Abstract
Over half of patients with BRCA1-deficient cancers do not respond to treatment with poly(ADP-ribose) polymerase (PARP) inhibitors. In this study, we report that a combination of 53BP1 and BRCA1 may serve as a biomarker of PARP inhibitor sensitivity. Based on the mRNA levels of four homologous recombination repair (HR) genes and PARP inhibitor sensitivity, we selected BRCA1-deficient MDA-MB-436 cells to conduct RNA interference. Reducing expression of 53BP1, but not the other three HR genes, was found to lower simmiparib sensitivity. Additionally, we generated 53BP1−/−/BRCA1−/− clonal variants by the transcription activator-like effector nuclease (TALEN) technique and found that depleting 53BP1 impaired PARP inhibitor sensitivity with a 36.7-fold increase in their IC50 values. Consistent with its effect on PARP inhibitor sensitivity, 53BP1 loss alleviated cell cycle arrest and apoptosis and partially restored HR function. Importantly, 53BP1 depletion dramatically reduced the ability of PARP inhibitors to suppress tumor growth in vivo. The inhibition rate of simmiparib was 74.16% for BRCA1-deficient MDA-MB-436 xenografts, but only 7.79% for 53BP1/BRCA1-deficient xenografts. Re-expressing 53BP1 in the dual-deficient cells restored PARP inhibitor sensitivity and the levels of HR regulators. Considering that at least 10% of BRCA1-deficient breast and ovarian cancers have reduced expression of 53BP1, using a combination of 53BP1 with BRCA1 as a biomarker for patient selection should reduce the number of patients undergoing futile treatment with PARP inhibitors.
Keywords: homologous recombination repair defects, breast cancer, ovarian cancer, TALEN, combined biomarker, simmiparib, olaparib
Introduction
The first poly(ADP-ribose) polymerase (PARP) inhibitor olaparib was approved for cancer therapy in December 20141. Since then, an additional inhibitor, rucaparib, has been accepted by the US FDA for priority review of its ability to treat advanced ovarian cancer associated with BRCA mutations2. Additionally, niraparib has demonstrated unexpected therapeutic efficacy and enhanced progression-free survival (PFS) in patients with BRCA-deficient ovarian cancer3. These important advances offer hope that cancers harboring homologous recombination repair (HR) defects can be cured. However, the objective response rates (ORR) to PARP inhibitors are only 30%–50%, even for patients with ovarian cancer associated with BRCA mutations1,2. These data imply that approximately over half of such patients will not respond to treatment and have a poor prognosis. Therefore, it is necessary to find other factors, in addition to BRCA defects, that participate in determining the sensitivity to PARP inhibitors. Additionally, because of the poor clinical response of BRCA-deficient breast cancer to PARP inhibitors1, it is vital to identify factor(s) that can be used to predict the therapeutic response to PARP inhibitors on the background of BRCA defects.
Aberrations in components of the HR pathway have been reported to affect PARP inhibitor sensitivity, including the HR factors Rad51, structural maintenance of chromosome 1 (SMC1), Pax transactivation domain-interacting protein (PTIP) and p53-binding protein 1 (53BP1)1,4,5,6,7,8,9,10. 53BP1 inhibited HR activity and its loss partially restored HR function in BRCA1-deleted mouse embryonic stem cells4 and conferred olaparib resistance to BRCA1-deficient mouse mammary tumors5. PTIP is a phospho-53BP1 binding protein that is required for 53BP1-mediated HR inhibition6. However, loss of PTIP did not restore HR activity but still caused PARP-1 inhibitor resistance in BRCA2-deficient B lymphocytes and embryonic stem cells7. Independent of 53BP1 signaling, SMC1 binds to BRCA1, and its reduction by RNA interference led to an approximate 3-fold increase in the sensitivity of triple-negative breast cancer (TNBC) cells to the PARP inhibitor veliparib8. In addition, Rad51, a core component of the HR pathway, was found to be upregulated in BRCA1-deficient breast tumors9, and loss of RAD51C resulted in increased sensitivity to olaparib10. However, few studies have explored the combinatorial impact of two or more such HR factors on the PARP inhibitor sensitivity of human cancer cells.
In this study, we examined the mRNA levels of Rad51, SMC1, PTIP and 53BP1 in seven human cancer cell lines harboring known HR defects. The BRCA1-deficient breast cancer line MDA-MB-436 had high levels of mRNA for all four genes and was highly sensitive to PARP inhibitors. Therefore, we selected this line for RNA interference to evaluate the impact of HR factors on sensitivity to simmiparib, a new PARP inhibitor undergoing clinical trials in China11,12. Silencing 53BP1 in MDA-MB-436 cells was more effective than silencing other HR factors at reducing simmiparib sensitivity, so we generated 53BP1 and BRCA1 dual-deficient MDA-MB-436 variants using the transcription activator-like effector nuclease (TALEN) technique. We tested the sensitivity of these variants to PARP inhibitors and measured their HR activity in vitro and in vivo. Our data indicate that 53BP1 profoundly affects the in vitro and in vivo HR activity and sensitivity to PARP inhibitors on a background of BRCA1 defects. These results suggest that a combination of 53BP1 and BRCA1 can serve as a biomarker for PARP inhibitor sensitivity and may help predict the therapeutic response of cancers to PARP inhibitors.
Materials and methods
Cell culture
Human normal breast epithelial MCF10A cells and human cancer cells, including MDA-MB-436 (breast), MDA-MB-231 (breast), UWB1.289 (ovarian), DoTc-4510 (cervical), Capan-1 (pancreatic), SK-ES-1 (Ewing's sarcoma), RD-ES (Ewing's sarcoma), and U87MG (glioma), were purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured according to the suppliers' instructions and periodically authenticated by morphologic inspection and tested for Mycoplasma contamination.
Drugs and reagents
Simmiparib was prepared as previously reported11. Talazoparib, niraparib, rucaparib and olaparib were purchased from Selleck Chemicals (Shanghai, China). All drugs were dissolved in dimethyl sulfoxide (DMSO), aliquoted, stored at -20 °C and diluted to desired concentrations in normal saline immediately prior to each experiment.
