SUMMARY
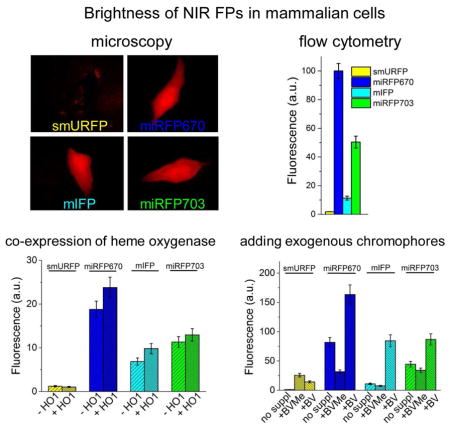
Numerous near-infrared (NIR) fluorescent proteins (FPs) were recently engineered from bacterial photoreceptors but lack of their systematic comparison makes researcher’s choice rather difficult. Here we evaluated side-by-side several modern NIR FPs, such as blue-shifted smURFP and miRFP670, and red-shifted mIFP and miRFP703. We found that among all NIR FPs, miRFP670 had the highest fluorescence intensity in various mammalian cells. For instance in common HeLa cells miRFP703, mIFP and smURFP were 2-, 9- and 53-fold dimmer than miRFP670. Either co-expression of heme oxygenase or incubation of cells with heme precursor weakly affected NIR fluorescence, however, in the latter case elevated cellular autofluorescence. Exogenously added chromophore substantially increased smURFP brightness but only slightly enhanced brightness of other NIR FPs. mIFP showed intermediate while monomeric miRFP670 and miRFP703 exhibited high binding efficiency of endogenous biliverdin chromophore. This feature makes them easy to use as GFP-like proteins for spectral multiplexing with FPs of visible range.
Keywords: heme oxygenase, 5-ALA, biliverdin, phytochrome, iRFP
Graphical Abstract
A systematic side-to-side comparison of the modern near-infrared fluorescent proteins in live mammalian cells was performed. Shemetov et al. found that the quality of the initial molecular engineering of the proteins is the key factor that determines their fluorescence in cells.
INTRODUCTION
Non-invasive in vivo imaging techniques demand for near-infrared (NIR) fluorescent probes. Engineering of fluorescent proteins (FP) from bacterial phytochrome photoreceptors (BphPs) has advanced deep-tissue imaging (Shcherbakova et al., 2015b; Shcherbakova et al., 2015c). Major advantages of BphP-derived FPs over far-red FPs of the green fluorescent protein (GFP)-like protein family are their excitation and emission spectra, which lie within a NIR tissue transparency “optical window” (~650–900 nm), which is characterized by low cellular autofluorescence, reduced light scattering, and minimal absorption of hemoglobin, melanin and water (Weissleder and Ntziachristos, 2003). NIR FPs were used to study protein-protein interaction (Filonov and Verkhusha, 2013), to design chimeric luciferases for multimodal imaging (Rumyantsev et al., 2016), and as probes for multiparameter photoacoustic tomography (Krumholz et al., 2014).
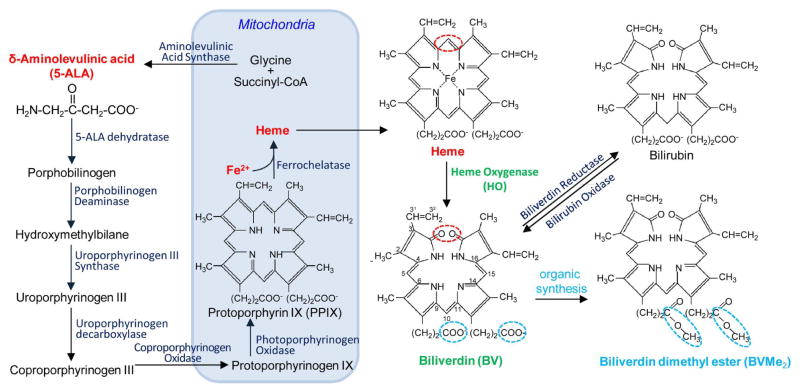
NIR spectral properties of BphP-based FPs originate from the ability of these proteins to utilize a product of the heme metabolism, linear tetrapyrrole biliverdin IXα (BV), as a chromophore. BV is abundantly present in all mammalian cells (Tran et al., 2014). In mammalian cells heme synthesis and degradation occurs in cytosol and mitochondria (Ponka, 1999) (Figure 1). Initially, small linear δ-aminolevulinic acid (5-ALA) forms protoporphyrin precursors. Then, heme formation occurs in mitochondria as a result of binding of Fe2+ to protoporphyrin IX (PPIX). Under oxidative stress, heme does not reach its primary binding proteins, such as hemoglobin or cytochrome c, and instead undergoes enzymatic oxidation by heme oxygenase (HO). In cells BV exists in the equilibrium with bilirubin, which is maintained by biliverdin reductase and bilirubin oxidase. BV and bilirubin act as a part of the protective system against cellular stress (Jozkowicz et al., 2007).
Figure 1. Scheme of biliverdin formation in mammalian cells.
Biosynthesis of a heme starts with the synthesis of 5-aminolevulinic acid (5-ALA) in mitochondria, and then 5-ALA is transferred to the cytosol. There it is merging in a chain of several enzymatic reactions through the formation of porphobilinogen (simple pyrrole), hydroxymethylbilane (linear tetrapyrrole) and uroporphyrinogen III (cyclic tetrapyrrole). Decarboxylation of uroporphyrinogen III results in a formation of coproporphyrinogen III, which is converted by coproporphyrinogen III oxidase, located in the mitochondrial membrane. Formed protoporphyrinogen IX is further oxidized to protoporphyrin IX (PPIX). PPIX is loaded by Fe2+ ions with help of ferrochelatase, which results in a formation of heme. Heme can be further converted to biliverdin IXα (BV) by heme oxygenase (HO), which cleaves the heme ring at the α-methene bridge (red ring in both heme and BV structures). In this reaction NADPH is used as the reducing agent. Molecular oxygen enter to the reaction resulted in carbon monoxide (CO) production, and the iron is released from the BV molecule. BV can be further converted into bilirubin with biliverdin reductase. Biliverdin dimethyl ester (BVMe2) can be produced using organic synthesis by esterification of carboxylic groups of BV (blue rings in BV and BVMe2 structures).
Recent FP engineering efforts have resulted in three types of the next-generation NIR FPs, which differ by their wild-type templates, spectral and biochemical characteristics. First, a monomeric NIR FP, named mIFP, based on BrBphP from Bradyrhizobium sp. was reported (Yu et al., 2015). Then, several monomeric miRFPs with different spectral properties developed from RpBphP1 of Rhodopseudomonas palustris were published (Shcherbakova et al., 2016). All these NIR FPs share a common structural fold and consist of two BphP domains, known as PAS and GAF, of 34.5 kDa in total. Lastly, a NIR FP, termed smURFP, that is a dimer of 32.0 kDa was developed from an allophycocyanin α-subunit (APCα) of Trichodesmium erythraeum cyanobacteria (Rodriguez et al., 2016). Although wild-type APCa binds phycocyanobilin (PCB) chromophore, the directed evolution of smURFP made it capable of binding BV.
