ABSTRACT
Nuclear bodies are cellular compartments that lack lipid bilayers and harbor specific RNAs and proteins. Recent proposals that nuclear bodies form through liquid-liquid phase separation leave the question of how different nuclear bodies maintain their distinct identities unanswered. Here we investigate Cajal bodies (CBs), histone locus bodies (HLBs) and nucleoli – involved in assembly of the splicing machinery, histone mRNA 3′ end processing, and rRNA processing, respectively – in the embryos of the zebrafish, Danio rerio. We take advantage of the transcriptional silence of the 1-cell embryo and follow nuclear body appearance as zygotic transcription becomes activated. CBs are present from fertilization onwards, while HLB and nucleolar components formed foci several hours later when histone genes and rDNA became active. HLB formation was blocked by transcription inhibition, suggesting nascent histone transcripts recruit HLB components like U7 snRNP. Surprisingly, we found that U7 base-pairing with nascent histone transcripts was not required for localization to HLBs. Rather, the type of Sm ring assembled on U7 determined its targeting to HLBs or CBs; the spliceosomal Sm ring targeted snRNAs to CBs while the specialized U7 Sm-ring localized to HLBs, demonstrating the contribution of protein constituents to the distinction among nuclear bodies. Thus, nucleolar, HLB, and CB components can mix in early embryogenesis when transcription is naturally or artificially silenced. These data support a model in which transcription of specific gene loci nucleates nuclear body components with high specificity and fidelity to perform distinct regulatory functions.
KEYWORDS: Cajal body, coilin, fibrillarin, liquid phase separation, histone locus body, zebrafish, zygotic genome activation
Introduction
Recent evidence has highlighted the importance of cellular compartments that lack lipid bilayers yet concentrate specific molecular components and enhance their interactions.1-3 These nuclear bodies and cytoplasmic granules are often RNA-rich and are characterized by protein constituents that have intrinsically disordered or low complexity regions. An emerging model for their assembly is that these protein constituents undergo liquid-liquid phase separation (LLPS) and thereby form droplet-like, round objects in cells that can move, split, and fuse.2,4 Numerous recent in vitro studies have begun to catalog the proteins and their domains that are capable of undergoing LLPS. While these studies are essential to our understanding of biochemical and biophysical mechanisms, the simultaneous presence of multiple bodies and granules in vivo – each containing numerous specific protein and RNA constituents – raises the question of how such cellular compartments maintain their integrity from one another. For example, nucleoli – the sites of rDNA transcription and rRNA processing – arise as rDNA transcription is activated, and it is well-known that multiple small nucleoli fuse into a bigger nucleolus as the cell cycle proceeds.5,6 However, nucleoli do not fuse over time with other distinct nuclear bodies. How does specificity among membraneless compartments arise? How is integrity maintained?
The nucleolus, the Cajal body (CB) and the histone locus body (HLB) are well-studied nuclear bodies that have distinct functions in ribosome biogenesis, spliceosomal small nuclear ribonucleoprotein (snRNP) assembly, and histone mRNA 3′ end formation, respectively.7 CBs are scaffolded by the low complexity protein, coilin, and concentrate snRNAs, snoRNAs, and protein components of both snRNPs and snoRNPs.3,7-9 CBs are the sites of snRNP assembly and knock down of coilin is lethal in zebrafish due to a splicing deficit.10,11 HLBs contain the machinery required for 3′ end cleavage of replicative histone mRNAs: U7 snRNP with a specialized Sm ring containing Lsm10 and Lsm11 proteins, stem loop binding protein (SLBP) and 2 low complexity proteins – FLASH and NPAT – that also have roles in transcription.12-15 Interestingly, the protein and RNA constituents of CBs and HLBs sometimes overlap; for example, U7 snRNA is sometimes present in CBs.7 In transformed tissue culture cells like HeLa, CBs and HLBs are usually combined in a single body.16 However, in non-transformed cells – ES cells, embryonic cells of flies and fish, and in fly tissues – CBs and HLBs are separate spatially and vary independently in number and size.15,17-20 Thus, the overlap between HLBs and CBs in transformed cells may indicate a tendency for association that might take place under certain, possibly abnormal, physiological conditions. For example, nucleoli under stress accumulate CB components.21 Taken together, the data indicate that components of these distinct nuclear bodies can indeed mix.
Here we take advantage of the early zebrafish embryo to investigate how CBs, HLBs and nucleoli are established as separate entities. We investigate the hypothesis that transcription of genes encoding the RNAs recognized by nuclear body components could serve as nucleation sites that potentially drive separation of related nuclear bodies. Previous studies have shown that nucleoli, CBs and HLBs are localized at transcription sites and at least partially transcription-dependent in tissue culture cells.1,15,16,22,23 Indeed, ectopic expression of histone genes and U2 gene arrays in HeLa cells are sufficient for nucleation of additional CBs and HLBs, respectively.16,24,25 In the embryo, we investigate the architecture of the nucleus before, during and after zygotic genome activation (ZGA), which entails the transcriptional activation of the genome after a period of maternal control and transcriptional silence.26 The 1-cell embryo can be injected with inhibitors of transcription, nucleotide analogs, fluorescent RNAs and other reporters; our previous study utilizing metabolic labeling of the first zygotic transcripts has identified the precise cell cycle at which transcription begins.27 This system enables us to investigate the question of nuclear body integrity in the context of normal and disrupted cellular function during vertebrate development.
Results and discussion
Fibrillarin resides in CBs until rDNA transcription activation
We sought to investigate the behavior of shared components of nucleoli and CBs with respect to rDNA transcription in early zebrafish embryos. About 700 human proteins co-purify with nucleoli,28 including many protein components – such as fibrillarin – of snoRNPs. Interestingly, hundreds of snoRNAs as well as fibrillarin pass through CBs en route to the nucleolus where they guide modifications of rRNA.9,29,30 Thus, both nucleoli and HLBs contain major components that also display minor localization to CBs in tissue culture cells. Nucleoli are transcription-dependent in somatic cells23; they form at the end of mitosis, coalesce into fewer and larger nucleoli in S-phase, and disassemble before M phase. The oocyte is packed with maternally provided rRNA and ribosomes,26,31 and rDNA transcription is undetectable at the 512-cell stage.27 However, when nucleoli form and when rDNA becomes active in the zebrafish embryo is unknown.