Antibodies against β-Actin, γH2AX, 53BP1, ATM, p-ATM, PTIP, Chk1, FANCD2 and RAD51 were from Santa Cruz Biotechnology (Santa Cruz, CA, USA) and SMC1, p-Chk1, Chk2, p-Chk2, cdc2, p-cdc2, BRCA2 and RPA32 were from Cell Signaling Technology (Danvers, MA, USA).
Proliferation inhibition assays
Cells were seeded into 96-well plates, allowed to attach overnight and treated for six days with graded concentrations of the indicated agents. Then, the proliferation of cells was determined using the Cell Counting Kit 8 (CCK8; Dojindo, Kumamoto, Japan) assays, as described previously13,14. The inhibition rate (%) was calculated as [1−(A450treated/A450control)]×100%. Average IC50 values (mean±SD) were determined from three independent experiments using the Logit method.
Western blotting
Cells were treated with the indicated agents for the indicated times. Western blotting was used to measure the cellular levels of the indicated proteins, as previously described15,16.
RNA interference
Cells were seeded into 96-well plates and allowed to attach overnight. Specific siRNA duplexes (100 nmol/L) against 53BP1(si53BP1: 5′-CCUGAUGCUUUCCGAUCUA-3′), RAD51 (siRad51: 5′-GUUGCCUAUGCGCCAAAGA-3′), PTIP (siPTIP: 5′-CGCGUAUGCACAGGCAAUA-3′) or SMC1 (siSMC1: 5′-GUAGGAGGUUCUUCUGAGU-3′), or a scrambled siRNA (siCtrl: 5′-UUCUCCGAACGUGUCACGU-3′) (RayBiotech, Guangzhou, China) were transfected with the RNAiMAX Transfection Reagent (Invitrogen, CA, USA) according to the manufacturer's instructions.
qRT-PCR
Total RNA was extracted with TRIzol (Life Technologies, CA, USA). cDNA was prepared with the PrimeScript RT Master Mix (Takara, Tokyo, Japan) from 500 ng of RNA and amplified with a SYBR Premix EX TaqII Kit (TaKaRa, Tokyo, Japan) in a 7500 Fast Real-Time PCR System (Applied Biosystems, CA, USA). The primers were from Sangon (Shanghai, China) with the sequences as follows: 5′-GCTGTCTTGGGTGCATTGGA-3′ (forward) and 5′-AAGGGACTTCCTGTAACAATGCA-3′ (reverse) for β-actin; 5′-TGAGCAGTTACCTCAGCCAAA-3′ (forward) and 5′-AAGGGAATGTGTAGTATTGCCTG-3′ (reverse) for 53BP1; 5′-CAACCCATTTCACGGTTAGAGC-3′ (forward) and 5′-TTCTTTGGCGCATAGGCAACA-3′ (reverse) for RAD51; 5′-ACAATGCACTAGCCTCACACA-3′ (forward) and 5′-ACACTGAACGGACAGAATCAC-3′ (reverse) for PTIP; 5′-CATCAAAGCTCGTAACTTCCTCG-3′ (forward) and 5′-CCCCAGAACGACTAATCTCTTCA-3′ (reverse) for SMC1; and 5′-GCTGTCTTGGGTGCATTG GA-3′ (forward) and 5′-GCATGATCTTAAAGGCTACCAGG-3′ (reverse) for RPA32.
Stable knockout of 53BP1 with the TALEN technique
To deplete 53BP1 expression, TALEN13,17 specifically targeting 53BP1 was performed with a FASTALE TALEN kit (SiDanSai Biotechnology, Shanghai, China). MDA-MB-436 cells were transfected with positive TALEN plasmids and incubated with 2 μg/mL puromycin. After puromycin screening, surviving cells were selected for 53BP1-deficient monoclonal cells. The positive clones were verified by DNA sequencing in order to generate stable 53BP1-knockout cell lines.
Transfection with 53BP1 plasmids
The plasmid N-myc-53BP1 WT pLPC-Puro (#19836; expressing wild-type 53BP1) was purchased from Addgene (Cambridge, MA, USA). Stable 53BP1-knockout cells were transfected with this plasmid using Lipofectamine 3000 (Invitrogen, CA, USA) according to the manufacturer's instructions.
Immunofluorescence microscopy
Cells were seeded onto glass coverslips, cultured overnight and then exposed to medium containing drugs. Treated cells were fixed with methanol at -20 °C for 20 min, blocked with 3% bovine serum albumin (BSA) for 15 min, stained with primary antibodies for 1 h and fluorescence-conjugated secondary antibodies for 30 min. Finally, the cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and imaged with an Olympus confocal microscope (Olympus, Tokyo, Japan) with a 100×objective.
Fluorescence-activated cell sorting (FACS)
FACS was performed to detect cell cycle distribution and apoptosis, as previously described13. For cell cycle assays, the treated cells were stained with 10 μg/mL propidium iodide (PI) in the dark for 10 min. Apoptosis was detected by staining the cells with an AnnexinV-PI apoptosis detection kit from Keygen (Nanjing, China). Fluorescence of the cells was measured immediately with a FACS Calibur flow cytometer (BD Biosciences, NJ, USA).
Assaying caspase 3/7 activity
Cells were seeded into 96-well plates, cultured overnight and treated with the indicated concentrations of simmiparib for 5 d. The caspase 3/7 activity of the treated cells was measured using a Caspase-Glo® 3/7 Assay kit (Promega, WI, USA) according to the manufacturer's instructions.
Assaying in vivo anticancer activity
MDA-MB-436 and 53BP1-deficient MDA-MB-436 xenografts were established by inoculating 5×106 cells subcutaneously into nude mice. After 1–2 passages, well-grown xenografts were cut into 1.5-mm3 fragments that were transplanted subcutaneously into the right flank of nude mice. When xenografts reached a volume of 100–200 mm3, the mice were randomized into control and experimental groups and received the indicated treatments. The size of the xenografts and the body weight of the mice were measured individually twice a week. The xenografts were measured for the maximal width (X) and length (Y) and the volume (V) was calculated as (X2Y)/2. Relative tumor volume (RTV) was calculated as Vt/V0, where V0 and Vt were the volume before and after treatments, respectively. The in vivo anticancer activity of each treatment was judged by its T/C (%) value. T/C (%) was calculated as (TRTV/CRTV)×100%, where TRTV and CRTV represented the RTV of the treatment and the vehicle group, respectively. The experiments abided by the institutional ethical guidelines of the Animal Care and Use Committee in Shanghai Institute of Materia Medica.
