 0 | ||||||
|
|
|||||||
| 461 | 0 | ||||||
|
| |||||||
| 5 | 1 | Del 5q23.1(LOC51334) | |||||
|
|
|||||||
| 186 | 1 | ||||||
|
|
|||||||
| 271 | 1 | ||||||
|
|
|||||||
| 272 | 1 | ||||||
|
|
|||||||
| 313 | 1 | Trisomy 9 | |||||
|
| |||||||
| 330 | 1 | 9p | |||||
|
|
|||||||
| 374 | 1 | ||||||
|
| |||||||
| MMM | 29 | 0 | 1p | RB del | S204F | ||
|
|
|||||||
| 56 | 0 | ||||||
|
|
|||||||
| 84 | 0 | 1p | RB del | Dup 8q21.3 | W515L | ||
|
|
|||||||
| 138 | 0 | ||||||
|
|
|||||||
| 183 | 0 | Dup 9q21.11-qter, Dup 4q28.1-qter | |||||
|
| |||||||
| 196 | 0 | ||||||
|
|
|||||||
| 325 | 0 | 1p | W515L | ||||
|
|
|||||||
| 459 | 0 | ||||||
|
|
|||||||
| 485 | 0 | NF1 del | Del 12p13 | ||||
|
|
|||||||
| 109 | 1 | 9p | Del 4q24 | ||||
|
| |||||||
| 120 | 1 | 9p | |||||
|
|
|||||||
| 122 | 1 | 9p | |||||
|
|
|||||||
| 191 | 1 | 9p | |||||
|
|
|||||||
| 287 | 1 | 9p | RB del | ||||
|
|
|||||||
| 442 | 1 | 9p | |||||
|
| |||||||
| 457 | 1 | 9p | |||||
UPD: uniparental disomy, Del: deletion, Dup: duplication.
- both wild-type and mutated alleles of JAK2 were detected;
- only mutated allele of JAK2 was detected.
Subpopulation had UPD which was detected by allele-specific gene dosage analysis.
Mutational status of MPL gene (see Result Section)
Seven of 16 samples of PMF had loss and/or gain of genomic material. Notably, five of the nine cases of PMF with no JAK2 mutations had genomic alterations including deletion involving RB1 (13q14) (PMF case #29, #84 and #287); deletion involving NF1 (17q11) (PMF case #485), duplication of 8q21.3, duplication of 9q21.11-qter, duplication of 4q28.1-qter, deletion of 12p13 and deletion of 4q24. Deletion of 4q24 (PMF case #109) was also detected by conventional cytogenetics confirming the SNP-chip results (data not shown). Deletions of NF1 and RB1 were also confirmed by quantitative genomic PCR (Supplement Figure).
Only two of 17 ET cases had genomic alterations; those with wild-type JAK2 (10 cases) had a normal genomic pattern (Table 1). One case of ET had deletion at 5q23.1 involving a single gene, LOC51334. We analyzed nucleotide sequences of all exons of the remaining allele, as well as, methylation status of the promoter region of this gene in this case; and found neither methylation nor mutation of the gene (data not shown).
9p UPD and JAK2 point mutations in MPD
One of the advantages of SNP-chip analysis is the ability to detect UPD [17]. This novel analysis can detect UPD even when clones with UPD are not dominan [17]. We found UPD in the 9p region that involved JAK2 in five PV samples, in which the clones with the UPD were dominant (Fig. 2A and Table 1) since large regions of loss of heterozygosity (LOH) without loss of DNA copy number were detected. Each of these cases showed homozygous JAK2V617F mutations by allele specific PCR (Table 1).
Figure 2. Uniparental disomy of 9p in MPD samples.
A: UPD in dominat clones. Green bars indicate heterozygous SNPs. Regions with pink lines show those regions with loss of heterozygosity but with normal DNA copy number (uniparental disomy). Position of JAK2 gene is indicated by an arrow. Case numbers are shown to the right of the chromosomes. B: UPD in non-dominant clone. Representative cases having non-dominant clones with 9p UPD are shown. In each panel, a blue line in the top indicates level of total gene dosage. Green bars under the chromosome indicates heterozygous SNP sites detected by SNP-chip analysis. Green and red lines in the bottom in each panel indicate levels of parental gene dosage. UPD regions have decrease of one of the parental gene dosage (green line) and incrase of gene dosage of the other parental allele (red line).
Further, we identified 11 cases with 9p UPD (3 PV, 1 ET, and 7 PMF), in which the clones with the UPD were not dominant (Table 1). Although LOH regions of 9p in these cases were quite as obvious, allele-specific gene dosage analysis clearly indicated that dosage level of one of the parental alleles was lower than normal level, and the other parental allele was higher (Fig. 2B).