Since fluorescence of the NIR FPs depends on a BV chromophore, an efficient and specific binding of endogenous BV is required to provide high fluorescence of mammalian cells. Cellular brightness (also termed as effective brightness) of NIR FPs in mammalian cells depends on molecular brightness, which is a product of molar extinction coefficient and quantum yield, on protein expression level, folding and stability, as well as on cellular BV level and its binding characteristics (Shcherbakova et al., 2015b; Yu et al., 2014). The difference in the BV binding by NIR FPs results in their various effective brightness.
It has been shown that the BV interaction with BphPs and BphP-derived NIR FPs consists of two steps (Shcherbakova et al., 2015a). First, a BV molecule enters a pocket formed in the GAF domain, resulting in a non-covalent binding with the apoform of NIR FP. This step determines the specificity of NIR FP to BV. Second, a covalent bond between the C32 atom of the ring A of BV and S atom of the cysteine residue occurs. This step determines a possibility of the NIR FP holoform formation. If an NIR FP has a low specificity to BV, the other intracellular tetrapyrroles, primarily PPIX, compete for the binding to its apoform (Lehtivuori et al., 2013; Wagner et al., 2008). Since the PPIX complex with the apoform does not fluoresce in the NIR region, this results in lower brightness of mammalian cells expressing the sub-optimal NIR FP. Addition of exogenous BV can increase the brightness of cells expressing a NIR FP with the low specificity to the BV chromophore, such as the first-generation Deinococcus radiodurans DrBphP-based proteins, IFP1.4 (Shu et al., 2009) and IFP2.0 (Yu et al., 2014). smURFP has been shown to be able binding more hydrophobic and thus more membrane permeable BV analog, biliverdin dimethyl ester (BVMe2) (Figure 1).
Another approach to upregulate a BV chromophore level in mammalian cells is to increase the holoform amount of an NIR FP with the low BV specificity by a heterologous co-expression of HO (Rodriguez et al., 2016; To et al., 2015; Yu et al., 2014). HO catalyzes the first step of heme degradation and plays an important role in oxidative stress defense and recycling of iron (Maines, 1988, 1997). HO cleaves the α-meso carbon bridge of heme, yielding equimolar quantities of carbon monoxide (CO), iron ions and BV (Figure 1). HO seems to be ubiquitously present in photosynthetic bacteria and plants as a key enzyme for the synthesis of photon-accepting chromophores (Beale, 1994; Davis et al., 1999; Migita et al., 2003). In mammals, HO has three major isoforms of which HO1 is inducible by oxidative stress, HO2 is constitutive, and HO3 is a pseudotranscript of HO2 (Abraham and Kappas, 2008). It has been reported that co-expression of either Synechocystis cyanobacterial HO (Rodriguez et al., 2016) or human HO (To et al., 2015; Yu et al., 2014) via either T2A peptide or internal ribosome entry site (IRES) notably increased fluorescence of several NIR FPs in mammalian cells. It has been also suggested to enhance the effect of HO co-expression with NIR FP by addition of a precursor of heme synthesis 5-ALA and iron ions (Figure 1). The latter approach led to a considerable increase of smURFP fluorescence in cells (Rodriguez et al., 2016). Overall, a large volume of data on the enhancement of the effective brightness of BphP-derived and APCα-derived NIR FPs in cultured cells and in vivo was accumulated. However, to date there is no direct comparison of the NIR FPs, similar to that performed for the GFP-like FP family (Bindels et al., 2017; Goedhart et al., 2011).
Here we characterized side-by-side four the most recently developed NIR FPs in live mammalian cells. Among them, smURFP and miRFP670 are blue-shifted NIR FPs with fluorescence maxima at 670 nm (Table 1). Another NIR FP pair consists of red-shifted mIFP and miRFP703 with fluorescence maxima at around 703 nm. We first compared the NIR FPs in different mammalian cell types without addition of exogenous chromophore. We then tested how a co-expression of human HO1 enzyme via either IRES2 sequence or T2A peptide affects the cellular brightness of NIR FPs. We next studied the brightness of NIR FPs in cells supplemented with exogenous BV and BVMe2. Lastly, we examined an effect of cell treatment with a precursor of heme synthesis 5-ALA and simultaneous co-expression of HO1.
Table 1.
Spectral properties of NIR FPs and their effective brightness in various mammalian cells. See also Figures 2, S1 and Table S1.
| NIR FP | Ex, nm | Em, nm | Extinction coefficient, M−1cm−1 | Quantum yield, % | NIR FP brightness in live cells relative to miRFP670: co-expression with EGFP downstream of T2A from the same plasmid (co-expression with EGFP from two separate plasmids), % | ||||
|---|---|---|---|---|---|---|---|---|---|
| HeLa | HEK293 | U87 | COS-7 | U-2 OS | |||||
| smURFP | 642 | 670 | 180,000 | 18 | 1.9 (0.7) | 0.8 (0.8) | 1.2 (2.4) | 1.7 (1.6) | 0.6 (0.9) |
| miRFP670 | 642 | 670 | 87,400 | 14 | 100 (100) | 100 (100) | 100 (100) | 100 (100) | 100 (100) |
| mIFP | 683 | 704 | 82,000 | 8.4 | 11 (26) | 12 (29) | 16 (28) | 14 (44) | 15 (38) |
| miRFP703 | 674 | 703 | 90,900 | 8.6 | 50 (68) | 50 (69) | 57 (58) | 67 (71) | 62 (89) |
RESULTS
Effective brightness of NIR FPs in different cells without supplements
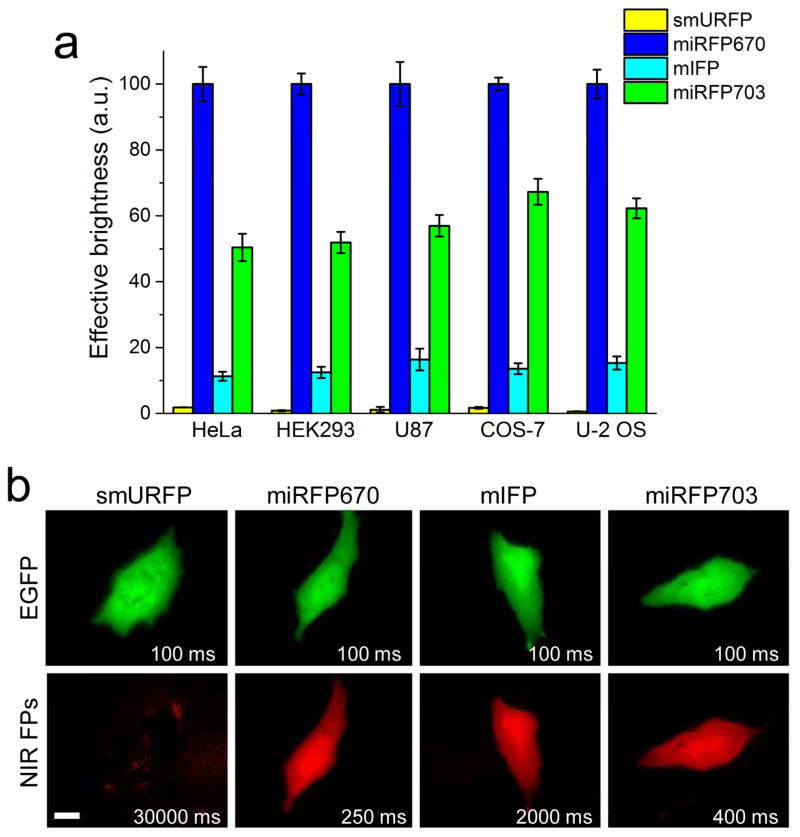
We studied effective brightness of smURFP, miRFP670, mIFP and miRFP703 in commonly used mammalian cell lines, such as HeLa, HEK293, COS-7, U87 and U-2 OS, using flow cytometry. NIR FPs were co-expressed via T2A self-cleavable peptide downstream of EGFP from the same plasmid to achieve the equal expression conditions for the NIR FPs. We found that miRFP670 exhibited the highest fluorescent signal among the NIR FPs in all tested live cells (Figure 2a, Table 1). The miRFP670 expressing cells were about 100-fold brighter than those with smURFP and 8–9-fold brighter than the mIFP cells. The cellular brightness of miRFP703 was lower than that of miRFP670 but substantially exceeded that of mIFP. Specifically, the miRFP703-expressing HeLa cells were 2-fold dimmer than the cells with miRFP670 but 4.4-fold brighter than the mIFP cells. Likely, some difference in endogenous BV levels between the cell types affected the observed effective brightness of NIR FPs. Although smURFP has the largest extinction coefficient and quantum yield (Table 1), it exhibited the lowest brightness in all tested mammalian cells, either because of the poor protein maturation or due to the weak chromophore binding efficiency. We further estimated the holoform amount of NIR FPs in mammalian cells (Table S1). Depending on the cell type, the amount of smURFP holoform was 102–103-fold lower than that of miRFP670.
