ABSTRACT
The assembly of specialized sub-nuclear microenvironments known as nuclear bodies (NBs) is important for promoting efficient nuclear function. In particular, the Cajal body (CB), a prominent NB that facilitates spliceosomal snRNP biogenesis, assembles in response to genomic cues. Here, we detail the factors that regulate CB assembly and structural maintenance. These include the importance of transcription at nucleating gene loci, the grouping of these genes on human chromosomes 1, 6 and 17, as well as cell cycle and biochemical regulation of CB protein function. We also speculate on the correlation between CB formation and RNA splicing levels in neurons and cancer. The timing and location of these specific molecular events is critical to CB assembly and its contribution to genome function. However, further work is required to explore the emerging biophysical characteristics of CB assembly and the impact upon subsequent genome reorganization.
KEYWORDS: Cajal bodies, coilin, chromatin, genome organization, histone locus bodies, nuclear bodies, oncogenesis, phase separation, RNA splicing, spliceosomal snRNPs
Introduction
It is evident that gene expression is controlled by the functional interplay between spatial genome organization, nuclear architecture and metabolic demand. To achieve optimal regulation of gene expression and RNA processing, the cell nucleus is non-randomly organized into a number of structurally-distinct regions, including chromosome territories, the nuclear lamina and a number of nuclear domains or microenvironments known as nuclear bodies (NBs).1-3 These domains are frequently associated with specific gene loci, the activities of which appear to contribute to their biogenesis. However, as the nuclear interior lacks defining membranes, NBs behave like liquid-phase droplets and are excluded from the surrounding nucleoplasm by concentration-dependent phase separation.4 As a result, these regions are enriched in key but frequently limited factors and enzymes essential to expediting specific molecular events. These processes would be energetically unfavorable to perform without temporal and spatial compartmentalization in a defined space within the nucleus.1,5-7 Recent evidence has indicated that these structures are functionally diverse but indispensable for cell viability and proliferation, although some NBs appear to be more essential than others.6 The nucleus is a highly dynamic and cooperative network and these structurally defined domains are not completely functionally independent. Thus, there are numerous regulatory feedback loops and physiological influences that trigger the nucleation of specific NBs and control their function.
Established NBs include the nucleolus, Cajal body (CB), histone locus body (HLB), PML nuclear body, nuclear speckle and paraspeckle and our knowledge of the specific functions of these domains is continuously improving.1,3,8 Additionally, a number of “orphan” NBs have been recognized that require further characterization of their resident proteins, RNAs and specific functions.9 However, the role of the CB is relatively well established. Assembly of the CB is an important contributing factor to efficient pre-mRNA processing (RNA splicing) by the multi-megadalton ribonucleoprotein complex known as the spliceosome.10 The enzymatic core of the spliceosome consists of small nuclear ribonucleoproteins (snRNPs), which are RNA-protein complexes, as well as a large number of accessory proteins that regulate their function.10,11 Briefly, RNA splicing involves the 5′ and 3′ definition of introns in target pre-mRNAs by U1 and U2 snRNPs, respectively, followed by binding of the U4/U6.U5 tri-snRNP to an intronic region known as the branchpoint sequence.11 The intron is removed by lariat formation and is targeted for degradation, or further processed to generate intron-encoded small nuclear and nucleolar RNAs (sn/snoRNAs), while the adjacent exons are joined together.12 Following the completion of each round of splicing, the snRNP components are individually liberated from the target pre-mRNA. U4/U6 di-snRNP and U4/U6.U5 tri-snRNP must then be reassembled and recycled to form competent splicing units for another round of RNA processing.
Spliceosomal snRNA and sn/snoRNP enrichment in CBs was first observed in the 1990s13-18 but splicing itself takes place predominantly in the space between the nuclear speckle periphery and adjacent chromatin domains.19 The CB augments spliceosomal U snRNA, intron-encoded snoRNA, and small CB-specific RNA (scaRNA) gene expression20,21 and the co-transcriptional processing of their 3′ extended end.22 Other CB-related processes include RNA base modifications, (site-specific pseudouridinylation and 2′-O-methylation guided by scaRNAs)23 snRNP assembly and maturation,24-26 as well as telomere maintenance.27 CB disassembly, or relocalization of several key CB components, has also been linked to the cellular response to DNA damage.28-31 Furthermore, the CB influences the stepwise assembly of U4/U6.U5 tri-snRNPs before and after each round of RNA splicing.32 These CB-enriched sn/snoRNAs also regulate RNA splicing,33 as well as pre-rRNA processing,34 and guide the base modification of RNA transcripts.35 However, despite the dependency of certain cell types on CBs for function, CBs have only been observed in a limited number of cells,36 including most neuronal cells, embryonic and fetal tissue,37 stem cells,38 and a large variety of aneuploid transformed cells. Thus, the CB is an important nuclear microenvironment that is likely to be a prototypical model example of the influence of synergistic cellular pathways in regulating nuclear structure assembly.
So what molecular events must occur for a CB to materialize in the nucleus? In plain language, the assembly of CBs appears to correlate with a “perfect storm” of high local threshold concentrations of key nucleation and scaffolding factors associated with specific gene regions of high transcriptional activity. Other regulators of this process include biochemical modifications of key CB structural components, cell cycle progression and metabolic demand.5 In the following sections we examine the reported details of CB nucleation, assembly steps and structural maintenance, and comment on some of the mysteries of CB formation that have yet to be resolved.
Temporal and spatial regulation of Cajal body assembly
Many, if not all, NBs assemble at specific genomic loci or chromosomal regions during defined cell cycle stages and times of specific metabolic need, stimuli or stress.3 It is important to note that the CB is likely the most canonical NB to assemble through heterogeneous nucleation, which requires an initial nucleation step using a seeding scaffold followed by stochastic CB self-assembly.39 CBs do not form through homogenous nucleation, which would be represented by the spontaneous formation of randomly positioned CBs in the nucleoplasm. In particular, the CB has long been known to be frequently associated with specific genomic loci enriched with U snRNA and intron-encoded snoRNA, scaRNA and histone gene loci.15,20,40-45 However, it has not been fully explored and genomically mapped whether the initiation of CB formation is restricted to a limited number of prominent primary nucleation sites (such as the highly expressed and prominent RNU1 and RNU2 gene arrays46 or histone gene clusters) or other highly expressed individual gene loci or repeats. It is unknown whether these must be positioned either in cis configurations on the same chromosome or in trans on different chromosomes to trigger CB formation.20 Also, there may not be a one-to-one relationship between a specific gene locus and the CB, as several of these transcriptionally active regions have been observed to be attracted to the CB periphery by simultaneous repositioning or at least in a semi-coordinated fashion.20,47 The CB exists and maintains a close relationship with the genome, as it arises at particular gene loci after mitosis in late G1 phase when widespread transcription is fully established, and CB number doubles during genome duplication in S phase.
The assembly and disassembly of all NBs is functionally linked to their inheritance during cell division. Some NBs, including CBs, persist though mitosis in the form of mitotic bodies that contain essential components in an inactive state.48 These mitotic NBs are equally distributed between daughter cells after cell division of the mother cell.49,50 Efficient NB reassembly at the end of mitosis ensures the full functionality of newly formed daughter cells. CBs remain intact through interphase and at the onset of mitosis. However, after the nuclear envelope breaks down they scatter throughout the cytoplasm and are physically separate from condensed chromosomes. The number of mitotic CBs remains the same from metaphase to telophase and functionally they are likely to be in an arrested state or at least contain high levels of retained intermediates. Indeed, these mitotic CBs contain spliceosomal snRNPs but target gene transcription and splicing is abolished during this stage of the cell cycle. Accordingly, the composition of these mitotic CBs is different from interphase CBs, as the essential CB component TCAB1/WRAP53 is absent from the domain51 but a thorough characterization of mitotic CB composition is necessary. Interestingly, as TCAB1/WRAP53 has been proposed to be the key element that targets sn/snoRNAs and scaRNAs to the CB,52 this may coincide with a redistribution of these small RNAs through the nucleoplasm. During mitosis, the key structural component of CBs, coilin,53 undergoes a change in phosphorylation status, which regulates its stability and leads to CB reassembly after mitosis.54-56 Once the nuclear envelope is reformed in daughter nuclei in early telophase, mitotic CBs rapidly disintegrate and coilin rapidly diffuses through the nuclear interior. Newly re-assembled post-mitotic CBs are formed later in G1 phase when global transcription is fully restored.
However, the CB does not exist in isolation within the nucleus. Frequent physical associations have been reported, primarily with the HLB,57 nucleolus,58,59 and PML nuclear body.60 Additionally, the current total number of reported target sn/snoRNA and scaRNA genes that are potentially targeted to, processed or sequestered in the CB is close to 1700 loci, although the exact number is under consideration.61 These gene loci are located across the genome on all human chromosomes) and more are frequently being discovered and annotated.62 We performed a simple but novel analysis by collecting the known locations of CB target genes from publically-accessible databases63,64 and plotting their enrichment across the genome in discrete 1 Mbp bins (Fig. 1). Several chromosomes, including 1, 6 and 17, appear to display higher local and total CB target gene density than other size-matched chromosomes (2, 7 and 18, respectively; Fig. 1a-b) and are reported to be frequently proximal to CBs.20,64,65 This may contribute to CB nucleation and their sub-nuclear positions. Given the known genomic organizational properties of the CB and the frequent but not universal presence of this sub-nuclear structure in most cell types, including a high variety of transformed cancer cells, the CB represents a unique model system for the study of the role in NB formation.