One PV and one ET had both wild-type JAK2 and JAK2V617F with trisomy 9. Another PV had both wild-type JAK2 and JAK2V617F with duplication of 9p, but we were unable to determine if the mutant or wild type allele was duplicated in these cases.
Point mutations of the MPL gene in MPD with 1p UPD
SNP-chip analysis showed 1p UPD in two cases of PMF (case # 29 and # 84) (Fig 3A), suggesting the possibility of a transforming allele in this region. Analysis of the common region of UPD on 1p of these two cases of PMF showed two tyrosine kinase (FGR and TIE1) genes and the thrombopoietin receptor (MPL) gene; each is known to be expressed in normal hematopoietic cells 18–20. We sequenced all coding exons of the FGR, TIE1, and MPL genes in these two PMF cases. No mutations in FGR and TIE1 genes were detected (data not shown). However, we found a point mutation in the MPL gene that changed codon 204 from TCT to TTT (S204F) in case # 29 (Fig 3B) and changed codon 515 from TGG to TTG (W515L) in case # 84 (Fig 3C). Each sample had no wild type allele as shown by direct sequencing analysis (Figs 3B and C).
Figure 3. UPD of 1p and point mutations of MPL in PMF.
A: Two cases of PMF with clear UPD in 1p region are shown. Green bars indicate heterozygous SNPs. Regions with pink lines show those regions with loss of heterozygosity but with normal DNA copy number (uniparental disomy). Position of MPL gene is indicated by an arrow. Point mutations of MPL found in 2 PMF cases are shown: S204F (B) and W515L (C). The sequences were determined by direct sequencing. Sequences of normal alleles are shown in upper panels.
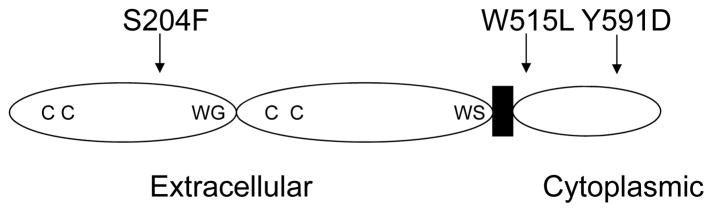
In another sample of PMF (case # 325) and PV (case # 298), 1p UPD was detected in subpopulation of the tumor cells (Fig. 4). We sequenced all exons of the MPL gene in these two cases and found point mutations of the MPL gene (W515L and Y591D) (Fig. 4). In case # 325 of PMF, TGG changed to TTG at amino-acid 515, leading to substitution of leucine for tryptophan (Fig. 4A); in case # 298 of PV, TAC changed to GAC at amino-acid position 591, leading to substitution of asparatate for tyrosine (Fig. 4B). In these cases, wild-type sequences were detected as well, suggesting that clones with these mutations were not predominant. All point mutations were also confirmed after subcloning of the PCR products into plasmids (data not shown). Positions of the MPL mutations are shown in Figure 5; S204F mutation is in the extracellular region; W515L and Y591D are in the cytoplasmic domain.
Figure 4. Non-dominant clones with 1p UPD in MPD were detected by SNP-chip.
Top panels: Allele-specific gene dosage analysis. Blue lines above the chromosomal panels indicate levels of total gene dosage (0 indicates 2N, normal diploid). Green and red lines below the chromosomal panels indicate respective parental allele gene dosage. Higher level of the red line and lower level of the green line indicates the regions of UPD. Middle and bottom panels: normal (middle) and mutated (bottom) nucleotide sequences of the MPL gene encoding amino-acid positions at 515 and 591; PMF case 325 (A) had W515L mutation and PV case 298 (B) had Y591D mutation.
Figure 5. Position of point mutations of the MPL gene found in MPD.
Schema of MPL protein. Position of the point mutations are indicated by arrows. CC: conserved cysteine residues; WG: WGXWS motif; WS: WSXWS motif; extracellular: extracellular domain; cytoplasmic: cytoplasmic domain; black box depicts the transmembrane domain.
Discussion
Recently, we developed a new algorithms for SNP-chip analysis which reduced noises (false-positive and false-negative) and make the data reliable. We have previously validated the sensitivity and specificity of the SNP-chip analysis using genomic real time PCR, FISH, nucleotide sequencing. This platform and our novel algorithm allow detect very small genomic abnormalities at very high sensitivity.
SNIP-chip is a powerful tool for genome-wide analysis of genetic abnormalities [14, 15]. It allows for detection of small deletions and amplification that cannot be detected by standard karyotyping techniques [14, 15]. Using this approach, commonly deleted and/or amplified regions of the genome can be defined to facilitate the identification of target tumor suppressor genes and/or oncogenes in these loci. For example, we identified deleted regions of PMF that include either the RB1 (13q14) or NF1 (17q11) locus.