To determine the localization of nucleolar and CB components before, during, and after ZGA, early zebrafish embryos were co-immunostained for fibrillarin and coilin (Fig. 1A). We found that fibrillarin exclusively colocalized with coilin until early gastrulation stages (6hpf), indicating its association with CBs rather than independent nucleoli. Similarly, we found that fluorescently-labeled U8 snoRNA colocalizes with U85 scaRNA, another CB marker ([19] and data not shown). To determine whether rDNA was transcriptionally active at these time points, RT-qPCR was employed to quantify the levels of unprocessed pre-rRNA over 1-hour intervals of early development (Fig. 1B). The data indicate no maternally supplied unprocessed pre-rRNA; detection of unprocessed pre-rRNA above background levels was only possible with samples prepared from embryos at early gastrulation. De novo transcription of rDNA at 6hpf is consistent with our observation that fibrillarin is present in CBs and a second nuclear body – likely the nucleolus – at 6hpf (Fig. 1A). We conclude that activation of rDNA transcription in zebrafish takes place several hours after RNA pol II becomes transcriptionally active and that canonical nucleoli are absent from pre-gastrula zebrafish embryos. Instead fibrillarin resides inside CBs during pre-gastrulation stages.
Figure 1.
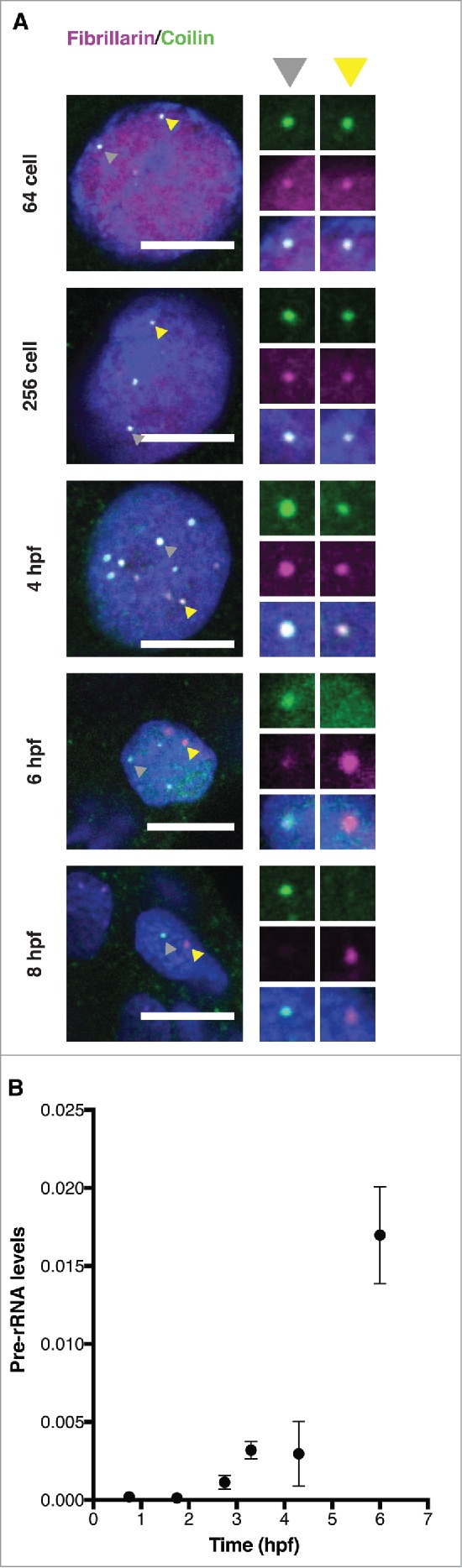
Activation of rDNA transcription recruits fibrillarin from the CB. (A) Immunostaining for the CB marker coilin (green) and the snoRNP component fibrillarin (magenta) over a developmental time course from the 64-cell stage to 8 hours post-fertilization (hpf). Insets magnify (2x) 2 representative bodies in the same nucleus (indicated by gray and yellow arrowheads). All scale bars, 10 μm. A Gaussian blur filter was applied for display reasons. (B) Detection of the onset of rDNA transcription through RT-qPCR amplification of pre-rRNA. Primers complementary to the 5′ external transcribed spacer (ETS) of 18S RNA were used to amplify pre-rRNA; signal (y-axis) was normalized to RT-qPCR values for pre-mitochondrial RNA, which was previously shown to be constant over these time points.27
Zygotic transcription is required for HLB formation
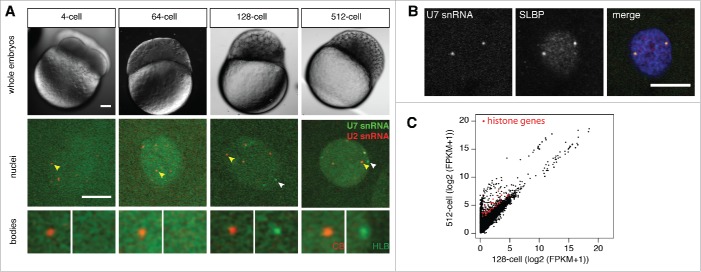
To visualize and distinguish between nuclear bodies in the early embryo, probes for fluorescent imaging are necessary. Injection of GFP-tagged constructs is not well suited for imaging nuclear bodies before ZGA, because 2.5 hours are required for translation and maturation of the GFP.19 Previously, we and others have employed injection of in vitro transcribed, fluorescent snRNAs and snoRNAs into tissue culture cells and 1-cell zebrafish embryos.11,19,32-34 Co-injection of mRNA encoding mRFP-Lsm11 and U7 snRNA results in colocalization in a discrete number of foci separate from coilin, suggesting that CBs and HLBs are separate entities at 3 hours post-fertilization (hpf), several hours after ZGA begins.19 Similarly, co-injected fluorescent U2 and U7 snRNAs localize to distinct CBs and HLBs at the 128- and 512-cell stages (Fig. 2A) and after zygotic genome activation (Fig. S1). Moreover, we could validate that the nuclear bodies marked by WT U7 are HLBs by co-staining with α-SLBP and are clearly separated from CBs (Fig. 2B, Fig. S1); SLBP binds to the stem loop of the nascent histone transcripts and exhibits diffuse nuclear staining as well as colocalization with HLBs in mammalian tissue culture cells.15
Figure 2.
