Abstract
In an effort to develop a general thermodynamic model from first principles to describe mixing behavior of lipid membranes we examined lipid mixing induced by targeted binding of small (Green Fluorescent Protein (GFP)) and large (Nanolipoprotein Particles (NLPs)) structures to specific phases of phase separated lipid bilayers. Phases were targeted by incorporation of phase-partitioning iminodiacetic acid (IDA) functionalized lipids into ternary lipid mixtures consisting of DPPC, DOPC and cholesterol. GFP and NLPs, containing histidine tags, bound the IDA portion of these lipids via a metal, Cu2+, chelating mechanism. In giant unilamellar vesicles (GUVs), GFP and NLPs bound to the Lo domains of bilayers containing DPIDA, and bound to the Ld region of bilayers containing DOIDA. At sufficiently large concentrations of DPIDA or DOIDA, lipid mixing was induced by bound GFP and NLPs. The validity of the thermodynamic model was confirmed when it was found that the statistical mixing distribution as a function of crowding energy for smaller GFP and larger NLPs collapsed to the same trend line for each GUV composition. Moreover, results of this analysis show that the free energy of mixing for a ternary lipid bilayer consisting of DOPC, DPPC, and cholesterol varied from 7.9×10−22 to 1.5×10−20 Joules/lipid at the compositions observed, decreasing as the relative cholesterol concentration was increased. It was discovered that there appears to be a maximum packing density, and associated maximum crowding pressure, of the NLPs, suggestive of circular packing. A similarity in mixing induced by NLP1 and NLP3 despite large difference in projected areas was analytically consistent with monovalent (one histidine) vs. divalent (two histidine) surface interactions, respectively. In addition to GUVs, binding and induced mixing behavior of NLPs was also observed on planar, supported lipid multibilayers. The mixing process was reversible, with Lo domains reappearing after addition of EDTA for NLP removal.
Graphical Abstract
INTRODUCTION
Phospholipids self-assemble in aqueous environments to form bilayers that are capable of existing in either liquid or solid phases.1–4 The type of phase formed depends on various properties and conditions, such as head group structure, length/degrees of unsaturation of the acyl tails, and temperature.4–5 The addition of cholesterol to a binary liquid-solid phospholipid mixture can result in a tertiary mixture consisting of two immiscible, coexisting liquid phases. Specifically, liquid-ordered (Lo) and liquid-disordered (Ld) phases are formed.1–4 These coexisting phases have distinct compositions and are able to selectively partition fluorescent probes and functionalized lipids.6–7 The selective partitioning of fluorescent probes allows for identification of phases via fluorescence microscopy, while functionalized lipids allow for phase-specific protein targeting. Phase-specific adsorption and binding of proteins has been of interest for the development of biomaterials for various applications, such as small-scale fluidic devices, biosensors, and high-density arrays.8–12
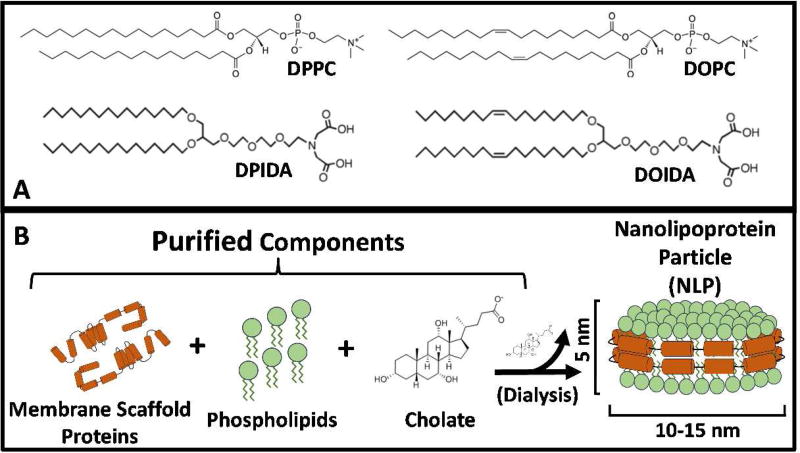
Various mechanisms for attachment of proteins to functionalized head groups have been developed, such as disulfide coupling of thiols,13 linkages via single-stranded DNA,14 tethering via glycan-phosphatidyl inositol,15 and biotinylation.16 In addition to these, metal chelation can also serve as a method for effectively adsorbing proteins to lipid bilayers.17 In this system, the lipid head group can be functionalized with either nitrilotriacetic acid (NTA) or iminodiacetic acid (IDA) groups. These groups coordinate divalent transition metal ions such as Zn2+, Ni2+, and Cu2+ through four coordination sites, leaving two sites exposed to bind to poly-histidine tags that are covalently attached to a protein of interest.18 IDA membranes are more favorable for switchable surfaces than NTA membranes, as NTA binds much more tightly to histidine tags; IDA requires a 1000-fold lower EDTA concentration than NTA to remove bound proteins.17, 19–20 The two functionalized lipids used in this study, dipalmitoyl iminodiacetic acid (DPIDA) and dioleoyl iminodiacetic acid DOIDA, have been shown to partition into dipalmitoyl phosphocholine(DPPC)-rich Lo and dioleoyl phosphocholine DOPC-rich Ld lipid bilayer phases, respectively21 (structures shown in Fig. 1A). This is due in part because of similarity of tail structure of DPIDA to DPPC (a gel phase lipid) and DOIDA to DOPC (a liquid-crystalline phase