Recently, UPD that is not detectable by conventional techniques, has been reported in human cancers [14, 21, 22]. Most investigators have assayed for LOH using microsatellite markers, combined with fluorescence in situ hybridization (FISH) to detect UPD [22]. A region was defined as having UPD when FISH displayed an intact chromosome pattern and the LOH analysis demonstrated allelic imbalance at the locus [22]. However, this approach is both time- and labor-intensive compared with use of SNP-chip for detection of UPD. For example, we detected five PV cases with 9p UPD. Each 9p UPD case had two JAK2 mutated alleles as confirmed by allele-specific PCR. As previously reported, UPD is a common mechanism for duplication of the mutant JAK2V617F allele in PV [6], presumably through mitotic recombination and subsequent positive selection for clones harboring two mutant alleles. We previously developed a new algorithm allowing us to evaluate allele-specific gene dosage (gene dosages of paternal and maternal alleles) without matched control samples [17]. This new algorithm can very sensitively and accurately detect UPD regions even though the clones with UPD are as small as 20% of whole tissues [17]. We previously found that 9p UPD in non-dominant clones were frequent events in MPD [17].
Using this method, we could detect non-dominant clones with 9p and/or 1p UPD, which may be overlooked by standard algorithm used for SNP-chip analysis. UPD was a very frequent event in PMF; 3 cases had 1p UPD and 7 cases had 9p UPD. In contrast, only one of the ET cases in this study had UPD. In ET, we only found one sample with deletion of 5q23.1 and another with trisomy 9, as well as, 7 cases with a JAK2 mutation. This observation could be explained by either lack of involvement of the granulocytic lineage (our source of DNA) in these ET patients, or by point mutations or other genetic abnormalities in these samples that were not detected by SNP-chip analysis. Clearly, the pathogenesis leading to ET, especially ET without a JAK2 mutation, remains to be elucidated.
We detected a novel 1p UPD in samples from three PMF and one PV individuals. In each case, we found a point mutation of the MPL gene; two samples involved the well-know mutation, W515L [10, 11], and the other two were novel mutations (S204F and Y591D). We screened 100 DNA samples from normal individuals to determine if these latter nucleotide substitutions were polymorphic; we could not detect these nucleotide substitutions in normal DNA (data not shown). We screened all of our remaining MPD cases for these mutations (S204F, W515L and Y591D) and found none (data not shown). Amino-acid substitution of MPL was found in all cases with 1p UPD, suggesting that these substitutions are mutations, not rare polymorphisms, and MPL gene is a target gene of 1pUPD. 1p UPD may be a hallmark for a MPL mutation in MPD. The precise function of novel MPL mutants (S204F and Y591) remains to be explored.
To elucidated functional significance of these novel mutations of the MPL gene, we expressed them in the IL3-dependent murine B-cell line, BaF/3 cells. Mutant S204F or Y591D MPL did not allow the cells to grow independently of IL3, whereas W515L MPL did (data not shown). How these novel MPL mutants contribute to development of PMF is unclear. Interestingly, one PV (case 298) had a point mutation of both a JAK2 and MPL (Y591D) suggesting that mutations of these two molecules may independently contribute to development of MPD even though they are involved in a same Mpl/Jak/Stat signal transduction pathway.
Myelofibrosis, not thrombocytosis, is a prominent feature in PMF [1]. It is not clear that an activating point mutation of MPL by itself can induce myelofibrosis. Interestingly, two cases with MPL mutations had deletion of the RB1 gene. Mutation of MPL and deletion of RB1, may cooperate in development of PMF; or loss of RB1 may potentiate acquisition of mutations of MPL.
In summary, we found recurrent genetic abnormalities including 9p UPD/JAK2 mutations, 1p UPD/MPL mutations, as well as, deletions of the RB and the NF1 genes in MPD. We found two novel point mutations of the MPL gene (S204F and Y591D) in PMF and PV samples with 1p UPD. SNP-chip analysis is a robust tool to detect genetic abnormalities, especially small deletions and UPD in MPD.