Statistical analyses
All the data are presented as the mean±SD. The student's t-test was used to determine significant differences between the treatment and control groups. P<0.05 was considered to be statistically significant.
Results
Reduction of 53BP1 decreases the sensitivity of BRCA1-deficient MDA-MB-436 cells to PARP inhibitors
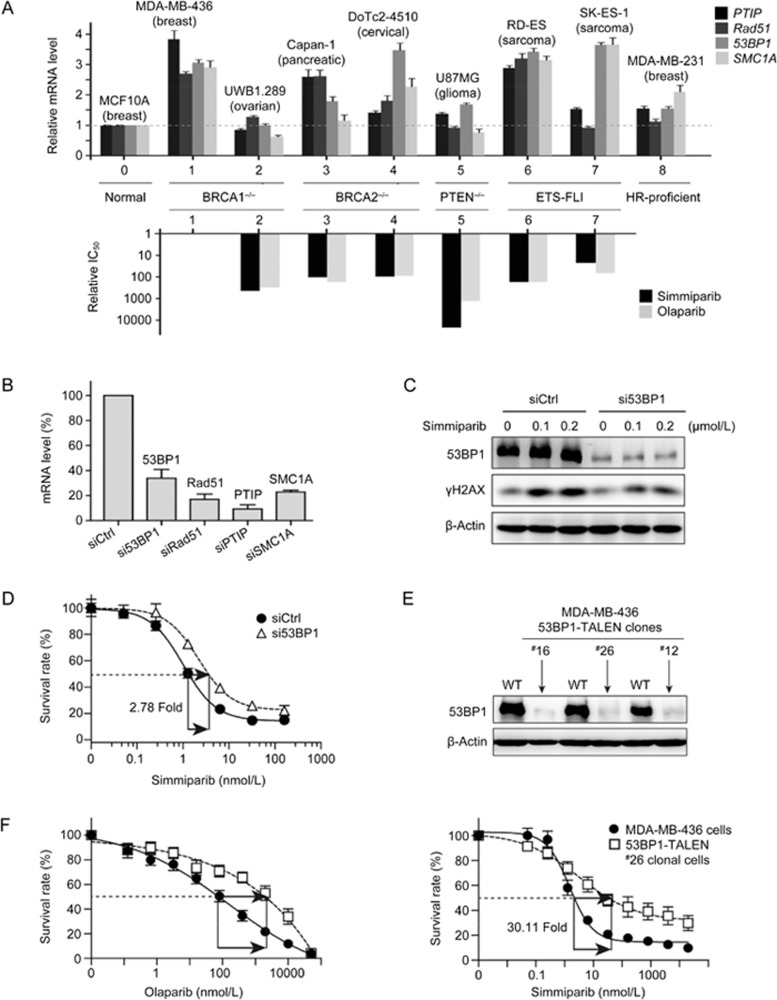
53BP1, PTIP, Rad51 and SMC1 participate in HR and have been proposed as sensitivity markers for PARP inhibitors1,4,5,6,7,8,9,10. To investigate whether these factors affect the PARP inhibitor sensitivity of cancer cells that harbor known HR defects, we measured the basal mRNA levels of these four factors in 7 HR-deficient (BRCA1−/−, BRCA2−/−, PTEN−/− or ETS-FL1) cancer cell lines, 1 HR-proficient cancer cell line, and 1 normal cell line. The basal mRNA levels of these factors did not correlate with either the HR defects or cellular sensitivity to the PARP inhibitors simmiparib and olaparib (Figure 1A)11. Additionally, their mRNA levels were not associated with BRCA1 status. The basal mRNA levels of all four genes were high in the breast cancer MDA-MB-436 cell line but low in the ovarian cancer UWB1.289 cell line, although both cell lines carry BRCA1 defects (Figure 1A). In our previous studies, MDA-MB-436 cells were more sensitive than UWB1.289 cells to PARP inhibitors, displaying 298- and 450-fold higher sensitivity to olaparib and simmiparib, respectively11. Therefore, we selected the BRCA1-deficient MDA-MB-436 cell line for subsequent studies.
Figure 1.
Reduction or loss of 53BP1 lowers the sensitivity of BRCA1-deficient MDA-MB-436 cells to PARP inhibitors. (A) The relationships between the relative mRNA levels of four genes and the relative IC50 of PARP inhibitors in seven HR-deficient cells. Upper, qRT-PCR for mRNA levels in eight human cancer cell lines. Levels are expressed as relative values with levels in the normal human breast epithelial cells normalized to 1; lower, the IC50 values of simmiparib and olaparib in No 2–7 cells expressed as relative values with IC50 values in MDA-MB-436 cells normalized to 1. The original IC50 values were previously determined11. (B) mRNA levels of four genes detected by qRT-PCR in MDA-MB-436 cells transfected with the indicated siRNAs for 24 h. (C) Protein levels of 53BP1 and γH2AX detected by Western blotting in siRNA-transfected MDA-MB-436 cells treated with simmiparib for 72 h. (D) The change in simmiparib sensitivity induced by 53BP1 reduction by siRNAs. MDA-MB-436 cells were transfected with the indicated siRNAs, treated with simmiparib for 6 d and then subjected to CCK8 assays. (E) The protein levels of 53BP1 detected by Western blotting in different 53BP1-TALEN clonal variants of MDA-MB-436 cells. WT, MDA-MB-436 cells. (F) The change in PARP inhibitor sensitivity induced by 53BP1 loss by TALEN. The cells were treated with olaparib or simmiparib for 6 d and then subjected to CCK8 assays. All the data were from three independent experiments.
We used specific siRNAs to silence 53BP1, PTIP, Rad51 or SMC1 in MDA-MB-436 cells and examined how this affected sensitivity to the PARP inhibitor simmiparib. RNA interference caused >60% reduction of both mRNA and protein levels of 53BP1, PTIP, Rad51 and SMC1 (Figure 1B and 1C; Supplementary Figure S1A). si53BP1 decreased the sensitivity of MDA-MB-436 cells to simmiparib by 2.78-fold (Figure 1D), but siPTIP and siSMC1 had little effect, and siRad51 actually increased the cellular sensitivity by 1.89-fold (Figure S1B). Consistent with these findings, treatment with simmiparib enhanced the levels of γH2AX, a DSB marker, in siRad51 and siCtrl cells but caused a more modest increase in γH2AX levels in si53BP1 cells (Figure S1A). These data suggest that, among the four tested HR factors, 53BP1 has the greatest impact on the cellular sensitivity to PARP inhibitors, specifically on a background of BRCA1 defects.