lipid). In the presence of CuCl2, the phase-targeted binding of proteins to DPIDA and DOIDA, as well as other IDA-functionalized lipids has been observed extensively by Sasaki and coworkers.20–23 Recombinant histidine-tagged Green Fluorescent Protein (His-GFP) was particularly convenient for identifying targeted binding through the appearance of green fluorescence associated with the Lo phase in Lo-Ld phase separated giant unilamellar vesicles (GUVs). Significant changes to membrane shape and morphology of GUVs, such as membrane bending and tubule formation, was observed upon various proteins binding to Lo domains.21–23 This behavior was attributed to the steric confinement of the bound proteins in combination with highly localized binding which resulted in the transformation from flat domains to highly curved structures. This phenomenon was most frequent when the lipid diphytanoyl phosphocholine (DPhPC) was incorporated into the membrane.
Figure 1.
(A) Chemical structures for phospholipids and IDA-lipids. (B) Schematic of nanolipoprotein particle (NLP) synthesis and its structure.
Scheve et al. examined protein binding in the absence of DPhPC and focused on Lo-Ld phase separated GUVs composed primarily of the ternary mixture DPPC, DOPC, and cholesterol.24 DPIDA, which partitions into the Lo phase, was included in the lipid mixture used to make the GUVs, in order to achieve targeted binding of His-GFP to the Lo phase. Instead of membrane deformation and tubule formation, they reported mixing of the Lo and Ld phases. This phenomenon was attributed to sufficiently large steric pressure being exerted by the localized protein binding in Lo domains. This behavior was observed for several different proteins ranging in size from 8 – 180 kDa. There is an inherent free energy of mixing associated with phase separated lipid bilayers.25 Therefore, Scheve et al. used a thermodynamic approach to model the system by comparing the enthalpy of mixing between the immiscible phases to the steric interactions exerted by bound proteins. Though accurate for their data, the approach and methodology was somewhat empirical and specific to their system.
In this present work, we expand upon the work of Scheve et al. to develop a thermodynamic model from first principles that not only describes mixing behavior, but can be used for a wide variety of similar systems with minimal modification. From this model, free energies of mixing can be determined for a given lipid mixture composition. Steric pressure-induced mixing of Lo and Ld phases in GUVs is observed by fluorescence microscopy after the phase-targeted binding of recombinant histidine-tagged GFP and significantly larger histidine-tagged nanolipoprotein particles (NLPs).
NLPs are disc-shaped patches of lipid bilayer that have two amphiphilic membrane scaffold proteins (MSPs) surrounding the outer periphery, thus making the entire particle water soluble (structure shown in Fig. 1B). The length of the MSP controls the diameter of the NLP.26–27 Recombinant histidine-tagged MSPs of various length were previously developed by Sligar et al. and are commercially available.28–29 Two different types of recombinant histidine-tagged MSP (MSP1 and MSP3) were used to synthesize two different populations of His-NLPs, each incorporating Oregon Green-DHPE lipid for visualization. Use of MSP1 resulted in a His-NLP population with an average diameter of 9 nm (NLP1), while MSP3 resulted in a His-NLP population with an average diameter of 14 nm (NLP3).30 We investigate His-NLPs to test the size-dependence of steric-pressure induced mixing in our model since His-NLPs are approximately 10X larger in cross-sectional area in comparison to His-GFP. In addition, NLPs are able to solubilize integral membrane proteins (IMPs).31–32 Though IMPs are not used in this work, the proposed architecture provides foundation for a methodology of IMP adsorption that was previously not feasible.
Our results indicate that His-NLPs are capable of selectively binding to either Lo or Ld regions of lipid bilayers, inducing mixing of the two phases, and refraining from rapid lipid exchange/fusion to the bilayer surface. Binding was observed for both giant unilamellar vesicles and supported lipid multibilayers on mica. In addition to this, our thermodynamic model shows that the free energy of mixing for lipid bilayers can be determined for various compositions, and that the values obtained follow a rationalized trend. Lastly, we report a newly observed phenomenon, where reversibility of pressure induced mixing was observed.
