Abstract
Objective
The purpose of this study was to identify a panel of novel serum tumor antigen-associated autoantibody (TAAb) biomarkers for the diagnosis of high-grade serous ovarian cancer.
Methods
To detect TAAb we probed high-density programmable protein microarrays (NAPPA) containing 10,247 antigens with sera from patients with serous ovarian cancer (n = 30 cases/ 30 healthy controls) and measured bound IgG. We identified 735 promising tumor antigens and evaluated these with an independent set of serous ovarian cancer sera (n = 30 cases/ 30 benign disease controls/ 30 heathy controls). Thirty-nine potential tumor autoantigens were identified and evaluated using an orthogonal programmable ELISA platform against a total of 153 sera samples (n = 63 cases/ 30 benign disease controls/ 60 healthy controls). Sensitivities at 95% specificity were calculated and a classifier for the detection of high-grade serous ovarian cancer was constructed.
Results
We identified 11-TAAbs (ICAM3, CTAG2, p53, STYXL1, PVR, POMC, NUDT11, TRIM39, UHMK1, KSR1, and NXF3) that distinguished high-grade serous ovarian cancer cases from healthy controls with a combined 45% sensitivity at 98% specificity.
Conclusion
These are potential circulating biomarkers for the detection of serous ovarian cancer, and warrant confirmation in larger clinical cohorts.
Keywords: Ovarian Cancer, Biomarker, Autoantibody, Proteomics, Diagnostics
1. Introduction
Ovarian cancer remains a leading cause of death from gynecologic malignancy, with over 21,290 new cases/year and an estimated 14,180 deaths in the United States in 2014 [1]. The 5 –year survival rate for stage I ovarian cancer is over 80%, compared with a survival rate of only 11% for stage IV ovarian cancer [2]. Early diagnosis is also associated with improved morbidity [3]. Unfortunately, over 60% of patients have advanced disease at the time of diagnosis. This is likely related to several factors, including an aggressive tumor biology that causes rapid, early dissemination through the peritoneum, as well as limitations of current biomarkers and imaging modalities for early detection.
A number of screening strategies and biomarkers have been proposed to improve the early detection of ovarian cancer. Two biomarkers, Cancer Antigen 125 (CA 125) and Human Epididymis Protein 4 (HE4), as well as two algorithms, OVA1 and Risk of Ovarian Malignancy Algorithm (ROMA), have been approved by the US FDA for risk assessment and management of ovarian cancer [4]. The most common approaches used in screening are sequential testing using serum CA 125 and transvaginal ultrasound (TVUS), which have been shown to have a high specificity (99.9%) and positive predictive value (PPV; 26.8%) [5]. Overall, these strategies are limited by the low prevalence of the disease in the general population, interpatient variability of CA 125 testing, false positive CA 125 levels from benign ovarian tumors and diverticulitis, as well as the limited specificity of pelvic ultrasound for detection of early ovarian cancers. Thus, there is an urgent need to identify biomarkers to complement CA 125 and HE4 for the early identification of ovarian cancer.
Tumor antigen-associated autoantibodies (TAAb) represent a well-documented source of potential early diagnostic biomarkers for ovarian cancer [6–10]. TAAbs are generated in response to protein overexpression or mutations in cancer patients. Our group as well as others have identified panels of TAAbs for the detection of breast [11, 12], prostate [13], colorectal [14], lung [15], and ovarian cancers [16]. In addition to CA 125 and HE4, serum antibodies to p53 are the most frequently studied TAAb in ovarian cancer [4, 17]. Mutations in TP53 occur early in high-grade serous ovarian carcinogenesis, are present in over 80% of serous ovarian cancer, and are strongly associated with the presence of p53-TAAb. We detected p53-TAAb in the sera of 6–7% of patients with limited-stage ovarian cancer, and 41% of women with high-grade serous ovarian carcinomas, including 30% of women with false-negative CA 125 levels [16, 18]. However, the false-positive rate of 8% limits its application for screening as a stand-alone biomarker [18]. We have reported a screen of 4,988 human proteins, and identified two additional TAAb (PTPRA and PTGFR) with modestly improved sensitivity (23.3%) at high specificity (98.3%). The specificity was maintained when compared to benign ovarian disease [16]. Taken together, these data suggest that the addition of other TAAb biomarkers beyond p53 may improve the detection of ovarian cancer.
In this study, we predicted that technological improvements in protein array production, protein expression, and an increased protein library (>5,000 new full-length proteins) would lead to the identification of novel TAAb biomarkers beyond p53, PTPRA, and PTGFR that could increase the sensitivity of early detection of serous ovarian cancer. To achieve this objective, we performed a large-scale proteomic analysis using custom Nucleic Acid Programmable Protein Microarrays (NAPPA), and an orthogonal Rapid Antigenic Protein In situ Display (RAPID) ELISA assay for validation [18–20]. In this system, full-length human proteins are expressed using in vitro mammalian cell lysate and captured on a solid support using epitope fusion tags [21]. We used a sequential screening strategy to identify candidate TAAb biomarkers and limit the false discovery rate (Figure 1). First, we developed custom protein microarrays expressing 10,247 full-length candidate human proteins and profiled the serum of 30 cases of high-grade serous ovarian cancer and 30 matched healthy controls (Discovery). Second, we selected 735 antigens for further validation using an independent serum set consisting of 30 cases of high-grade serous ovarian cancer, 30 benign ovarian disease controls, and 30 healthy controls (Training) and identified 39 potential candidate TAAb biomarkers. Third, using an orthogonal ELISA assay to display these antigens, we retested the sera from the training set. Finally, using the same ELISA platform we displayed 11 antigens and screened an independent serum set consisting of 34 cases of high-grade serous ovarian cancer and 32 healthy controls (Validation). Here we present the sensitivity and specificity of each individual biomarker, as well as the biomarker panel.
Figure 1.