Generation of 53BP1-BRCA1 dual-deficient MDA-MB-436 variants and their changes in drug sensitivity
RNA interference did not completely abolish expression of 53BP1, and the remaining 53BP1 protein (∼40%) was likely to be functional. The TALEN technique for genome editing and genetic modifications has been used to eliminate protein expression of specific genes13,17. Therefore, we used TALEN to completely block expression of 53BP1 in BRCA1-deficient MDA-MB-436 cells in order to verify the effect of 53BP1 on PARP inhibitor sensitivity revealed by RNA interference. As shown in Figure S2, we designed five 53BP1 TALEN recognition sequences and used them to construct 6 TALEN plasmid combinations. Following transfection into BRCA1-deficient MDA-MB-436 cells, the L1+R3 plasmid combination produced the highest 53BP1 gene targeting efficiency. Through monoclonal selection of the cells transfected with this combination, we obtained three 53BP1/BRCA1 dual-deficient clonal variants, ie, #12, #16 and #26 clonal cell lines. DNA sequencing results showed that both the #16 and #26 variant had four bases deleted from the 53BP1 cDNA, while the #12 variant had seven deleted bases (Figure S2). These base deletions depleted 53BP1 protein in all three clonal variants (Figure 1E).
Relative to the BRCA1-deficient parental MDA-MB-436 cells, the 53BP1/BRCA1 dual-deficient variants displayed up to 36.7-fold reduced sensitivity of various PARP inhibitors, including olaparib, simmiparib, talazoparib, niraparib and rucaparib (Figure 1F; Table 1). Moreover, the sensitivity of the #26 clonal cells to the Top1 inhibitor SN38 was reduced by 5.19-fold, although their sensitivity to other conventional anticancer drugs with different mechanisms of action was only slightly altered (Table S1). These results further indicate that the HR factor 53BP1 contributes to the sensitivity of BRCA1-deficient MDA-MB-436 cells to PARP inhibitors.
Table 1. The sensitivity of 53BP1-knockout cells to PARP inhibitors in comparison with that of the parental MDA-MB-436 cells.
| PARP inhibitors | MDA-MB-436 | IC50 (mean±SD) (nmol/L) |
|||||
|---|---|---|---|---|---|---|---|
| #26 | RF | #16 | RF | #12 | RF | ||
| Olaparib | 169.80±12.29 | 5530.75±926.37 | 32.6 | 3644.29±1117.53 | 34.1 | 895.44±134.00 | 19.69 |
| Simmiparib | 2.78±0.52 | 83.60±12.51 | 30.1 | 76.95±0.122 | 27.7 | ND | ND |
| Rucaparib | 200.23±28.12 | 7357.5±419.27 | 36.7 | 4076.10±483.17 | 20.4 | 608.66±126.77 | 26.2 |
| Niraparib | 324.15±28.39 | 4317.66±527.84 | 13.3 | 1842.17±71.98 | 15.1 | 1601.39±2.31 | 4.94 |
| Talazoparib | 1.14±0.14 | 36.62±7.16 | 32.1 | 18.10±0.68 | 15.8 | ND | ND |
Notes: #12, #16 and #26 represent three different clonal cell lines derived from MDA-MB-436 cells by using the TALEN technique to knock out 53BP1. RF was calculated as (IC50 from #12, #16 or #26 cells)/(IC50 from MDA-MB-436 cells). ND, not determined.
Notably, DNA sequencing revealed clear single peaks adjacent to the deleted bases in the 53BP1 cDNA for both the #16 and #26 variant but continuous dual peaks for the #12 variant (Figure S2). These data indicate that both copies of the 53BP1 gene were effectively disrupted in the #16 and #26 variants but possibly not in the #12 variant. Therefore, the former variants were selected for subsequent studies.
Loss of 53BP1 alleviates cell cycle arrest and apoptosis induced by PARP inhibitors
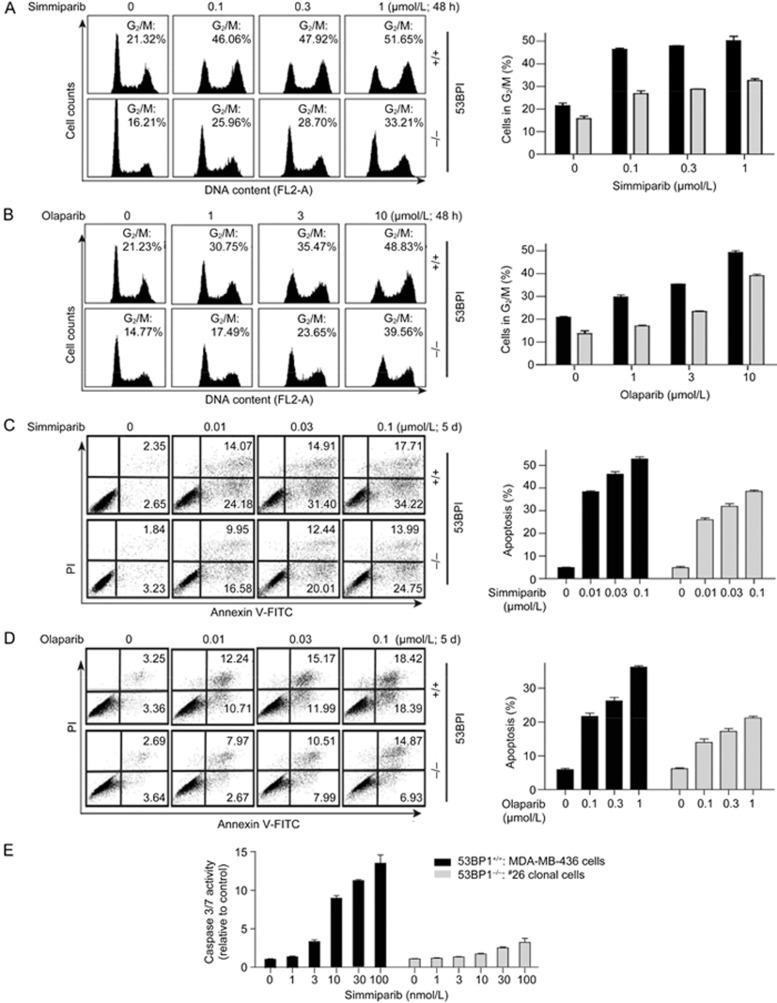
Both simmiparib (Figure 2A) and olaparib (Figure 2B) caused a dose-dependent G2/M arrest in the BRCA1-deficient parental MDA-MB-436 cells. This arrest was partially blocked in the 53BP1/BRCA1 dual-deficient #26 variant (Figure 2A and B). Similarly, loss of 53BP1 decreased apoptosis triggered by treatment with either simmiparib (Figure 2C) or olaparib (Figure 2D). Consistent with the FACS results, the simmiparib-induced activation of the apoptotic effector enzymes caspase 3 and 7 was much weaker in the 53BP1-deficient cells than in the 53BP1-proficient cells (Figure 2E). These data further support the conclusion that 53BP1 contributes to the PARP inhibitor sensitivity of BRCA1-deficient cells.