Figure 1.
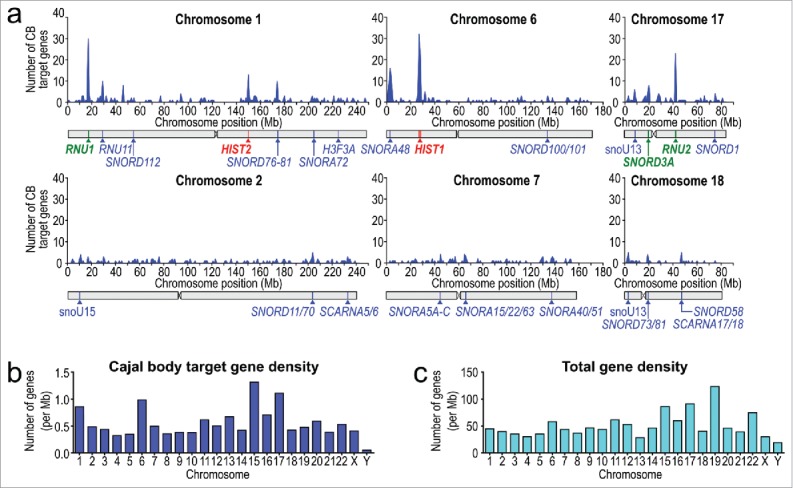
Cajal bodies nucleate at chromatin regions with high target gene density. (a) CB target gene density for human chromosomes containing major sn/snoRNA gene arrays and size-matched chromosomes. Genes were binned into 106 base pair (1 Mbp) windows. The positions of potential major CB nucleation sites and other notable intron-encoded small RNAs are indicated with pointers. List of U sn/snoRNA genes were acquired from publically-accessible databases and potentially includes pseudogenes and other intron-encoded sn/sno/scaRNAs that are neither expressed nor sensitive to CB disassembly by knockdown of essential CB components by siRNA. CB target gene loci include spliceosomal U snRNA, SNORD, SNORA, SCARNA and histone genes. (b) CB target gene density and (c) total gene density for all human chromosomes, normalized per 106 base pairs indicates that CB target gene density is independent of total gene density (Pearson coefficient = 0.49).
Seeding RNA by transcription to grow Cajal bodies
The CB is an excellent example of a dynamic but structurally stable nuclear entity that forms at sites and times of target gene transcription or RNA processing. Inhibition of RNA Polymerase II transcription initially results in redistribution of snRNPs away from the CB,66 which is followed by CB structural disassembly after several hours.58,67 Additionally, high levels of transcription have been associated with increased CB numbers per cell.68 Recent studies have indicated that several nuclear bodies, including the CB, the nucleolus, HLBs, nuclear speckles, paraspeckles and nuclear stress bodies require a specific RNA scaffold for an initial nucleation step.69,70 Therefore, it is likely that transcription plays a critical role in regulating CB assembly.
However, CB-positioning does not coincide with transcriptional activity at random gene loci. Long-established sites of CB association include the major and minor replication-dependent histone gene clusters on chromosomes 6 and 1, respectively (HIST1 and HIST2),42 as well as major (U1, U2, U4, U5, U6) and minor (U4atac, U6atac, U11, U12) spliceosomal U snRNA genes.40,41,43,71-73 These are all RNA polymerase II-transcribed genes, except RNU6, which is transcribed by RNA polymerase III, and several are situated in large clusters (RNU1 and RNU2). These possess independent and structurally unique promoters that are activated by specific transcription factors, including the pentameric snRNA-activating protein complex (SNAPc).74,75 Furthermore, transcription of snRNA genes requires the Little Elongation Complex, whose components are present in both the nucleoplasm and CB,76 for initiation and elongation.77 The Integrator complex is responsible for co-transcriptional cleavage of the unique 3′-end of pre-U snRNA transcripts78 but only one subunit is currently known to be present in CBs.79 A number of intron-encoded sn/snoRNA and scaRNA genes have also been identified as frequent CB-interacting regions.20 These genomic regions are targeted to the CB periphery in an active repositioning process.47 This occurs after formation of the structure and their RNA products are sequestered and maturated in the CB microenvironment.62 However, it is unclear how many of these specific gene loci represent primary CB nucleation hubs, or if all CB target gene loci are capable of nucleating the structure to some degree. Regardless, the nucleation of the CB can be summarized into 3 main steps: i) Transcription and nucleation, ii) Phase separation, and iii) Genome clustering (summarized in Fig. 2).
Figure 2.
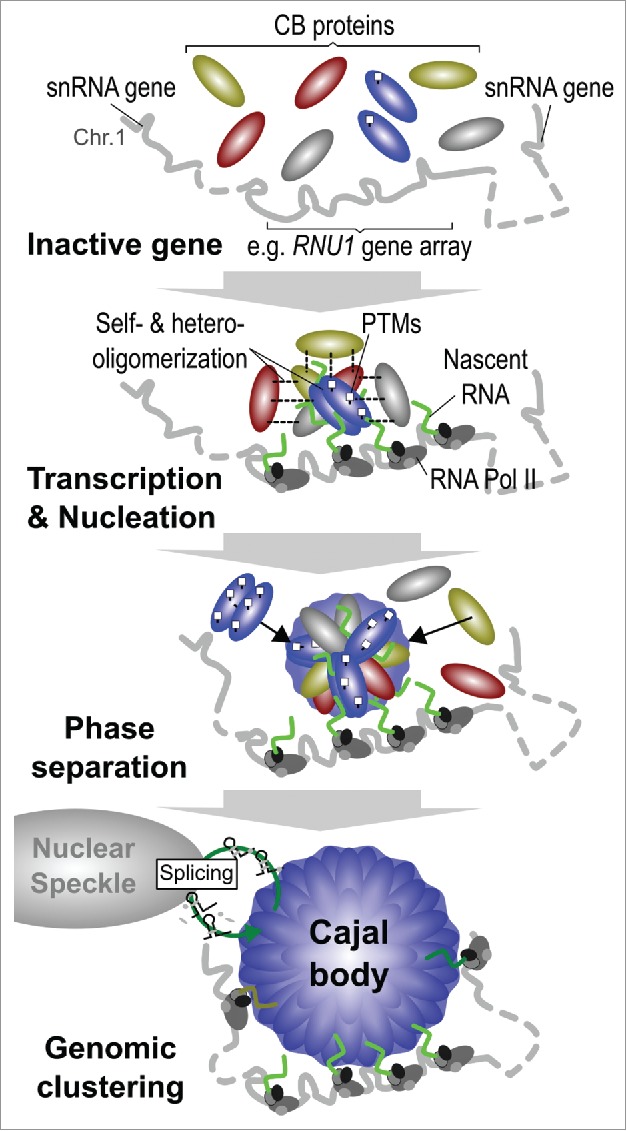
The stages of Cajal body assembly. Canonical Cajal bodies do not form at inactive genes, or spontaneously at random gene loci in the nucleoplasm. Instead, CB components are attracted to specific gene loci, such as the U1 snRNA gene array on chromosome 1, upon transcriptional activation by RNA polymerase II (RNA pol II) and the Integrator complex during G1 phase of the cell cycle. This involves a large number of homo- and hetero-association steps between multivalent hub proteins and other components that are modulated by post-translational modifications (PTMs). The CB becomes phase separated from the surrounding nucleoplasm, enriching specific CB factors within the microenvironment. As the CB grows, other active target gene loci are attracted to the periphery of the domain, which create gene clusters in both cis and trans configurations. Mature snRNPs produced by the CB are exported to the nuclear speckle for spliceosome assembly and mRNA processing prior to recycling in the CB.