HLB assembly coincides with the onset of zygotic transcription. (A) Embryonic nuclei imaged at the indicated stages after injection of fluorescent U2 snRNA (red) to mark CBs and U7 snRNA (green) to mark HLBs. Upper panel: DIC image from in vivo imaging of whole zebrafish embryos with 10x magnification. Scale bar equals 100μm. Middle panel: Single confocal sections of in vivo imaging of zebrafish embryos labeled with microinjected U7 and U2 snRNA. Scale bar equals 10μm. Arrowheads point to the nuclear body shown 2x magnified in bottom panel. A Gaussian blur filter was applied for display reasons. (B) Immunostaining for SLBP (red in the merged panel) shows the concentration of SLBP in the same foci as those marked by injected U7 snRNA (green). Scale bar equals 10 μm (C) Scatterplot showing the levels in FPKM of individual transcripts metabolically labeled by 4s-UTP injection into 1-cell embryos and purified from 128-cell (x-axis) and 512-cell (y-axis) embryos; histone genes with a fold change in transcript level of log2 ≥ 2 are displayed in red, showing that transcription of histone genes begins between the 128-cell and 512-cell stage. Data taken from ref. 27.
To investigate the relationship of the HLB to transcriptional activity during the first rapid cell cycles, early time points were examined (Fig. 2A). Confirming that CBs are present in the embryo in large numbers from fertilization onwards,19 CBs were readily detected from the 4-cell stage onwards. The first detection of U7 snRNA foci in our assay was at the 128-cell stage, coinciding with the transcriptional activity of zebrafish embryos.27 Specific examination of metabolically labeled histone mRNA transcripts during the 128- to 512-cell stage, confirms that histone genes are transcriptionally activated during this time window (Fig. 2C). Detection of U7 snRNA foci at later stages was more robust, suggesting that increasing histone gene transcriptional activity during ZGA enhances HLB formation. Taken together, our data indicate that the HLB forms at the onset of zygotic transcription.
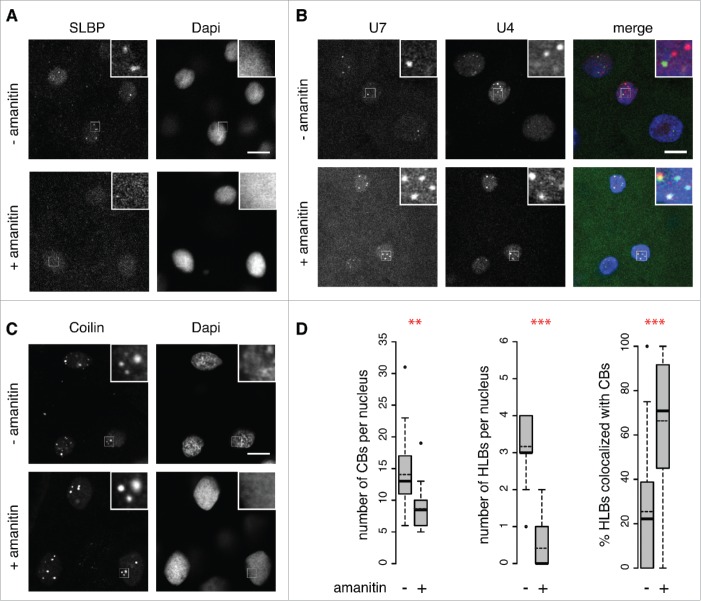
In contrast to HLBs, CBs are present in large numbers just after fertilization and before ZGA ([19] and Fig. 2). To determine whether the HLB is indeed transcription-dependent we blocked RNA Pol II activity in the embryo by α-amanitin injection and immunostained for coilin and SLBP (Fig. 3). Note that coilin is a definitive marker of CBs, which we have characterized extensively in previous studies.11,19 Although CBs were still present in transcription blocked embryos, SLBP foci were undetectable. To test if other HLB components also disperse or fail to coalesce, microinjected U7 snRNA and U4 snRNA foci were examined with and without transcription block. Upon loss of transcription, U7 snRNA colocalizes significantly more often with CBs marked by U4 snRNA (Fig. 3B). Note that α-amanitin treatment leads to developmental arrest.35 However, the strength of transcription block might differ between individual cells of the embryo, which may explain why re-localization is not 100%. All of the data are quantitated in Fig. 3D, which shows a modest effect of α-amanitin treatment on CB numbers (13/cell vs 9/cell), dramatic effects on HLB numbers (3/cell vs 0) and a significant increase in the overlap between CB and HLB components (22% vs 71%). Taken together these data imply that – in contrast to CBs – HLBs in early embryos are transcription-dependent and their formation coincides with histone gene transcription. Furthermore, inhibition of transcription causes U7 snRNA to relocalize to CBs, despite the presence of its HLB targeting sequence and Sm-ring.
Figure 3.