THERMODYNAMIC MODEL OF STERIC PRESSURE-INDUCED MIXING
The bimodal distribution between the two states of a GUV lipid bilayer (mixed and unmixed) can be modelled with a Boltzmann distribution33 determined by the free energy contributions from mixing, ΔFmix, and steric pressure, ΔFp as shown in Equation 1. Nmix and Nunmix correspond to the number of GUVs in mixed and unmixed states, respectively, while kB is the Boltzmann constant, and T is temperature.
| (1) |
The differential free energy (F) for a reversible process subjected to pressure-volume (P-V) work (W) at constant temperature is given by Equation 2. A more detailed derivation can be found in the Supporting Information.
| (2) |
Vc corresponds to the volume occupied by protein, or crowding agent, bound to the surface of the vesicle. Upon mixing, the area occupied by phase-targeted protein (Ac) expands in 2-dimensions along the surface of a giant vesicle. Therefore, Equation 2 can be rewritten in terms of a differential area multiplied by height (h) of the protein as shown in Equation 3.
| (3) |
Integration of Equation 3 results in Equation 4, which is the total contribution to free energy from steric pressure (ΔFp).
| (4) |
Af and Ai correspond to the final and initial areas occupied by the bound layer of protein, respectively. The Carnahan-Starling equation of state is an appropriate equation of state to use for this system. It accurately describes the pressure generated by particles packed within a finite space and has been previously used for calculating steric pressures generated by protein crowding.23–24, 34 The equation of state is shown in Equation 5.
| (5) |
N represents the number of bound protein molecules or NLPs. The variable η represents the fractional surface coverage and is shown in Equation 6.
| (6) |
Ap corresponds to the projected area of a protein or NLP. By rearranging Equations 5 and 6, and appropriately substituting them into Equation 4, Equation 7 is obtained.
| (7) |
In the integral limits, ηi represents the fractional coverage of domains (i.e. an unmixed state), while ηf represents the fractional coverage of the entire GUV (i.e. when the vesicle becomes mixed). Details for how these values were determined are outlined in the Supporting Information. By plotting experimentally measured values of Nmix/Nunmix vs calculated values of ΔFp, a value of ΔFmix can be found by regressing Equation 1 such that the error between the experimental data and a theoretical curve is minimized. The Boltzmann distribution can utilize energy on a “per lipid” basis (Joules/lipid) or a “per vesicle” basis (Joules/vesicle) when determining ΔFmix. Both types will be used in our analysis.
For a “per lipid” basis, the free energy terms must be divided by the total number of lipids (NL) in a vesicle. This corresponds to the average free energy per lipid, which is comparable in magnitude to kBT. This modification to Equation 1 is shown in Equation 8. This model forces an exponential fit through the origin (i.e. 0% mixed vesicles) when ΔFmix is sufficiently large. However, due to compositional differences among vesicles,35 actual systems will not truly exhibit this 0% mixed vesicle phenomena and Nmix/Nunmix will always have an experimentally measured value greater than 0. This results in a slight underestimation by this theoretical model at low values of ΔFp.
| (8) |
For the “per vesicle” basis, the ratio of free energy to kT can be as large as 10 orders of magnitude. This causes the exponential term to diverge. Therefore, Equation 9 is used to find ΔFmix in terms of Joules/vesicle. As with Equation 8, Equation 9 also results in underestimation of regression values at low ΔFp.
| (9) |
Equation 9 is the linearized version of Equation 1. Plotting the left-hand side against ΔFp can be fit to a line with a slope of (−1). The intercept of this line corresponds to − ΔFmix.
According to Equation 7, the steric pressure contribution to free energy diverges to infinity as η approaches 1. This would correspond to Nmix/Nunmix also diverging to infinity in Equations 8 and 9. However, an infinite steric pressure can be avoided if (Nmix/Nunmix) approaches a finite, asymptotic value due to the presence of a maximal packing density (i.e. ηmax), as observed in phenomena such as circular packing36. Therefore, data points with theoretical η values higher than ηmax should not be used for determining ΔFmix,.