(i) (a) Transcription and Nucleation – sn/snoRNA genes. The major U1 snRNA (RNU1) gene cluster, which spans 0.5 Mbp on the 5′ end of the chromosome 1 p-arm, appears to possess a unique relationship with CB formation as it displays the highest association frequency with the CB in HeLa cell colocalization studies.20 We hypothesize that once U1 snRNA gene expression at this array reaches a critical local threshold concentration, the CB begins to nucleate. This attracts and accumulates key CB components, including the structural multivalent CB component coilin, which has the capability to interact with numerous CB components and form homo- and hetero-oligomers.80,81 Coilin accumulation enhances NB assembly and stabilizes transient interactions among other CB components. However, only approximately 40% of CBs are closely associated with the U1 snRNA gene array on chromosome 1 (60% of all CBs were associated with chromosome 1) in HeLa cells.20 Therefore, it is likely that other genomic loci located on other chromosomes also contribute to global CB nucleation. Based upon sn/snoRNA gene density (Fig. 1a-c) and expression levels it is highly likely that the U2 and SNORD3A (U3) sn/snoRNA gene arrays on chromosome 17 are also essential CB-nucleating regions. Indeed, chromosome 17 is the second most frequently CB-associated region identified in HeLa cells, followed by chromosomes 15 and 6, the latter of which contains the major histone gene cluster (HIST1).20
(i) (b) Transcription and Nucleation – Histone genes. As mentioned above, CBs are frequently physically associated with HLBs. Approximately 42% of HeLa cells and 88% of MCF10A cells, which are spontaneously immortalized human breast epithelial cells, possess CBs and HLBs in physical contact to one another (Fig. 3a-d).57 In support of this observation, tethering of unprocessed human histone pre-mRNAs on an engineered genomic locus82 resulted in de novo formation of a CB with a physically associated HLB.70 These data indicate that a high local concentration of unprocessed histone pre-mRNA at a gene locus is able to act as seeding scaffold to nucleate a de novo CB. It is tempting to speculate upon which components are shared between these 2 NBs and might be responsible for bring CB and HLB to physical association. We hypothesize that this frequent CB-HLB associations arises due to 5 members of the heptameric Sm ring that are shared by both the CB-enriched major spliceosomal U snRNPs and the HLB-enriched histone-specific U7 snRNP. These Sm proteins display the highest de novo CB nucleation efficiency in HeLa cells tethering experiments.83 Thus, through physical association, HLBs can benefit from the ability of the CB to topologically reshape genomic regions and target specific CB-associated gene loci, including HLB-regulated replication-dependent histone gene clusters,84 to highly transcriptionally active regions located outside of chromosome territories.
Figure 3.
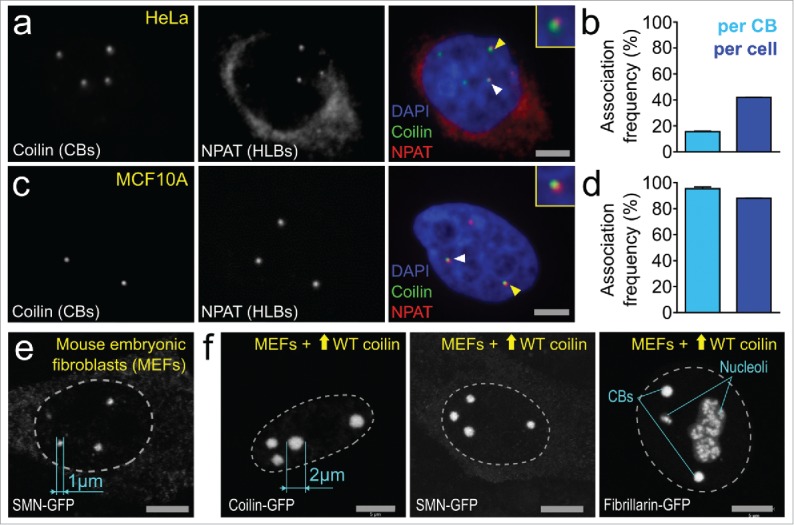
Cajal bodies are dynamic structures. (a-d) HeLa cells and human breast epithelial cell line MCF10A cultured for 3 d and fixed using 4% paraformaldehyde were stained with antibodies to detect CBs (CB marker protein coilin, green) and HLBs (histone transcription factor NPAT, far red) and imaged using the Opera 5020 high-content automated confocal microscopy system (PerkinElmer, Waltham MA). Briefly, nuclear segmentation and automated spot detection was performed using proprietary PerkinElmer Acapella software. CB-HLB distances were exported and a threshold (4 pixels between spot centers) was applied to quantify CB-HLB associations. N = 2, approx. Five,000 cells. Associations were normalized on a per-CB and per-cell basis. (e) Maximal intensity projection image of mouse embryonic fibroblasts (MEFs) expressing endogenous WT coilin transfected with plasmids containing SMN-GFP. CB size was measured using Zeiss Zen 2 software. (f) Maximal intensity projection images of MEFs transfected with plasmids containing WT (untagged) mouse coilin plus SMN-GFP, mouse coilin-GFP or fibrillarin-GFP (to co-stain both CBs and nucleoli). CB size was measured using Zeiss Zen 2 software.
(i) (c) Transcription and Nucleation – Summary. Comparison of the linear genomic clustering of CB target genes, including sn/snoRNA and histone gene arrays, as well intron-encoded small RNAs, revealed that the density of these CB-interacting regions is far higher on chromosomes 1, 6 and 17 compared to size-matched pairs (chromosomes 2, 7, and 18, Fig. 1a-b). We hypothesize that, similar to the proposed nucleation of Polycomb bodies,85 the linear clustering of transcriptionally active target CB genes in close genomic proximity may be important for initial CB assembly. However, it should be noted that interesting small RNA genes are also found on less CB target gene-dense chromosomes (which is independent of total gene density, Pearson correlation coefficient = 0.49 between total and snRNA somatic chromosome gene densities, Fig. 1b-c). In summary, CB formation is most likely to occur at the prominent sites of target gene expression with high gene density and/or transcription levels, including the U1, U2 and SNORD3A (U3) sn/snoRNA gene arrays and the histone gene clusters.
ii) Phase Separation. Following transcription, the CB condenses from the surrounding nucleoplasm by concentration-dependent phase separation. Briefly, phase separation describes the process by which a single-phase system (e.g. nucleoplasm) transitions to a system containing multiple coexisting liquid phases (e.g., nucleoplasm + CB). Modeling of NB nucleation and growth in vivo under different experimental conditions has confirmed that NBs are indeed phase separated droplet organelles.86,87 As a result, NB domains are compositionally distinct, but not impermeable,88 to the surrounding interchromatin space leading to optimized regulation of their specific molecular functions. This sub-nuclear phase transition may be dependent upon coilin's ability to act as a molecular hub and interact with numerous CB components via hetero-oligomerization, which stabilizes the developing structure.81,89 Nucleation of similar RNP-based droplets occurs via phase separation and is dependent upon both RNA and a rapid local accumulation of RNA binding proteins, supporting this hypothesis.90 When these CB components reach a local threshold concentration, liquid phase separation occurs and forms the microenvironment.4,91 This coincides with further concentration of specific essential and frequently limited factors that boost CB-dependent transcription of spliceosomal snRNA genes, the processing of their 3′ extended end and maturation of snRNPs by macromolecular crowding.6,92 Intriguingly, the frequent association between CBs and chromatin may further enhance phase separation and body formation.93 Thus, the CB potentially becomes a more efficient processing microenvironment, which substantially contributes to the elevated levels of RNA splicing in highly proliferative cells and histones needed for packing newly replicated DNA into nucleosomes. It is unknown at what stage (and to what extent) snRNA processing and snRNP maturation is accelerated during CB assembly. However, in theory, as soon as there is an increased local concentration of CB factors (compared to the surrounding nucleoplasm) this should be sufficient to ensure that snRNA and snRNP processing is augmented.
iii) Genomic Clustering. Finally, the growing CB attracts other target genes in cis and trans configurations to its physical proximity.20,44,71 It is unclear whether these target gene loci may possess their own smaller CBs (that are sub-microscopic) prior to amalgamation into a larger structure,94 or if there are only moderate local concentrations of coilin and the various sn/snoRNA transcriptional and processing complexes. Nevertheless, the CB induces a topological rearrangement of the genome and is seen to frequently associate with several target gene loci simultaneously, while pushing away nearby non-target genomic regions.20,44 The biological consequences of this cluster formation around the CB is not apparent and further work is required to describe the stability and benefit of these higher-order gene association events. Several studies have indicated that CBs are capable of both fusion and fission, but it is unknown with which genomic loci these CBs are associated.47,95-98 Following fusion, CBs were observed to increase in volume but this process appears to be limited by a size criticality in most cell lines as theoretically-possible CBs of more than 1.5 µm are not reported in the literature.8 This potentially indicates that the size of the CB is a reflection of the number of simultaneous gene associations that form with targets across the genome, global chromatin organization or the accumulation of sn/snoRNA transcripts that are sequestered within the domain. There are few reported exceptions to this rule, including overexpression of SMN99 and snRNPs,100 which induces small increases in human CB volume, as well as in plants where overexpression of a coilin homolog resulted in substantially larger CBs.101 However, using mouse embryonic fibroblast (MEF) cells, we have observed CBs of an increased size (2 µm diameter or more) without an increase in the number of CBs following overexpression of WT untagged coilin alongside co-expression of GFP-tagged CB components such as SMN (Fig. 3e-f). This phenotype is not observed in cells overexpressing coilin-GFP only.102 We believe that tagging of the N- or C-terminal regions of coilin by GFP potentially inhibits the enlargement of CBs in these embryonic cells, possibly by disrupting coilin's self-association ability. These data indicate that attaching fluorescent tags to either the C- or N-termini of coilin inhibits full function in ectopic overexpression studies and that the previously reported size criticality may not be universally applicable across all mammalian cell types.
