ABSTRACT
Aside from nucleoli, Cajal bodies (CBs) are the best-characterized organelles of mammalian cell nuclei. Like nucleoli, CBs concentrate ribonucleoproteins (RNPs), in particular, spliceosomal small nuclear RNPs (snRNPs) and small nucleolar RNPs (snoRNPs). In one of the best-defined functions of CBs, most of the snoRNPs are involved in site-specific modification of snRNAs. The two major modifications are pseudouridylation and 2′-O-methylation that are guided by the box H/ACA and C/D snoRNPs, respectively. This review details the modifications, their function, the mechanism of modification, and the machineries involved. We dissect the different classes of noncoding RNAs that meet in CBs, guides and substrates. Open questions and conundrums, often raised and appearing due to experimental limitations, are pointed out and discussed. The emphasis of the review is on mammalian CBs and their function in modification of noncoding RNAs.
KEYWORDS: 2′-O-methyl, Cajal body, C/D RNA, H/ACA RNA; pseudouridine, telomerase, spliceosomal, snRNA, snoRNA, scaRNA
Introduction
Although many functions have been ascribed to Cajal bodies (CBs), perhaps the best-established function is that as sites of modification of spliceosomal small nuclear RNAs (snRNAs). Whereas other potential functions are covered in other sections of this Special Focus on CBs, this review will concentrate on the role of CBs in the modification of noncoding RNAs. Other aspects of CBs are covered in other sections.
Over the past few years, RNA modification has garnered some limelight mainly due to technical improvements of detection of the modifications enabling genome-wide approaches. For example, the identification of enzymes that remove N6-methyladenosines from mRNAs showed the modification to be reversible and led to the development of methods identifying this modification genome-wide.18,30,63 This precipitated exciting findings on the function and mechanism of this most abundant internal modification of mRNA.49 In turn, this bounty of information stimulated interest in developing techniques for identification of other modifications that were long known in abundant noncoding RNAs, but technically inaccessible in mRNA. Thus, 4 independent studies identified pseudouridines in yeast and mammalian mRNAs showing that this modification is much more widespread than previously appreciated.8,48,51,74 Despite the considerable interest and potential impact of these findings in mRNAs,47 pseudouridines (and 2′-O-methyl groups) in noncoding RNAs far outnumber those in mRNAs for 2 reasons: first, ribosomal RNAs (rRNAs), snRNAs, and small nucleolar RNAs (snoRNAs) are orders of magnitude more abundant than mRNAs and, second, they each carry multiple modifications. It is perhaps for this reason that the sites of modification of these RNAs occupy specific organelles in the nucleus, nucleoli and CBs. Unless specified otherwise, this review focuses on modification events in mammalian CBs.
Overview
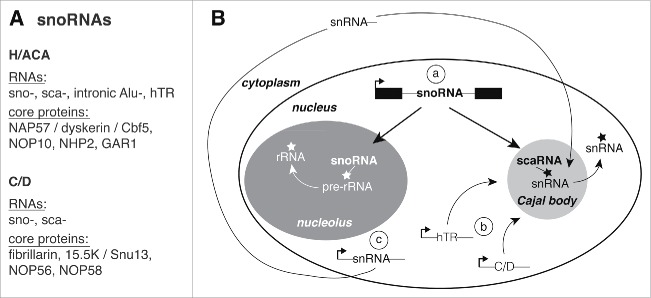
This brief overview is intended to make the subject more accessible to the uninitiated and serve as reference for the remainder of the review. Based on common sequence motifs and secondary structures, snoRNAs form 2 major families, box H/ACA and box C/D snoRNAs (Fig. 1A).14,39 H/ACA and C/D RNAs each associate with their own set of the same 4 core proteins (Fig. 1A) to generate the functional units, small nucleolar ribonucleoproteins (snoRNPs). As the names suggest, the most abundant RNAs function in nucleoli (snoRNAs) and in CBs (small CB-specific, scaRNAs), where they guide the modification of rRNAs and snRNAs, respectively (Fig. 1B). A large, but low-abundant class of H/ACA RNAs is that of the AluACA RNAs, derived from intronic Alu elements (Fig. 1A).29 Whereas most snoRNAs are expressed from introns of mainly housekeeping genes (Fig. 1B, a), a prominent H/ACA RNA, human telomerase RNA (hTR), and 5 abundant C/D RNAs (U3, U8, U13, mgU2-25/61, mgU12-22/U4-8) are expressed from their own RNA polymerase II promoters (Fig. 1B, b).64,70,80,81,98 These independently expressed snoRNAs traffic through CBs, where apparently their 5′-cap is hypermethylated. The major modification targets in CBs, the spliceosomal snRNAs, are also independently expressed (Fig. 1B, c). However, most snRNAs then embark on an obligate cytoplasmic journey during which their 5′-cap is hypermethylated, their 3′-end is trimmed, and they assimilate Sm proteins for reimport into nuclei and trafficking to CBs (Fig. 1B). Armed with this information, we can now delve into the nuts and bolts of CB function in RNA modification.
Figure 1.
SnoRNAs and life cycle. (A) List of small nucleolar RNAs (snoRNAs) and ribonucleoprotein (RNP) core proteins. Abbreviations: small Cajal body (CB)-specific (sca) and human telomerase RNA (hTR). (B) Schematic of snoRNA mode of expression, trafficking, and sites of action and that of its main target in CBs, spliceosomal small nuclear RNAs (snRNAs). See text for full explanation. Pseudouridylation and 2′-O-methylation modifications are indicated (asterisks). Note proteins were left off for simplicity and clarity.