MATERIALS AND METHODS
Materials
Lyophilized Green Fluorescent Protein (GFP) containing a histidine-tag was purchased from Thermo Fisher Scientific. Lyophilized Membrane Scaffold Proteins (MSPs) used in the synthesis of NLPs also each contained a histidine-tag. Two different types of MSP were purchased; MSP1 (sold as his-MSP1D1, 217 amino acids, 25.3 kDa) was purchased from Cube Biotech, Inc. and MSP3 (sold as his-MSP1E3D1, 277 amino acids, 32.6 kDa) was purchased from Sigma-Aldrich, Inc. Imidazole (≥ 99%), copper (II) chloride (≥ 99%), sodium chloride (≥ 99%), methanol (≥ 99%), and sodium cholate (≥ 99%) were also purchased from Sigma-Aldrich, Inc. Chloroform and hexane were purchased from Fisher Scientific International, Inc. DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), and cholesterol were purchased from Avanti Polar Lipids, Inc. Texas Red® DHPE (Texas Red® 1,2,-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) and Oregon Green® 488 DHPE (Oregon Green® 488 1,2,-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) were purchased in a lyophilized state from Thermo Fisher Scientific. DPIDA (1,2-dipalmitoyl-sn-glycero-triethyleneoxy-iminodiacetic acid)21 and DOIDA (1,2-dioleoyl-sn-glycero-3-triethylenoxy-iminodiacetic acid)37 were synthesized according to previously reported protocols. Ni-NTA agarose was purchased from 5 PRIME, Inc. Tris(hydroxymethyl)aminomethane (MB Grade) and hydrochloric acid (12.1 N) were purchased from USB Corporation and Fisher Scientific International, Inc., respectively. All water used in these experiments was purified using a Barnstead Nanopure System (Barnstead Thermolyne, Dubuque, IA) with a resistivity of 17.9 MΩ•cm or greater.
Preparation of GFP and NLPs
GFP was dissolved in water at a concentration of 0.1 mg/mL, aliquoted into small centrifuge tubes, and frozen at −20°C. Individual aliquots were thawed immediately prior to binding experiments. NLPs were synthesized as previously described,30, 38 with appropriate stoichiometric modifications for use with DOPC. Briefly, DOPC was doped with trace amounts of Oregon Green® DHPE (0.1 mol%) in chloroform, dried using nitrogen gas, then subjected to further drying under vacuum for 2 hours. The lipid film was rehydrated in Tris Buffer (20 mM Tris, 100 mM NaCl, pH 7.4) containing 40 mM sodium cholate and combined with either MSP1D1 or MSP1E3D1 in lipid:protein molar ratios of 60:1 or 120:1, respectively. The mixture was then dialyzed over the course of 2 days to remove cholate and promote NLP synthesis. The final product was purified using Ni-NTA resin and eluted using 400 mM imidazole. The eluate was then dialyzed for a 24 hour period to remove imidazole and the NLPs subsequently stored in Tris Buffer at 4°C.
Lipid Multibilayer and GUV Binding and Imaging Experiments
GUVs were synthesized using a standard electroformation method described in the Supporting Information. For binding experiments, a well consisting of polylactic acid (PLA) rectangular walls on a microscope slide was used to contain each sample. Chamber walls with dimensions of 1.5 cm × 1.0 cm and a depth of 0.2 cm were prepared using a 3D printer. To each well, the following was added: 300 µL of Tris Buffer (20 mM Tris, 100 mM NaCl, pH 7.4), 2.5 µL of 16 mM CuCl2 (in water), and 20 µL of GFP or NLP solution. GFP and NLP stock solutions were prepared such that their final concentrations were approximately 3.8 µM. After mixing the contents of the wells, 18 uL of GUVs in sucrose was added to the well. Samples were allowed to set for 10 minutes to allow sufficient time for GUV settling and protein binding.
Planar lipid multibilayers (MBLs) were prepared by a standard spin-coating technique described in the Supporting Information. For NLP binding, 30 µL of 16 mM CuCl2 was added to petri dish. Afterward, NLP stock solution was added to reach a final concentration of 0.2 µM. To remove bound NLPs, 16 uL of 0.5 M EDTA was added to the petri dish. All mixing was done using a 1 mL pipette and in a manner such that the region in the immediate vicinity of the supported MBLs was minimally disturbed.
Imaging was performed using a 60X water immersion lens on a Nikon TE400 fluorescence microscope. The filter set used was comprised of FITC and Texas Red filters (Chroma Technology, Bellows Falls, VT). FITC filters were used to visualize GFP and Oregon Green-DHPE-containing NLPs, while Texas Red Filters were used to visualize Texas Red containing GUVs. With the concentrations of dye and protein used, no visual overlap was observed between the two filters. Simultaneous use of the filters allowed for visualization of domain formation and protein binding. For determining domain percentages in GUVs, 90–200 vesicles were counted during each of the triplicate trials.