Pulling itself together: Oligomerization and Cajal body assembly
An important dynamic property of NBs is their overall structural stability, as the CB itself persists for up to 10 hours.47 This is maintained by the continuous flux of components into and out of the domain.88 The typical residence time of components within a CB is in the range of seconds to a few minutes, although coilin and survival motor neuron (SMN) protein appear to exchange between the CB and the surrounding nucleoplasm at slower rates than other CB components.8,103-105 It has been established that the assembly and structural maintenance of NBs is predominantly influenced by several prominent architectural proteins that are able to self-associate to form homo- and hetero-oligomers.80,87 Many CB proteins have been identified as structurally essential for CB integrity, but little is known about the biochemical properties and regulation of each factor. The majority of our knowledge regarding the proteins essential for CB assembly regards coilin and SMN, which are the classical CB marker proteins, as well as a limited number of other proteins.
Coilin is the most well-characterized obligate multivalent protein factor in the CB assembly process.81 The protein is densely phosphorylated and contains a self-interaction domain on its N-terminus. Coilin is capable of forming homo-oligomers through these N-terminal domains that are predicted to be one of the early events CB assembly and a major stabilization factor. However, it seems that this self-association is influenced by strict regulation of phosphorylation events on residues located on the C-terminus domain. In human primary cells, which typically lack CBs, coilin is hyperphosphorylated on C-terminal phosphoserine residues and this is proposed to reduce its ability to self-interact.102 By contrast, in transformed cells, which typically have a high number of CBs, coilin is hypophosphorylated at these sites and able to pull the body together. To date, 18 distinct human coilin phosphorylation residues have been reported, with several known regulators of their modification, including the phosphatase PPM1G.106 Cell cycle-dependent phosphorylation, potentially by the Serine-Threonine kinase VRK1,55,107 has also been linked to structural integrity of the CB. Differential coilin phosphorylation may mislocalize CBs to the intra-nucleolar space, potentially resulting in aberrant snRNP processing.108,109 Furthermore, PRMT5-dependent symmetrical arginine dimethylation (sDMA) in the coilin arginine/glycine (RG) box mediates coilin's association with SMN.110 Ablation of sDMA modification induces the physical separation of coilin-positive residual CBs and SMN-positive foci (Gems).88,111 Interestingly, it has been suggested that coilin shares many similarities with other organelle forming proteins, including an enrichment of intrinsically disordered domains112 which may be crucial for nucleating the CB phase transition.113 Also, coilin is predicted to bind DNA and RNA, which may influence protein oligomerization. In particular, binding of U1 and U2 snRNA to coilin are reported to change the protein conformation. This may enhance self-association, but a defined binding region within coilin has not been identified114 (although the C-terminal Tudor domain is predicted to bind nucleic acids115). Thus, many aspects of CB biology are regulated by coilin oligomerization and post-translational modifications.
The SMN protein is another major CB component and forms the oligomeric core of a multiprotein complex that is involved in the biogenesis of spliceosomal snRNPs.116 SMN forms stable dimers,117 which in turn self-associate to form tetramers and octamers. Nearly half of the missense mutations found in Spinal Muscular Atrophy (SMA) (a genetic disorder in infants whose phenotype includes abnormal CB formation, caused by deletions or mutations in the SMN1 gene) patients are located in the C-terminal region of SMN within a highly conserved oligomerization domain termed the YG box.118 Some SMA patient YG box mutations have been shown to result in a decrease in the ability of SMN to self-oligomerize and associate with other interactive partners. As an enriched protein within the CB, it is predicted that these SMN self-oligomerization events help to drive CB nucleation and growth, resulting in enrichment of spliceosomal snRNPs within the CB.
The function of several other proteins have also been suggested to regulate CB integrity. Depletion of TCAB1/WRAP53,119 USPL176 or hCINAP120 (among others,121-124 summarized in Table 1) results in CB disassembly and disruption of CB target processes. Of note, TCAB1/WRAP53 appears to mediate the formation of functional CBs by bridging 2 large and mostly independent protein-based complexes that are centered on multivalent proteins, coilin and SMN, respectively.125,126 However, the functional role of other proteins in CB formation and maintenance is unclear. These proteins may regulate coilin and SMN self-association, or another aspect of CB assembly. Further work is required to categorize these proteins as essential assembly or maintenance factors and describe their explicit CB-based functions. Finally, full characterization of other, non-phosphorylation, coilin and SMN post-translational modifications that are reported to influence CB stability is required.127,128
Table 1.
List of selected proteins reported to be required for CB structural maintenance. For simplification, disruption was determined based upon localization of the universally used CB marker protein coilin (or SMN following coilin depletion).
| Protein | Cajal body disruption* |
|---|---|
| AIDA1c | Partial |
| Coilin | Partial114 |
| Fam118B | Complete |
| hCINAP | Complete |
| hTGS1 | Partial |
| ICE1 | Complete |
| INTS4 | Complete |
| PHAX | Partial |
| SMN | Partial |
| TCAB1/WRAP53 | Complete |
| TDP-43 | Partial |
| TOE1 | Complete |
| USPL1 | Complete |
| VRK1 | Complete |
Cajal bodies and a cellular game of “Hide-and-go-seek”
A long-unanswered problem within the CB field is why CBs are present within specific cell types but not others, leading researchers to hunt for the structure in many cell types and various physiological or experimental conditions.129 Indeed, for an evolutionarily-conserved structure it is puzzling to observe CBs within the brain and fetal tissue, as well as transformed cancer cells, but few normal primary adult diploid cells. The presence of CBs in neuronal cells, including terminally differentiated neurons that are incapable of cell division, is potentially indicative of a lineage-specific factor that positively or negatively regulates CB component oligomerization, such as neuronal growth factor (NGF) or fibroblast growth factor (FGF).130,131 Alternatively the appearance of CBs in these cells is part of a metabolic response. Specifically, the presence of CBs in brain tissue could be a cellular response to high splicing demands, as neuronal tissues have among the highest rates of alternative splicing in humans.132 As previously shown, neuritogenesis itself is a splicing-intensive process and requires high-levels of U snRNPs and component recycling.133 Thus, higher levels of snRNA transcription is required, as well as snRNP turnover after each round of splicing, resulting in CB formation.
Much of our knowledge to date regarding CB biology has been acquired using model cell lines, such as the widely used aneuploid HeLa cervical carcinoma cells (as well as essential work in model organisms, such as Xenopus,134 Drosophila135 and Zebrafish136). What makes mammalian aneuploid cancer cells unique compared to diploid controls cells derived from the same tissue of origin that renders them capable of assembling CBs? A particularly appealing idea is that CBs form in response to specific metabolic and processing demands. Like neuronal cells, cancers exhibit high levels of alternative splicing.137 Interestingly, activation of Ras, a known oncogene, is associated with differential regulation of Serine-Arginine (SR)-proteins, which are essential splicing factors.138 The CB has been suggested as important for leukemogenesis through its association with elongation factor for RNA polymerase II (ELL), a component of the little elongation complex.139 This hypothesis, in which CB formation correlates with splicing demand, is consistent with recent studies in which specific splicing events76 and genome-wide splicing fidelity20 were disrupted following CB disassembly. More studies are required to define the extent to which the CB is important during oncogenic transformation.
Finally, it has been reported that CBs are able to form in human primary diploid cells that previously lacked them following inhibition of U4.U6/U5 tri-snRNP assembly by SART3 siRNA knockdown, which is a U4/U6 di-snRNP recycling factor.140 These CBs may form as a stress response to incomplete snRNP assembly and insufficient spliceosome formation as they were enriched with snRNP assembly intermediate structures. It would be interesting to know if these intermediate CBs primarily form at sites of histone gene and spliceosomal snRNA gene transcription or if these CBs nucleate at other, still uncharacterized, specific genomic locations. This would provide revealing information regarding any potential heterogeneity in CB assembly sites. Thus, the formation of the CB can be presumed to be linked to supporting proper spliceosome formation during stress. These primary data and studies suggest that CB formation is a metabolic response to increased and essential splicing demands, including oncogenesis, which requires the potentially more efficient spliceosomal snRNP biogenesis pathway provided by the CB.
What can we learn from other Nuclear bodies and in vitro studies?
Many studies have also been published that have characterized the formation of other NBs using various cell models and technical approaches, which may assist researchers to better understand the dynamics of CB assembly. Of interest is the potential identification of an essential structural long non-coding RNA (lncRNA). Indeed, an important determinant of splicing speckle141 and paraspeckle142 formation is the presence of a seeding lncRNA that regulates body formation.143 However, to date, no lncRNA has been successfully identified that is structurally important for CB assembly. This indicates that CBs do not require a stabilizing lncRNA, the RNA component of snRNPs is sufficient to fulfil this role or that a lncRNA remains to be identified. This is similar to nucleolar assembly, which is dependent upon ribosomal rDNA transcription by RNA polymerase I.144 A recent coilin interactome study did not suggest any candidate lncRNAs for this role, suggesting that the former scenario is more likely to be true.62 For the nucleolus and the CB, it can be assumed that the high local expression of highly structured RNAs (the ribosomal rRNAs and snRNAs, respectively) may compensate for the lack of a singular structural lncRNA.