The modifications
Two modifications predominate in snRNAs and have been associated with CBs, pseudouridylation and 2′-O-methylation. Pseudouridylation is the isomerization of uridine to pseudouridine by breaking the N-glycosidic bond, followed by a 180° rotation of the base, and the formation of a C–C-glycosidic bond (Fig. 2A). 2′-O-methylation is the methylation of the 2′-hydroxyl group of the ribose moiety of any ribonucleoside (Fig. 2A). Both modifications change the biophysical properties of the RNA, even if to a minor degree. Compared to uridine, pseudouridine stabilizes base stacking, rigidifies the backbone (through coordination of a water molecule by the additional amino group of the base), and is more polar.1,4,13 2′-O-methylation changes the hydration shell around the oxygen and protects the RNA against alkaline hydrolysis.5,23 As a consequence, modifications fine-tune the function of substrate RNAs.
Figure 2.
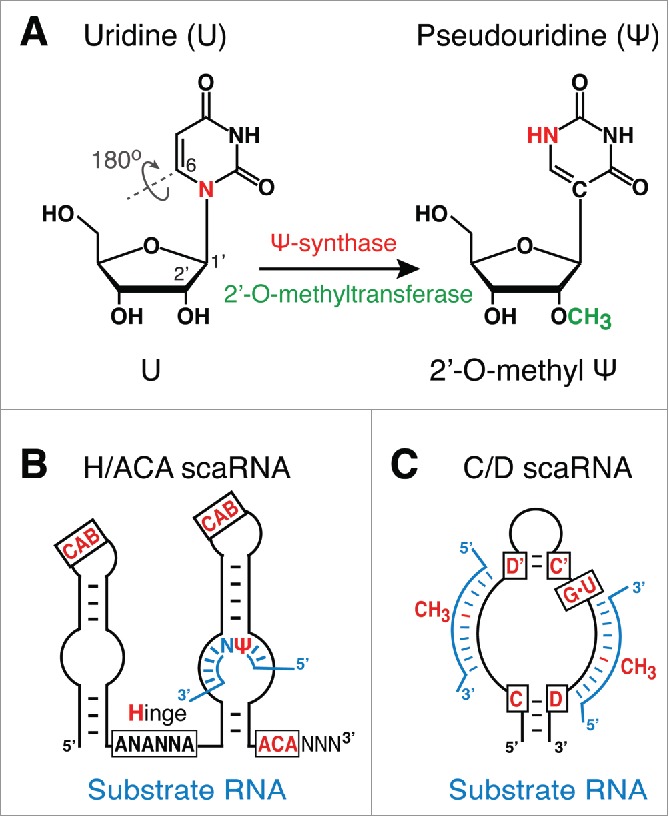
Modifications and scaRNAs. (A) Schematic of pseudouridylation (red) and 2′-O-methylation (green). Note although indicated on the same nucleoside, these are independent modification reactions. (B) H/ACA scaRNA with CB localizing elements (CAB) and a substrate RNA in one of the 2 pseudouridylation pockets. Note CAB boxes and guide elements can reside in one or the other hairpin or both. (C) C/D scaRNA with a CB localizing G•U/U•G wobble stem (G•U) and 2 substrate RNAs. Note the fifth nucleotide of the substrate from boxes D and/or D′ is targeted for 2′-O-methylation (CH3).
Spliceosomal snRNAs harbor additional modifications. Except snRNA U6 and U6atac, all snRNAs possess a trimethylguanosine cap structure followed by two 2′-O-methylated residues that are modified during cap formation. One or two N6-methyladenosines and 2-methylguanosines have also been identified in snRNAs.58 However, these modifications do not occur in CBs. For example, cap hypermethylation occurs during the cytosolic step of U snRNP maturation (Fig. 1B).61 Surprisingly, the cap methylase Tgs1 is also concentrated in CBs, where it may specifically hypermethylate the cap of independently expressed snoRNAs (Fig. 1B, b).21,85 Similar to Tgs1, the survival of motor neurons protein (SMN), which functions in the cytoplasm to assemble Sm rings on snRNAs, also concentrates in CBs despite snoRNAs being devoid of Sm proteins.50 Obviously, some aspects of the snRNP lifecycle and CB function remain to be elucidated.