Figure 2. Effective brightness of NIR FPs co-expressed with EGFP downstream of T2A self-cleavable peptide in different mammalian cells.
(a) NIR fluorescence intensity of cells transiently transfected with smURFP, miRFP670, mIFP and miRFP703 (in the EGFP-T2A-NIRFP constructs) were analyzed using flow cytometry 48 h after transfection. Mean NIR fluorescence intensity was normalized to mean EGFP fluorescence intensity to account for cell transfection efficiency. NIR effective brightness of miRFP670 was assumed to be 100%. Error bars, s.e.m. (n=3; transfection experiments). (b) Representative fluorescence images of NIR FPs in live HeLa cells. Acquisition time for each image is indicated. Scale bar, 10 μm. See also Figure S1, Tables 1 and S1.
We next studied performance of NIR FPs using fluorescence microscopy. Fluorescence imaging required distinct acquisition times for live HeLa cells with different NIR FPs to achieve comparable image quality (Figure 2b). All three BphP-derived NIR FPs exhibited homogenous intracellular distributions and absence of intracellular aggregates. However, mIFP cells required 5–8-fold longer exposure time as compared to that of miRFP670 and miRFP703 cells. Intracellular distribution of smURFP was difficult to determine even at the 30 s exposure time because its fluorescence signal was lower than cellular autofluorescence.
We also compared NIR FPs using a dual plasmid transfection approach in which NIR FPs and EGFP were co-expressed from two different plasmids, as previously described (Shcherbakova et al., 2016; Shcherbakova and Verkhusha, 2013). We found that miRFP670 again overperformed other NIR FPs in all cell types (Figure S1a and Tables 1, S1). Effective brightness of miRFP703 was slightly higher as compared to that with co-expression with EGFP via T2A from a single plasmid. Effective brightness of mIFP was 4-fold higher in HeLa cells and reached 25% and 40% of that for miRFP670 and miRFP703, respectively, which is in agreement with published resulted (Shcherbakova et al., 2016). Fluorescence microscopy confirmed the flow cytometry results and showed the decrease in exposure time for mIFP (Figure S1b). Likely, additional amino acid residues at the N-terminus of mIFP originated from the T2A peptide affected its expression level. Effective brightness of smURFP was not changed and remained the lowest in all tested mammalian cells. Thus, the relative effective brightness of the NIR FPs did not change, meaning that the both EGFP and NIR FP co-expression approaches, either from a single plamid or from two separate plasmids, can be equally used to compare various NIR FPs.
Co-expression of NIR FPs with human HO1 and treatment with heme precursor
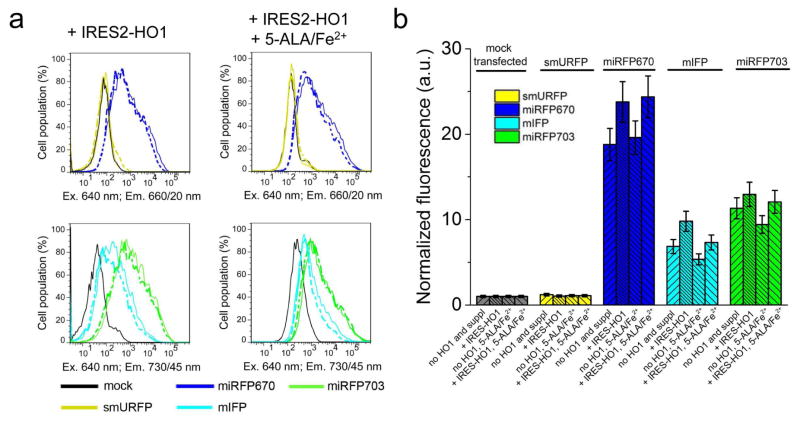
Since some NIR FPs showed low fluorescence in mammalian cells we next studied ways to increase it by co-expressing HO, as was suggested by others (Rodriguez et al., 2016; To et al., 2015; Yu et al., 2014). We co-expressed human HO1 encoded via IRES2 element in the same plasmid as NIR FPs, resulting in the EGFP-T2A-NIRFPs-IRES2-HO1 constructs. As a control, we similarly co-expressed catalytically inactive HO1 containing two amino acid substitutions, such as H25A (Wilks et al., 1995) and D140F (Fujii et al., 2001). We found that the co-expression with HO1 slightly increased NIR fluorescence of all cells, except for those with smURFP whose fluorescence was not affected (Figure 3). However, the increase in the effective brightness of miRFP670, miRFP703 and mIFP did not exceed 25% over that observed with the co-expression of catalytically inactive HO1. Likely, the low fluorescence increase was because the produced BV molecules remained bound to hHO1. It has been shown that the release of BV product is a rate-limiting step in a reaction catalyzed by HOs, including hHO1 (Liu and Ortiz de Montellano, 2000). Structural analysis of hHO1 revealed that its active site is adopted for interaction with a planar substrate, and that the formed linear BV product further moves to an internal pocket of the enzyme (Lad et al., 2004). Spontaneous dissociation of BV molecule from the internal pocket was shown to be very slow and could be only accelerated in the presence of biliverdin reductase that interacted with hHO1 (Liu and Ortiz de Montellano, 2000). Thus, the HO1 co-expression just slightly enhanced the effective brightness of the BphP-based NIR FPs and had no effect on the smURFP cellular brightness.