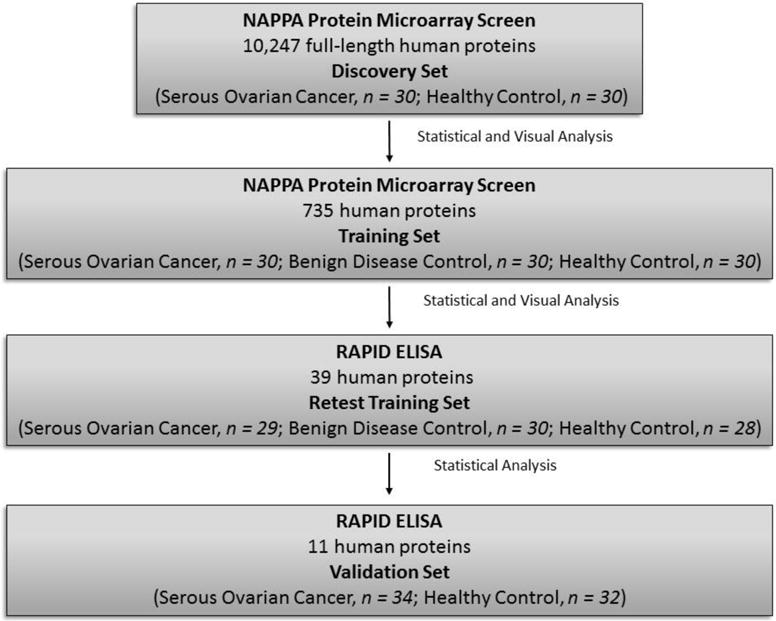
Overview of study design.
2. Methods
2.1 Patient sera
Sera from patients with serous ovarian cancer were obtained from the Brigham and Women’s Hospital. Sera derived from ovarian cancer patients were obtained at the time of presentation prior to surgery. Sera from age-, gender-, and location-matched general population control women were obtained using a standardized serum collection protocol and stored at −80°C until use. Cases and matched controls were processed simultaneously. Women with a personal history of cancer (other than non-melanoma skin cancer) were excluded as controls (Table 1). These sera have been reported in [16, 22, 23]. Written consent was obtained from all subjects under institutional review board approval.
Table 1.
Characteristics of Cases and Controls
| Discovery Set | Training Set | Validation Set | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||
| Controls | Serous Cases | Chi-square | Controls | Benign Controls | Serous Cases | Chi-square | Controls | Serous Cases | Chi-square | |
| p-value* | p-value* | p-value* | ||||||||
| (N=30) N (%) |
(N=30) N (%) |
(N=30) N (%) |
(N=30) N (%) |
(N=30) N (%) |
(N=32) N (%) |
(N=34) N (%) |
||||
| Age | ||||||||||
| <50 | 6 (20.0%) | 6 (20.0%) | 0.99 | 12 (40.0%) | 12 (40.0%) | 11 (36.7%) | 0.99 | 1 (3.1%) | 2 (5.9%) | 0.85 |
| 50–60 | 12 (40.0%) | 12 (40.0%) | 11 (36.7%) | 11 (36.7%) | 11 (36.7%) | 10 (31.2%) | 11 (32.4%) | |||
| >60 | 12 (40.0%) | 12 (40.0%) | 7 (23.3%) | 7 (23.3%) | 8 (26.7%) | 21 (65.6%) | 21 (61.8%) | |||
| Menopausal status | ||||||||||
| Pre | 8 (26.7%) | 6 (20.0%) | 0.54 | 13 (43.3%) | 10 (33.3%) | 10 (33.3%) | 0.65 | 3 (9.4%) | 3 (8.8%) | 0.94 |
| Post | 22 (73.3%) | 24 (80.0%) | 17 (56.7%) | 20 (66.7%) | 20 (66.7%) | 29 (90.6%) | 31 (91.2%) | |||
| Race | ||||||||||
| White | 30 (100.0%) | 30 (100.0%) | – | 30 (100.0%) | 27 (90.0%) | 23 (88.5%) | 0.17 | 30 (93.8%) | 32 (97.0%) | 0.54 |
| Non-white | 0 (0%) | 0 (0%) | 0 (0%) | 3 (10.0%) | 3 (11.5%) | 2 (6.2%) | 1 (3.0%) | |||
| OC use | ||||||||||
| Ever | 10 (33.3%) | 13 (43.3%) | 0.43 | 10 (33.3%) | 11 (36.7%) | 10 (38.5%) | 0.92 | 18 (56.2%) | 20 (60.6%) | 0.72 |
| Never | 20 (66.7%) | 17 (56.7%) | 20 (66.7%) | 19 (63.3%) | 16 (61.5%) | 14 (43.8%) | 13 (39.4%) | |||
| Parity | ||||||||||
| Nulliparous | 4 (13.3%) | 6 (20.7%) | 0.45 | 5 (16.7%) | 9 (30.0%) | 7 (26.9%) | 0.46 | 3 (9.4%) | 3 (8.8%) | 0.94 |
| Parous | 26 (86.7%) | 23 (79.3%) | 25 (83.3%) | 21 (70.0%) | 19 (73.1%) | 29 (90.6%) | 31 (91.2%) | |||
| Year of specimen collection | ||||||||||
| 2001–2002 | 10 (33.3%) | 8 (26.7%) | 0.4 | 8 (26.7%) | 10 (33.3%) | 3 (10.0%) | 0.19 | 10 (31.2%) | 11 (32.4%) | 0.91 |
| 2003–2005 | 8 (26.7%) | 13 (43.3%) | 13 (43.3%) | 11 (36.7%) | 12 (40.0%) | 15 (46.9%) | 17 (50.0%) | |||
| 2006–2010 | 12 (40.0%) | 9 (30.0%) | 9 (30.0%) | 9 (30.0%) | 15 (50.0%) | 7 (21.9%) | 6 (17.6%) | |||
| Length of storage | ||||||||||
| <5.4 years | 13 (43.3%) | 9 (30.0%) | 0.07 | 11 (36.7%) | 10 (33.3%) | 17 (56.7%) | 0.25 | 8 (25.0%) | 8 (23.5%) | 0.84 |
| 5.44–7.6 years | 84(13.3%) | 12 (40.0%) | 9 (30.0%) | 10 (33.3%) | 9 (30.0%) | 11 (34.4%) | 14 (41.2%) | |||
| >7.6 years | 13 (43.3%) | 9 (30.0%) | 10 (33.3%) | 10 (33.3%) | 4 (13.3%) | 13 (40.6%) | 12 (35.3%) | |||
2.2 Sample characteristics and biomarker selection
The primary goal of this study was to identify serum TAAb biomarkers that would distinguish serous ovarian cancer from both benign disease and healthy controls in order to improve the sensitivity of current biomarkers and guide clinical decisions. We performed a sequential screening strategy, described in Figure 1, in order to identify a panel of TAAb biomarkers from 10,247 full-length human proteins. All the discovery and validation case and control sera were gender- and age-matched. Sera from cases, primarily stage III/IV (95%), grade 3 (93.7%), was collected prior to surgery under standard collection protocols [16]. Table I details the age distribution, menopausal status, race, oral contraceptive (OC) use, parity, grade, year of sample collection, and length of storage.