Guide RNAs
To appreciate the role of CBs in RNA modification, it is important to understand the mechanism of modification. Although pseudouridylation and 2′-O-methylation can be catalyzed by single-protein enzymes, the modification of mammalian snRNAs appears exclusively accomplished by snoRNPs. Some of the snoRNPs have distinctive features and names (see below).32,52,60,91 SnoRNPs each consist of a short, function-defining guide RNA and 4 core proteins including the pseudouridine synthase or the methyltransferase (Fig. 1A). Conserved sequence motifs characterize the guide RNAs, boxes H (ANANNA) and ACA in pseudouridylation guides and boxes C/C' (RUGAUGA) and D/D' (CUGA) in methylation guides (Fig. 2B and C). Base pairing of the guide RNAs with the substrate RNAs determines the nucleotides to be modified (Fig. 2B and C). Accordingly, each modification site possesses at least one complementary guide RNA. A large number of guide RNAs thus mirrors the large number of modification sites. Presently, some 700 snoRNAs are expressed at significant levels.32 As the name suggests, the most abundant RNAs are localized in nucleoli where they function in the modification of rRNA, which contains about 100 of each modification that are all guided by snoRNAs.56
Guide RNPs
Each snoRNA is stabilized by 4 core proteins, box C/D RNAs by Nop56, Nop58, 15.5K, and the methylase fibrillarin and box H/ACA RNAs by NHP2, NOP10, GAR1, and the pseudouridine synthase NAP57, also known as dyskerin and Cbf5.52,60,87 Each RNP contains 2 sets of the core proteins, one for each kink-turn motif formed by boxes C/D and C'/D' and one for each hairpin of H/ACA RNAs. Although most individual snoRNAs are low-abundant and consequently difficult to detect in cells, collectively, they are readily identified through their core proteins by indirect immunofluorescence. In fact, the 2 enzymes of the RNPs, fibrillarin and NAP57, were among the first proteins identified in CBs, colocalizing with the CB marker coilin.2,62,69
Substrate RNAs
The identification of coilin afforded the immunolocalization in CBs of trimethylguanosine capped RNAs, Sm proteins, and the U1 snRNP.68,76 These RNAs corresponded to the spliceosomal snRNAs U1, U2, U4, and U5 whose concentration in nuclear foci was visualized around the same time by RNA fluorescent in situ hybridization (FISH).10 After a cytoplasmic maturation phase – where they acquire a heptameric ring of Sm proteins, their cap is hypermethylated, and their 3′-end is trimmed – mammalian snRNAs reenter the nucleus and shuttle to CBs, possibly with the help of the cap hypermethylase Tgs1 and the Sm assembly protein SMN.61,66,79,84,86 In addition to snRNAs, the snoRNAs U3 and U8 target to CBs.42,43,44,65,73 All these noncoding RNAs are subject to pseudouridylation and 2′-O-methylation. Spliceosomal snRNAs collectively contain 28 pseudouridines and 18 2′-O-methyl groups (Table 1).33,58,93 At least in the case of U2 snRNA, which contains 13 pseudouridines alone, the modifications are essential for snRNP formation and splicing.92 Further, the perhaps most prominent H/ACA RNA, human telomerase RNA (hTR), accumulates in CBs.27,97 The hTR of active telomerase RNPs is pseudouridylated and its modification affects the structure of the RNA and the function of telomerase.36 Apparently, all snoRNA substrates are independently expressed and it remains to be determined if intronic snoRNAs are also modified.
Table 1.
Number of human internal snRNA modifications.
| snRNA | U1 | U2 | U4 | U5 | U6 | U4atac | U6atac | U12 | Total |
|---|---|---|---|---|---|---|---|---|---|
| Pseudouridines* | 2 | 13 | 3 | 3 | 3 | 1 | 1 | 2 | 28 |
| 2′-O-methyl groups* | 1 | 7 | 2 | 2 | 5 | 1 | 18 |
Numbers are from.33,58,93
Small CB-Specific RNAs – scaRNAs
The guide RNAs responsible for snRNA modification are specialized snoRNAs in CBs, the small CB-specific RNAs (scaRNAs). Although they possess all the features of common box H/ACA and C/D RNAs, they harbor additional short sequence motifs that are responsible for CB localization, in case of H/ACA RNAs, it is the CAB box (ugAG) and in that of C/D RNAs, it is the G•U/U•G wobble stem (Fig. 2B and C).57,71 The CAB box is recognized by the WD40 repeat protein Wdr79 (aka Wrap53 and TCAB1) that is required for localization of scaRNPs to CBs.82,83 Although C/D scaRNAs lack a CAB box, Wdr79 is also involved in their targeting to CBs even if it recognizes these scaRNAs with a G•U/U•G wobble stem about 20-fold less than H/ACA scaRNAs with a CAB box.82
In addition to these sequence motifs required for CB localization, some scaRNAs show remarkable features not seen in other snoRNAs. They can occur as tandem snoRNAs with 4 potential guide sequences instead of 2 and they can form hybrid snoRNAs, wherein an H/ACA RNA with a CAB box is inserted into the loop of a C/D snoRNA giving rise to 2 potential pseudouridylation pockets and 2 methyl guide sequences (Table 2).11,26,38 Currently 29 scaRNAs have been described, 17 H/ACA, 2 C/D, 1 tandem H/ACA, 4 tandem C/D, and 5 hybrid C/D-H/ACA scaRNAs (Table 2).32 Additionally, hTR, which carries a CAB box and ends in an H/ACA domain, is a scaRNA running up the current number of scaRNAs to 30.27,64 If we consider the expression levels of the scaRNAs reported in the ENCODE sRNA-seq data,32 then the H/ACA motifs outnumber the C/D motifs over 20-fold. Consequently, box H/ACA core proteins should outnumber box C/D core proteins more than 20-fold in CBs because scaRNAs concentrate in CBs and the snoRNP core proteins assemble proportionately around each motif. Indeed, relative to nucleoli, the indirect immunofluorescence signal for NAP57 is higher in CBs, whereas that for fibrillarin is consistently lower than in nucleoli.62,69 However, it should be noted that some of the scaRNAs have reported targets in both, CBs and nucleoli (Table 2). Although, the targets need to be experimentally verified, these scaRNAs are therefore likely residents of both nuclear bodies. Thus it seems that even in the case of scaRNAs, there is no absolute separation between nuclear organelles.
Table 2.