Figure 2.
Loss of 53BP1 alleviates cell cycle arrest and apoptosis. MDA-MB-436 cells (53BP1+/+) and the #26 clonal variant (53BP1−/−) were treated with simmiparib or olaparib for 48 h (A, B) or 5 d (C–E). Treated cells were then subjected to PI-staining flow cytometry analyses for cell cycle arrest (A, B) and PI/Annexin V-FITC double-staining flow cytometry analyses for apoptosis (C, D). Left, representative images with values representing the percentage of apoptotic cells; right, mean±SD from three independent experiments. Caspase 3/7 activity was determined by Caspase-Glo® 3/7 assays and the data expressed as the mean±SD from three independent experiments (E).
Loss of 53BP1 partially restores the HR function in BRCA1-deficient MDA-MB-436 cells
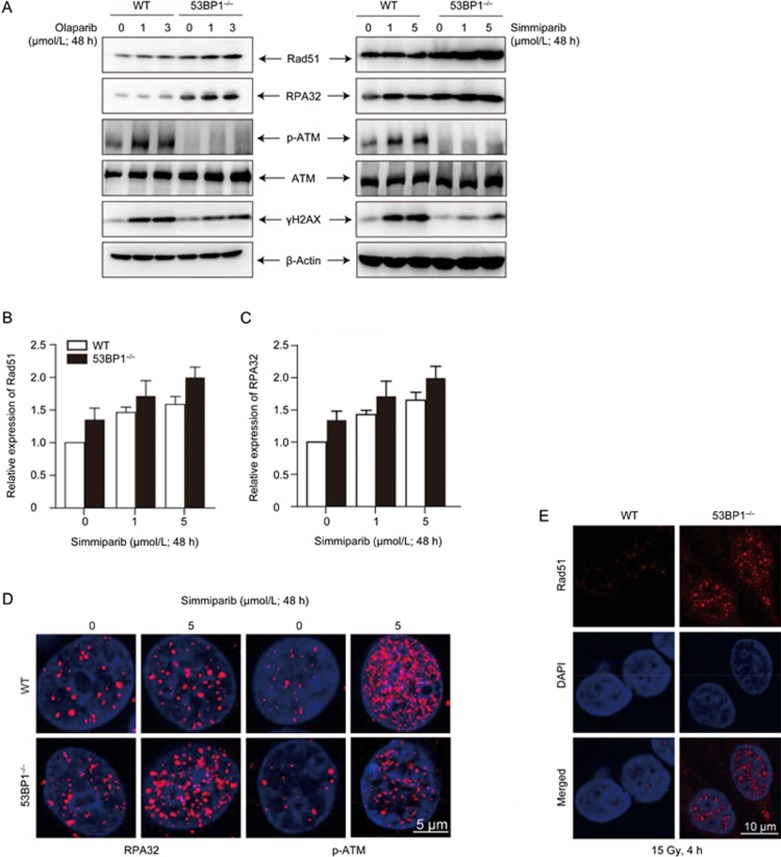
To explore how the loss of 53BP1 reduces the sensitivity of BRCA1-deficient cells to PARP inhibitors, we compared the HR activity of a 53BP1/BRCA1 dual-deficient variant to the activity of the parental BRCA1-deficient MDA-MB-436 cells. The basal protein (Figure 3A) and mRNA (Figure 3B and 3C) levels of the HR factors Rad51 and RPA32 were higher in the #26 clonal cells than in the parental MDA-MB-436 cells. In contrast, the basal levels of phosphorylated ATM (p-ATM) and γH2AX were reduced in the dual-deficient variant (Figure 3A). Treatments with olaparib or simmiparib enhanced these differences in a concentration-dependent manner (Figure 3A–3C) with more Rad51 and RPA32 but fewer p-ATM and γH2AX in #26 clonal cells than in parental cells. Consistent with these results, treatment with simmiparib (Figure 3D) or ionizing radiation (Figure 3E) resulted in the formation of more RPA32 (Figure 3D) and Rad51 foci (Figure 3E) but fewer p-ATM (Figure 3D) foci in #26 clonal cells than in parental cells. The enhanced formation of Rad51 foci following the loss of 53BP1 suggests that 53BP1 depletion partially restores HR in BRCA1-null MDA-MB-436 cells.
Figure 3.
Loss of 53BP1 partially restores HR in BRCA1-deficient MDA-MB-436 cells. (A–D) MDA-MB-436 cells (WT) and the #26 clonal variant (53BP1−/−) were treated with simmiparib or olaparib for 48 h for qRT-PCR and confocal microscopy or for 72 h for Western blotting. Western blotting (A), qRT-PCR (B, C) or confocal microscopy (D) of the treated cells. (E) Confocal images of immunofluorescent cells 4 h after irradiation with 15 Gy. The data are expressed as the mean±SD (B, C) or representative images (A, D, E) from three independent experiments.