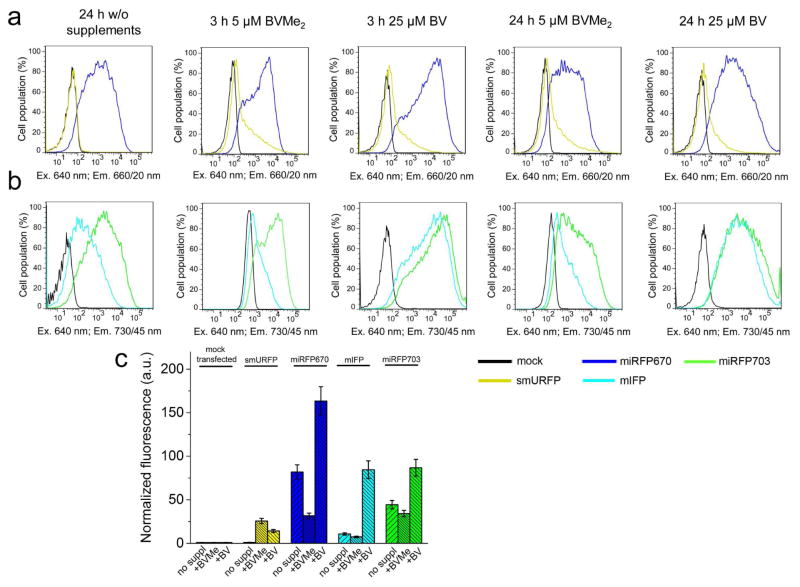
Figure 3. Effective brightness of NIR FPs in presence of HO1 and 5-ALA with FeSO4.
(a) Fluorescence intensity distribution of live HeLa cells transfected with smURFP, miRFP670, mIFP and miRFP703 co-expressed with HO1 downstream of IRES2 element (in EGFP-T2A-NIRFP-IRES2-HO1 constructs) with or without 250 μM 5-ALA and 40 μM FeSO4 was analyzed using flow cytometry. The supplements were added for 24 h period. Fluorescence distribution of cells co-expressing NIR FPs with catalytically inactive HO1 (H25A/D140F) mutant as a control is shown with dashed lines. (b) Quantification of the data presented in (a). Mean NIR fluorescence intensity was normalized to mean green fluorescence intensity of EGFP co-expressed via T2A peptide upstream of NIR FPs (to account for transfection efficiency) and to mean fluorescence intensity of mock transfected cells in the 660/20 nm channel (for miRFP670 and smURFP) or in the 730/45 nm channel (for miRFP703 and mIFP) (to account for cellular autofluorescence). Error bars, s.e.m. (n = 3; transfection experiments). See also Figure S2 and Table 2.
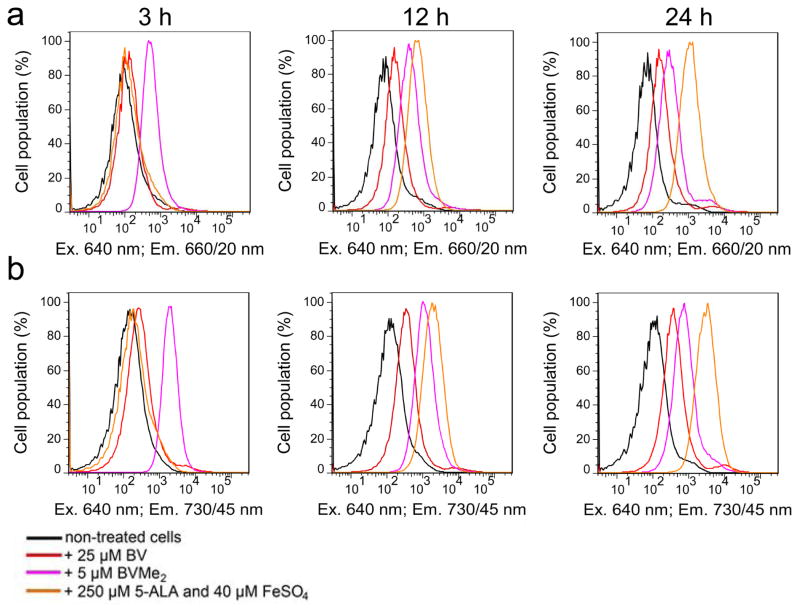
Earlier, it has been suggested to elevate intracellular level of heme, a substrate for HO, by adding of heme precursors (Rodriguez et al., 2016). Therefore, we next supplied HeLa cells transfected with EGFP-T2A-NIRFPs-IRES2-HO1 with a precursor of heme synthesis 5-ALA (Figure 1) and FeSO4. Incubation of the control mock-transfected cells with 5-ALA for 12 h resulted in a 6-fold increase of autofluorescence in the 660/20 nm channel (used for imaging of smURFP and miRFP670) and a 12-fold increase of autofluorescence in the 730/45 nm channel (used for imaging of mIFP and miRFP703) (Figure 4). After 24 h of incubation with 5-ALA, the cells exhibited up to 21-fold higher NIR autofluorescence. This indicated that HeLa cells actively metabolized 5-ALA and likely accumulated the produced fluorescent PPIX (Figure 1), as observed in cancer cell lines (Krieg et al., 2002; Nemes et al., 2016). After normalization of NIR FP fluorescence to the increased autofluorescence we found that the real effective brightness of all NIR FPs has not changed and remained the same as without 5-ALA (Figure 3b). Moreover, it was reported that PPIX competes with BV for binding of NIR FP apoforms, resulting in non-fluorescent protein-PPIX complexes (Lehtivuori et al., 2013; Wagner et al., 2008). Thus, the treatment of mammalian cells with heme precursors is the inefficient way to increase cellular brightness of suboptimally engineered NIR FPs.
Figure 4. Autofluorescence of mammalian cells with exogenous chromophores or heme precursor.
Fluorescence intensity distribution of live HeLa cells transfected with a mock plasmid in (a) the 660/20 nm channel and (b) the 730/45 nm channel without (black line) or with 25 μM BV (red line), 5 μM BVMe2 (magenta line), or 250 μM 5-ALA and 40 μM FeSO4 (orange line) was analyzed using flow cytometry 3, 12 and 24 h after the supplementation.
We next studied an effect of the HO1 co-expression on the NIR FP brightness using the dual plasmid approach in which cells co-expressed NIR FP and HO1 encoded by two separate plasmids. We also compared this dual plasmid co-expression with a single plasmid transfection whereby proteins were co-expressed via either T2A peptide or IRES element. Similar analysis of co-expression of two GFP-like FPs was performed previously (Goedhart et al., 2011). Co-expression of miRFP703 and HO1 from two separate plasmids with both proteins encoded under CMV promoters slightly increased NIR cell fluorescence (Figure S2), likely due to elevated BV production. However, the increase in the effective brightness did not exceed 1.2-fold as compared to that of miRFP703 expressed alone. The similar result was obtained in the case of the co-expression of miRFP703 with HO1 downstream of IRES2 element (Figure 3b). We then co-expressed HO1 downstream of T2A peptide in the same plasmid with miRFP703 to obtain equimolar quantities of NIR FP and enzyme. Transfection with this plasmid resulted in 3.1-fold decrease of miRFP703 fluorescence as compared to the co-expression of miRFP703 and HO1 using two plasmids (Figure S2a). The 2A peptides, also known as CHYSELs (cis-acting hydrolase elements), consist of two parts: a non-conserved amino acid sequence with a strong α-helical propensity and a downstream conserved D(V/I)EXNPGP motif. It has been shown that the 2A peptide-like sequences can interact with the ribosomal exit tunnel and, consequently, act as arrest sequences for the translation termination (Donnelly et al., 2001; Muto et al., 2006; Nakatogawa and Ito, 2001). Likely, in our experiments T2A peptide slowed down the translation termination of an upstream gene, such as miRFP703, resulting in the decrease of the protein expression and, consequently, in the lower NIR cellular brightness as compared to expression of miRFP703 from a separate plasmid.