List of scaRNAs.
| Name | Alt. Name | ID | Type | Target 1 | Target 2 | Target 3 | Target 4 |
|---|---|---|---|---|---|---|---|
| SCARNA7 | U90 | snoID_0598 | CD-SCARNA | 5.8S-76 | U1.1-70 | ||
| SCARNA28 | snoID_0620 | CD-SCARNA | U2.2-47 | NA | |||
| SCARNA1 | ACA35 | snoID_0603 | HACA-SCARNA | NA | 18S-1441 | ||
| SCARNA3 | HBI-100 | snoID_0596 | HACA-SCARNA | NA | U6.6-40 | ||
| SCARNA4 | ACA26 | snoID_0595 | HACA-SCARNA | U2.3-41 | U2.3-39 | ||
| SCARNA8 | U92 | snoID_0601 | HACA-SCARNA | U2.3-34 | U2.3-44 | ||
| SCARNA11 | ACA57 | snoID_0610 | HACA-SCARNA | NA | U5.3-41 | ||
| SCARNA14 | U100 | snoID_0612 | HACA-SCARNA | NA | U1.1-72 | ||
| SCARNA15 | ACA45 | snoID_0604 | HACA-SCARNA | NA | U2.3-39 | ||
| SCARNA16 | ACA47 | snoID_0602 | HACA-SCARNA | NA | U1.4-5 | ||
| SCARNA18 | U109 | snoID_0609 | HACA-SCARNA | NA | U1.4-6 | ||
| SCARNA18B | snoID_0707 | HACA-SCARNA | NA | U1.4-6 | |||
| SCARNA19 | hTR/TERC | snoID_1118 | HACA-SCARNA | telomeres | |||
| SCARNA20 | ACA66 | snoID_0592 | HACA-SCARNA | NA | U12.1-27 | ||
| SCARNA21B | snoID_0577 | HACA-SCARNA | U12.1-18 | 28S-4426 | |||
| SCARNA22 | ACA11 | snoID_0611 | HACA-SCARNA | NA | NA | ||
| SCARNA23 | ACA12 | snoID_0594 | HACA-SCARNA | NA | U6.6-40 | ||
| SCARNA26A | snoID_0618 | HACA-SCARNA | U4.2-79 | NA | |||
| SCARNA26B | snoID_0625 | HACA-SCARNA | U4.1-79 | NA | |||
| SCARNA27 | snoID_0614 | HACA-SCARNA | NA | NA | |||
| SCARNA5 | U87 | snoID_0597 | Hybrid | U5.1-39 | 18S-595 | 18S-1530 | U4.1-65 |
| SCARNA6 | U88 | snoID_0613 | Hybrid | U5.1-39 | 18S-1628 | 28S-2861 | 28S-1530 |
| SCARNA10 | U85 | snoID_0608 | Hybrid | 18S-283 | U5.1-44 | 1818S-101 | U5.1-43 |
| SCARNA12 | U89 | snoID_0607 | Hybrid | NA | 18S-917 | 18S-556 | 18S-464 |
| SCARNA21 | ACA68 | snoID_0599 | Hybrid | U12.1-17 | U12.1-18 | U6atac-83 | 28S-4426 |
| SCARNA2 | HBII-382 | snoID_0593 | Tandem-CD | U2.1-25 | NA | 18S-1363 | 28S-1963 |
| SCARNA9 | mgU2-19/30 | snoID_0605 | Tandem-CD | U2.1-19 | NA | NA | U2.1-30 |
| SCARNA9L | snoID_0600 | Tandem-CD | U2.1-19 | NA | NA | U2.1-30 | |
| SCARNA17 | U91 | snoID_0591 | Tandem-CD | U12-21 | NA | U4.1-8 | U2.1-43 |
| SCARNA13 | U93 | snoID_0606 | Tandem-HACA | NA | U7-7 | U5.1-51 | U2.3-54 |
Where substrate and guide RNAs meet – A function for CBs
Given the congregation of modification machinery and substrate RNAs in CBs, it was a small step to predict that CBs are the sites of snRNA modification – guilty by association. Demonstration of this theory however was not straightforward. The elegant work of the Kiss group pointed the way.28 First, they showed that a mutant U2 snRNA that is unable to reenter the nucleus indeed failed to be modified. Only when targeted to CBs, but not to nucleoli, was U5 snRNA modified documenting that CBs are the sites of snRNA modification. For other potential functions of CBs, such as RNP assembly, the reader is referred to other reviews of this Special Focus issue. Modification of snRNAs, however, can also occur in the absence of CBs, i.e. in coilin knockout cells, which are left with 2 types of remnants of CBs, one that accumulates the scaRNAs and snRNAs and one that accumulates independently transcribed snoRNAs and the snoRNP chaperone Nopp140, hinting at a separation of snRNA and snoRNA modification.28,79 That CB structures per se are not required for snRNA modification is further supported by data from fly, which, when lacking CBs and Wdr79 still contain fully modified snRNAs.15,16 Moreover, snRNA modification can occur in the absence of SMN.17 Finally, even mammalian cells often do not contain visible CBs, yet their pre-mRNA splicing seems unperturbed.76 Obviously, snRNAs and scaRNPs can get together without the environs of CBs.
On the other hand, the number and size of CBs correlates positively with the metabolic rate of the cell including transcription and with it pre-mRNA splicing and snRNA synthesis and modification.40,45,75,77 Altogether, these findings support a model whereby, through concentration of snRNAs and scaRNPs, CBs promote snRNA modification, which can also occur outside of these bodies even if at lesser efficiency.