53BP1/BRCA1 dual-deficient xenografts derived from #26 clonal cells are resistant to PARP inhibitors
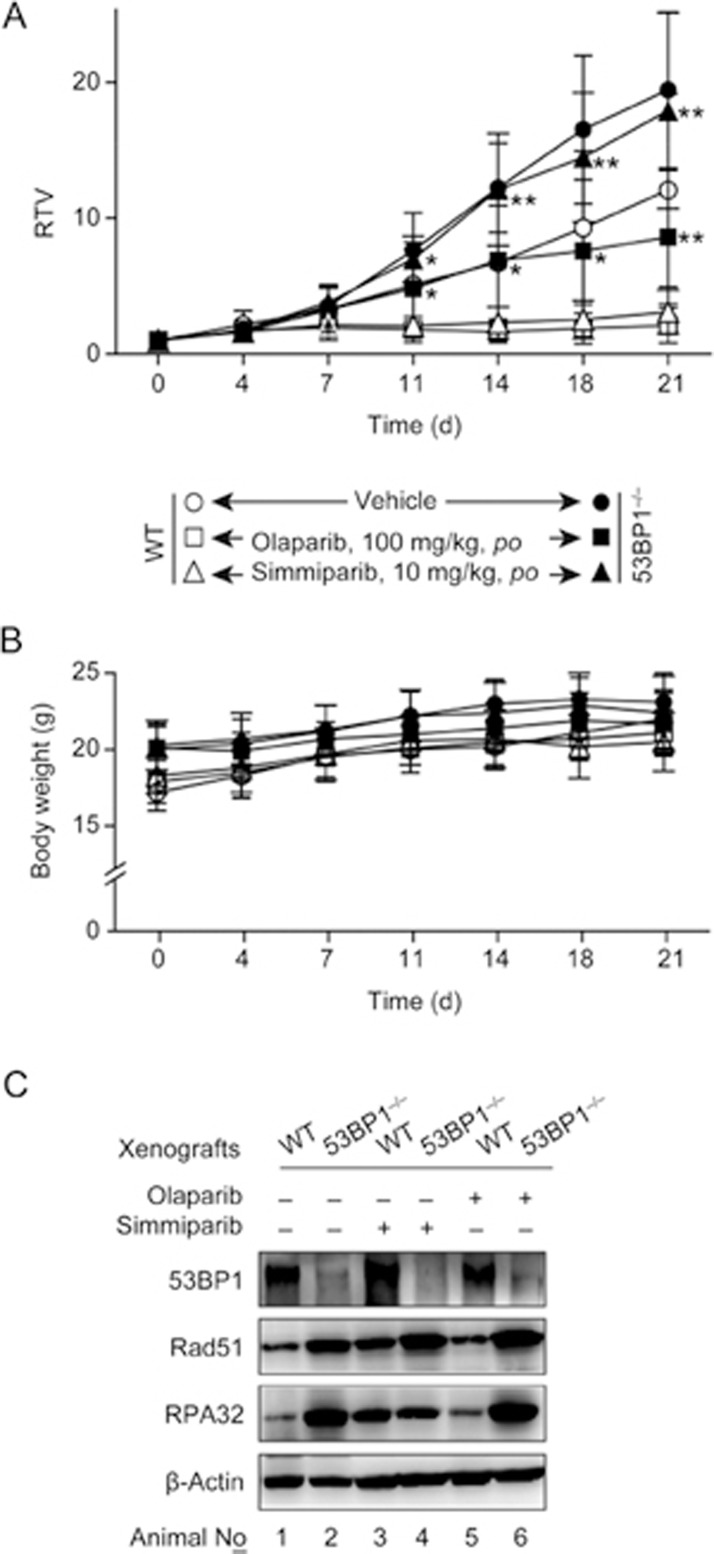
To test whether 53BP1 affects in vivo sensitivity to PARP inhibitors, we used the #26 clonal cells and the parental cells to establish xenograft models in nude mice. The #26 clonal cell-derived xenografts grew much faster than the parental cell-derived xenografts (Figure 4A). Oral administration of simmiparib or olaparib inhibited the growth of #26 tumor xenographs by 7.79% and 55.78%, respectively, and inhibited parental tumor xenographs by 74.16% and 82.49%, respectively (Figure 4A). Neither PARP inhibitor affected body weight (Figure 4B). These data demonstrate that on a BRCA1-deficient background, disruption of 53BP1 can lead to a partial or nearly complete loss of in vivo sensitivity to PARP inhibitors.
Figure 4.
Loss of 53BP1 lowers the response of MDA-MB-436 xenografts to PARP inhibitors in nude mice. Simmiparib or olaparib were orally administered to nude mice bearing xenografts derived from MDA-MB-436 cells (WT) or the #26 clonal variant (53BP1−/−), for 21 d. (A and B) Changes in the relative tumor volume (RTV) (A) and the animal body weight (B) over the treatment time; n=6 for the treatment groups and 12 for the control groups. (C) Protein levels in the indicated treated xenografts, measured by Western blotting. In each group, three xenografts were detected (because some xenografts were very small so that not enough materials could be obtained for the detection, particularly in the WT treatment group). Representative images are shown. *P<0.05, **P<0.01 vs WT.
In addition, Western blotting revealed that 53BP1, Rad51 and RPA32 had an expression profile in the xenografts (Figure 4C) similar to those in their corresponding cell lines (Figure 3A).