Separation of CBs and HLBs is transcription-dependent. Zygotic transcription was blocked by injection of α-amanitin into 1-cell embryos. Single Z-stacks of confocal series from fixed blastula stage embryos. Untreated (- amanitin) and treated (+ amanitin) embryos are labeled with antibodies specific for SLBP (A) or coilin (C); co-injection of U4 (red) and U7 snRNAs (green) provides an independent view of CBs and HLBs, respectively (B). White square indicates area shown magnified in each inset (4x magnification). Scale bars equal 10 μm. (D) Quantification of CBs per nucleus (coilin immunostaining, as in C; data from n=18 embryos from 3 independent experiments at 4hpf), HLBs per nucleus (SLBP, as in A; data from n = 18 untreated and n = 17 α-amanitin injected embryos from 3 independent experiments at 4hpf), and of the overlapping signals between HLBs and CBs (U4 and U7 snRNAs, as in B; n = 31 nuclei from untreated embryos and n = 16 nuclei from α-amanitin injected embryos at 3hpf). Distribution, mean number (dashed line) and median number (bold line) are shown. Asterisks indicate statistical significance (Welch 2-sample t-test, P = 0.004702 for CBs, P = 1.043e-10 for HLBs and P = 7.208e-05 for U4/U7 snRNA).
U7 snRNP targeting to HLBs depends on the Sm-ring
The accurate sorting of snRNAs to CBs and HLBs can also be appreciated in Fig. 4A, where fields of nuclei labeled with U2 and U7snRNAs are shown in 3 dimensional reconstructions at 4hpf. How does snRNA localization to either specific body occur? The results of the previous section suggest that U7 snRNA may be targeted to HLBs through base-pairing to nascent histone transcripts. The 5′ region of U7 snRNA is complementary to downstream sequences in canonical histone genes and base pairs with newly transcribed, still uncleaved histone RNA.12,36 To determine whether base pairing with nascent histone RNA is required to target U7 snRNA to HLBs, we generated mutated U7 snRNAs in vitro and tested their localization after injection. Two mutant U7 snRNAs harboring alterations in the sequence of the histone RNA base-pairing region were constructed (Fig. 4B): the HDE mutant – replacing most of the uracils with other nucleotides – and the A-mutant – replacing a stretch of adenines with uracils. We would expect these mutants to be dispersed throughout the nucleus if base-pairing is important for HLB targeting. However, we observe colocalization of the A-mutant and the HDE-mutant with U7 WT in HLBs (Fig. 4D and E), indicating that U7 snRNA is recruited to HLBs without base-pairing to nascent histone RNA.
Figure 4.
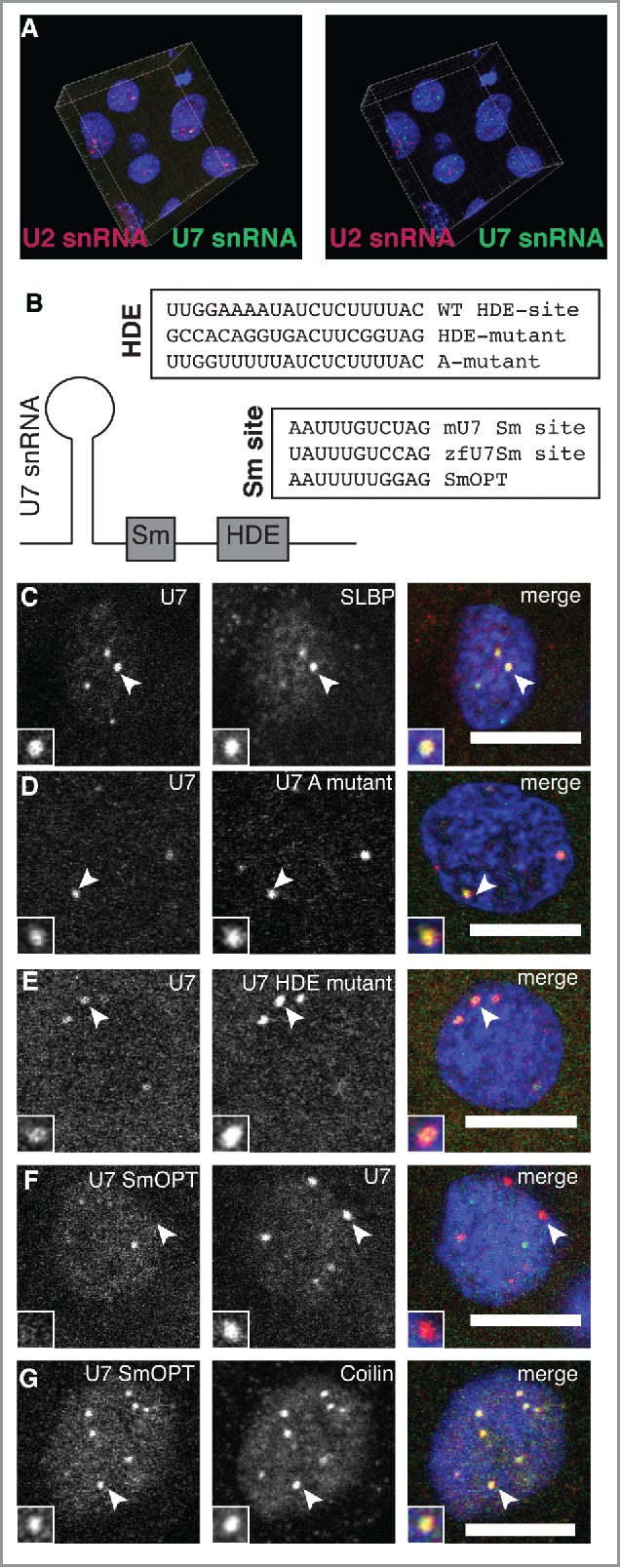
Sm ring composition specifies snRNA localization to CBs and HLBs. (A) Co-injection of U2 (magenta) and U7 (green) snRNAs shows independence of CBs and HLBs in 3 dimensions. Projection of the fluorescent data collected by confocal imaging is shown in the left panel; rendering of CB and HLB centers of mass, obtained through the use of Imaris analysis software, is shown in the right panel. (B) Scheme for testing the contribution of base-pairing to histone transcripts through mutational analysis of the U7 snRNA HDE site is shown. To test the contribution of the Sm-ring, the specialized Sm site of mouse U7 (mU7) was mutated to Sm-OPT, previously shown to target the spliceosomal Sm ring to U7 snRNA.47 (C-G) Confocal sections showing the localization patterns of wild-type U7 snRNA compared to other markers and mutants, as indicated. Scale bars equal 10 μm. Arrowheads point to foci magnified 2x in the insets.