Alternative methods of modification and consequences
Although snRNA modification in mammalian cells appears exclusively catalyzed by scaRNPs, in yeast there are 2 mechanisms, an RNA-guided and an RNA-independent, i.e., protein-only, mechanism.53,54 Whether the latter exists in mammalian cells is not clear, though a recombinant Pus7 homolog is capable of pseudouridylating the uridine in position 34 of human U2 snRNA and a redundant modification mechanism was indicated.33,96 Alternatively, the absence of a true CB (and with it scaRNAs) in yeast could account for the difference. It will be interesting to determine if mammalian snRNA modification can be induced at novel sites as demonstrated for yeast snRNAs and if such modification would also occur in CBs.89 These are important questions, as modification of snRNAs can have serious consequences. Thus, modification is generally required for snRNP assembly and pre-mRNA splicing.92 And specifically, pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly and a pseudouridine in U6 snRNA is part of the filamentous growth program in yeast.7,88 In mRNA, pseudouridines can turn nonsense codons into sense codons diversifying the cellular proteome?34
Open questions
Though the function of CBs as sites of snRNA modification is firmly established, many questions and puzzles remain. For example, are all snRNAs modified in CBs, what about U6, which lacks a trimethylguanosine cap, does not transit through the cytoplasm, and which was proposed to be modified in the nucleolus?20,94
Regardless, the pseudouridylation of at least one of its uridines is catalyzed by a scaRNP – scaRNA23 guides the pseudouridylation at position 40 of U6 snRNA.37
What about snoRNAs, do all traffic through CBs? Unlike most other snoRNAs, U3 and U8 are highly abundant and thus could be isolated and their modification directly demonstrated,35,70 but no guide RNAs have been identified so far.32,46 Whether other, less abundant snoRNAs are modified is unknown. After transfection or microinjection, U3, U8, and U14 snoRNAs indeed traffic to or through CBs, but, except for U3 snoRNA,31,67 endogenous molecules have so far escaped detection in CBs.9,10,65,66,73 In contrast, the endogenous scaRNAs U85, U88, U91, and U92 have been visualized in CBs.11
Regardless, the identification of most snoRNAs among RNAs UV-crosslinked to coilin would suggest that most snoRNAs, if not all, traffic through CBs.55 Indeed, after microinjection some of these snoRNAs are detected in CBs before accumulating in nucleoli, their place of action. Nevertheless, whereas coilin is highly concentrated in CBs, some 70% of it is present in the nucleoplasm, even if much more dilute.3,9,41,59 Therefore, it cannot be excluded that some of the snoRNA associations with coilin occur outside CBs. If CBs play a role in the biogenesis of all snoRNPs, then why are none of the snoRNP maturation factors, except Nopp140, present in CBs?12,22,24 Importantly, assembly of most core proteins, at least in the case of H/ACA RNPs, does not occur in CBs but at the site of snoRNA transcription.6,12,19,72,90 Finally, it is unclear how all the snoRNPs involved in rRNA modification find their way into CBs without specific localizing motifs.
To what degree are snRNAs modified? The simple fact that snRNA modifications were recognized early on suggests that most of the snRNAs are fully modified at individual sites. Indeed, this has been verified for the case of yeast rRNA using a quantitative mass spectrometric approach showing that some 84% of the 112 modified nucleotides are nearly fully modified.78 This is in stark contrast to genome-wide RNAseq based approaches that identified much lower levels of, e.g., pseudouridylation.95 Thus, it can be safely assumed that most modification positions in snRNAs are modified to a high degree.
Furthermore, there is the conundrum of how the different types of RNAs find their way into CBs. While there seems to be a role for Tgs1 and SMN in targeting of snRNAs and for PHAX in targeting of snoRNA substrates to CBs,84 how the remainder of the snoRNPs find their way into CBs remains unclear. In contrast, accumulation of scaRNAs clearly depends on their CAB box that is recognized by Wdr79, but CAB boxes are also present in the over 300 AluACA RNAs that are present in the nucleoplasm but not CBs.29 Therefore, although necessary, Wdr79 may not be sufficient for targeting of scaRNAs but require additional factors, perhaps such as the nucleolar and CB protein Nopp140.25 Obviously, these are only some of the questions that remain to be clarified, leaving plenty of CB modification work ahead.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowlegments
I thank Jonathan Bizarro for critical reading of the manuscript.
Funding
The work in the author's laboratory was supported by a grant from the National Institutes of Health (GM097752 to U.T.M.).