Supplementary Material
References
- 1.Tefferi A. Myelofibrosis with myeloid metaplasia. N Engl J Med. 2000;342:1255–65. doi: 10.1056/NEJM200004273421706. [DOI] [PubMed] [Google Scholar]
- 2.Spivak JL. Diagnosis of the myeloproliferative disorders: resolving phenotypic mimicry. Semin Hematol. 2003;40(1 Suppl 1):1–5. doi: 10.1053/shem.2003.50026. [DOI] [PubMed] [Google Scholar]
- 3.Thiele J, Kvasnicka HM, Orazi A. Bone marrow histopathology in myeloproliferative disorders--current diagnostic approach. Semin Hematol. 2005;42:184–95. doi: 10.1053/j.seminhematol.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 4.Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet. 2005;365:1054–1061. doi: 10.1016/S0140-6736(05)71142-9. [DOI] [PubMed] [Google Scholar]
- 5.James C, Ugo V, Le Couedic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434:1144–1148. doi: 10.1038/nature03546. [DOI] [PubMed] [Google Scholar]
- 6.Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med. 2005;352:1779–1790. doi: 10.1056/NEJMoa051113. [DOI] [PubMed] [Google Scholar]
- 7.Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 8.Zhao R, Xing S, Li Z, et al. Identification of an acquired JAK2 mutation in polycythemia vera. J Biol Chem. 2005;280:22788–22792. doi: 10.1074/jbc.C500138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott LM, Tong W, Levine RL, Scott MA, Beer PA, Stratton MR, Futreal PA, Erber WN, McMullin MF, Harrison CN, Warren AJ, Gilliland DG, Lodish HF, Green AR. JAK2 exon 12 mutations in polycythemia vera and idiopathic erythrocytosis. N Engl J Med. 2007;356:459–68. doi: 10.1056/NEJMoa065202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pikman Y, Lee BH, Mercher T, McDowell E, Ebert BL, Gozo M, Cuker A, Wernig G, Moore S, Galinsky I, DeAngelo DJ, Clark JJ, Lee SJ, Golub TR, Wadleigh M, Gilliland DG, Levine RL. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med. 2006;3:e270. doi: 10.1371/journal.pmed.0030270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pardanani AD, Levine RL, Lasho T, Pikman Y, Mesa RA, Wadleigh M, Steensma DP, Elliott MA, Wolanskyj AP, Hogan WJ, McClure RF, Litzow MR, Gilliland DG, Tefferi A. MPL515 mutations in myeloproliferative and other myeloid disorders: a study of 1182 patients. Blood. 2006;108:3472–6. doi: 10.1182/blood-2006-04-018879. [DOI] [PubMed] [Google Scholar]
- 12.Levine RL, Belisle C, Wadleigh M, et al. X-inactivation-based clonality analysis and quantitative JAK2V617F assessment reveal a strong association between clonality and JAK2V617F in PV but not ET/MMM, and identifies a subset of JAK2V617F-negative ET and MMM patients with clonal hematopoiesis. Blood. 2006;107:4139–4141. doi: 10.1182/blood-2005-09-3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garraway LA, Widlund HR, Rubin MA, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 14.Kawamata N, Ogawa S, Zimmermann M, Kato M, Sanada M, Hemminki K, Yamatomo G, Nannya Y, Koehler R, Flohr T, Miller CW, Harbott J, Ludwig WD, Stanulla M, Schrappe M, Bartram CR, Koeffler HP. Molecular allelokaryotyping of pediatric acute lymphoblastic leukemias by high resolution single nucleotide polymorphism oligonucleotide genomic microarray. Blood. 2007 doi: 10.1182/blood-2007-05-088310. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann S, Ogawa S, Raynaud SD, Sanada M, Nannya Y, Ticchioni M, Bastard C, Akagi T, Kawamata N, Koeffler HP. Molecular allelokaryotyping of early stage untreated chronic lymphocytic leukemia. Cancer. 2007 doi: 10.1002/cncr.23270. in press. [DOI] [PubMed] [Google Scholar]
- 16.Nannya Y, Sanada M, Nakazaki K, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, Koeffler HP, Ogawa S. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–26. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Batard P, Sansilvestri P, Scheinecker C, et al. The Tie receptor tyrosine kinase is expressed by human hematopoietic progenitor cells and by a subset of megakaryocytic cells. Blood. 1996;87:2212–2220. [PubMed] [Google Scholar]
- 19.Lannutti BJ, Shim MH, Blake N, Reems JA, Drachman JG. Identification and activation of Src family kinases in primary megakaryocytes. Exp Hematol. 2003;31:1268–1274. doi: 10.1016/j.exphem.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Kaushansky K. The molecular mechanisms that control thrombopoiesis. J Clin Invest. 2005;115:3339–3347. doi: 10.1172/JCI26674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raghavan M, Lillington DM, Skoulakis S, et al. Genome-wide single nucleotide polymorphism analysis reveals frequent partial uniparental disomy due to somatic recombination in acute myeloid leukemias. Cancer Res. 2005;65:375–378. [PubMed] [Google Scholar]
- 22.Kralovics R, Guan Y, Prchal JT. Acquired uniparental disomy of chromosome 9p is a frequent stem cell defect in polycythemia vera. Exp Hematol. 2002;30:229–236. doi: 10.1016/s0301-472x(01)00789-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.