Re-expression of 53BP1 in the 53BP1/BRCA1 dual-deficient cells restores PARP inhibitor sensitivity
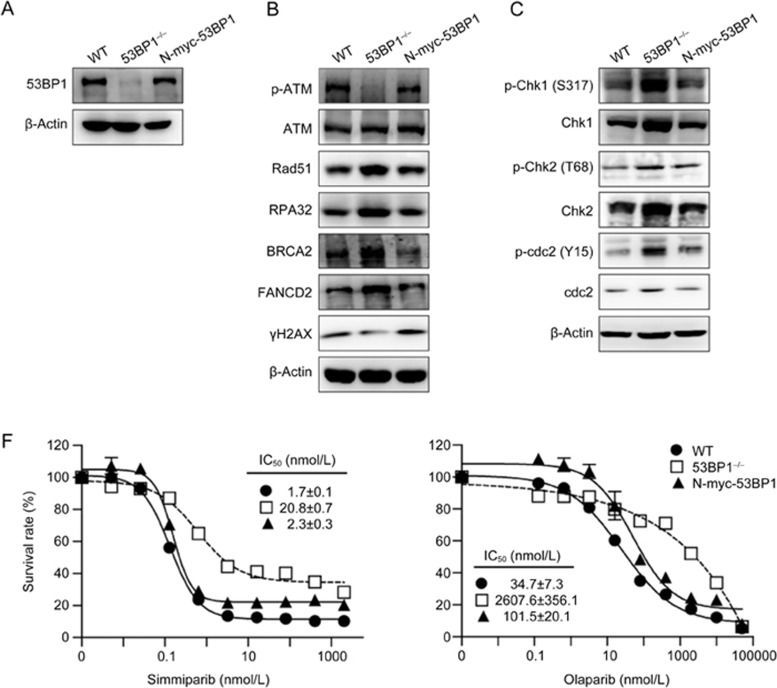
To further validate the impact of 53BP1 on PARP inhibitor sensitivity, we used full-length 53BP1 cDNA contained in the N-myc-53BP1 plasmid to restore 53BP1 protein levels in the #26 clonal cells to the levels found in the MDA-MB-436 parental cells (Figure 5A). Notably, restoring expression of the 53BP1 protein also restored the expression profiles of the indicated HR-related factors and cell-cycle regulators (Figure 5B and 5C). Consequently, 53BP1 re-expression almost completely restored sensitivity to simmparib and partially restored sensitivity to olaparib (Figure 5D).
Figure 5.
Re-expression of 53BP1 in the 53BP1/BRCA1 dual-deficient MDA-MB-436 cells leads to restoration of PARP inhibitor sensitivity. 53BP1/BRCA1 dual-deficient #26 clonal cells (53BP1−/−) derived from the BRCA1-deficient MDA-MB-436 cells (WT) were transfected with full-length 53BP1 cDNA leading to the stable re-expression of 53BP1 (N-myc-53BP1). (A–C) Protein levels of 53BP1 (A), other HR factors (B) and cell cycle regulators (C) in the three cell types. (D) Cells were treated with PARP inhibitors for 6 d and then assayed by CCK8. The survival curves from three separate experiments were plotted (mean±SD).
Discussion
In this study, we show that the basal mRNA levels of four HR-related genes, Rad51, SMC1, PTIP and 53BP1, do not correlate with either the HR defects or PARP inhibitor sensitivity of 7 human cancer cell lines. Therefore, expression of these genes cannot be used to predict PARP inhibitor sensitivity of cells with differing HR defects, although these genes were proposed as potential biomarkers for PARP inhibitor sensitivity in previous reports1,4,5,6,7,8,9,10. There was a potential correlation between the mRNA levels of the four HR genes and the PARP inhibitor sensitivity in two BRCA1-deficient cell lines: the breast cancer MDA-MB-436 and ovarian cancer UWB1.289 lines. MDA-MB-436 cells have high basal mRNA levels of the four genes and are highly sensitive to both olaparib and simmiparib, but UWB1.289 cells have low mRNA levels and are insensitive to both drugs. Hence, mRNA levels of these genes might correlate with the sensitivity of BRCA1-deficient cancer cells to PARP inhibitors. However, when RNAi was used to individually target these four genes, only reduced 53BP1 expression impaired simmiparib sensitivity. This impairment was exacerbated (from 2.78 to 30.11 times) following TALEN-mediated depletion of the 53BP1 protein. Additionally, 53BP1/BRCA1 dual-deficient xenografts were less sensitive than parental BRCA1-deficient-MB-436 xenografts in an in vivo model for olaparib or simmiparib sensitivity. These data further support the conclusion that 53BP1 contributes to the sensitivity of cancer cells harboring BRCA1 defects to PARP inhibitors.
53BP1 was demonstrated to be required for non-homologous end joining repair (NHEJ) but dispensable for HR in mice18. However, 53BP1 depletion alleviated proliferation arrest and partially restored HR function in BRCA1- but not BRCA2-deleted mouse embryonic stem cells4. Supporting this, the loss of 53BP1 resulted in resistance of BRCA1-deficient mouse mammary tumors to the PARP inhibitor olaparib5. Knockdown of 53BP1 mediated by specific shRNAs partially reduced the olaparib sensitivity of BRCA1-deficient but not BRCA1-proficient cells19. In the current study, we eliminated 53BP1 in BRCA1-deficient MDA-MB-436 cells and tested different 53BP1/BRCA1 dual-deficient variants for their in vitro and in vivo sensitivity to PARP inhibitors. The observed reduction of PARP inhibitor sensitivity due to 53BP1 depletion was consistent with the changes observed in cell cycle arrest, apoptosis and HR activity. Based on this study and previous reports, the combination of 53BP1 with BRCA1 is a promising biomarker for predicting PARP inhibitor sensitivity.
Importantly, 53BP1 aberrations (loss or reduction) have been found in approximately 14% (5/35) of BRCA1-mutated breast cancer4. Low levels of 53BP1 protein were also observed in approximately 72% (13/18) of BRCA1-promoter-methylated human breast cancers20. Thus, it is possible that more that 10% of patients with BRCA1-deficient breast cancer have aberrant 53BP1, and these patients may not respond to therapy with PARP inhibitors. Notably, similar results were seen in BRCA1-mutated ovarian cancer with decreased 53BP1 expression found in 12% (7/59) of sampled cases21. Therefore, for cancer patients with BRCA1 defects, the expression level of 53BP1 may be used to optimize therapies, thus improving the ORR to PARP inhibitors and reducing futile treatments. In addition, a recent report showed that the loss of 53BP1 lowered PARP inhibitor sensitivity in breast cancer cells harboring defects in the HR factor and BRCA1 kinase, ATM22. This result further supports the conclusion that both 53BP1 and BRCA1 should be assayed to promote the appropriate use of PARP inhibitors for cancer therapy.
Author contribution
Zhong-min YANG and Xue-mei LIAO contributed to the conception and design of the study. Zhong-min YANG, Jin-xue HE, and Ze-hong MIAO were involved in data collection. Yi CHEN, Yan-yan SHEN, Xin-ying YANG, Yi SU, Yi-ming SUN, Ying-lei GAO, Jian DING, and Ao ZHANG participated in the coordination of the study. Zhong-min YANG and Jin-xue HE conducted the statistical analysis. All authors contributed to the interpretation of the results, provided critical revision on the draft manuscript for important intellectual content, and approved the final version of the manuscript.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No 81573450, 81373446, 81321092, 81603160 and 81402961), the Chinese Academy of Sciences (Hundred Talents Project, XDA12020104, XDA12020205, CASIMM0120152003 and CASIMM0120153005), the Science and Technology Commission of Shanghai Municipality (No 16JC1406300) and the State Key Laboratory of Drug Research (No SIMM1601ZZ-03).