References
- 1.Agris PF. The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Prog Nucleic Acid Res Mol Biol 1996; 53:79-129; PMID:8650309; https://doi.org/ 10.1016/S0079-6603(08)60143-9 [DOI] [PubMed] [Google Scholar]
- 2.Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med 1991; 173:1407-19; PMID:2033369; https://doi.org/ 10.1084/jem.173.6.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA 1993; 90:1947-51; PMID:8446613; https://doi.org/ 10.1073/pnas.90.5.1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 1994; 33:7560-67; PMID:8011621; https://doi.org/ 10.1021/bi00190a008 [DOI] [PubMed] [Google Scholar]
- 5.Auffinger P, Westhof E. Rules governing the orientation of the 2′-hydroxyl group in RNA. J Mol Biol 1997; 274:54-63; PMID:9398515; https://doi.org/ 10.1006/jmbi.1997.1370 [DOI] [PubMed] [Google Scholar]
- 6.Ballarino M, Morlando M, Pagano F, Fatica A, Bozzoni I. The cotranscriptional assembly of snoRNPs controls the biosynthesis of H/ACA snoRNAs in Saccharomyces cerevisiae. Mol Cell Biol 2005; 25:5396-403; PMID:15964797; https://doi.org/ 10.1128/MCB.25.13.5396-5403.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basak A, Query CC. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep 2014; 8:966-73; PMID:25127136; https://doi.org/ 10.1016/j.celrep.2014.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515:143-6; PMID:25192136; https://doi.org/ 10.1038/nature13802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carmo-Fonseca M, Ferreira J & Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis–evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 1993; 120:841-52; PMID:7679389; https://doi.org/ 10.1083/jcb.120.4.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J 1991; 10:195-206; PMID:1824936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 2002; 21:2746-56; PMID:12032087; https://doi.org/ 10.1093/emboj/21.11.2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 2006; 173:207-18; PMID:16618814; https://doi.org/ 10.1083/jcb.200601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis DR. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 1995; 23:5020-6; PMID:8559660; https://doi.org/ 10.1093/nar/23.24.5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Decatur WA, Fournier MJ. RNA-guided nucleotide modification of ribosomal and other RNAs. J Biol Chem 2003; 278:695-8; PMID:12431975; https://doi.org/ 10.1074/jbc.R200023200 [DOI] [PubMed] [Google Scholar]
- 15.Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 2009; 20:5250-9; PMID:19846657; https://doi.org/ 10.1091/mbc.E09-09-0777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deryusheva S, Gall JG. Novel small Cajal-body-specific RNAs identified in Drosophila: probing guide RNA function. RNA 2013; 19:1802-14; PMID:24149844; https://doi.org/ 10.1261/rna.042028.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deryusheva S, Choleza M, Barbarossa A, Gall JG, Bordonné R. Post-transcriptional modification of spliceosomal RNAs is normal in SMN-deficient cells. RNA 2012; 18:31-6; PMID:22124016; https://doi.org/ 10.1261/rna.030106.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201-6; PMID:22575960; https://doi.org/ 10.1038/nature11112 [DOI] [PubMed] [Google Scholar]
- 19.Fatica A, Dlakić M, Tollervey D. Naf1 p is a box H/ACA snoRNP assembly factor. RNA 2002; 8:1502-14; PMID:12515383; http://doi.org/ 10.1017/S1355838202022094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganot P, Jády BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol 1999; 19:6906-17; PMID:10490628; https://doi.org/ 10.1128/MCB.19.10.6906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Girard C, Verheggen C, Neel H, Cammas A, Vagner S, Soret J, Bertrand E, Bordonné R. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J Biol Chem 2008; 283:2060-9; PMID:18039666; https://doi.org/ 10.1074/jbc.M704209200 [DOI] [PubMed] [Google Scholar]
- 22.Grozdanov PN, Roy S, Kittur N & Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA 2009; 15:1188-97; PMID:19383767; https://doi.org/ 10.1261/rna.1532109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 2006; 34:721-33; PMID:16452298; https://doi.org/ 10.1093/nar/gkj471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoareau-Aveilla C. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 2006; 12:832-40; PMID:16601202; https://doi.org/ 10.1261/rna.2344106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol 1998; 142:319-29; PMID:9679133; https://doi.org/ 10.1083/jcb.142.2.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jády BE, Kiss T. A small nucleolar guide RNA functions both in 2′-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J 2001; 20:541-51; https://doi.org/ 10.1093/emboj/20.3.541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jády BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 2004; 164:647-52; https://doi.org/ 10.1083/jcb.200310138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 2003; 22:1878-88; https://doi.org/ 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jády BE, Ketele A, Kiss T. Human intron-encoded Alu RNAs are processed and packaged into Wdr79-associated nucleoplasmic box H/ACA RNPs. Genes Dev 2012; 26:1897-910; https://doi.org/ 10.1101/gad.197467.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, He C. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885-7; PMID:22002720; https://doi.org/ 10.1038/nchembio.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiménez-García LF, Segura-Valdez ML, Ochs RL, Rothblum LI, Hannan R, Spector DL. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell 1994; 5:955-66; PMID:784152327174936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR. An updated human snoRNAome. Nucleic Acids Res 2016; 44:5068-82; PMID:27174936; http://doi.org/ 10.1093/nar/gkw386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karijolich J, Yu YT. Spliceosomal snRNA modifications and their function. RNA Biol 2010; 7:192-204; PMID:20215871; http://doi.org/ 10.4161/rna.7.2.11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011; 474:395-8; PMID:21677757; http://doi.org/ 10.1038/nature10165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kato N, Harada F. Nucleotide sequence of nuclear 5.4 S RNA of mouse cells. Biochim Et Biophys Acta (BBA) - Gene Struct Expression 1984; 782:127-31; http://doi.org/ 10.1016/0167-4781(84)90015-0 [DOI] [PubMed] [Google Scholar]