An alternative determinant of snRNA localization is the Sm-ring. Assembly of a heptameric ring of Sm-proteins onto a single-stranded Sm-site within snRNAs stabilizes snRNAs within snRNPs.37 The canonical Sm-ring on spliceosomal snRNPs binds to coilin and may be required for targeting to CBs,33,34,38 suggesting that the specific type of Sm-ring assembled on the snRNA might determine targeting specificity. Previous studies have shown that knockdown of spinal motor neuron protein (SMN) that participates in Sm-ring assembly causes loss of CBs19,39,40; this could either be due to reduced snRNP biogenesis or to a direct role for SMN in CB assembly. A requirement for the Sm ring in CB and HLB localization has not been rigorously tested. Sequence alignments of U7 snRNA with other snRNAs revealed that U7 snRNA harbors an atypical Sm-ring binding site.41 This unique Sm-ring binding site promotes the assembly of a specialized Sm-ring where the 2 proteins SmD1/D2 are substituted with Lsm10 and Lsm11.42,43 Lsm11 binds the HLB component FLASH, suggesting a possible mechanism for U7 snRNP localization to HLBs.12,44
To determine whether the identity of the Sm ring determines nuclear body localization, we next changed the special Sm-ring site of U7 to the consensus sequence of the spliceosomal snRNAs termed SmOPT.45,46 U7 SmOPT RNA was shown to assemble the Sm-proteins found on the spliceosomal snRNAs U1, U2, U4, U5 and to render the U7 snRNP inactive for histone mRNA 3′ end processing.12,46,47 If the Sm-ring determines the localization of U7 snRNA, we would expect U7 SmOPT to show distinct localization compared to WT U7 snRNA. Indeed, co-injections of U7 SmOPT and U7 Sm-WT revealed that those 2 RNAs indeed localize to distinct nuclear foci (Fig. 4F). To test if U7 SmOPT localizes to CBs, as expected for spliceosomal snRNAs,11,19 injected embryos were co-stained with α-coilin. Indeed, U7 SmOPT colocalizes with coilin, a marker of CBs (Fig. 4G). We conclude that the Sm-ring identity determines the specificity of snRNA localization to CBs and HLBs.
Model for the formation and maintenance of discrete nuclear bodies during embryogenesis
Our current findings allow us to generate a comprehensive picture of nuclear architecture in a developing vertebrate embryo, incorporating the role that transcription plays in establishing nuclear compartments (Fig. 5). During transcriptional silence before ZGA, the nucleus harbors about 20 CBs that also contain the nucleolar factors, like fibrillarin. HLBs are absent at these initial stages and emerge as histone transcription starts at ZGA. Localization of the snRNAs to CBs or HLBs depends on the kind of Sm-ring assembled on the Sm site. If transcription is blocked, HLBs disperse and U7 snRNA re-localizes to CBs. We speculate that U7 snRNP may join CBs due to the shared protein components of both types of Sm ring, which have affinity for coilin.9,38 Because U7 snRNA does not concentrate in CBs before transcription onset, we presume that another HLB component – perhaps FLASH which binds the Lsm-ring component Lsm1112-14– may buffer a tendency for U7 to localize to CBs early or alternatively promote colocalization late. As there are many components of CBs and HLBs to test with regard to this activity; unfortunately, most antibodies specific for HLB components like FLASH are non-reactive in zebrafish. In Drosophila, fibrillarin positive bodies form during cleavage stages, but these are variable in size and number before stabilization by transcription.5 In zebrafish, fibrillarin foci – spatially distinct from CBs – form much later during gastrulation, reflecting the late onset of rDNA transcription and likely nucleolus formation. Taken together, these observations suggest that transcription of histone and rDNA genes drives the nucleation of specific nuclear bodies at their respective chromosomal sites. Nuclear body components are then recruited away from previous, less stable sites of segregation, such as the zebrafish embryonic CB. It is currently unclear how segregation into one nuclear body and resorting into a distinct nuclear body is compatible with LLPS. Alternatively, newly translated protein must perform this function. However, previous work has shown that the molecular components of nuclear bodies are exchangeable with nucleoplasm, providing opportunities for re-sorting after nuclear body formation.48-50
Figure 5.
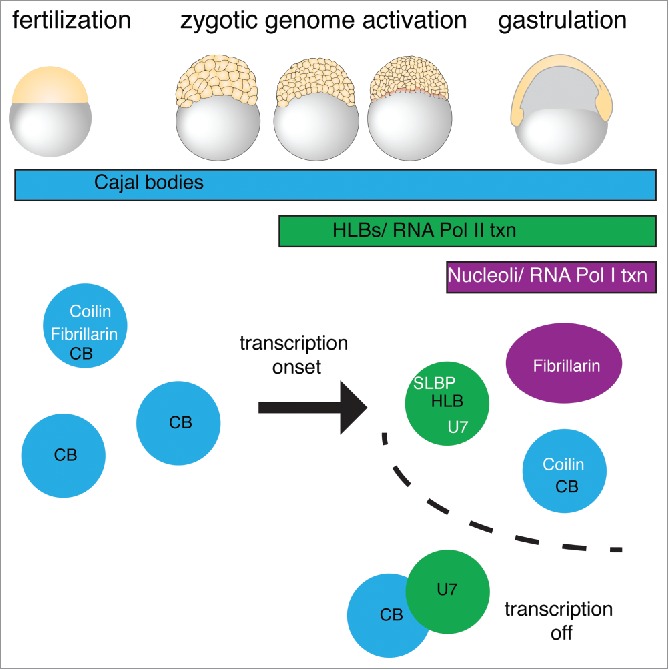
Transcription-dependent changes in embryonic nuclear architecture. Working model showing that between fertilization and ZGA, fibrillarin initially concentrates in CBs while U7 snRNA is not present in foci. Upon transcription activation of histone loci, U7 and SLBP are recruited to HLBs that are separate from CBs in zebrafish embryos. If Pol II transcription is inhibited, U7 accumulates in CBs. Fibrillarin remains associated with CBs until activation of rDNA transcription at 6hpf. Thus, the integrity and composition of each nuclear body depends on transcription.