2.3 Protein microarray production
Sequence-verified, full-length cDNA expression plasmids in the T7-based mammalian expression vector pANT7_cGST were obtained from Arizona State University’s Biodesign Institute Plasmid Repository and are publicly available through DNASU (http://dnasu.asu.edu/DNASU/). The high-throughput preparation of high-quality supercoiled DNA for cell-free protein expression was performed as described [21]. Protein arrays displaying 10,247 human proteins were distributed evenly over five slides. Quality control was performed to determine inter and intra assay consistency by measuring DNA and protein expression (Figure 2 and Supplemental Table 1).
Figure 2.
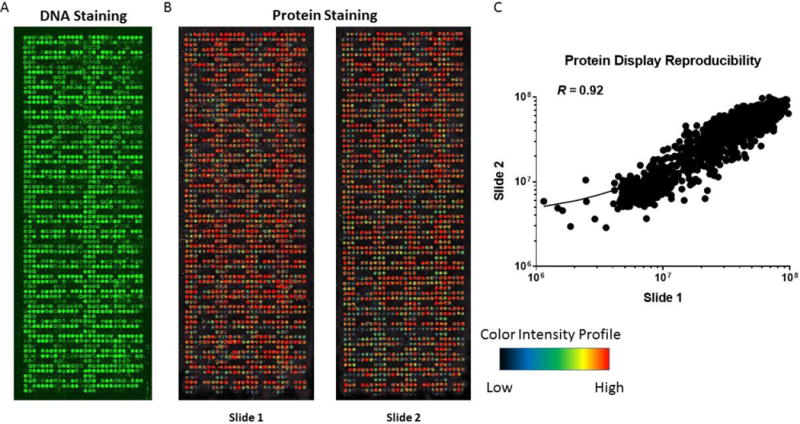
Quality control of protein microarrays. A. Quality control of DNA printing with picogreen. B. Quality control of in vitro protein production with anti-GST Mab staining. C. Reproducibility of protein signal intensity of two protein arrays produced in a single print batch.
2.4 Protein array screening protocol
The slides were initially blocked with 1x Superblock® (Pierce, MA, USA) for 1 hour at 30°C. In vitro transcription translation (IVTT) was performed using HeLa lysate, Accessory Proteins, and Reaction Mix from Thermo Scientific (Carlsbad, CA, USA). Per the manufacturer’s recommendations, Hela Lysate was slowly injected onto the slide and the expression of the proteins was automated in two steps: 1. Hybridization at 30°C for 90 minutes with no agitation; 2. Hybridization at 15°C for 30 minutes with no agitation using an EchoTherm Chilling Incubator (Torrey Pines Scientific, Inc. Carlsbad, CA, USA). Following the IVTT step the slides are washed with 0.2% PBST then with with 5% milk in 0.2% PBST [21].
2.5 Image analysis and quantification
Image analysis and quantification were performed as previously described [11]. Briefly, slides were scanned using a PowerScanner™ (Tecan, Mannedorf, Switzerland) and the spot intensity was measured using ArrayPro Analyzer (MediaCybernetics, Rockville, MD, USA). Raw intensity values were normalized by the following steps: 1. Remove background by subtracting non-spot control using the first quartile value (Background); 2. Determine the median intensity of every gene, excluding the control spots (Median Protein Intensity); 3. Subtract the raw signal intensity from the background and divide it over the median protein intensity minus the background. In addition, the visual intensity of captured antibodies was evaluated by at least two researchers to identify and confirm positive responses, previously described in [24]. Visually, antigens were selected for further validation if a ring was present in two or more cases and not identified in controls.
2.6 Antigen selection criteria for validation protein arrays
Using the normalized values from the discovery array, for each protein antigen we determined the sensitivity at 95% and 98% specificity, area under receiver operating characteristics curve (AUC), partial AUC above 95% and 98% specificity (pAUC), including the Welch’s t test P value. We also used the K-statistic as previously described in [11]. Briefly, the K-statistic measures the separation of the top 20 percentile of antigens by evaluating qcases(0.975) - qcases(0.800) divided by qcontrols(0.975) – qcontrols(0.025). A higher K value indicates a greater separation of the antibody reactivity of positive cases and negative controls. Antibodies with the same sensitivity as other markers but a higher K-statistic were selected for further validation.
Custom validation NAPPA arrays were composed of antigens that met at least one of the following criteria: (a) ≥10% sensitivity at 95% specificity, (b) ≥0.500 AUC, (c) ≥0.700 pAUC, (d) significant P value of Welch t test, (e) K≥0.9, and (f) antigens with a greater prevalence in cases than in controls by visual analysis. In total, 735 full-length proteins were included for further validation (Supplemental Table 2). These antigens were manufactured as described above and printed in duplicate on a single microarray. The protein array data discussed in this manuscript have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) and are accessible through GEO Super Series accession number GSE79517 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79517).