RESULTS
Steric Pressure-induced Mixing by Binding to Lo Phase in GUVs
We investigated the binding of GFP and two types of NLPs (NLP1 and NLP3) to the liquid ordered (Lo) regions of GUVs. GUVs were composed of a ternary mixture containing DOPC, DPPC, and cholesterol. DPIDA was incorporated for Lo phase targeting of histidine-tagged GFP and NLPs in the presence of CuCl2. The DOPC:(DPPC+DPIDA) molar ratio was held constant at 3:2, while the cholesterol content was varied from 18 mol% to 30 mol%. The diameters of NLP1 and NLP3 were determined by dynamic light scattering to be 9.0±1.4 nm and 14.2±1.3 nm, respectively (see Supporting Information). The doping of GUVs with 0.1 mol% Texas-Red DHPE – a lipid that partitions into the Ld phase - allowed for visualization of phase separation of the GUVs by fluorescence microscopy. Higher cholesterol content (32 mol% and greater) resulted in no visible phase separation, indicating a critical/mixing point was present for this composition between 30 and 32 mol% cholesterol at 22°C. For compositions used in our experiments, DPIDA was found to have minimal influence on the phase behavior of a true DOPC/DPPC/cholesterol ternary system due to its very similar features to DPPC (i.e. carbon tail structure and head group size) – see the Supporting Information.
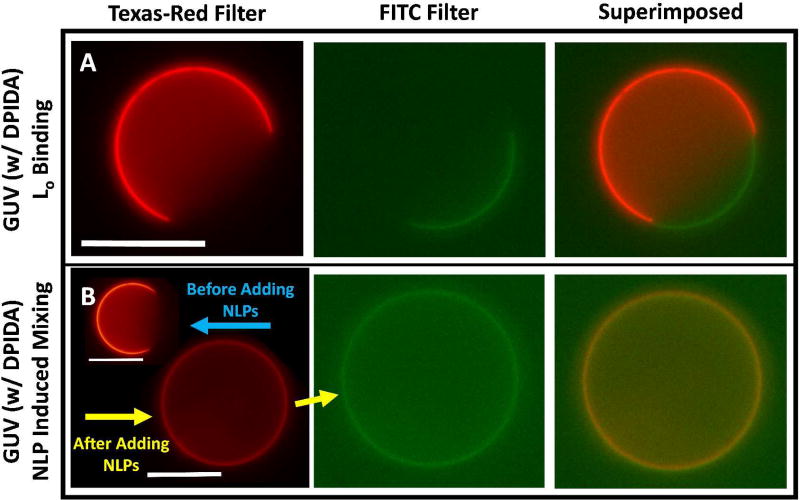
Texas-Red and FITC filters were used to visualize the Ld regions of GUVs and bound GFP or Oregon Green-DHPE (OG-DHPE)-containing NLPs, respectively. Fig. 2A shows a representative phase-separated GUV containing 2 mole % DPIDA with NLP1 bound to the Lo region. Mixing was not induced as indicated by the enhanced signal observed in the FITC filter overlapping with the missing signal from the Texas-Red filter. Fig. 2B shows a GUV containing 10 mole % DPIDA that underwent mixing due to the crowding effect of bound NLP1. This is confirmed by the complete overlap of signal from both filters. These phenomena were also observed for bound GFP and NLP3. In the absence of the chelating agent, Cu2+, there was no binding to the GUVs – see Supporting Information.
Figure 2.
(A) A representative GUV in the presence of NLP1 exhibiting targeted binding to the Lo domain. GUVs in this sample were synthesized with a lipid composition consisting of 50% DOPC, 30% DPPC, 2% DPIDA, and 18% cholesterol. (B) A representative GUV before and after NLP1 binding exhibiting crowding induced mixing. GUVs in this sample were synthesized with a lipid composition consisting of 50% DOPC, 22% DPPC, 10% DPIDA, and 18% cholesterol. Scale bars are 20 µm.