- 36.Kim N-K, Theimer CA, Mitchell JR, Collins K, Feigon J. Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res 2010; 38:6746-56; PMID:20554853; http://doi.org/ 10.1093/nar/gkq525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kiss AM, Jády BE, Bertrand E, Kiss T. Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 2004; 24:5797-5807; PMID:15199136; http://doi.org/ 10.1128/MCB.24.13.5797-5807.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss AM, Jády BE, Darzacq X, Verheggen C, Bertrand E, Kiss T. A Cajal body-specific pseudouridylation guide RNA is composed of two box H/ACA snoRNA-like domains. Nucleic Acids Res 2002; 30:4643-9; PMID:12409454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 2002; 109:145-8; PMID:12007400 [DOI] [PubMed] [Google Scholar]
- 40.Lafarga M, Berciano MT, Garcia-Segura LM, Andres MA, Carmo-Fonseca M. Acute osmotic/stress stimuli induce a transient decrease of transcriptional activity in the neurosecretory neurons of supraoptic nuclei. J Neurocytol 1998; 27:205-17; PMID:10640180 [DOI] [PubMed] [Google Scholar]
- 41.Lam YW, Lyon CE, Lamond AI. Large-scale isolation of Cajal bodies from HeLa cells. Mol Biol Cell 2002; 13:2461-73; PMID:12134083; http://doi.org/ 10.1091/mbc.02-03-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange TS, Borovjagin A, Maxwell ES, Gerbi SA. Conserved boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J 1998a; 17:3176-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lange TS, Borovjagin AV, Gerbi SA. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA 1998b; 4:789-800; http://doi.org/ 10.1093/emboj/17.11.3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange TS, Ezrokhi M, Borovjagin AV, Rivera-León R, North MT, Gerbi SA. Nucleolar localization elements of Xenopus laevis U3 small nucleolar RNA. Mol Biol Cell 1998c; 9:2973-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Lührmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 2006; 17:3221-31; PMID:16687569; http://doi.org/ 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lestrade L, Lestrade L, Weber MJ, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 2006; 34:D158-62; PMID:16381836; http://doi.org/ 10.1093/nar/gkj002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Ma S, Yi C. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol 2016; 33:108-16; PMID:27348156; http://doi.org/ 10.1016/j.cbpa.2016.06.014 [DOI] [PubMed] [Google Scholar]
- 48.Li X, Zhu P, Ma S, Song J, Bai J, Sun F & Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015; 11:592-7; PMID:26075521; http://doi.org/ 10.1038/nchembio.1836 [DOI] [PubMed] [Google Scholar]
- 49.Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol 2016; 23:98-102; PMID:26840897; http://doi.org/ 10.1038/nsmb.3162 [DOI] [PubMed] [Google Scholar]
- 50.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J 1996; 15:3555-65; PMID:8670859 [PMC free article] [PubMed] [Google Scholar]
- 51.Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: Pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 2014; 9:e110799; PMID:25353621; http://doi.org/ 10.1371/journal.pone.0110799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lui L, Lowe T. Small nucleolar RNAs and RNA-guided post-transcriptional modification. Essays Biochem 2013; 54:53-77; PMID:23829527; http://doi.org/ 10.1042/bse0540053 [DOI] [PubMed] [Google Scholar]
- 53.Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT. Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 2005; 24:2403-13; PMID:15962000; http://doi.org/ 10.1038/sj.emboj.7600718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 2003; 22:1889-97; PMID:12682021; http://doi.org/ 10.1093/emboj/cdg191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The Coilin interactome identifies hundreds of small noncoding RNAs that traffic through cajal bodies. Mol Cell 2014; 56:389-99; PMID:25514182; http://doi.org/ 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 56.Maden BE. The numerous modified nucleotides in eukaryotic ribosomal RNA. Prog Nucleic Acid Res Mol Biol 1990; 39:241-303; PMID:2247610; http://doi.org/ 10.1234/12345678 [DOI] [PubMed] [Google Scholar]
- 57.Marnef A, Richard P, Pinzón N, Kiss T. Targeting vertebrate intron-encoded box C/D 2′-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res 2014; 42:6616-29; PMID:24753405; http://doi.org/ 10.1093/nar/gku287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massenet S, Mougin A, Branlant C. Posttranscriptional modifications in the U small nuclear RNAs. In modification and editing of RNA. Am Soc Microbiol 1998; 201-27; http://doi.org/ 10.1128/9781555818296.ch11 [DOI] [Google Scholar]
- 59.Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem 1998; 70:181-92; PMID:9671224; http://doi.org/ 10.1038/nrm2124 [DOI] [PubMed] [Google Scholar]
- 60.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 2007; 8:209-20; PMID:17318225; http://doi.org/ [DOI] [PubMed] [Google Scholar]
- 61.Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 1986; 46:905-11; PMID:2944599; http://doi.org/ 10.1016/0092-8674(86)90072-3 [DOI] [PubMed] [Google Scholar]
- 62.Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol 1994; 127:1505-14; PMID:7798307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, Jaffrey SR. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012; 149:1635-46; PMID:22608085; http://doi.org/ 10.1016/j.cell.2012.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitchell JR, Cheng J, Collins K. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 1999; 19:567-76; PMID:9858580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell 1999; 10:2131-47; PMID:10397754; http://doi.org/ 10.1091/mbc.10.7.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pradet-Balade B, Girard C, Boulon S, Paul C, Azzag K, Bordonné R, Bertrand E, Verheggen C. CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J 2011; 30:2205-18; PMID:21522132; http://doi.org/ 10.1038/emboj.2011.128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J Cell Biochem 1995; 59:11-26; PMID:8530530 [DOI] [PubMed] [Google Scholar]