Then, we tested the effect of co-expression of HO1 encoded via IRES2 element in the same plasmid with miRFP703. This bicistronic hHO1 co-expression resulted in the 1.8-fold decrease of the miRFP703 cellular brightness (Figure S2). Treatment of cells with tin-PPIX, an inhibitor of HOs, did not increase their brightness, indicating that the observed decrease of NIR cell fluorescence is not related to the HO1 enzymatic activity. Likely, this decrease was caused by the reduced expression of miRFP703 itself. Bicistronic IRES2-containing mRNA molecule encoding an NIR FP and HO1 is twice longer than mRNA encoding the NIR FP alone. This may result in the lower number of miRFP703-encoding mRNAs and, consequently, in the lower number of the NIR FP molecules.
Effective brightness of NIR FPs in presence of exogenous chromophores
Next, we studied the effect of supply of the NIR FP expressing cells with exogenous chromophores, such as 25 μM of BV or 5 μM of BVMe2. Here, HeLa cells were transfected with EGFP-T2A-NIRFPs constructs. The addition of BV for 3 h slightly increased fluorescence of miRFP670 and miRFP703 (Figure 5) while the incubation with BV for 24 h resulted in 2-fold increase of their fluorescence. In contrast, mIFP fluorescence increased 7-fold after the incubation with BV. smURFP fluorescence elevated 10-fold after 24 h incubation with the excess of BV chromophore, suggesting much lower specificity of this protein to BV. In contrast to BV, treatment of the NIR FP expressing cells with BVMe2 led to the substantial redistribution of the smURFP cell population and a notable increase of effective brightness of three other NIR FPs. However, BVMe2 also caused a fluorescence increase of the control mock-transfected cell population (Figure 4). Incubation with BVMe2 for 3 h increased autofluorescence of the control cells 4.2-fold in the 660/20 nm channel and 12.8-fold in the 730/45 nm channel. In contrast, 3 h incubation with BV resulted in a 1.9-fold and a 3.4-fold fluorescence increase in the 660/20 nm and 730/45 nm channels, respectively. The brightness of HeLa cells expressing NIR FPs in presence of BV or BVMe2, normalized to autofluorescence of the mock transfected cells supplemented with the same chromophores (Figure 5c), revealed the real fluorescence changes. Only smURFP showed the substantial fluorescence increase after the incubation with both chromophores for 24 h: 10-fold with BV and 24-fold with BVMe2 (Table 2). In contrast, exogenous BV increased the brightness of the miRFP670 and miRFP703 expressing cells less than 2-fold. Importantly, the addition of BVMe2 decreased the effective brightness of three other NIR FPs 1.5–3-fold (Table 2), suggesting that the BphP-derived proteins do not bind esterified BV derivatives.
Figure 5. Effect of exogenous chromophores on fluorescence of NIR FPs in mammalian cells.
Fluorescence intensity distribution of live HeLa cells transfected with (a) smURFP or miRFP670 and (b) mIFP or miRFP703 was analyzed using flow cytometry in presence of 25 μM BV or 5 μM BVMe2 in the 660/20 nm channel (a) or in the 730/45 nm channel (b), respectively. Chromophores were added for 24 h period. Entire constructs were EGFP-T2A-NIRFPs. (c) Quantification of the data represented in panels (a) and (b). Mean NIR fluorescence intensity was normalized to mean green fluorescence intensity of EGFP co-expressed via T2A peptide upstream of NIR FPs (to account for transfection efficiency) and to mean fluorescence intensity of mock transfected cells in the 660/20 nm channel (for smURFP and miRFP670) or the 730/45 nm channel (for mIFP and miRFP703) (to account for cellular autofluorescence). Error bars, s.e.m. (n = 3; transfection experiments). See also Table 2.
Table 2.
Effect of supplements on the brightness of mammalian cells expressing NIR FPs. See also Figures 3 and 5.
| NIR FP | Effective brightness in live HeLa cells relative to miRFP670, % | |||
|---|---|---|---|---|
| without supplements | with 5 μM BVMe2 for 24 h | with 25 μM BV for 24 h | with 250 μM 5-ALA for 24 h and co-expressed HO1 via IRES | |
| smURFP | 1.3 | 31 | 17 | 4.6 |
| miRFP670 | 100 | 39 | 199 | 102 |
| mIFP | 13 | 9 | 102 | 30 |
| miRFP703 | 54 | 42 | 106 | 51 |
DISCUSSION
Our side-by-side comparison of the most recent BphP-derived miRFP670, mIFP and miRFP703 proteins demonstrated their high fluorescence brightness in various types of mammalian cells. The highest effective brightness in cells was observed for miRFP670. In contrast, the engineered from the APCα photoreceptor smURFP protein exhibited the lowest brightness in all tested cell lines and conditions. miRFP670 and miRFP703 notably outperformed mIFP in terms of the cellular brightness (Figures 2 and S1). An additional advantage of miRFPs is that they can be combined in two-color NIR imaging (Shcherbakova et al., 2016).
Co-expression of all NIR FPs with human HO1 encoded via IRES element provided rather limited enhancement of their cellular brightness (Figure 3), which can be explained by binding of BV by the enzyme. We also found the notable decrease of the protein expression levels when modern co-expression approaches were applied, such as co-expression via T2A and via IRES2 sequences, as compared to the dual plasmid co-transfection (Figure S2). We hypothesized that the larger size of bicistronic mRNA might decrease its amount, consequently, decreasing the level of the translated NIR FPs. To circumvent the negative effect of 2A peptide and IRES element on expression of the downstream protein, other approaches to co-introduce several genes into cells should be considered, for instance, a CRISPR/Cas9 system (Wright et al., 2016).
Incubation of mammalian cells with exogenous chromophores, such as BV or BVMe2, significantly enhanced the smURFP effective brightness. At the same time, the addition of the exogenous chromophores substantially increased cellular autofluorescence (Figure 4). In contrast, cellular fluorescence of miRFP670 and miRFP703 increased 2-fold and only with BV, whereas BVMe2 caused the decrease of their effective brightness. The latter is likely because the carboxylic groups of BV should be unmodified for efficient interaction with the pocket of the BphP-based NIR FPs. In contrast, the homology crystallographic modeling of smURFP showed lack of the BV carboxylates recognition, which enables BVMe2 binding (Rodriguez et al., 2016).