Recent phase separation studies have greatly improved our understanding of NB assembly and structural organization. Several groups have shown that multivalent proteins enriched with highly disordered domains are capable of forming liquid droplets in vitro that display many characteristic features of NBs, including fusion and fission events.86,87,90,91,145 It has also been suggested that separation of nucleoli, paraspeckles146 and Nuage bodies formed by DEAD-box helicase 4 (Ddx4) protein86,147 is driven by these disordered domains-containing proteins. Coilin and its homologs are highly disordered but Arabidopsis thaliana-derived coilin is stabilized upon snRNA binding leading to multimerization in vitro,114 which can be assumed to contribute to CB nucleation. Intriguingly, it is proposed that the nucleolus and droplets in vitro produced using nucleolar proteins may consist of multiple, immiscible phases that are separated by different surface tensions.147 This is also true of Tat-activating regulatory DNA-binding protein-43 (TDP-43) nuclear foci, which display NB characteristics in vivo and possess nucleoplasm-rich inclusions.148 This may result in a molecular production-line by coupling transcription with spatially segregated downstream RNA and RNP processing steps. Similarly, multiple coexisting but mutually-exclusive coilin-rich and SMN-rich phases within the CB have been identified by super resolution microscopy (although the HLB does not display a similar internal structure).140,149 At what point during CB assembly these multiple coexisting phases arise and whether they influence nucleation is unclear. However, these data indicate that intra-NB biochemistry may be more discrete than previously believed and NB sub-structure is compartmentalized to efficiently catalyze multiple parallel molecular processes.
A similar but distinct biophysical property of NBs is increased macromolecular crowding compared to the surrounding nucleoplasm. This phenomenon arises following the accumulation of large macromolecules in limited spaces and is an underappreciated contributing factor to biochemical and assembly reactions.150 This leads to the exclusion of water thereby lowering entropy and energetic barriers within the NB. Crowded environments display enhanced reaction rates, slower diffusion and better protein/RNA folding compared to non-crowded volumes.92,151-153 In total, these effects may explain the proposed acceleration of molecular processes conferred by NBs, including transcription and RNA processing, as well as the early stages of coilin and SMN oligomerization.154 Indeed, crowding effects within picoliter droplets containing DNA and polymerases in vitro enhances the retention of transcriptional machinery and enhanced RNA synthesis at transcription sites.155 This may be an important contributing factor to the early stages of CB nucleation by enhancing snRNA gene transcription.
These studies indicate that the crowded environment within NBs is necessary for various specific molecular processes and this coordination may be more highly optimized than previously believed. However, considering the number of NBs and the range of biological functions that they are linked to, inferences from studies that do not directly examine CB function must be treated with caution. Regardless, they provide noteworthy suggestions for future investigations into the biology of CB assembly and structural maintenance.
Conclusions
The CB is found in a limited number of cells and tissues but appears to play an important role in boosting and supporting high spliceosome turnover when present. We propose that assembly of this specialized nuclear microenvironment that improves cellular efficiency is a dynamic response to defined genomic and cellular cues (Fig. 4). Upon transcription of target gene loci and arrays, such as RNU1 and HIST1, coilin and other resident CB architectural proteins undergo numerous post-translational modifications. This initiates the formation of a stabilized and phase-separated macromolecular hub that attracts additional target gene loci to form specific clusters in both cis- and trans-chromosomal configurations. Protein and RNA factors become enriched in the crowded CB environment, which theoretically expedites spliceosomal snRNP production. CB formation also appears to correlate with the level of mRNA processing in a cell and lineage-specific factors. However, it is imperative that the relationship between phase separation, snRNP processing and gene association is fully elucidated. In particular, uncovering the mechanisms that define 3D genome reorganization following CB assembly and their biological significance is essential. Together, these data will improve our understanding of the functional benefit of CB-dependent snRNP biogenesis compared to that which occurs in the nucleoplasm of CB-deficient cells.
Figure 4.
Factors that influence Cajal body assembly and target processes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Greg Matera for providing Mouse Embryonic Fibroblasts and Dr. Philip Oberdoerffer for MCF10A cells. We also thank Dr. Mark Olson, Dr. Jiri Bartek and Dr. Eric Wagner for stimulatory discussions. High-content confocal microscopy images were acquired using equipment in the High-Throughput Imaging Facility (HiTIF) at the National Cancer Institute (Bethesda, MD) and confocal microscopy images were acquired at the NCI Core Fluorescence Imaging Facility in Building 41, National Institutes of Health (Bethesda, MD).
Funding
The authors are supported by a National Institutes of Health, National Institute of General Medical Sciences grant (R01GM090156, awarded to M.D.) and the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research (G.L.H.).
References
- 1.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genetics 2011; 27:295-306; PMID:21680045; https://doi.org/ 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol 1999; 9:302-9; PMID:10407409; https://doi.org/ 10.1016/S0962-8924(99)01606-2 [DOI] [PubMed] [Google Scholar]
- 3.Dundr M. Nuclear bodies: multifunctional companions of the genome. Curr Opin Cell Biol 2012; 24:415-22; PMID:22541757; https://doi.org/ 10.1016/j.ceb.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 2015; 112:E5237-45; PMID:26351690; https://doi.org/ 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 2009; 17:639-47; PMID:19922869; https://doi.org/ 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawyer IA, Dundr M. Nuclear bodies: Built to boost. J Cell Biol 2016; 213:509-11; PMID:27241912; https://doi.org/ 10.1083/jcb.201605049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatomer DC, Terzo E, Curry KP, Salzler H, Sabath I, Zapotoczny G, McKay DJ, Dominski Z, Marzluff WF, Duronio RJ. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J Cell Biol 2016; 213:557-70; PMID:27241916; https://doi.org/ 10.1083/jcb.201504043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000711; PMID:21068152; https://doi.org/ 10.1101/cshperspect.a000711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca M, Berciano MT, Lafarga M. Orphan nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000703; PMID:20610547; https://doi.org/ 10.1101/cshperspect.a000703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014; 15:108-21; PMID:24452469; https://doi.org/ 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Case AM, Sawyer IA, Dundr M, Hastings ML. Pre-mRNA Splicing: Function and Dysfunction In: Bradshaw RA, Stahl PD, eds. Encyclopedia of Cell Biology. Waltham: Academic Press, 2016:503-11; https://doi.org/10.1016/B978-0-12-394447-4.10053-7 [Google Scholar]
- 12.Hesselberth JR. Lives that introns lead after splicing. Wiley Interdisciplinary Rev RNA 2013; 4:677-91; PMID:23881603; https://doi.org/10.1002/wrna.1187 [DOI] [PubMed] [Google Scholar]
- 13.Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell 1992; 3:555-69; PMID:1535243; https://doi.org/ 10.1091/mbc.3.5.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J 1991; 10:195-206; PMID:1824936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915-9; PMID:7597053; https://doi.org/ 10.1073/pnas.92.13.5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bellini M, Gall JG. Coilin can form a complex with the U7 small nuclear ribonucleoprotein. Mol Biol Cell 1998; 9:2987-3001; PMID:9763457; https://doi.org/ 10.1091/mbc.9.10.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sleeman J, Lyon CE, Platani M, Kreivi JP, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res 1998; 243:290-304; PMID:9743589; https://doi.org/ 10.1006/excr.1998.4135 [DOI] [PubMed] [Google Scholar]
- 18.Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 2002; 21:2736-45; PMID:12032086; https://doi.org/ 10.1093/emboj/21.11.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girard C, Will CL, Peng J, Makarov EM, Kastner B, Lemm I, Urlaub H, Hartmuth K, Luhrmann R. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun 2012; 3:994; PMID:22871813; https://doi.org/ 10.1038/ncomms1998 [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. Cajal bodies are linked to genome conformation. Nat Commun 2016; 7:10966; PMID:26997247; https://doi.org/ 10.1038/ncomms10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schul W, van Driel R, de Jong L. Coiled bodies and U2 snRNA genes adjacent to coiled bodies are enriched in factors required for snRNA transcription. Mol Biol Cell 1998; 9:1025-36; PMID:9571237; https://doi.org/ 10.1091/mbc.9.5.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci 2012; 125:166-75; PMID:22250197; https://doi.org/ 10.1242/jcs.090837 [DOI] [PubMed] [Google Scholar]