2.7 RAPID ELISA
Rapid Antigenic Protein In situ Display (RAPID) ELISA was performed as previously described [25], with the following modifications. All GST fusion proteins were expressed from plasmids using 1-Step Human Coupled in vitro Expression system (Thermo Scientific) at 30°C for 1.5 hours. Following in vitro protein expression, the GST-fused proteins were diluted 1:100 in 5% milk in 0.2% PBST and added to the GST coated 96 well plates at 100 μl/well and shaken at room temperature at 500 rpm for 1 hour. Plates were then washed 5 times with 0.2% PBST. During incubation the patient serum was diluted 1:500 in 50% E. coli lysate and 5% Milk in 0.2% PBST. Plates were then incubated with 100 μl/well diluted patient serum, with shaking at RT at 500 rpm for 1 hour, and washed 5 times with 0.2% PBST. Secondary HRP conjugated goat anti-human IgG (Jackson ImmunoResearch Laboratories) to detect serum TAAb were diluted 1:10,000 in 5% milk in 0.2% PBST, and the GST positive control secondary HRP sheep anti-Mouse IgG (Jackson ImmunoResearch Laboratories) was diluted 1:6250. The secondary Abs were shaken at RT at 500 rpm for 1 hour. Plates were washed 5 times in 0.2% PBST prior to the addition of the developing buffer, Supersignal ELISA Femto Chemiluminescent Substrate (Thermo Scientific). Relative luminescence units (RLU) were measured on a Glomax 96 Microplate Luminometer (Promega, Madison, WI) using a wavelength of 425 nm. All assays were performed in duplicate, and values are plotted as mean values. RLU ratios were calculated using the RLU of a specific antigen divided by the RLU of the control GST-protein. The sensitivity and specificities of TAAb levels was determined by applying a cutoff value of the mean of the controls plus two standard deviations. We performed a two-tailed t-test to determine significance.
2.8 Statistical Analysis
ROC analysis was performed utilizing normalized values from our orthogonal RAPID ELISA assay without feature selection, using a binary logistic regression analysis. We selected an 11-TAAb panel (ICAM3, CTAG2, p53, STYXL1, PVR, POMC, NUDT11, TRIM39, UHMK1, KSR1, and NXF3) to develop a classifier based on their individual sensitivity and specificity. The binary logistic regression analysis classifier was trained on normalized RAPID ELISA values from Training Set and tested on normalized RAPID ELISA values from Validation Set. Samples were selected as positive if they exceed antigen specific cutoffs of two standard deviations the mean of the benign ovarian disease controls. Antigen specific cutoffs were set to achieve 95% specificity by only selecting antigens for the classifier if they met ≥10% sensitivity at ≥95% specificity (Table 2).
Table 2.
Training set and validation set test statistics for potential serous ovarian cancer autoantibody biomarkers. Sensitivity, specificity, and cutoff values were determined for both the healthy and benign disease controls in training set and the healthy controls in validation set.
| Antigen | Training Set - RAPID ELISA | Validation Set - RAPID ELISA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| (Serous Ovarian Cancer, n = 29; Benign Disease Control, n = 30; Healthy Control, n = 28) | (Serous Ovarian Cancer, n = 34; Healthy Control, n = 32) | ||||||||
|
| |||||||||
| Benign vs Cases | Healthy vs Cases | Healthy vs Cases | |||||||
|
| |||||||||
| Sensitivity | Specificity | Cutoffs (2SD) | Sensitivity | Specificity | Cutoffs (2SD) | Sensitivity | Specificity | Cutoffs (2SD) | |
| CTAG2 | 0.103 | 0.967 | 2.088 | 0.103 | 0.967 | 1.882 | 0.235 | 0.968 | 3.465 |
| ICAM3 | 0.034 | 1.000 | 1.711 | 0.034 | 1.000 | 1.900 | 0.176 | 0.968 | 1.801 |
| KSR1 | 0.137 | 0.933 | 2.605 | 0.000 | 0.933 | 3.224 | 11.8 | 0.968 | 2.001 |
| NUDT11 | 0.275 | 0.967 | 2.436 | 0.275 | 0.967 | 2.734 | 0.324 | 1.000 | 2.345 |
| NXF3 | 0.034 | 0.900 | 2.085 | 0.034 | 0.900 | 1.473 | 0.088 | 0.968 | 3.686 |
| POMC | 0.068 | 0.933 | 2.250 | 0.172 | 0.933 | 1.649 | 0.118 | 0.937 | 2.804 |
| PVR | 0.137 | 0.933 | 1.468 | 0.207 | 0.933 | 1.366 | 0.176 | 0.968 | 2.734 |
| STYLX1 | 0.138 | 0.933 | 1.813 | 0.275 | 0.933 | 1.583 | 0.088 | 0.968 | 4.878 |
| TP53 | 0.069 | 0.967 | 3.171 | 0.067 | 0.967 | 2.310 | 0.441 | 0.968 | 6.012 |
| TRIM39 | 0.241 | 0.967 | 2.477 | 0.034 | 0.967 | 19.130 | 0.147 | 0.968 | 6.912 |
| UHMK1 | 0.034 | 0.900 | 2.006 | 0.138 | 0.900 | 1.539 | 0.118 | 0.968 | 2.387 |
|
| |||||||||
| Combined Sensitivity | > 2 antigens = 11/29 = 37.9% | > 2 antigens = 9/29 = 31% | > 2 antigens = 16/34 = 47.1% | ||||||
|
| |||||||||
| Combined Specificity | > 2 antigens = 2/30 = 93.3% | > 2 antigens = 2/28 = 92.8% | > 2 antigens =2/32 = 93.7% | ||||||
3 Results
3.1 Tumor Antigen-Associated Autoantibody screen of 10,247 human antigens using NAPPA protein microarrays
In the discovery phase, we performed a comprehensive analysis of 10,247 full-length human proteins against 30 patients with serous ovarian cancer and 30 healthy controls to identify potential serum IgG TAAb biomarkers associated with serous ovarian cancer (Figure 1 and Supplemental Table 1). The entire cohort consisted of 94 women with newly diagnosed serous ovarian cancer, 92 healthy control women, and 30 women with benign ovarian disease (Table 1). For each set, cases were intentionally matched to controls for age and gender. The median age at diagnosis was 60 years of age and were >85% white. Because the training set included women with benign ovarian disease, all cases and controls in that set were younger, on average, than the other sets (median age 54 vs 62.5). As seen in Table 1, cases and controls were balanced for menopausal status, oral contraceptive use, parity, and year of sample collection. The discovery set cases were stored longer than controls (p=0.07) but this variable was evenly distributed for the remaining datasets.
The goal of the discovery screen was to eliminate uninformative antigens as well as reducing the false positive rate and lower the overall cost of the screen. Sample protein arrays are shown in Figure 2. The batch-to-batch variation of DNA printing was R = 0.97 and protein expression, as measured by anti-GST monoclonal antibody binding, was R = 0.92 (Figure 2). The interassay reproducibility of serum screening is shown in Figure 2C, and was R = 0.92. In order to identify candidate TAAbs among the 10,247 antigens that distinguish serous ovarian cancer from healthy controls, we selected antigens based on statistical significance and visual analysis (see Methods).