The reported data on the effect of exogenous chromophores on the effective brightness of NIR FPs in vivo are rather contradictory. For instance, it was reported that an intravenous administration of BV significantly increased fluorescence of IFP1.4 expressed in the liver of mice (Shu et al., 2009). In contrast, the attempts to visualize the growth of IFP1.4-expressing glioblastoma cells in the brain with BV injections have been unsuccessful (Jiguet-Jiglaire et al., 2014). Similarly, an intravenous injection of BV did not increase fluorescence of HT1080 tumor xenografts expressing smURFP(Rodriguez et al., 2016). Importantly, BV has a broad range of biological activities and besides of protecting from cellular oxidative stress (Satoh et al., 2003) it also affects cell proliferation (Nuhn et al., 2009), cell survival, and activation of several signaling pathways (Gibbs and Maines, 2007; Miralem et al., 2012; Mölzer et al., 2013). Therefore, an exogenous supply of tetrapyrroles, such as BV and BVMe2, to cultured cells and in vivo should be performed with caution to avoid affecting metabolism.
Because the BphP-derived miRFP670, mIFP and miRFP703 already exhibit the efficient binding of endogenous chromophore, future engineering efforts on these NIR FPs should focus on an increase of their quantum yields, e.g., via reduction of various excited-state deactivation pathways (Hontani et al., 2016). In contrast, the APCα-derived smURFP should be further evolved to substantially enhance its efficiency and specificity to bind endogenous BV chromophore in mammalian cells.
NIR FPs provide several advantages over GFP-like FPs of visible range for multicolor imaging of biological processes across scales, from super-resolution fluorescence microscopy to deep-tissue optical tomography. Currently, miRFPs are the best available NIR FPs because of their highest effective brightness and spectral separation making them exceptional among already heavy and crowded FP toolbox (Shcherbakova et al., 2016). The use of the same NIR FP at the cellular and the organismal levels should become a common approach in cell and developmental biology, cancer studies, immunology, neuroscience, and biomedicine.
SIGNIFICANCE
The comprehensive side-to-side comparison of the modern monomeric miRFP670, mIFP and miRFP703 NIR FPs has revealed that they exhibit the efficient binding of endogenous BV chromophore in mammalian cells and can be readily applied to cellular and in vivo imaging. Several approaches that were tested in order to further increase the NIR cellular fluorescence, including co-expression HOs via T2A or IRES, and incubation with heme precursor, in fact, did not increase the NIR FP effective brightness and, moreover, in the latter case substantially elevated cellular autofluorescence. The supply of exogenous chromophore resulted in rather limited cellular enhancement of miRFP670 and miRFP703 fluorescence, which was in contrast to the significant fluorescence increase observed for smURFP. mIFP showed intermediate performance among BphP-based FPs but was substantially brighter than smURFP. Thus, a quality of the initial molecular engineering rather than an insufficient amount of the endogenous BV chromophore is the key factor determining the NIR FP brightness in mammalian cells.
STAR METHODS
KEY RESOURCES TABLE
CONTACT FOR REAGENT AND RESOURCE REQUEST
Further information and requests for reagents should be directed to the Lead contact, Vladislav Verkhusha (vladislav.verkhusha@einstein.yu.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines
HeLa, HEK293, U87, COS-7 and U-2 OS cells were obtained from ATCC and grown in high-glucose DMEM medium (HyClone) containing 10% (v/v) FBS (Gemini Bio-Products), 0.5% (v/v) penicillin-streptomycin (Mediatech). All cell lines were grown at 37°C in a humidified 5% CO2 atmosphere. Transfection of plasmids was performed with an Effectene reagent (Qiagen) according to the manufacturer’s protocol. Cell medium was changed 8 h after the transfection. If necessary, 25 μM of BV (Frontier Scientific), 5 μM BVMe2 (Frontier Scientific) or 250 μM 5-ALA (Acros Organics) with 40 μM of FeSO4 (Sigma-Aldrich) were added 24 h after the transfection, and cell fluorescence was analyzed using flow cytometry at indicated times.
METHODS DETAILS
Design of plasmids
To generate a pCMV-MCS-T2A-hHO1 plasmid, a pcDNA3.1-iCasper-T2A-HO1 vector (Addgene #64278) (To et al., 2015) was treated with BspTI and SgsI restriction endonucleases (Thermo Scientific) to cut out iCasper gene and then MCS (multiple cloning site) was inserted by ligation of MCS_HO1_sense and MCS_HO1_asense oligonucleotides there. To generate a pEGFP-T2A-HO1 plasmid, an EGFP gene was amplified with EGFP_HindIII_fw and EGFP_XhoI_rv oligonucleotides, treated with HindIII and XhoI restriction endonucleases (Thermo Scientific) and inserted into the pCMV-MCS-T2A-HO1 plasmid digested with the same enzymes. To generate a pIRES2-HO1 plasmid, an AgeI restriction site was removed from HO1 gene in the pcDNA3.1-iCasper-T2A-HO1 vector by quick-change mutagenesis using HO1_AgeI_del_fw and HO1_AgeI_del_rv oligonucleotides and PrimeSTAR DNA Polymerase (Takara Bio). Before transformation into E.coli TOP10 (Invitrogen), the PCR product was treated with DpnI restriction endonuclease. Resulted HO1 gene without AgeI recognition site was amplified with HO1_AgeI_fw and HO1_NotI_rv oligonucleotides, treated with BshTI and NotI restriction endonucleases (Thermo Scientific) and inserted into the pIRES2-EGFP plasmid digested with the same enzymes to cut out an EGFP gene. EGFP-T2A-HO1 DNA fragment was amplified from pEGFP-T2A-HO1 with EGFP_SacI_fw and HO1_XmaI_rv oligonucleotides, digested with SacI and Cfr9I restriction endonucleases (Thermo Scientific) and inserted into a pEGFP-N1 plasmid (Clontech) treated with the same enzymes. Then, a SpeI restriction site was added before a HO1 gene by quick-change mutagenesis using HO1_SpeI_add_fw and HO1_SpeI_add_rv oligonucleotides and PrimeSTAR DNA Polymerase (Takara Bio). The miRFP670 and the miRFP703 genes (Shcherbakova et al., 2016) was PCR-amplified with miRFP_SpeI_fw and miRFP_XmaI_rv oligonucleotides. The mIFP gene was PCR-amplified with mIFP_SpeI_fw and mIFP_XmaI_rv oligonucleotides from a pmIFP-N1 plasmid (Addgene #54620) (Yu et al., 2015). The smURFP gene was PCR-amplified with smURFP_SpeI_fw and smURFP_XmaI_rv oligonucleotides from a pcDNA3-smURFP-IRES-HO1 plasmid (Addgene #80345) (Rodriguez et al., 2016). Then, the genes of all NIR FPs were treated with BcuI and Cfr9I restriction endonucleases (Thermo Scientific) and inserted into the pEGFP-T2A-HO1 plasmid (with SpeI recognition site added previously) digested with the same enzymes. An EGFP gene was cut out with BshTI and NotI restriction endonucleases, the plasmids were treated with T4 DNA Polymerase (Thermo Scientific) and self-ligated with T4 DNA Ligase (Thermo Scientific). In this way, the pEGFP-T2A-NIR FP plasmids were generated where NIR FP is miRFP670, miRFP703, mIFP or smURFP. To generate a pEGFP-T2A-NIR FP-IRES2-HO1 plasmid, the pIRES2-HO1 vector was treated with Cfr9I and NotI restriction endonucleases (Thermo Scientific) and then the IRES2-HO1 fragment was inserted into the pEGFP-T2A-NIR FP plasmids digested with the same enzymes. To obtain pEGFP-T2A-NIR FP-IRES2-HO1 plasmids with inactive HO1, a two-step quick-change mutagenesis was performed to introduce the H25A and D140F substitutions.