Materials and methods
Zebrafish maintenance
Wildtype AB strain zebrafish embryos were raised in E3 embryo medium (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, 10-5 % Methyl blue in ddH2O) and staged as previously described.51
In vitro transcription of RNA
In vitro transcription of RNA was done according to published methods.11,19 In brief, the MEGAshortscript™ T7 Kit was used for the production of snRNAs according to manufacturer instructions. For the production of fluorescent RNA modified ribonucleotides (Alexa488-UTP, Alexa546-UTP or Cy5-UTP) were used at a ratio of 1:4 to unmodified UTP. The reactions for U2, U4 and U7 snRNA (and variants) were supplemented with m7G(5′)ppp(5′)G cap analog (Ambion) at a final concentration of 1 mM (1:4 ratio of rGTP: m7G(5′)ppp(5′)G). For zfU7 snRNA, mMU7 snRNA, U7-A mutant and U7-HDE mutant an oligo with the reverse complementary sequence of the T7 promoter fused to the reverse complementary of the respective sequence was annealed to a primer with the T7 promoter sequence. For U7smOPT an oligo with the reverse T7 promoter sequence and the reverse mU7 sequence, where the Sm-binding site was changed to the U7smOPT sequence was annealed with a complementary oligo and used as the IVT template. Human U2 and U4 snRNA IVT template were generated by PCR with hU2 snRNA or hU4 snRNA primers from a vector containing the human U2 snRNA or U4 snRNA gene respectively. See Table S1 for all primers.
Indirect Immunofluorescence
Staining was performed as previously described19with minor modifications. In brief, embryos were manually dechorionated and fixed in small containers containing 4%PFA in 0.2 M Pipes pH6.9 for 20min or 30min (SLBP or Coilin/Fibrillarin detection respectively). All incubation steps were performed in a wet chamber under agitation at room temperature. After fixation embryos were washed 3 times with MgPBS (10 mM MgCl in PBS). Permeabilization of membranes was achieved by incubation for 20 or 30 min (SLBP/Fibrillarin or Coilin detection respectively) in MgPBS supplemented with 0.2% Triton™ X-100. Blocking of unspecific binding was realized by incubation with 5% BSA supplemented with 0.2% β-Glycerol phosphate disodium salt pentahydrate for 15min. After a quick single MgPBS wash step embryos were incubated for one hour with the respective primary antibodies (anti-SLBP S-18 (Santa Cruz sc-26522) in a 1:100 dilution or anti-Fibrillarin (monoclonal autoantibody 72B952) in a 1:100 dilution or anti-Coilin 9EA2–319 in a 1:2000 dilution) in 3% BSA supplemented with 0.2% β-Glycerol phosphate disodium salt pentahydrate. For double staining, embryos were fixed for 30min and incubated simultaneously with both primary antibodies. Next, embryos were washed 3 times with MgPBS and incubated with secondary antibodies (anti–goat conjugated with FITC, anti-mouse conjugated with FITC and anti-rabbit conjugated with TRITC, Jackson ImmunoResearch Laboratories for SLBP, Fibrillarin or Coilin respectively) diluted at 1:200 in 3% BSA supplemented with 0.2% β-Glycerol phosphate disodium salt pentahydrate. Before mounting to microscope glass slides embryos were again washed twice with MgPBS and once with ddH2O and then left to dry on the glass slide. For mounting 80% Glycerol supplemented with 25 mg/ml DABCO and 2μg/ml Hoechst 33342 was applied and cover slips placed on top.
Microinjections
Embryos were injected into the cell at 1-cell stage (fluorescent RNAs) or into the yolk at 1-to 2-cell stage (drugs). The injected material was solved in Danieu buffer (0.4 mM MgSO4, 0.6 mM CaCl2, 0.7 mM KCl, 58 mM NaCl, 5 mM Hepes pH = 7 .6) One to 4 nl were injected into the embryo. Between 100 pg and 300 pg per embryo of fluorescent RNA. To block transcription 2 nl (0.4 mg/ml) of α-amanitin were injected. For co-injection with fluorescent U7 snRNA and U4 snRNA 2 nl of α-amanitin (0.2 mg/ml) were injected.
Microscopy
Indirect immunofluorescence microscopy slides, in vivo imaging of embryos and fixed specimens were imaged on the Olympus FluoView1000 confocal microscope. The following 2 objectives were used: UPLSAPO 10X NA:0.40 (in vivo whole embryo images) and UPLSAPO 60X Oil NA:1.35.
Image processing
Microscopic images were processed using Fiji.53 The image was analyzed with the 3-D object counter to detect 3-D objects by a 2-passes connexity analysis with manual threshold setting.54 For counting of CBs immunostained by coilin the threshold was set to 3-times the background in the nucleoplasm and objects filtered for a diameter of at least 0.5 μm. Data of at least 2–3 nuclei were averaged per embryo for 18 embryos from 3 independent experiments. HLBs immunostained by SLBP were manually counted. Data of 2–3 nuclei were averaged per embryo for 18 untreated and 17 α-amanitin injected embryos from 3 independent experiments. For colocalization the objects maps produced of the 3-D object counter were overlaid and colocalization of pixels assigned to objects in the 2 channels manually inferred. Display figures from fluorescent RNA injections or immunostainings are representative single z-stacks. Images were contrast and brightness adjusted for display purposes.