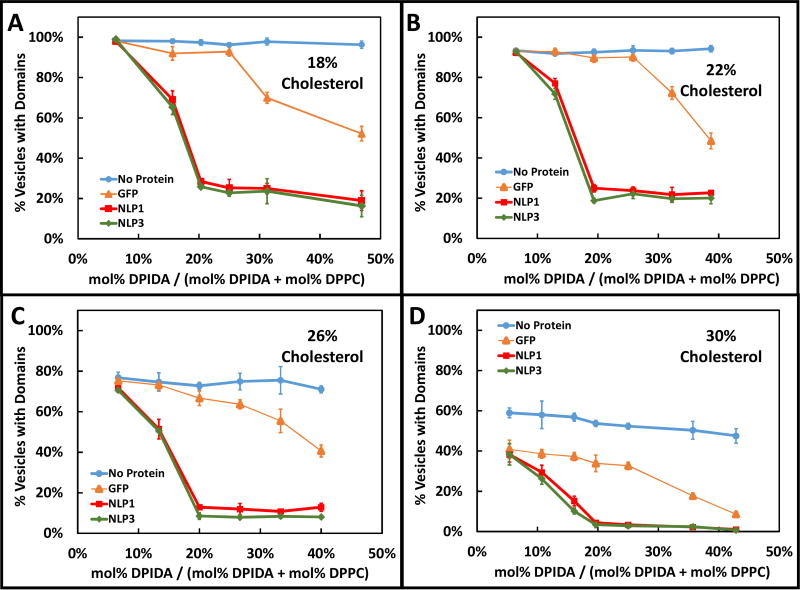
Variation of the DPIDA percentage in the domains and binding to smaller GFP or larger NLPs allowed for control of the steric pressure contribution to domain mixing. Variation of the cholesterol content controlled the mixing energy contribution. The percentage of GUVs displaying phase-separated Lo domains in the presence (or absence) of GFP, NLP1, or NLP3, as DPIDA molar percentage in the domains was varied is shown in Fig. 3. Cholesterol content was varied from 18 mol% (Fig. 3A), 22 mol% (Fig. 3B), 26 mol% (Fig. 3C) to 30 mol% (Fig. 3D). In the absence of protein, less than 100% of the vesicles exhibited the presence of visible domains over the range of DPIDA concentrations. With the addition of GFP, the percentage of GUVs displaying domains decreased as the relative DPIDA content increased. The magnitude of the decrease became significant when the DPIDA concentration reached 25–30%. When NLP1 or NLP3 were used, the percentage of GUVs with visible domains decreased significantly when the DPIDA concentration was 6–16%. The decrease appeared to reach a plateau once the DPIDA concentration reached 20% for cholesterol contents below 30%. As the cholesterol content was increased, the percentage of vesicles with domains, over all experimental conditions, shifted to lower values.
Figure 3.
The percentage of GUVs with visible domains observed in the absence of protein as well as the presence of GFP, NLP1, or NLP3, for various levels of DPIDA and cholesterol content. DPIDA content is represented as relative doping concentration with DPPC. The DOPC:(DPPC+DPIDA) ratio was held constant at 3:2. Vertical error bars represent the standard deviation of triplicate measurements.
For a more detailed description of Fig. 3 and the method of obtaining mixing statistics see the Supplemental Information.
Curve Fitting and Determination of ΔFmix
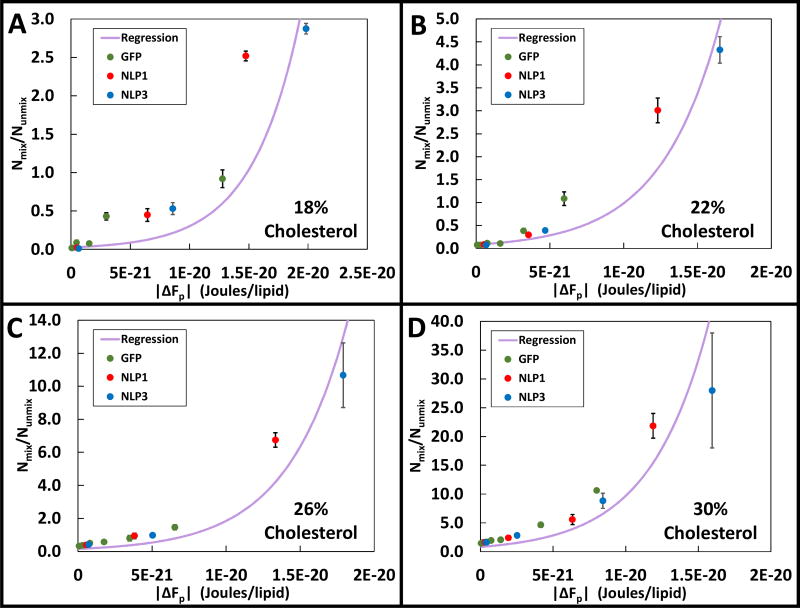
To determine ΔFmix, Equation 8 was used to regress, by method of weighted least squares, experimentally observed GUV populations plotted against calculated steric pressure contributes to free energy (ΔFp) as shown in Figure 4. In our ΔFp calculations, we assume that the projected area of GFP, which had a single histidine tag, was 10 nm2. We uniformly assumed 2.5 DPIDAs were bound per tag as in the work of Stachowiak.24 Projected areas for NLP1 and NLP3 ranged from ~45–65 nm2 and ~75–150 nm2, respectively, depending on their orientation after binding (i.e. upright rectangular or flat circular relative to the bilayer surface). Also, each NLP contained two histidine tags. Whether one or both histidine tags were able bind to DPIDA was unclear. Thus for each NLP, there were four general binding possibilities: flat or upright, with one or two tags attached. Based on the similarities in crowding behavior exhibited by NLP1 and NLP3 in Fig. 3, we used in the thermodynamic model an area of 65 nm2 with one bound tag for NLP1 and two energetically equivalent cases for NLP3 – area of 75 nm2 with one bound tag and an area of 150 nm2 with two bound tags (see Supporting Information). Fig. 4 shows regressed plots using Equation 8 with cholesterol content increasing from 18% to 30%. In all of these plots, the experimental data was regressed against GFP, NLP1, and NLP3 samples simultaneously. Experimental data points within the “plateau” region (see Fig. 3) were not used. The limiting surface coverage (ηmax) was determined from onset of the plateau regions to be 0.92 ± 0.01 for NLP1 and 0.94 ± 0.01 for NLP3.