Fluorescence microscopy
Live HeLa, HEK293, U87, COS-7 and U-2 OS cells were imaged with an Olympus IX81 inverted epifluorescence microscope 48 h after the transfection. The microscope was equipped with a 200 W metal halide arc lamp (Lumen220PRO, Prior), a 60× 1.35 numerical aperture (NA) oil objective lens (UPlanSApo, Olympus) and an opiMOS sCMOS camera (QImaging). During imaging, HeLa cells were incubated in a cell imaging medium (Life Technologies-Invitrogen) and kept at 37°C. The microscope was operated with a SlideBook v.6.0.8 software (Intelligent Imaging Innovations).
Flow cytometry
Flow cytometry analysis was performed using a BD LSRII flow cytometer. All NIR FPs were excited with a 640 nm solid-state laser. Fluorescence of smURFP and miRFP670 was detected with a 660/20 nm emission filter, and fluorescence of mIFP and miRFP703 was detected with a 730/45 nm emission filter. EGFP was excited with a 488 nm solid-state laser, and its fluorescence was detected with a 530/40 nm emission filter. To determine an NIR FP effective (cellular) brightness, a mean NIR fluorescence intensity of the entire EGFP-positive cell population was divided by a mean fluorescence intensity in the EGFP channel, thus normalizing to the transfection efficiency. To normalize for a cellular autofluorescence, this value was further divided by the similarly obtained value for the mock transfected cells. Minimally 3×105 live cells were analyzed in each cell sample. Data were analyzed using a FACSDiva v.8.0.1 and a FlowJo v.7.6.2 software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data fitting and statistical analysis were performed using an OriginPro v.9.2.196 software (OriginLab). Statistical values including the exact n and statistical significance are reported in Figure legends.
Supplementary Material
Highlights.
miRFP670 is the brightest monomeric NIR FP in mammalian cells.
co-expression of NIR FPs with heme oxygenase does not affect cellular brightness.
precursor of biliverdin chromophore does not increase cellular brightness of NIR FPs.
endogenous chromophore suffices for miRFP670, mIFP and miRFP703 to fluoresce.
Acknowledgments
We thank Konstantin Lukyanov (Institute of Bioorganic Chemistry, Russia), Andrii Kaberniuk and Daria Shcherbakova (both from Albert Einstein College of Medicine) for the valuable discussions. This work was supported by GM122567 and NS099573 grants from the US NIH extramural program and by ERC-2013-ADG-340233 grant from the EU FP7 program.
Footnotes
AUTHOR CONTRIBUTIONS
A.A.S. and O.S.O. acquired the data. A.A.S., O.S.O. and V.V.V. analyzed and interpret the data. A.A.S., O.S.O. and V.V.V. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham NG, Kappas A. Pharmacological and clinical aspects of heme oxygenase. Pharmacol Rev. 2008;60:79–127. doi: 10.1124/pr.107.07104. [DOI] [PubMed] [Google Scholar]
- Beale SI. Biosynthesis of open-chain tetrapyrroles in plants, algae, and cyanobacteria. Ciba Found Symp. 1994;180:156–168. doi: 10.1002/9780470514535.ch9. discussion 168–171. [DOI] [PubMed] [Google Scholar]
- Bindels DS, Haarbosch L, van Weeren L, Postma M, Wiese KE, Mastop M, Aumonier S, Gotthard G, Royant A, Hink MA, et al. mScarlet: a bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 2017;14:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Kurepa J, Vierstra RD. The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc Natl Acad Sci U S A. 1999;96:6541–6546. doi: 10.1073/pnas.96.11.6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly ML, Hughes LE, Luke G, Mendoza H, ten Dam E, Gani D, Ryan MD. The ‘cleavage’ activities of foot-and-mouth disease virus 2A site-directed mutants and naturally occurring ‘2A-like’ sequences. J Gen Virol. 2001;82:1027–1041. doi: 10.1099/0022-1317-82-5-1027. [DOI] [PubMed] [Google Scholar]
- Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol. 2013;20:1078–1086. doi: 10.1016/j.chembiol.2013.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H, Zhang X, Tomita T, Ikeda-Saito M, Yoshida T. A role for highly conserved carboxylate, aspartate-140, in oxygen activation and heme degradation by heme oxygenase-1. J Am Chem Soc. 2001;123:6475–6484. doi: 10.1021/ja010490a. [DOI] [PubMed] [Google Scholar]
- Gibbs PE, Maines MD. Biliverdin inhibits activation of NF-kappaB: reversal of inhibition by human biliverdin reductase. Int J Cancer. 2007;121:2567–2574. doi: 10.1002/ijc.22978. [DOI] [PubMed] [Google Scholar]
- Goedhart J, van Weeren L, Adjobo-Hermans MJ, Elzenaar I, Hink MA, Gadella TW. Quantitative co-expression of proteins at the single cell level--application to a multimeric FRET sensor. PLoS One. 2011;6:e27321. doi: 10.1371/journal.pone.0027321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontani Y, Shcherbakova DM, Baloban M, Zhu J, Verkhusha VV, Kennis JT. Bright blue-shifted fluorescent proteins with Cys in the GAF domain engineered from bacterial phytochromes: fluorescence mechanisms and excited-state dynamics. Sci Rep. 2016;6:37362. doi: 10.1038/srep37362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiguet-Jiglaire C, Cayol M, Mathieu S, Jeanneau C, Bouvier-Labit C, Ouafik L, El-Battari A. Noninvasive near-infrared fluorescent protein-based imaging of tumor progression and metastases in deep organs and intraosseous tissues. J Biomed Opt. 2014;19:16019. doi: 10.1117/1.JBO.19.1.016019. [DOI] [PubMed] [Google Scholar]
- Jozkowicz A, Was H, Dulak J. Heme oxygenase-1 in tumors: is it a false friend? Antioxid Redox Signal. 2007;9:2099–2117. doi: 10.1089/ars.2007.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg RC, Messmann H, Rauch J, Seeger S, Knuechel R. Metabolic characterization of tumor cell-specific protoporphyrin IX accumulation after exposure to 5-aminolevulinic acid in human colonic cells. Photochem Photobiol. 2002;76:518–525. doi: 10.1562/0031-8655(2002)076<0518:mcotcs>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Krumholz A, Shcherbakova DM, Xia J, Wang LV, Verkhusha VV. Multicontrast photoacoustic in vivo imaging using near-infrared fluorescent proteins. Sci Rep. 2014;4:3939. doi: 10.1038/srep03939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad L, Friedman J, Li H, Bhaskar B, Ortiz de Montellano PR, Poulos TL. Crystal structure of human heme oxygenase-1 in a complex with biliverdin. Biochemistry. 2004;43:3793–3801. doi: 10.1021/bi035451l. [DOI] [PubMed] [Google Scholar]