Detection of pre-rRNA by RT-qPCR
Groups of 5 staged embryos were quickly washed 3 times with PCR-grade water, homogenized in Trizol and total RNA was extracted according to manufacturer's instructions. Residual DNA was removed with TurboDNAse. Total RNA was converted into cDNA with Superscript III and random hexamers as primers, including a no Reverse Transcriptase (RT) control. For Pre-rRNA quantification qRT-PCRs were performed on a MxPro3000 Stratagene system with the Absolute qPCR SYBR Green Mix. All primers used (Table S1) were optimized for a working concentration of 100% (+/- 5%) efficiency of amplification. The experiment was performed twice in 3 biological replicates per developmental time point. The mean Pre-rRNA levels (2-(ΔCT) of 5′ETS-18S-rRNA normalized to ND3-ND4I mtRNA) and standard deviations were calculated and plotted with Prism 6.
Analysis of histone gene transcription onset
To analyze the onset of histone gene transcription we intersected annotated canonical histone genes with the first zygotically transcribed genes as published in Heyn et al., 2014. The normalized expression levels (as in Heyn et al., 2014) were compared between the 128-cell and 512-cell stage and histone genes that showed an increase in expression greater or equal to log2 of 2 were highlighted.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Nick Watkins for providing the Fibrillarin antibody. We are grateful to the Neugebauer lab and Nadine Vastenhouw for helpful discussions and support during the course of this work. We thank Vladimir Despic for helpful comments on the manuscript and Jonah Pearl for technical assistance.
Funding
We are grateful for funding from MPI-CBG in Dresden (to K.M.N.), a grant from the Deutsche Forschungsgemeinschaft (NE909/2–2 to K.M.N.) and funding from the Dresden International Graduate School for Biomedicine and Bioengineering (DIGS-BB fellowship to P.H.).
References
- 1.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends in genetics : TIG 2011; 27(8):p. 295-306; PMID:21680045; https://doi.org/ 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J 2016; 35(15):p. 1603-12; PMID:27357569; https://doi.org/ 10.15252/embj.201593517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Developmental cell 2009; 17(5):p. 639-47; PMID:19922869; https://doi.org/ 10.1016/j.devcel.2009.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berry J, Weber SC, Vaidya N, Haataja M, Brangwynne CP. RNA transcription modulates phase transition-driven nuclear body assembly. Proc Natl Acad Sci U S A 2015; 112(38):p. E5237-45; PMID:26351690; https://doi.org/ 10.1073/pnas.1509317112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol 2016; 26(3):p. 277-85; PMID:26776729; https://doi.org/ 10.1016/j.cub.2015.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pederson T. The nucleolus. Cold Spring Harb Perspect Biol 2011; 3(3): a000638; PMID:21106648; https://doi.org/23042601 10.1101/cshperspect.a000638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley interdisciplinary reviews. RNA 2013; 4(1):p. 17-34; PMID:23042601; https://doi.org/ 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- 8.Machyna M, Neugebauer KM, Stanek D. Coilin: The first 25 years. RNA Biol 2015; 12(6):p. 590-6; PMID:25970135; https://doi.org/ 10.1080/15476286.2015.1034923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through cajal bodies. Mol Cell 2014; 56(3):p. 389-99; PMID:25514182; https://doi.org/ 10.1016/j.molcel.2014.10.004 [DOI] [PubMed] [Google Scholar]
- 10.Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 2004; 166(7):p. 1015-25; PMID:15452143; https://doi.org/ 10.1083/jcb.200405160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nature structural & molecular biology 2010; 17(4):p. 403-9; PMID:20357773; https://doi.org/ 10.1038/nsmb.1783 [DOI] [PubMed] [Google Scholar]
- 12.Romeo V, Schumperli D. Cycling in the nucleus: regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol 2016; 40:p. 23-31; PMID:26895140; https://doi.org/ 10.1016/j.ceb.2016.01.015 [DOI] [PubMed] [Google Scholar]
- 13.Tatomer DC, Terzo E, Curry KP, Salzler H, Sabath I, Zapotoczny G, McKay DJ, Dominski Z, Marzluff WF, Duronio RJ. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J Cell Biol 2016; 213(5):p. 557-70; PMID:27241916; https://doi.org/ 10.1083/jcb.201504043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9(11):p. 843-54; PMID:18927579; https://doi.org/ 10.1038/nrg2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci U S A 2008; 105(44):p. 16964-9; PMID:18957539; https://doi.org/ 10.1073/pnas.0809273105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol 1999; 9(3):p. 126-35; PMID:10021385; https://doi.org/ 10.1016/S0960-9822(99)80066-9 [DOI] [PubMed] [Google Scholar]
- 17.Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. J Cell Physiol 2009; 220(1):p. 129-35; PMID:19277982; https://doi.org/ 10.1002/jcp.21740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol 2006; 172(6):p. 875-84; PMID:16533947; https://doi.org/ 10.1083/jcb.200511038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strzelecka M, Oates AC, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 2010; 1(1):p. 96-108; PMID:21327108; https://doi.org/ 10.4161/nucl.1.1.10680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 2008; 7(15):p. 2357-67; PMID:18677100; https://doi.org/ 10.4161/cc.6344 [DOI] [PubMed] [Google Scholar]
- 21.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell 2010; 40(2):p. 216-27; PMID:20965417; https://doi.org/ 10.1016/j.molcel.2010.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carmo-Fonseca M, Pepperkok R, Carvalho MT, Lamond AI. Transcription-dependent colocalization of the U1, U2, U4/U6, and U5 snRNPs in coiled bodies. J Cell Biol 1992; 117(1):p. 1-14; PMID:1532583; https://doi.org/ 10.1083/jcb.117.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007; 8(7):p. 574-85; PMID:17519961; https://doi.org/ 10.1038/nrm2184 [DOI] [PubMed] [Google Scholar]