Figure 4.
The regression curves used to determine ΔFmix from Equation 8 for various concentrations of cholesterol. The DOPC:(DPPC+DPIDA) ratio was held constant at 3:2. Vertical error bars represent the standard deviation of triplicate measurments.
As shown in Figure 4A, for GUVs containing 18 mol% cholesterol as ΔFp was increased from 7.7×10−23 to 1.9×10−20 Joules/lipid, the ratio of mixed to unmixed GUVs exponentially increased from 0.01 to 2.9. The regression curve fit to this data yielded a ΔFmix value of (1.5 ± 0.4) × 10−20 Joules/lipid, as shown in Table 1. In Figs. 4B, 4C, and 4D cholesterol contents of 22, 26, and 30 mol% were used, respectively. In these figures, ΔFp was increased from 8.0×10−23 - 1.6×10−20, 8.3×10−23 - 1.7×10−20, and 5.7×10−23 - 1.5×10−20 Joules/lipid, respectively. The ratio of mixed to unmixed vesicles increased from 0.08–4.3, 0.3–10.7, and 1.4–27.9, respectively, in exponential patterns. The regression curve fit to these samples yielded values for ΔFmix of (1.0 ± 0.2) × 10−20, (7.5± 1.8) × 10−21, and (7.9 ± 5.4) × 10−22 Joules/lipid, respectively, as shown in Table 1. Use of equation 9 to evaluate the data in Fig. 4 yielded comparable values for ΔFmix but with significantly larger error bars (see Table 1 and Supporting Information).
Table 1.
Values of ΔFmix for various cholesterol concentrations. The DOPC:(DPPC+DPIDA) ratio was held constant at 3:2.
| Cholesterol Concentration (mol%) |
ΔFmix (from Eqn 8) (Joules/Lipid) |
ΔFmix (from Eqn 9) (Joules/Lipid) |
ΔFmix (from Eqn 9) (Joules/Vesicle) |
|---|---|---|---|
| 18% | (1.5 ± 0.4) × 10−20 | (1.3 ± 0.5) × 10−20 | (3.5 ± 1.5) × 10−11 |
| 22% | (1.0 ± 0.2) × 10−20 | (8.6 ± 5.6) × 10−21 | (2.4 ± 1.5) × 10−11 |
| 26% | (7.5 ± 1.8) × 10−21 | (3.4 ± 2.4) × 10−21 | (9.3 ± 6.5) × 10−12 |
| 30% | (7.9 ± 5.4) × 10−22 | (8.9 ± 8.8) × 10−22 | (2.5 ± 2.4) × 10−12 |
Steric pressure-induced Mixing by Binding to Ld Phase in GUVs
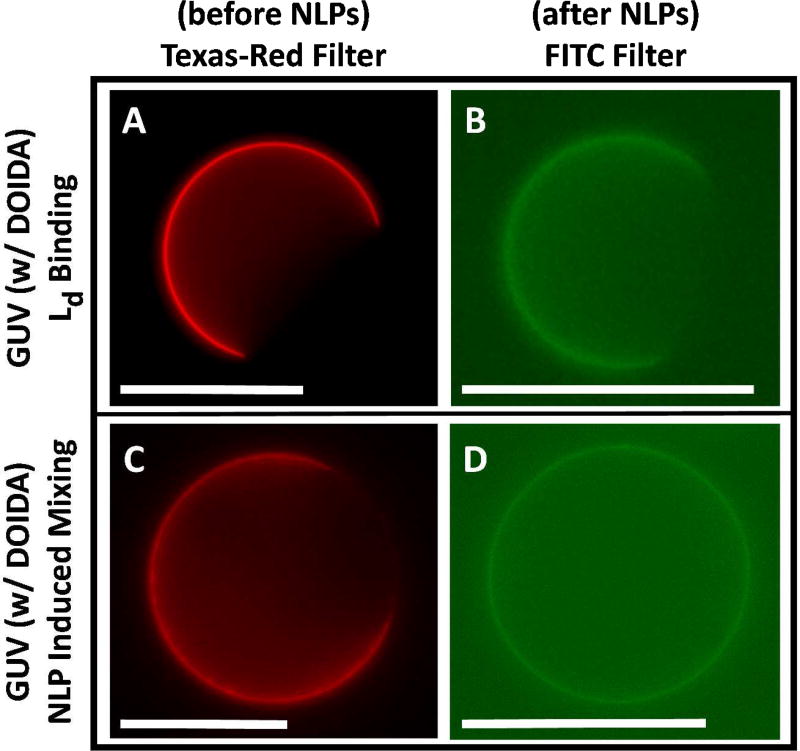
For investigation of targeted binding of GFP, NLP1, and NLP3 to the liquid-disordered (Ld) region of GUVs, DOIDA was used. DOIDA was incorporated into the DOPC/DPPC/cholesterol mixtures, as DOIDA will partition primarily into the Ld phase. To prevent significant differences from the phase behavior of a true DOPC/DPPC/cholesterol system, DOIDA concentrations in GUVs were limited to 40 mol%. Fig. 5A shows a representative phase-separated vesicle synthesized using 0.1 mol% Texas-Red DHPE and 5% DOIDA (mol% DOIDA/[mol% DOIDA + mol% DOPC]) in the Ld region. Fig. 5B shows another vesicle synthesized with an identical composition minus