3.2 Selection of antigens for training of biomarker panel
We selected 735 antigens for training and printed each antigen in duplicate for testing with sera form the Training Set. These arrays consisted of 649 antigens with sensitivities above 10% at 95% specificity and/or K>0.9 in our Discovery Set. Visually, we also selected 86 antigens to be included in our focused 735 antigen array. The selected 735 antigens were screened with the separate sera Training Set consisting of 30 cases of serous ovarian cancer, 30 benign disease controls, and 30 matched healthy controls (Supplementary Table 2). The inclusion of sera from women with benign ovarian disease at this early stage of biomarker discovery is critical to limit false positive detection and for ultimate clinical applicability. From this data, 39 antigens were selected based on sensitivity by visual and/or statistical analysis. Statistically, they had to meet two of the following criteria for selection, >10% sensitivity at 95% specificity, AUC ≥0.44, and a K-statistic ≥0.9.
3.3 RAPID ELISA test of 39 antigen biomarkers associated with serous ovarian cancer
We performed an orthogonal RAPID ELISA assay to validate our 39 candidate TAAb biomarkers. The RAPID ELISA assay is highly reproducible and readily amenable for screening large numbers of sera [16]. First, we rescreened the 39 antigens against our training set serum samples in order to establish cutoff values, sensitivity, and specificity for each antigen in comparison to benign disease and healthy controls (Table 2 and Supplemental Table 3). All 39 antigens were assessed using cutoff values defined by the benign disease control samples. In the training set screen, antibodies against NUDT11 (27.5%), TRIM39 (24.1%), KSR1 (13.7%), and PVR (13.7%) demonstrated the best discrimination of serous ovarian cancer from benign disease controls at 93.3% and 96.7% specificity (Table 2). We selected a panel of the top 11 antigens (CTAG2, ICAM3, KSR1, NUDT11, NXF3, POMC, PVR, STYXL1, p53, TRIM39, and UHMK1) based on their sensitivity (3.4 to 27.5%) and specificity (90 to 100%). Using this panel, we calculated the overall sensitivity and specificity of training set, defining a patient as positive if they express ≥2 antigens. The combined overall sensitivity of the top 11 antigens for discriminating benign disease (37.9%) and healthy (31%) controls at 93% specificity, are presented in Table 2. In addition, all of the cases and benign disease controls were screened for CA125 prior to treatment. There were 5 cases that were falsely negative (below 35 U/mL) for CA125. Using our final panel of 11-AAb, all 5 cases were classified as positive. Similarly, there were 6 benign disease controls with false positive CA125 values (above 35 U/mL) and using our 11-AAb panel the specificity of CA125 improved to 2 false positives.
3.4 Validation of biomarkers and development of the 11 autoantibody panel
We screened the top 11 antigens against an independent validation 2 sera set consisting of 34 cases of serous ovarian cancer and 32 healthy controls (Figures 1 and 3, Table 2). In validation set, TAAb against p53, CTAG2, NUDT11, PVR, and ICAM3 had sensitivities ≥17.6% at specificities of ≥96.8%; with a combined sensitivity of 47.1% at 93.7% specificity using the same criteria as our training set (Table 2).
Figure 3.
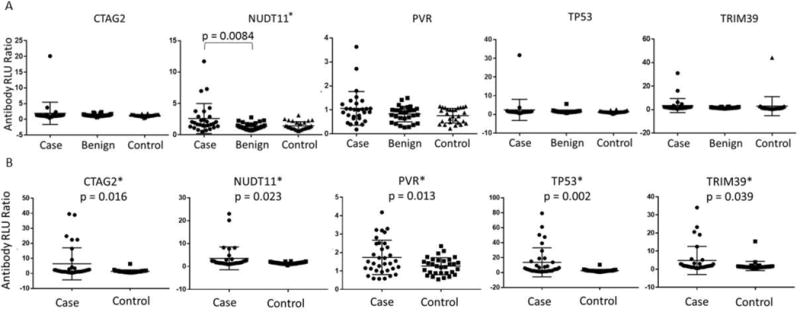
Autoantibody responses in serous ovarian cancer A. training set and B. validation set. P-value was calculated by a two-tail t test and * indicates statistically significant antigens (p < 0.05). Data is presented as antibody Relative Light Unit (RLU) ratio (Antigen: GST control ratio).
A receiver operating characteristic (ROC) analysis was computed using a binary logistic regression analysis (see Material and Methods). The 11 TAAb classifier was trained using antibody relative light unit (RLU) ratios from our training set comparing serous ovarian cases against benign disease and healthy controls with an area under the curve (AUC) of 0.807 and a sensitivity of 32% at 100% specificity (Figure 4A, dotted line, Supplemental Table 4). We tested our classifier using antibody RLU ratios from our validation set comparing serous ovarian cases against healthy controls with an ROC of 0.719 at a sensitivity of 45% at >99% specificity (Figure 4A, solid line). In comparison, we generated a separate classifier utilizing the same training and test samples for the top two TAAb to serous ovarian cancer, p53 and CTAG2 (Figure 4B). While this could be due to the unexpected improvement in sensitivity from the training set to the validation set; this suggests that p53 and CTAG2 alone have a higher sensitivity and AUC when discriminating serous ovarian cases from healthy controls but on their own do not discriminate serous ovarian cases from benign disease controls. In addition, all cases were screened for CA125 prior to treatment. In our validation sample set there were 5 cases that were false negative for CA125. Using our 11-AAb panel, four of the five false negative CA125 cases were classified as positive, improving the sensitivity of the CA125 assay.
Figure 4.