- 24.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011; 13(2):p. 167-73; PMID:21240286; https://doi.org/ 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- 25.Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154(3):p. 499-509; PMID:11489914; https://doi.org/ 10.1083/jcb.200105084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development 2009; 136(18):p. 3033-42; PMID:19700615; https://doi.org/ 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- 27.Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, Neugebauer KM. The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell reports 2014; 6(2):p. 285-92; PMID:24440719; https://doi.org/ 10.1016/j.celrep.2013.12.030 [DOI] [PubMed] [Google Scholar]
- 28.Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics 2010; 9(3):p. 457-70; PMID:20026476; https://doi.org/ 10.1074/mcp.M900429-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonné R, et al.. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16(5):p. 777-87; PMID:15574332; https://doi.org/ 10.1016/j.molcel.2004.11.013 [DOI] [PubMed] [Google Scholar]
- 30.Narayanan A, Speckmann W, Terns R, Terns MP. Role of the box C/D motif in localization of small nucleolar RNAs to coiled bodies and nucleoli. Mol Biol Cell 1999; 10(7):p. 2131-47; PMID:10397754; https://doi.org/ 10.1091/mbc.10.7.2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Farrell PH. Growing an embryo from a single cell: a hurdle in animal life. Cold Spring Harb Perspect Biol 2015; 7:a019042; PMID:26254311; https://doi.org/ 10.1101/cshperspect.a019042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins NJ, Dickmanns A, Luhrmann R. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol Cell Biol 2002; 22(23):p. 8342-52; PMID:12417735; https://doi.org/ 10.1128/MCB.22.23.8342-8352.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 2006; 17(12):p. 4972-81; PMID:16987958; https://doi.org/ 10.1091/mbc.E06-06-0513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerbi SA, Borovjagin AV, Odreman FE, Lange TS. U4 snRNA nucleolar localization requires the NHPX/15.5-kD protein binding site but not Sm protein or U6 snRNA association. J Cell Biol 2003; 162(5):p. 821-32; PMID:12939253; https://doi.org/ 10.1083/jcb.200301071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mishima Y, Tomari Y. Codon Usage and 3′ UTR Length Determine Maternal mRNA Stability in Zebrafish. Mol Cell 2016; 61(6):p. 874-85; PMID:26990990; https://doi.org/ 10.1016/j.molcel.2016.02.027 [DOI] [PubMed] [Google Scholar]
- 36.Marz M, Mosig A, Stadler BM, Stadler PF. U7 snRNAs: a computational survey. Genomics Proteomics Bioinformatics 2007; 5(3-4):p. 187-95; PMID:18267300; https://doi.org/ 10.1016/S1672-0229(08)60006-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddy R, Busch H. Small nuclear RNAs and RNA processing. Prog Nucleic Acid Res Mol Biol 1983; 30:p. 127-62; PMID:6198692; https://doi.org/ 10.1016/S0079-6603(08)60685-6 [DOI] [PubMed] [Google Scholar]
- 38.Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schümperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 2005; 114(3):p. 155-66; PMID:16003501; https://doi.org/ 10.1007/s00412-005-0003-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Lührmann R. Ongoing U snRNP Biogenesis Is Required for the Integrity of Cajal Bodies. Mol Biol Cell 2006; 17(7):p. 3221-31; PMID:16687569; https://doi.org/ 10.1091/mbc.E06-03-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 2005; 102(48):p. 17372-7; PMID:16301532; https://doi.org/ 10.1073/pnas.0508947102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gruber A, Soldati D, Burri M, Schümperli D. Isolation of an active gene and of two pseudogenes for mouse U7 small nuclear RNA. Biochim Biophys Acta 1991; 1088(1):p. 151-4; PMID:1989694; https://doi.org/ 10.1016/0167-4781(91)90167-K [DOI] [PubMed] [Google Scholar]
- 42.Pillai RS, Will CL, Lührmann R, Schümperli D, Müller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J 2001; 20(19):p. 5470-9; PMID:11574479; https://doi.org/ 10.1093/emboj/20.19.5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, Schümperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev 2003; 17(18):p. 2321-33; PMID:12975319; https://doi.org/ 10.1101/gad.274403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang XC, Sabath I, Dębski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol Cell Biol 2013; 33(1):p. 28-37; PMID:23071092; https://doi.org/ 10.1128/MCB.00653-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grimm C, Stefanovic B, Schumperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J 1993; 12(3):p. 1229-38; PMID:8458335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stefanovic B, Wittop Koning TH, Schumperli D. A synthetic histone pre-mRNA-U7 small nuclear RNA chimera undergoing cis cleavage in the cytoplasm of Xenopus oocytes. Nucleic Acids Res 1995; 23(16):p. 3152-60; PMID:7667091; https://doi.org/ 10.1093/nar/23.16.3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci 2004; 61(19-20):p. 2560-70; PMID:15526162; https://doi.org/ 10.1007/s00018-004-4190-0 [DOI] [PubMed] [Google Scholar]
- 48.Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol 2004; 164(6):p. 831-42; PMID:15024031; https://doi.org/ 10.1083/jcb.200311121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, Misteli T. A kinetic framework for a mammalian RNA polymerase in vivo. Science 2002; 298(5598):p. 1623-6; PMID:12446911; https://doi.org/ 10.1126/science.1076164 [DOI] [PubMed] [Google Scholar]
- 50.Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009; 324(5935):p. 1729-32; PMID:19460965; https://doi.org/ 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- 51.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203(3):p. 253-310; PMID:8589427; https://doi.org/ 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 52.Reimer G, Raska I, Tan EM, Scheer U. Human autoantibodies: probes for nucleolus structure and function. Virchows Arch B Cell Pathol Incl Mol Pathol 1987; 54(3):p. 131-43; PMID:2894087; https://doi.org/ 10.1007/BF02899205 [DOI] [PubMed] [Google Scholar]
- 53.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al.. Fiji: an open-source platform for biological-image analysis. Nature methods 2012; 9(7):p. 676-82; PMID:22743772; https://doi.org/ 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 2006; 224(Pt 3):p. 213-32; PMID:17210054; https://doi.org/ 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.