- 23.Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 2003; 22:1878-88; PMID:12682020; https://doi.org/ 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 2004; 166:1015-25; PMID:15452143; https://doi.org/ 10.1083/jcb.200405160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 2003; 160:505-16; PMID:12578909; https://doi.org/ 10.1083/jcb.200210087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 2008; 19:2534-43; PMID:18367544; https://doi.org/ 10.1091/mbc.E07-12-1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stern JL, Zyner KG, Pickett HA, Cohen SB, Bryan TM. Telomerase recruitment requires both TCAB1 and Cajal bodies independently. Mol Cell Biol 2012; 32:2384-95; PMID:22547674; https://doi.org/ 10.1128/MCB.00379-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilder AS, Do PM, Carrero ZI, Cosman AM, Broome HJ, Velma V, Martinez LA, Hebert MD. Coilin participates in the suppression of RNA polymerase I in response to cisplatin-induced DNA damage. Mol Biol Cell 2011; 22:1070-9; PMID:21289084; https://doi.org/ 10.1091/mbc.E10-08-0731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henriksson S, Rassoolzadeh H, Hedstrom E, Coucoravas C, Julner A, Goldstein M, Imreh G, Zhivotovsky B, Kastan MB, Helleday T, et al.. The scaffold protein WRAP53beta orchestrates the ubiquitin response critical for DNA double-strand break repair. Genes Dev 2014; 28:2726-38; PMID:25512560; https://doi.org/ 10.1101/gad.246546.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol 2006; 175:401-13; PMID:17088425; https://doi.org/ 10.1083/jcb.200604099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rassoolzadeh H, Bohm S, Hedstrom E, Gad H, Helleday T, Henriksson S, Farnebo M. Overexpression of the scaffold WD40 protein WRAP53[β] enhances the repair of and cell survival from DNA double-strand breaks. Cell Death Dis 2016; 7:e2267; PMID:27310875; https://doi.org/ 10.1038/cddis.2016.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 2002; 21:2724-35; PMID:12032085; https://doi.org/ 10.1093/emboj/21.11.2724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bratkovic T, Rogelj B. The many faces of small nucleolar RNAs. Biochim Et Biophys Acta 2014; 1839:438-43; PMID:24735946; https://doi.org/ 10.1016/j.bbagrm.2014.04.009 [DOI] [PubMed] [Google Scholar]
- 34.Dupuis-Sandoval F, Poirier M, Scott MS. The emerging landscape of small nucleolar RNAs in cell biology. Wiley Interdisciplinary Rev RNA 2015; 6:381-97; PMID:25879954; https://doi.org/ 10.1002/wrna.1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karijolich J, Yi C, Yu YT. Transcriptome-wide dynamics of RNA pseudouridylation. Nat Rev Mol Cell Biol 2015; 16:581-5; PMID:26285676; https://doi.org/ 10.1038/nrm4040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol 2003; 4:975-80; PMID:14685175; https://doi.org/ 10.1038/nrm1262 [DOI] [PubMed] [Google Scholar]
- 37.Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res 2001; 265:252-61; PMID:11302690; https://doi.org/ 10.1006/excr.2001.5186 [DOI] [PubMed] [Google Scholar]
- 38.Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci U S A 2008; 105:16964-9; PMID:18957539; https://doi.org/ 10.1073/pnas.0809273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dundr M. Seed and grow: a two-step model for nuclear body biogenesis. J Cell Biol 2011; 193:605-6; PMID:21576389; https://doi.org/ 10.1083/jcb.201104087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith KP, Lawrence JB. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol Biol Cell 2000; 11:2987-98; PMID:10982395; https://doi.org/ 10.1091/mbc.11.9.2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA Genes in interphase HeLa cells but not with U6 and U7 Genes. Mol Biol Cell 1999; 10:1653-63; PMID:10233169; https://doi.org/ 10.1091/mbc.10.5.1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shopland LS, Byron M, Stein JL, Lian JB, Stein GS, Lawrence JB. Replication-dependent histone gene expression is related to Cajal Body (CB) association but does not require sustained CB contact. Mol Biol Cell 2001; 12:565-76; PMID:11251071; https://doi.org/ 10.1091/mbc.12.3.565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 1995; 59:473-85; PMID:8749717; https://doi.org/ 10.1002/jcb.240590408 [DOI] [PubMed] [Google Scholar]
- 44.Sawyer IA, Shevtsov SP, Dundr M. Spectral imaging to visualize higher-order genomic organization. Nucleus (Austin, Tex) 2016; 7:325-38; PMID:27167405; https://doi.org/10.1080/19491034.2016.1187344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schul W, Adelaar B, van Driel R, de Jong L. Coiled bodies are predisposed to a spatial association with genes that contain snoRNA sequences in their introns. J Cell Biochem 1999; 75:393-403; PMID:10536363; https://doi.org/ 10.1002/(SICI)1097-4644(19991201)75:3%3c393::AID-JCB5%3e3.0.CO;2-G [DOI] [PubMed] [Google Scholar]
- 46.Zieve GW, Sauterer RA. Cell biology of the snRNP particles. Critical Rev Biochem Mol Biol 1990; 25:1-46; PMID:2138975; https://doi.org/ 10.3109/10409239009090604 [DOI] [PubMed] [Google Scholar]
- 47.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007; 179:1095-103; PMID:18070915; https://doi.org/ 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dellaire G, Eskiw CH, Dehghani H, Ching RW, Bazett-Jones DP. Mitotic accumulations of PML protein contribute to the re-establishment of PML nuclear bodies in G1. J Cell Sci 2006; 119:1034-42; PMID:16492707; https://doi.org/ 10.1242/jcs.02817 [DOI] [PubMed] [Google Scholar]
- 49.Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol 1994; 126:11-23; PMID:8027171; https://doi.org/ 10.1083/jcb.126.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 1993; 120:841-52; PMID:7679389; https://doi.org/ 10.1083/jcb.120.4.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogan JM, Collins K. Dynamics of human telomerase holoenzyme assembly and subunit exchange across the cell cycle. J Biol Chem 2015; 290:21320-35; PMID:26170453; https://doi.org/ 10.1074/jbc.M115.659359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marnef A, Richard P, Pinzon N, Kiss T. Targeting vertebrate intron-encoded box C/D 2′-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res 2014; 42:6616-29; PMID:24753405; https://doi.org/ 10.1093/nar/gku287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell 2009; 20:1661-70; PMID:19158395; https://doi.org/ 10.1091/mbc.E08-05-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 2008; 105:10762-7; PMID:18669648; https://doi.org/ 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cantarero L, Sanz-Garcia M, Vinograd-Byk H, Renbaum P, Levy-Lahad E, Lazo PA. VRK1 regulates Cajal body dynamics and protects coilin from proteasomal degradation in cell cycle. Scientific Reports 2015; 5:10543; PMID:26068304; https://doi.org/ 10.1038/srep10543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hebert MD. Signals controlling Cajal body assembly and function. Int J Biochem Cell Biol 2013; 45:1314-7; PMID:23583661; https://doi.org/ 10.1016/j.biocel.2013.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nizami Z, Deryusheva S, Gall JG. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol 2010; 2:a000653; PMID:20504965; https://doi.org/ 10.1101/cshperspect.a000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Structural Biol 1990; 104:120-7; PMID:2088441; https://doi.org/ 10.1016/1047-8477(90)90066-L [DOI] [PubMed] [Google Scholar]
- 59.Bohmann K, Ferreira JA, Lamond AI. Mutational analysis of p80 coilin indicates a functional interaction between coiled bodies and the nucleolus. J Cell Biol 1995; 131:817-31; PMID:7490287; https://doi.org/ 10.1083/jcb.131.4.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grande MA, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort HT, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem 1996; 63:280-91; PMID:8913879; https://doi.org/ 10.1002/(SICI)1097-4644(19961201)63:3%3c280::AID-JCB3%3e3.0.CO;2-T [DOI] [PubMed] [Google Scholar]
- 61.Makarova JA, Kramerov DA. SNOntology: Myriads of novel snoRNAs or just a mirage? BMC Genomics 2011; 12:543; PMID:22047601; https://doi.org/ 10.1186/1471-2164-12-543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 2014; 56:389-99; PMID:25514182; https://doi.org/ 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 63.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 2006; 34:D158-62; PMID:16381836; https://doi.org/ 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al.. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22:1760-74; PMID:22955987; https://doi.org/ 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dunham I, Kundaje A, Aldred SF, Collins PJ, Davis CA, Doyle F, Epstein CB, Frietze S, Harrow J, Kaul R, et al.. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57-74; PMID:22955616; https://doi.org/ 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 1992; 117:1-14; PMID:1532583; https://doi.org/ 10.1083/jcb.117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol Biol Cell 2005; 16:2395-413; PMID:15758027; https://doi.org/ 10.1091/mbc.E04-11-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pena E, Berciano MT, Fernandez R, Ojeda JL, Lafarga M. Neuronal body size correlates with the number of nucleoli and Cajal bodies, and with the organization of the splicing machinery in rat trigeminal ganglion neurons. J Comparative Neurol 2001; 430:250-63; PMID:11135260; https://doi.org/ 10.1002/1096-9861(20010205)430:2%3c250::AID-CNE1029%3e3.0.CO;2-L [DOI] [PubMed] [Google Scholar]