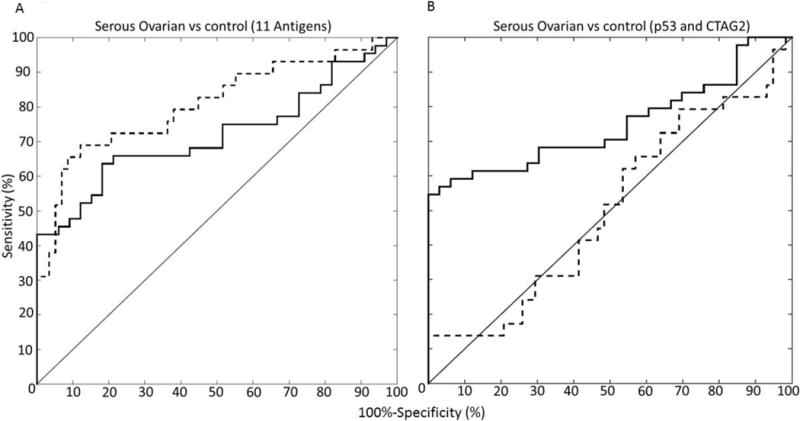
ROC classifier of the A. 11 TAAb classifier (CTAG2, ICAM3, KSR1, NUDT11, NXF3, POMC, PVR, STYXL1, TP53, TRIM39, and UHMK1) and B. p53 and CTAG2 classifier was calculated using a binary logistic regression analysis. The dotted line indicates the training set, consisting of serous ovarian cases, n = 29 vs benign disease and healthy controls, n = 58) and the solid line indicates the validation set (serous ovarian cases, n = 34 vs healthy controls, n = 32). We used the values obtained from our validation set to report our sensitivity and specificity.
4 Discussion
Sera samples from a total of 94 patients with serous ovarian cancer and 30 benign disease and 92 healthy control samples were screened for TAAb using our custom protein microarrays (Discovery and Training Sets). We identified an 11-AAb biomarker panel that distinguishes serous ovarian cancer from benign disease (32% sensitivity) and healthy controls (45% sensitivity) at 98% specificity (Figure 4A). Five of these markers have been previously identified in serous ovarian cancer (p53 and CTAG2) or related cancers (NUDT11, PVR, and TRIM39) as contributing to cancer progression. Three biomarkers, NUDT11, PVR, and TRIM39 were consistently selective for serous ovarian cancer with individual sensitivities ranging from 14.7% to 32.4% at >96% specificity. Previously identified biomarkers for serous ovarian cancer, p53 and CTAG2 displayed modest sensitivities 6.9% and 10.3% at 98% specificity discriminating serous ovarian cancer from benign disease controls but displayed higher sensitivities 44.1% and 23.5% at >96% specificity when compared to healthy controls.
Previous work on serous ovarian cancer associated TAAbs have focused on the detection of individual biomarkers with only recent attention being given to the development of a panel of TAAb [4]. Ovarian cancer is usually detected at late stages and with high mortality rates. CA 125 (MUC19) is a glycoprotein biomarker that detects 50% of early stage ovarian cancer and remains, to this day, the primary biomarker for ovarian cancer diagnosis in routine clinical use [3]. HE4 shows similar diagnostic properties as CA 125 [26]. Other potential biomarkers include MMP1 [27], cytokines [28], plasminogen activator receptor [29], osteopontin [9], MMP7 [30], B7-H4 [31], and kallikreins [32]. The use of multiple tumor markers may improve the performance characteristics of the tests [26]. In a phase III blinded validation study only serum concentrations of CA 125, HE4, and mesothelin have been shown to increase up to 3 years prior to clinical diagnosis [33]. This multimodal screening (MMS) strategy is further highlighted in the recent report from the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS), in which more than 200,000 women were followed for 14 year to assess the effects of early/annual screening on ovarian cancer mortality rates [34]. The finding, overall, suggests that annual long-term MMS reduces mortality rates by up to 28% compared to no screening. These two studies emphasize the importance of annual screening using a panel of biomarkers, multiple screening approaches, and detailed screening algorithms to reduce the mortality rates associated with ovarian cancer.
In our prior study using first generation NAPPA arrays we identified a panel of 12 TAAb with sensitivities ranging from 13% to 22% at >93% specificity [16]. Epitope presentation is critical for serological assays and it has been recently reported that over 90% of antibodies are generated against conformational epitopes, stressing the importance of protein folding and post-translational modifications for the identification of TAAb biomarkers [35]. Our first generation NAPPA arrays used rabbit reticulocyte lysate to perform IVTT [16]. Rabbit reticulocyte is a mammalian IVTT expression system with several drawbacks, including limited post-translation modifications and co-expression of off-target proteins, thus leading to high-background, batch-to-batch variability, and suboptimal expression conditions. In this study, we used improved second generation NAPPA arrays displaying >5,000 new full-length proteins as well as mammalian Hela cell lysate to express optimized and normalized cDNAs, providing higher protein yields. These changes have improved our reproducibility, lowered background, and increased assay sensitivity.
In addition to p53, we consistently identified TAAb to the previously reported CTAG2, as well as three novel ovarian autoantigens PVR, NUDT11, and TRIM39. While PVR was present on our first generation arrays both NUDT11 and TRIM39 are recent additions to our second generation arrays. We reconfirmed the presence of TAAbs to (p53, CTAG, and PVR) in serous ovarian cancer [16]. Tumor antigen-associated autoantibodies against p53 and CTAG2 proteins have been reported by our group as well as others as biomarkers for several tumor types such as serous ovarian [16], basal-like breast [11], and lung cancer [17]. Interestingly, TAAb against p53 and CTAG2 correlate with aggressive phenotypes of these cancers. The similar transcript expression profiles of ovarian, basal-like and lung cancer reported by The Cancer Genome Atlas (TCGA) suggest that these markers may have value in detecting multiple cancers [36].
Of the novel biomarkers we identified, several are associated with carcinogenesis. The family of nudix (NUDT) proteins, including NUDT11, are highly conserved among species and contribute to the hydrolysis of nucleoside diphosphate derivatives, hypothesized to control the cellular concentration of these compounds [37]. While the role of NUDT11 is unknown, there are studies supporting the role of NUDT1 driving cellular proliferation in breast, lung, colon, and prostate cancer [38]. Poliovirus receptor (PVR, also known as CD155) is an overexpressed cell surface protein in multiple tumor types that contributes to proliferation, invasion, and immune evasion [39]. PVR/ CD155 has been the recent target of oncolytic viral therapies, due to the abundant overexpression on tumor cells [40]. The TRIM family of proteins display both a positive and negative role in regulating oncogenesis by ubiquitinating key regulatory proteins such as p53 [41]. Although TRIM39 has not been directly implicated in tumor progression recent studies have demonstrated that TRIM39 can directly bind to and ubiquitinate p53, stabilizing p21 and leading to cell cycle progression in hepatocellular carcinoma [42].