Footnotes
Supplementary information is available at the website of Acta Pharmacologica Sinica.
Supplementary Information
The effect of reduction of PTIP, Rad51 or SMC1 on the sensitivity of BRCA1-deficient MDA-MB-436 cells to the PARP inhibitor simmiparib.
Generation of 53BP1/BRCA1 dual-deficient MDA-MB-436 variants.
The sensitivity of 53BP1-knockout cells to PARP inhibitors in comparison with that of the parental MDA-MB-436 cells.
References
- Wang YQ, Wang PY, Wang YT, Yang GF, Zhang A, Miao ZH. An update on poly(adp-ribose)polymerase-1 (PARP-1) inhibitors: opportunities and challenges in cancer therapy. J Med Chem 2016; 59: 9575–98. [DOI] [PubMed] [Google Scholar]
- Clovis Oncology, Inc. FDA accepts Clovis Oncology'S new drug application for rucaparib for priority review for the treatment of advanced mutant BRCA ovarian cancer. http://phx.corporate-ir.net/phoenix.zhtml?c=247187&p=irol-newsArticle_Print&ID=2196955.
- TESARO, Inc. TESARO's niraparib significantly improved progression-free survival for patients with ovarian cancer in both cohorts of the phase 3 NOVA trial. http://ir.tesarobio.com/releasedetail.cfm?ReleaseID=977524.
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol 2010; 17: 688–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspers JE, Kersbergen A, Boon U, Sol W, van Deemter L, Zander SA, et al. Loss of 53BP1 causes PARP inhibitor resistance in Brca1-mutated mouse mammary tumors. Cancer Discov 2013; 3: 68–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Li N, Li Y, Wang J, Gao M, Wang W, et al. Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discov 2015; 4: 15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Callen E, Ding X, Gogola E, Duarte AA, Lee JE, et al. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 2016; 535: 382–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S, Sehrawat A, Eroglu Z, Somlo G, Hickey R, Yadav S, et al. Role of SMC1 in overcoming drug resistance in triple negative breast cancer. PLoS One 2013; 8: e64338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RW, Orelli BJ, Yamazoe M, Minn AJ, Takeda S, Bishop DK. Bishop, RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res 2007; 67: 9658–65. [DOI] [PubMed] [Google Scholar]
- Min A, Im SA, Yoon YK, Song SH, Nam HJ, Hur HS, et al. RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther 2013; 12: 865–77. [DOI] [PubMed] [Google Scholar]
- Yuan B, Ye N, Song SS, Wang YT, Song Z, Chen HD, et al. Poly(ADP-ribose)polymerase (PARP) inhibition and anticancer activity of simmiparib, a new inhibitor undergoing clinical trials. Cancer Lett 2017; 386: 47–56. [DOI] [PubMed] [Google Scholar]
- Ye N, Chen CH, Chen T, Song Z, He JX, Huan XJ, et al. Design, synthesis, and biological evaluation of a series of benzo[de][1,7]naphthyridin-7(8H)-ones bearing a functionalized longer chain appendage as novel PARP1 inhibitors. J Med Chem 2013; 56: 2885–903. [DOI] [PubMed] [Google Scholar]
- Yi JM, Huan XJ, Song SS, Zhou H, Wang YQ, Miao ZH. Triptolide induces cell killing in multidrug-resistant tumor cells via CDK7/RPB1 rather than XPB or p44. Mol Cancer Ther 2016; 15: 1495–503. [DOI] [PubMed] [Google Scholar]
- He JX, Wang M, Huan XJ, Chen CH, Song SS, Wang YQ, et al. Novel PARP1/2 inhibitor mefuparib hydrochloride elicits potent in vitro and in vivo anticancer activity characteristic of high tissue distribution. Oncotarget 2017; 8: 4156–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi JM, Zhang XF, Huan XJ, Song SS, Wang W, Tian QT, et al. Dual targeting of microtubule and topoisomerase II by α-carboline derivative YCH337 for tumor proliferation and growth inhibition. Oncotarget 2015; 6: 8960–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzo SG, Zhou ZL, Wang YQ, Marinello J, He JX, Li YC, et al. Natural product triptolide mediates cancer cell death by triggering CDK7-dependent degradation of RNA polymerase II. Cancer Res 2012; 72: 5363–73. [DOI] [PubMed] [Google Scholar]
- Gaj T, Gersbach CA, Barbas CF, 3rd. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 2013; 31: 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsburn B, Escudero B, Prakash M, Gesheva S, Liu G, Huso DL, et al. Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1. Mol Cell Biol 2010; 30: 2341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oplustilova L, Wolanin K, Mistrik M, Korinkova G, Simkova D, Bouchal J, et al. Evaluation of candidate biomarkers to predict cancer cell sensitivity or resistance to PARP-1 inhibitor treatment. Cell Cycle 2012; 11: 3837–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacot W, Thezenas S, Senal R, Viglianti C, Laberenne AC, Lopez-Crapez E, et al. BRCA1 promoter hypermethylation, 53BP1 protein expression and PARP-1 activity as biomarkers of DNA repair deficit in breast cancer. BMC Cancer 2013; 13: 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennington KP, Wickramanayake A, Norquist BM, Pennil CC, Garcia RL, Agnew KJ, et al. 53BP1 expression in sporadic and inherited ovarian carcinoma: Relationship to genetic status and clinical outcomes. Gynecol Oncol 2013; 128: 493–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong R, Ma F, Zhang W, Yu X, Li Q, Luo Y, et al. 53BP1 depletion causes PARP inhibitor resistance in ATM-deficient breast cancer cells. BMC Cancer 2016; 16: 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The effect of reduction of PTIP, Rad51 or SMC1 on the sensitivity of BRCA1-deficient MDA-MB-436 cells to the PARP inhibitor simmiparib.
Generation of 53BP1/BRCA1 dual-deficient MDA-MB-436 variants.
The sensitivity of 53BP1-knockout cells to PARP inhibitors in comparison with that of the parental MDA-MB-436 cells.