- 69.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95-101; PMID:21170033; https://doi.org/ 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011; 13:167-73; PMID:21240286; https://doi.org/ 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- 71.Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol 1999; 9:126-35; PMID:10021385; https://doi.org/ 10.1016/S0960-9822(99)80066-9 [DOI] [PubMed] [Google Scholar]
- 72.Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154:499-509; PMID:11489914; https://doi.org/ 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res 1997; 25:4740-7; PMID:9365252; https://doi.org/ 10.1093/nar/25.23.4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.James Faresse N, Canella D, Praz V, Michaud J, Romascano D, Hernandez N. Genomic study of RNA polymerase II and III SNAPc-bound promoters reveals a gene transcribed by both enzymes and a broad use of common activators. PLoS Genet 2012; 8:e1003028; PMID:23166507; https://doi.org/ 10.1371/journal.pgen.1003028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hung KH, Stumph WE. Regulation of snRNA gene expression by the Drosophila melanogaster small nuclear RNA activating protein complex (DmSNAPc). Critical Rev Biochem Mol Biol 2011; 46:11-26; PMID:20925482; https://doi.org/ 10.3109/10409238.2010.518136 [DOI] [PubMed] [Google Scholar]
- 76.Hutten S, Chachami G, Winter U, Melchior F, Lamond AI. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J Cell Sci 2014; 127:1065-78; PMID:24413172; https://doi.org/ 10.1242/jcs.141788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takahashi H, Takigawa I, Watanabe M, Anwar D, Shibata M, Tomomori-Sato C, Sato S, Ranjan A, Seidel CW, Tsukiyama T, et al.. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat Commun 2015; 6:5941; PMID:25575120; https://doi.org/ 10.1038/ncomms6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen J, Wagner EJ. snRNA 3′ end formation: the dawn of the Integrator complex. Biochem Society Trans 2010; 38:1082-7; PMID:20659008; https://doi.org/ 10.1042/BST0381082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci 2012; 125:166-75; PMID:22250197; https://doi.org/ 10.1242/jcs.090837 [DOI] [PubMed] [Google Scholar]
- 80.Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 2000; 11:4159-71; PMID:11102515; https://doi.org/ 10.1091/mbc.11.12.4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Machyna M, Neugebauer KM, Stanek D. Coilin: The first 25 years. RNA Biol 2015; 12:590-6; PMID:25970135; https://doi.org/ 10.1080/15476286.2015.1034923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dundr M. Nucleation of nuclear bodies. Meth Mol Biol (Clifton, NJ) 2013; 1042:351-64; PMID:23980018; https://doi.org/18948503 10.1007/978-1-62703-526-2_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713-7; PMID:18948503; https://doi.org/ 10.1126/science.1165216 [DOI] [PubMed] [Google Scholar]
- 84.Romeo V, Schumperli D. Cycling in the nucleus: regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol 2016; 40:23-31; PMID:26895140; https://doi.org/ 10.1016/j.ceb.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 85.Pirrotta V, Li HB. A view of nuclear Polycomb bodies. Curr Opin Genet Dev 2012; 22:101-9; PMID:22178420; https://doi.org/ 10.1016/j.gde.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nott TJ, Petsalaki E, Farber P, Jervis D, Fussner E, Plochowietz A, Craggs TD, Bazett-Jones DP, Pawson T, Forman-Kay JD, et al.. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 2015; 57:936-47; PMID:25747659; https://doi.org/ 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banani Salman F, Rice Allyson M, Peeples William B, Lin Y, Jain S, Parker R, Rosen Michael K. Compositional control of phase-separated cellular bodies. Cell 166:651-63; PMID:27374333; https://doi.org/ 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol 2004; 164:831-42; PMID:15024031; https://doi.org/ 10.1083/jcb.200311121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mohamad N, Boden M. The proteins of intra-nuclear bodies: a data-driven analysis of sequence, interaction and expression. BMC Systems Biol 2010; 4:44; PMID:20388198; https://doi.org/ 10.1186/1752-0509-4-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang H, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges Andrew A, Brangwynne Clifford P, Gladfelter Amy S. RNA controls PolyQ protein phase transitions. Mol Cell 2015; 60:220-30; PMID:26474065; https://doi.org/ 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? Embo J 2016; 35:1603-12; PMID:27357569; https://doi.org/ 10.15252/embj.201593517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cho EJ, Kim JS. Crowding effects on the formation and maintenance of nuclear bodies: insights from molecular-dynamics simulations of simple spherical model particles. Biophys J 2012; 103:424-33; PMID:22947858; https://doi.org/ 10.1016/j.bpj.2012.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Oh I, Choi S, Jung Y, Kim JS. Phase separation of a Lennard-Jones fluid interacting with a long, condensed polymer chain: implications for the nuclear body formation near chromosomes. Soft Matter 2015; 11:6450-9; PMID:26179211; https://doi.org/ 10.1039/C5SM01096A [DOI] [PubMed] [Google Scholar]
- 94.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdisciplinary Rev RNA 2013; 4:17-34; PMID:23042601; https://doi.org/ 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- 95.Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol 2000; 151:1561-74; PMID:11134083; https://doi.org/ 10.1083/jcb.151.7.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pederson T. Dynamics and genome-centricity of interchromatin domains in the nucleus. Nat Cell Biol 2002; 4:E287-91; PMID:12461535; https://doi.org/ 10.1038/ncb1202-e287 [DOI] [PubMed] [Google Scholar]
- 97.Hutten S, Swift S, Lamond AI. Time-lapse imaging of nuclear bodies. Methods Mol Biol (Clifton, NJ) 2015; 1262:55-67; PMID:25555575; https://doi.org/12068306 10.1007/978-1-4939-2253-6_4 [DOI] [PubMed] [Google Scholar]
- 98.Platani M, Goldberg I, Lamond AI, Swedlow JR. Cajal body dynamics and association with chromatin are ATP-dependent. Nat Cell Biol 2002; 4:502-8; PMID:12068306; https://doi.org/ 10.1038/ncb809 [DOI] [PubMed] [Google Scholar]
- 99.Morris GE. The Cajal body. Biochim Et Biophys Acta 2008; 1783:2108-15; PMID:18755223; https://doi.org/ 10.1016/j.bbamcr.2008.07.016 [DOI] [PubMed] [Google Scholar]
- 100.Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 2001; 114:4407-19; PMID:11792806; https://doi.org/10.1.608.2151 [DOI] [PubMed] [Google Scholar]
- 101.Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of Cajal bodies in Arabidopsis. Mol Biol Cell 2006; 17:2942-51; PMID:16624863; https://doi.org/ 10.1091/mbc.E05-12-1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci 2009; 122:1872-81; PMID:19435804; https://doi.org/ 10.1242/jcs.044040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Misteli T. Physiological importance of RNA and protein mobility in the cell nucleus. Histochem Cell Biol 2008; 129:5-11; PMID:17994245; https://doi.org/ 10.1007/s00418-007-0355-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deryusheva S, Gall JG. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc Natl Acad Sci U S A 2004; 101:4810-4; PMID:15044688; https://doi.org/ 10.1073/pnas.0401106101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci 2003; 116:2039-50; PMID:12679382; https://doi.org/ 10.1242/jcs.00400 [DOI] [PubMed] [Google Scholar]
- 106.Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB'. Chromosoma 2010; 119:205-15; PMID:19997741; https://doi.org/ 10.1007/s00412-009-0249-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sanz-Garcia M, Vazquez-Cedeira M, Kellerman E, Renbaum P, Levy-Lahad E, Lazo PA. Substrate profiling of human vaccinia-related kinases identifies coilin, a Cajal body nuclear protein, as a phosphorylation target with neurological implications. J Proteomics 2011; 75:548-60; PMID:21920476; https://doi.org/ 10.1016/j.jprot.2011.08.019 [DOI] [PubMed] [Google Scholar]
- 108.Carrero ZI, Velma V, Douglas HE, Hebert MD. Coilin phosphomutants disrupt Cajal body formation, reduce cell proliferation and produce a distinct coilin degradation product. PLoS One 2011; 6:e25743; PMID:21991343; https://doi.org/ 10.1371/journal.pone.0025743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res 1997; 230:84-93; PMID:9013710; https://doi.org/ 10.1006/excr.1996.3380 [DOI] [PubMed] [Google Scholar]
- 110.Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA. PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD). Cell Cycle (Georgetown, Tex) 2015; 14:1716-29; PMID:25927346; https://doi.org/ 10.1080/15384101.2015.1033595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 2001; 15:2720-9; PMID:11641277; https://doi.org/ 10.1101/gad.908401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Frege T, Uversky VN. Intrinsically disordered proteins in the nucleus of human cells. Biochem Biophys Reports 2015; 1:33-51; https://doi.org/ 10.1016/j.bbrep.2015.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aguzzi A, Altmeyer M. Phase Separation: Linking Cellular Compartmentalization to Disease. Trends Cell Biol 2016; 26:547-58; PMID:27051975; https://doi.org/ 10.1016/j.tcb.2016.03.004 [DOI] [PubMed] [Google Scholar]