- 68.Raska I, Andrade LE, Ochs RL, Chan EK, Chang CM, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 1991; 195:27-37; PMID:2055273 [DOI] [PubMed] [Google Scholar]
- 69.Raska I, Ochs RL, Andrade LE, Chan EK, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol 1990; 104:120-7; PMID:2088441; http://doi.org/ 10.1002/jcb.240590103 [DOI] [PubMed] [Google Scholar]
- 70.Reddy R, Henning D, Busch H. Nucleotide sequence of nucleolar U3B RNA. J Biol Chem 1979; 254:11097-105; PMID:500626 [PubMed] [Google Scholar]
- 71.Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T. A common sequence motif determines the Cajal body-specific localization of box H/ACA scaRNAs. EMBO J 2003; 22:4283-93; PMID:12912925; http://doi.org/ 10.1093/emboj/cdg394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Richard P, Kiss AM, Darzacq X, Kiss T. Cotranscriptional recognition of human intronic box H/ACA snoRNAs occurs in a splicing-independent manner. Mol Cell Biol 2006; 26:2540-9; PMID:16537900; http://doi.org/ 10.1128/MCB.26.7.2540-2549.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Samarsky DA, Samarsky DA, Fournier MJ, Fournier MJ, Singer RH, Singer RH, Bertrand E, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J 1998; 17:3747-57; PMID:9649444; http://doi.org/ 10.1093/emboj/17.13.3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al.. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; http://doi.org/ 10.1016/j.cell.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 2001; 114:4407-19; PMID:11792806 [DOI] [PubMed] [Google Scholar]
- 76.Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell 1992; 3:555-69; PMID:1535243; https://doi.org/ 10.1091/mbc.3.5.555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Strzelecka M, Oates AC & Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus (Austin, Tex) 2010; 1:96-108; PMID:21327108; http://doi.org/ 10.4161/nucl.1.1.10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Taoka M, Nobe Y, Yamaki Y, Yamauchi Y, Ishikawa H, Takahashi N, Nakayama H, Isobe T. The complete chemical structure of Saccharomyces cerevisiae rRNA: partial pseudouridylation of U2345 in 25S rRNA by snoRNA snR9. Nucleic Acids Res 2016; 44(18):8951-61; PMID:27325748; http://doi.org/11470819 10.1093/nar/gkw564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 2001; 154:293-307; PMID:11470819; https://doi.org/ 10.1083/jcb.200104083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tyc K, Steitz JA. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J 1989; 8:3113-9; PMID:2531075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tycowski KT, Aab A, Steitz JA. Guide RNAs with 5′ caps and novel box C/D snoRNA-like domains for modification of snRNAs in metazoa. Curr Biol 2004; 14:1985-95; PMID:15556860; https://doi.org/ 10.1016/j.cub.2004.11.003 [DOI] [PubMed] [Google Scholar]
- 82.Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 2009; 34:47-57; PMID:19285445; https://doi.org/ 10.1016/j.molcel.2009.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for cajal body localization and telomere synthesis. Science 2009; 323:644-8; PMID:19179534; https://doi.org/ 10.1126/science.1165357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Verheggen C, Bertrand E. CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes. Nucleus (Austin, Tex) 2012; 3:132-7; PMID:22555597; http://doi.org/ 10.1093/emboj/20.19.5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Verheggen C, Lafontaine DLJ, Samarsky D, Mouaikel J, Blanchard JM, Bordonné R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 2002; 21:2736-45; PMID:12032086; https://doi.org/ 10.1093/emboj/21.11.2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Verheggen C, Mouaikel J, Thiry M, Blanchard JM, Tollervey D, Bordonné R, Lafontaine DL, Bertrand E. Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain. EMBO J 2001; 20:5480-90; PMID:11574480; https://doi.org/ 10.1093/emboj/20.19.5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; https://doi.org/ 10.1002/wrna.117 [DOI] [PubMed] [Google Scholar]
- 88.Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J 2016; 35:654-67; PMID:26873591; https://doi.org/ 10.15252/embj.201593113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 2011; 30:79-89; PMID:21131909; https://doi.org/ 10.1038/emboj.2010.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang PK, Hoareau C, Froment C, Monsarrat B, Henry Y, Chanfreau G. Cotranscriptional recruitment of the pseudouridylsynthetase Cbf5p and of the RNA binding protein Naf1p during H/ACA snoRNP assembly. Mol Cell Biol 2005; 25:3295-304; PMID:15798213; https://doi.org/ 10.1128/MCB.25.8.3295-3304.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine–pseudouridylation. RNA Biol 2014; 11:1483-94; PMID:25590339; https://doi.org/ 10.4161/15476286.2014.972855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre‐mRNA splicing. EMBO J 1998; 17:5783-95; PMID:9755178; https://doi.org/ 10.1093/emboj/17.19.5783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu YT, Scharl EC, Smith CM, Steitz JA. The growing world of small nuclear ribonucleoproteins In The RNA World, Gesteland RF, Cech TR, Atkins JF (eds). Cold Spring Harbor, NY; 1999; pp 487-524; http://doi.org/ 10.4161/15476286.2014.972855 [DOI] [Google Scholar]
- 94.Yu YT, Shu MD, Narayanan A, Terns RM, Terns MP, Steitz JA. Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J Cell Biol 2001; 152:1279-88; PMID:11257127; https://doi.org/ 10.1083/jcb.152.6.1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zaringhalam M, Papavasiliou FN. Pseudouridylation meets next-generation sequencing. Methods 2016; 107:63-72; PMID:26968262; https://doi.org/ 10.1016/j.ymeth.2016.03.001 [DOI] [PubMed] [Google Scholar]
- 96.Zhao X, Li Z-H, Terns RM, Terns MP, Yu YT. An H/ACA guide RNA directs U2 pseudouridylation at two different sites in the branchpoint recognition region in Xenopus oocytes. RNA 2002; 8:1515-25; PMID:12515384; http://doi.org/ 10.1017/S1355838202022537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 2003; 15:81-90; PMID:14528011; https://doi.org/ 10.1091/mbc.E03-07-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zieve G, Penman S. Small RNA species of the HeLa cell: Metabolism and subcellular localization. Cell 1976; 8:19-31; PMID:954090; https://doi.org/ 10.1016/0092-8674(76)90181-1 [DOI] [PubMed] [Google Scholar]