- Lehtivuori H, Rissanen I, Takala H, Bamford J, Tkachenko NV, Ihalainen JA. Fluorescence properties of the chromophore-binding domain of bacteriophytochrome from Deinococcus radiodurans. J Phys Chem B. 2013;117:11049–11057. doi: 10.1021/jp312061b. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ortiz de Montellano PR. Reaction intermediates and single turnover rate constants for the oxidation of heme by human heme oxygenase-1. J Biol Chem. 2000;275:5297–5307. doi: 10.1074/jbc.275.8.5297. [DOI] [PubMed] [Google Scholar]
- Maines MD. Heme oxygenase: function, multiplicity, regulatory mechanisms, and clinical applications. FASEB J. 1988;2:2557–2568. [PubMed] [Google Scholar]
- Maines MD. The heme oxygenase system: a regulator of second messenger gases. Annu Rev Pharmacol Toxicol. 1997;37:517–554. doi: 10.1146/annurev.pharmtox.37.1.517. [DOI] [PubMed] [Google Scholar]
- Migita CT, Zhang X, Yoshida T. Expression and characterization of cyanobacterium heme oxygenase, a key enzyme in the phycobilin synthesis. Properties of the heme complex of recombinant active enzyme. Eur J Biochem. 2003;270:687–698. doi: 10.1046/j.1432-1033.2003.03421.x. [DOI] [PubMed] [Google Scholar]
- Miralem T, Lerner-Marmarosh N, Gibbs PE, Tudor C, Hagen FK, Maines MD. The human biliverdin reductase-based peptide fragments and biliverdin regulate protein kinase Cδ activity: the peptides are inhibitors or substrate for the protein kinase C. J Biol Chem. 2012;287:24698–24712. doi: 10.1074/jbc.M111.326504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto H, Nakatogawa H, Ito K. Genetically encoded but nonpolypeptide prolyl-tRNA functions in the A site for SecM-mediated ribosomal stall. Mol Cell. 2006;22:545–552. doi: 10.1016/j.molcel.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Mölzer C, Pfleger B, Putz E, Roßmann A, Schwarz U, Wallner M, Bulmer AC, Wagner KH. In vitro DNA-damaging effects of intestinal and related tetrapyrroles in human cancer cells. Exp Cell Res. 2013;319:536–545. doi: 10.1016/j.yexcr.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. Secretion monitor, SecM, undergoes self-translation arrest in the cytosol. Mol Cell. 2001;7:185–192. doi: 10.1016/s1097-2765(01)00166-6. [DOI] [PubMed] [Google Scholar]
- Nemes A, Fortmann T, Poeschke S, Greve B, Prevedello D, Santacroce A, Stummer W, Senner V, Ewelt C. 5-ALA Fluorescence in Native Pituitary Adenoma Cell Lines: Resection Control and Basis for Photodynamic Therapy (PDT)? PLoS One. 2016;11:e0161364. doi: 10.1371/journal.pone.0161364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuhn P, Künzli BM, Hennig R, Mitkus T, Ramanauskas T, Nobiling R, Meuer SC, Friess H, Berberat PO. Heme oxygenase-1 and its metabolites affect pancreatic tumor growth in vivo. Mol Cancer. 2009;8:37. doi: 10.1186/1476-4598-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P. Cell biology of heme. Am J Med Sci. 1999;318:241–256. doi: 10.1097/00000441-199910000-00004. [DOI] [PubMed] [Google Scholar]
- Rodriguez EA, Tran GN, Gross LA, Crisp JL, Shu X, Lin JY, Tsien RY. A far-red fluorescent protein evolved from a cyanobacterial phycobiliprotein. Nat Methods. 2016;13:763–769. doi: 10.1038/nmeth.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumyantsev KA, Turoverov KK, Verkhusha VV. Near-infrared bioluminescent proteins for two-color multimodal imaging. Sci Rep. 2016;6:36588. doi: 10.1038/srep36588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Baba M, Nakatsuka D, Ishikawa Y, Aburatani H, Furuta K, Ishikawa T, Hatanaka H, Suzuki M, Watanabe Y. Role of heme oxygenase-1 protein in the neuroprotective effects of cyclopentenone prostaglandin derivatives under oxidative stress. Eur J Neurosci. 2003;17:2249–2255. doi: 10.1046/j.1460-9568.2003.02688.x. [DOI] [PubMed] [Google Scholar]
- Shcherbakova DM, Baloban M, Emelyanov AV, Brenowitz M, Guo P, Verkhusha VV. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun. 2016;7:12405. doi: 10.1038/ncomms12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Baloban M, Pletnev S, Malashkevich VN, Xiao H, Dauter Z, Verkhusha VV. Molecular Basis of Spectral Diversity in Near-Infrared Phytochrome-Based Fluorescent Proteins. Chem Biol. 2015a;22:1540–1551. doi: 10.1016/j.chembiol.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Baloban M, Verkhusha VV. Near-infrared fluorescent proteins engineered from bacterial phytochromes. Curr Opin Chem Biol. 2015b;27:52–63. doi: 10.1016/j.cbpa.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Shemetov AA, Kaberniuk AA, Verkhusha VV. Natural photoreceptors as a source of fluorescent proteins, biosensors, and optogenetic tools. Annu Rev Biochem. 2015c;84:519–550. doi: 10.1146/annurev-biochem-060614-034411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbakova DM, Verkhusha VV. Near-infrared fluorescent proteins for multicolor in vivo imaging. Nat Methods. 2013;10:751–754. doi: 10.1038/nmeth.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TL, Piggott BJ, Makhijani K, Yu D, Jan YN, Shu X. Rationally designed fluorogenic protease reporter visualizes spatiotemporal dynamics of apoptosis in vivo. Proc Natl Acad Sci U S A. 2015;112:3338–3343. doi: 10.1073/pnas.1502857112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran MT, Tanaka J, Hamada M, Sugiyama Y, Sakaguchi S, Nakamura M, Takahashi S, Miwa Y. In vivo image analysis using iRFP transgenic mice. Exp Anim. 2014;63:311–319. doi: 10.1538/expanim.63.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JR, Zhang J, von Stetten D, Günther M, Murgida DH, Mroginski MA, Walker JM, Forest KT, Hildebrandt P, Vierstra RD. Mutational analysis of Deinococcus radiodurans bacteriophytochrome reveals key amino acids necessary for the photochromicity and proton exchange cycle of phytochromes. J Biol Chem. 2008;283:12212–12226. doi: 10.1074/jbc.M709355200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R, Ntziachristos V. Shedding light onto live molecular targets. Nat Med. 2003;9:123–128. doi: 10.1038/nm0103-123. [DOI] [PubMed] [Google Scholar]
- Wilks A, Sun J, Loehr TM, Ortiz de Montellano PR. Heme Oxygenase His25Ala Mutant: Replacement of the Proximal Histidine Iron Ligand by Exogenous Bases Restores Catalytic Activity. J Am Chem Soc. 1995;177:2925–2926. [Google Scholar]
- Wright AV, Nuñez JK, Doudna JA. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- Yu D, Baird MA, Allen JR, Howe ES, Klassen MP, Reade A, Makhijani K, Song Y, Liu S, Murthy Z, et al. A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat Methods. 2015;12:763–765. doi: 10.1038/nmeth.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Gustafson WC, Han C, Lafaye C, Noirclerc-Savoye M, Ge WP, Thayer DA, Huang H, Kornberg TB, Royant A, et al. An improved monomeric infrared fluorescent protein for neuronal and tumour brain imaging. Nat Commun. 2014;5:3626. doi: 10.1038/ncomms4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.