There are several limitations of this study. First, there is evident variation in the frequency of TAAb detection between our training set and validation sets. We suspect that this may be due to the significantly older population in the validation set, which did not include benign disease controls (Table 1). These results warrant further evaluation with independent samples sets. We do not yet know how early these biomarkers are detected prior to ovarian cancer development, but p53-AAb have been detected up to 6 months prior to clinical disease [16, 18].
In summary, we performed the largest proteomic screen using programmable protein arrays to date on serous ovarian cancer and identified an 11-TAAb biomarker panel. Further validation of these markers is needed using independent sample sets. Early ovarian cancer diagnosis is limited by both the overall low incidence of the disease in the general population (1.5% lifetime risk) and the rapid development of peritoneal disease [4]. Routine screening with CA 125 serum levels, transvaginal ultrasound, and pelvic examinations has not been shown to improve morbidity or mortality from ovarian cancer [1]. Our analysis of TAAb associated with serous ovarian cancer represents promising markers for the early detection because their sensitivity is independent of TVUS, and blood tests can be performed often to determine the longitudinal changes in biomarker levels of high risk individuals without risk or expensive tests, providing a realistic approach for individuals who require more frequent testing. Early evaluation of these markers complement CA 125, improving sensitivity as well as improving the specificity of CA 125 for the evaluation of benign ovarian disease. Future work is needed to determine if these markers provide clinical value for diagnostic and/ or prognostic responses.
Supplementary Material
Supplementary Table 1. List of 10,247 antigens screened in our NAPPA discovery set, patients screened, and raw values.
Supplementary Table 2. List of 735 antigens screened in our NAPPA training set, patients screened, and raw values.
Supplementary Table 3. List of 39 antigens screened in our RAPID ELISA training set, patients screened, and GST normalized values.
Supplementary Table 4. List of 11 antigens screened in our RAPID ELISA validation set, patients screened, and GST normalized values.
Highlights.
Screened a total of 10,247 human antigens using customizable protein microarrays.
Performed orthogonal ELISA assay to validate 39 candidate autoantibody biomarkers.
Defined a panel of biomarkers with 45% sensitivity at >99% specificity to screen for serous ovarian cancer.
Acknowledgments
This study was funded by the NCI Early Detection Research Network U01 CA117374 (K.S.A and J.L.).
Abbreviations
- TAAb
tumor antigen-associated autoantibody
- Ab
Antibody
- SOC
serous ovarian cancer
- NAPPA
nucleic acid programmable protein arrays
- IVTT
In vitro transcription and translation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement
Dr. Anderson and Dr. LaBaer serve as members of the scientific advisory board for ProvistaDx. Dr. Anderson served as a consultant for ProvistaDx.
References
- 1.Society AC. Cancer Facts & Figures 2015. Atlanta: American Cancer Society; 2015. [Google Scholar]
- 2.Fast Stats: An interactive tool for access to SEER cancer statistics. Surveillance Research Program NCIhscgf.
- 3.Badgwell D, Bast RC., Jr Early detection of ovarian cancer. Dis Markers. 2007;23:397–410. doi: 10.1155/2007/309382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi JX, Qin JJ, Ye H, Wang P, Wang KJ, Zhang JY. Tumor associated antigens or anti-TAA autoantibodies as biomarkers in the diagnosis of ovarian cancer: a systematic review with meta-analysis. Expert review of molecular diagnostics. 2015;15:829–52. doi: 10.1586/14737159.2015.1035713. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2015 doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. Journal of proteome research. 2005;4:1123–33. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koziol JA, Zhang JY, Casiano CA, Peng XX, Shi FD, Feng AC, et al. Recursive partitioning as an approach to selection of immune markers for tumor diagnosis. Clin Cancer Res. 2003;9:5120–6. [PubMed] [Google Scholar]
- 8.Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, et al. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18:868–73. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee M, Mohapatra S, Ionan A, Bawa G, Ali-Fehmi R, Wang X, et al. Diagnostic markers of ovarian cancer by high-throughput antigen cloning and detection on arrays. Cancer Res. 2006;66:1181–90. doi: 10.1158/0008-5472.CAN-04-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson KS, Sibani S, Wong J, Hainsworth G, Mendoza EA, Eugene R, Raphael J, Logvinenko T, Ramachandran N, Godwin A, Marks J, Engstrom P, LaBaer J. Using custom protein microarrays to identify autoantibody biomarkers for the early detection of breast cancer. San Antonio Breast Cancer Symposium; San Antonio, TX: 2008. [Google Scholar]
- 11.Wang J, Figueroa JD, Wallstrom G, Barker K, Park JG, Demirkan G, et al. Plasma Autoantibodies Associated with Basal-like Breast Cancers. Cancer Epidemiol Biomarkers Prev. 2015;24:1332–40. doi: 10.1158/1055-9965.EPI-15-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katchman BA, Barderas R, Alam R, Chowell D, Field MS, Esserman LJ, et al. Proteomic Mapping of p53 Immunogenicity in Pancreatic, Ovarian, and Breast Cancers. Proteomics Clin Appl. 2016 doi: 10.1002/prca.201500096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Yu J, Sreekumar A, Varambally S, Shen R, Giacherio D, et al. Autoantibody signatures in prostate cancer. N Engl J Med. 2005;353:1224–35. doi: 10.1056/NEJMoa051931. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen JW, Gentry-Maharaj A, Fourkala EO, Dawnay A, Burnell M, Zaikin A, et al. Early detection of cancer in the general population: a blinded case-control study of p53 autoantibodies in colorectal cancer. British journal of cancer. 2013;108:107–14. doi: 10.1038/bjc.2012.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lubin R, Zalcman G, Bouchet L, Tredanel J, Legros Y, Cazals D, et al. Serum p53 antibodies as early markers of lung cancer. Nature medicine. 1995;1:701–2. doi: 10.1038/nm0795-701. [DOI] [PubMed] [Google Scholar]