- 114.Makarov V, Rakitina D, Protopopova A, Yaminsky I, Arutiunian A, Love AJ, Taliansky M, Kalinina N. Plant coilin: structural characteristics and RNA-binding properties. PLoS One 2013; 8:e53571; PMID:23320094; https://doi.org/ 10.1371/journal.pone.0053571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shanbhag R, Kurabi A, Kwan JJ, Donaldson LW. Solution structure of the carboxy-terminal Tudor domain from human Coilin. FEBS Lett 2010; 584:4351-6; PMID:20875822; https://doi.org/ 10.1016/j.febslet.2010.09.034 [DOI] [PubMed] [Google Scholar]
- 116.Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. The SMN complex: an assembly machine for RNPs. Cold Spring Harbor Symposia Quantitative Biol 2006; 71:313-20; PMID:17381311; https://doi.org/ 10.1101/sqb.2006.71.001 [DOI] [PubMed] [Google Scholar]
- 117.Gupta K, Martin R, Sharp R, Sarachan KL, Ninan NS, Van Duyne GD. Oligomeric properties of survival motor neuron Gemin2 Complexes. J Biol Chem 2015; 290:20185-99; PMID:26092730; https://doi.org/ 10.1074/jbc.M115.667279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martin R, Gupta K, Ninan NS, Perry K, Van Duyne GD. The survival motor neuron protein forms soluble glycine zipper oligomers. Structure 2012; 20:1929-39; PMID:23022347; https://doi.org/ 10.1016/j.str.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; https://doi.org/ 10.1371/journal.pbio.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang J, Zhang F, Zheng X. Depletion of hCINAP by RNA interference causes defects in Cajal body formation, histone transcription, and cell viability. Cell Mol Life Sci 2010; 67:1907-18; PMID:20186459; https://doi.org/ 10.1007/s00018-010-0301-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol 2013; 203:149-64; PMID:24127217; https://doi.org/ 10.1083/jcb.201303145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 2006; 17:3221-31; PMID:16687569; https://doi.org/ 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xu H, Hebert MD. A novel EB-1/AIDA-1 isoform, AIDA-1c, interacts with the Cajal body protein coilin. BMC Cell Biol 2005; 6:23; PMID:15862129; https://doi.org/ 10.1186/1471-2121-6-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li Y, Fong KW, Tang M, Han X, Gong Z, Ma W, Hebert M, Songyang Z, Chen J. Fam118B, a newly identified component of Cajal bodies, is required for Cajal body formation, snRNP biogenesis and cell viability. J Cell Sci 2014; 127:2029-39; PMID:24569877; https://doi.org/ 10.1242/jcs.143453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Söderberg O, Strömblad S, Wiman KG, Farnebo M. WRAP53 Is Essential for Cajal Body Formation and for Targeting the Survival of Motor Neuron Complex to Cajal Bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; https://doi.org/ 10.1371/journal.pbio.1000521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Poh YC, Shevtsov SP, Chowdhury F, Wu DC, Na S, Dundr M, Wang N. Dynamic force-induced direct dissociation of protein complexes in a nuclear body in living cells. Nat Commun 2012; 3:866; PMID:22643893; https://doi.org/ 10.1038/ncomms1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell 2002; 3:329-37; PMID:12361597; https://doi.org/ 10.1016/S1534-5807(02)00222-8 [DOI] [PubMed] [Google Scholar]
- 128.Han KJ, Foster D, Harhaj EW, Dzieciatkowska M, Hansen K, Liu CW. Monoubiquitination of survival motor neuron regulates its cellular localization and Cajal body integrity. Hum Mol Genet 2016; 25:1392-405; PMID:26908624; https://doi.org/ 10.1093/hmg/ddw021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gall JG. Cajal bodies: the first 100 years. Annual Rev Cell Dev Biol 2000; 16:273-300; PMID:11031238; https://doi.org/ 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- 130.Forthmann B, van Bergeijk J, Lee YW, Lubben V, Schill Y, Brinkmann H, Ratzka A, Stachowiak MK, Hebert M, Grothe C, et al.. Regulation of neuronal differentiation by proteins associated with nuclear bodies. PLoS One 2013; 8:e82871; PMID:24358231; https://doi.org/ 10.1371/journal.pone.0082871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Janevski J, Park PC, De Boni U. Changes in morphology and spatial position of coiled bodies during NGF-induced neuronal differentiation of PC12 cells. J Histochem Cytochem 1997; 45:1523-31; PMID:9358854; https://doi.org/ 10.1177/002215549704501109 [DOI] [PubMed] [Google Scholar]
- 132.Yeo G, Holste D, Kreiman G, Burge CB. Variation in alternative splicing across human tissues. Genome Biol 2004; 5:R74; PMID:15461793; https://doi.org/ 10.1186/gb-2004-5-10-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Navascues J, Berciano MT, Tucker KE, Lafarga M, Matera AG. Targeting SMN to Cajal bodies and nuclear gems during neuritogenesis. Chromosoma 2004; 112:398-409; PMID:15164213; https://doi.org/ 10.1007/s00412-004-0285-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wu Z, Murphy C, Gall JG. Human p80-coilin is targeted to sphere organelles in the amphibian germinal vesicle. Mol Biol Cell 1994; 5:1119-27; PMID:7532471; https://doi.org/ 10.1091/mbc.5.10.1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nizami ZF, Deryusheva S, Gall JG. Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harbor Symposia Quantitative Biol 2010; 75:313-20; PMID:21047905; https://doi.org/ 10.1101/sqb.2010.75.005 [DOI] [PubMed] [Google Scholar]
- 136.Strzelecka M, Oates AC, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus (Austin, Tex) 2010; 1:96-108; PMID:21327108; https://doi.org/10.416/nucl.1.1.10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016; 35:2413-27; PMID:26300000; https://doi.org/ 10.1038/onc.2015.318 [DOI] [PubMed] [Google Scholar]
- 138.Srebrow A, Kornblihtt AR. The connection between splicing and cancer. J Cell Sci 2006; 119:2635-41; PMID:16787944; https://doi.org/ 10.1242/jcs.03053 [DOI] [PubMed] [Google Scholar]
- 139.Polak PE, Simone F, Kaberlein JJ, Luo RT, Thirman MJ. ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia. Mol Biol Cell 2003; 14:1517-28; PMID:12686606; https://doi.org/ 10.1091/mbc.E02-07-0394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Novotný I, Malinová A, Stejskalová E, Matějů D, Klimešová K, Roithová A, Švéda M, Knejzlík Z, Staněk D. SART3-Dependent Accumulation of Incomplete Spliceosomal snRNPs in Cajal Bodies. Cell Reports 2015; 10:429-40; PMID:25600876 https://doi.org/17270048 10.1016/j.celrep.2014.12.030 [DOI] [PubMed] [Google Scholar]
- 141.Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 2007; 8:39; PMID:17270048; https://doi.org/ 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, et al.. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014; 25:169-83; PMID:24173718; https://doi.org/ 10.1091/mbc.E13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Et Biophys Acta 2016; 1859:139-46; PMID:26021608; https://doi.org/ 10.1016/j.bbagrm.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 144.Olson MOJ, Dundr M. Nucleolus: Structure and Function. eLS: John Wiley & Sons, Ltd, 2001 [Google Scholar]
- 145.Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 2015; 34:23-30; PMID:25942753; https://doi.org/ 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al.. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015; 210:529-39; PMID:26283796; https://doi.org/ 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. Coexisting liquid phases underlie nucleolar subcompartments. Cell 2016; 165:1-12; PMID:27212236; https://doi.org/27452472 10.1016/j.cell.2016.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Schmidt HB, Rohatgi R. In vivo formation of vacuolated multi-phase compartments lacking membranes. Cell Rep 2016; 16:1228-36; PMID:27452472; https://doi.org/ 10.1016/j.celrep.2016.06.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sawyer IA, Sturgill D, Sung M-H, Hager GL, and Dundr M (2016) Cajal body function in genome organization and transcriptome diversity. Bioessays, 38(12):1-12; PMID:27767214; 25517143https://doi.org/10.1002/bies.201600144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Richter K, Nessling M, Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim Et Biophys Acta (BBA) - Mol Cell Res 2008; 1783:2100-7; PMID:18723053; https://doi.org/25517143 10.1016/j.bbamcr.2008.07.017 [DOI] [PubMed] [Google Scholar]
- 151.ten Wolde PR, Mugler A. Chapter Twelve - Importance of Crowding in Signaling, Genetic, and Metabolic Networks. In: Ronald H, Kwang WJ, eds. International review of cell and molecular biology: Academic Press, 2014:419-42; PMID:24380601; 25517143https://doi.org/10.1016/B978-0-12-800046-5.00012-6 [DOI] [PubMed] [Google Scholar]
- 152.Mourao MA, Hakim JB, Schnell S. Connecting the dots: the effects of macromolecular crowding on cell physiology. Biophys J 2014; 107:2761-6; PMID:25517143; https://doi.org/ 10.1016/j.bpj.2014.10.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Christiansen A, Wang Q, Cheung MS, Wittung-Stafshede P. Effects of macromolecular crowding agents on protein folding in vitro and in silico. Biophys Rev 2013; 5:137-45; https://doi.org/ 10.1007/s12551-013-0108-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 2006; 17:4972-81; PMID:16987958; https://doi.org/ 10.1091/mbc.E06-06-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hansen MM, Meijer LH, Spruijt E, Maas RJ, Rosquelles MV, Groen J, Heus HA, Huck WT. Macromolecular crowding creates heterogeneous environments of gene expression in picolitre droplets. Nat Nanotechnol 2016; 11:191-7; PMID:26501750; https://doi.org/ 10.1038/nnano.2015.243 [DOI] [PMC free article] [PubMed] [Google Scholar]