- 16.Anderson KS, Cramer DW, Sibani S, Wallstrom G, Wong J, Park J, et al. Autoantibody signature for the serologic detection of ovarian cancer. Journal of proteome research. 2015;14:578–86. doi: 10.1021/pr500908n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desmetz C, Mange A, Maudelonde T, Solassol J. Autoantibody signatures: progress and perspectives for early cancer detection. J Cell Mol Med. 2011;15:2013–24. doi: 10.1111/j.1582-4934.2011.01355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson KS, Wong J, Vitonis A, Crum CP, Sluss PM, Labaer J, et al. p53 autoantibodies as potential detection and prognostic biomarkers in serous ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2010;19:859–68. doi: 10.1158/1055-9965.EPI-09-0880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, et al. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. Journal of proteome research. 2008;7:1490–9. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anderson KS. Multiplexed detection of antibodies using programmable bead arrays. Methods in molecular biology. 2011;723:227–38. doi: 10.1007/978-1-61779-043-0_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, et al. Next-generation high-density self-assembling functional protein arrays. Nature methods. 2008;5:535–8. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitonis AF, Titus-Ernstoff L, Cramer DW. Assessing ovarian cancer risk when considering elective oophorectomy at the time of hysterectomy. Obstet Gynecol. 2011;117:1042–50. doi: 10.1097/AOG.0b013e318212fcb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cramer DW, Vitonis AF, Welch WR, Terry KL, Goodman A, Rueda BR, et al. Correlates of the preoperative level of CA125 at presentation of ovarian cancer. Gynecol Oncol. 2010;119:462–8. doi: 10.1016/j.ygyno.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Barker K, Steel J, Park J, Saul J, Festa F, et al. A versatile protein microarray platform enabling antibody profiling against denatured proteins. Proteomics Clin Appl. 2013;7:378–83. doi: 10.1002/prca.201200062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong J, Sibani S, Lokko NN, LaBaer J, Anderson KS. Rapid detection of antibodies in sera using multiplexed self-assembling bead arrays. Journal of immunological methods. 2009;350:171–82. doi: 10.1016/j.jim.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore RG, McMeekin DS, Brown AK, DiSilvestro P, Miller MC, Allard WJ, et al. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol Oncol. 2009;112:40–6. doi: 10.1016/j.ygyno.2008.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertenshaw GP, Yip P, Seshaiah P, Zhao J, Chen TH, Wiggins WS, et al. Multianalyte profiling of serum antigens and autoimmune and infectious disease molecules to identify biomarkers dysregulated in epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2872–81. doi: 10.1158/1055-9965.EPI-08-0464. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Yu Y, Xu F, Berchuck A, van Haaften-Day C, Havrilesky LJ, et al. Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol Oncol. 2007;107:526–31. doi: 10.1016/j.ygyno.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henic E, Borgfeldt C, Christensen IJ, Casslen B, Hoyer-Hansen G. Cleaved forms of the urokinase plasminogen activator receptor in plasma have diagnostic potential and predict postoperative survival in patients with ovarian cancer. Clin Cancer Res. 2008;14:5785–93. doi: 10.1158/1078-0432.CCR-08-0096. [DOI] [PubMed] [Google Scholar]
- 30.Palmer C, Duan X, Hawley S, Scholler N, Thorpe JD, Sahota RA, et al. Systematic evaluation of candidate blood markers for detecting ovarian cancer. PLoS ONE. 2008;3:e2633. doi: 10.1371/journal.pone.0002633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon I, Liu Y, Krall KL, Urban N, Wolfert RL, Kim NW, et al. Evaluation of the novel serum markers B7-H4, Spondin 2, and DcR3 for diagnosis and early detection of ovarian cancer. Gynecol Oncol. 2007;106:112–8. doi: 10.1016/j.ygyno.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Yousef GM, Diamandis EP. The human kallikrein gene family: new biomarkers for ovarian cancer. Cancer Treat Res. 2009;149:165–87. doi: 10.1007/978-0-387-98094-2_8. [DOI] [PubMed] [Google Scholar]
- 33.Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, et al. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst. 102:26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945–56. doi: 10.1016/S0140-6736(15)01224-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy MA, O’Connell DJ, O’Kane SL, O’Brien JK, O’Toole S, Martin C, et al. Epitope presentation is an important determinant of the utility of antigens identified from protein arrays in the development of autoantibody diagnostic assays. Journal of proteomics. 2012;75:4668–75. doi: 10.1016/j.jprot.2012.02.031. [DOI] [PubMed] [Google Scholar]
- 36.Reuschenbach M, von Knebel Doeberitz M, Wentzensen N. A systematic review of humoral immune responses against tumor antigens. Cancer immunology, immunotherapy : CII. 2009;58:1535–44. doi: 10.1007/s00262-009-0733-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakabeppu Y. Cellular levels of 8-oxoguanine in either DNA or the nucleotide pool play pivotal roles in carcinogenesis and survival of cancer cells. International journal of molecular sciences. 2014;15:12543–57. doi: 10.3390/ijms150712543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grisanzio C, Werner L, Takeda D, Awoyemi BC, Pomerantz MM, Yamada H, et al. Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11252–7. doi: 10.1073/pnas.1200853109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kono T, Imai Y, Yasuda S, Ohmori K, Fukui H, Ichikawa K, et al. The CD155/poliovirus receptor enhances the proliferation of ras-mutated cells. International journal of cancer Journal international du cancer. 2008;122:317–24. doi: 10.1002/ijc.23080. [DOI] [PubMed] [Google Scholar]
- 40.Brown MC, Dobrikova EY, Dobrikov MI, Walton RW, Gemberling SL, Nair SK, et al. Oncolytic polio virotherapy of cancer. Cancer. 2014;120:3277–86. doi: 10.1002/cncr.28862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatakeyama S. TRIM proteins and cancer. Nature reviews Cancer. 2011;11:792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Mei Y, Fu NY, Guan L, Xie W, Liu HH, et al. TRIM39 regulates cell cycle progression and DNA damage responses via stabilizing p21. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20937–42. doi: 10.1073/pnas.1214156110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. List of 10,247 antigens screened in our NAPPA discovery set, patients screened, and raw values.
Supplementary Table 2. List of 735 antigens screened in our NAPPA training set, patients screened, and raw values.
Supplementary Table 3. List of 39 antigens screened in our RAPID ELISA training set, patients screened, and GST normalized values.
Supplementary Table 4. List of 11 antigens screened in our RAPID ELISA validation set, patients screened, and GST normalized values.