the use of Texas-Red DHPE, and then exposed to NLP1. The Oregon Green fluorescence from NLP1 covering a similar area of the vesicle surface, suggests that NLP1 bound to the Ld phase but did not induce mixing. Fig. 5C illustrates a phase-separated GUV prepared with 0.1% Texas-Red DHPE and 40% DOIDA in the Ld region. Fig. 5D shows a GUV with the same composition (minus Texas-Red DHPE) and then exposed to NLP1. Oregon Green fluorescence constitute all of the vesicle surface indicating NLP1 bound to the Ld phase and induced mixing. We gathered mixing statistics in 50% (DOPC+DOIDA), 32% DPPC, and 18% cholesterol GUVs in the absence of protein, as well as in the presence of GFP, NLP1, and NLP3, changing the DOIDA content from 5% to 40% (see Supporting Information for a histogram). With no added protein, the percentage of vesicles with Lo domains such as in Fig. 5A changed from (95 ± 3)% to (97 ± 1)% respectively. In the presence of GFP, the GUVs displaying dark Lo domains such as in Fig. 5B changed (94 ± 3)% to (50 ± 2)% respectfully. NLP1, and NLP3 once again induced virtually identical mixing (95 ± 2)% to (20 ± 3)%, and (92 ± 3%) to (18 ± 2)%, respectively. Very similar mixing behavior was observed for GUV samples incubated with NLP1 and NLP3 absent of (OG-DHPE), which showed that Texas Red-DHPE homogeneously distributed throughout the GUV membrane. Note that GFP and NLP binding could not be observed for Ld targeted vesicles whenever Texas-Red DHPE was present in the Ld phase which we believe is due to electrostatic repulsion (See Supporting Information).
Figure 5.
Representative 50% (DOPC+DOIDA), 32% DPPC, 18% cholesterol GUVs. (A) GUV with 5% DOIDA/(DOPC+DOIDA) either containing 0.1% Texas Red-DHPE displaying a red Ld domain or (B) after addition of OG-DHPE NLP1 displaying a green domain resulting from binding to an Ld region. (C) GUV with 40% DOIDA/(DOPC+DOIDA) either containing 0.1% Texas Red-DHPE displaying a red Ld domain or (D) after addition of OG-DHPE NLP1 displaying uniform green fluorescence resulting from mixing. Scale bars are 20 µm.
Reversibility of Steric Pressure-induced Mixing Demonstrated in Lipid Multibilayers
Binding of GFP, NLP1, and NLP3 to supported lipid multibilayers (MBLs) on mica was investigated. We utilized Lo domain targeting for these experiments. In Fig. 6, binding of NLP1 to Lo domains was observed. The molar composition of the MBL was 50% DOPC, 30% DPPC, 2% DPIDA and 18% cholesterol. In the Texas-Red filter channel, the uppermost layer of the MBL was observed to contain circular dark Lo domains with diameters on the order of 1–20 µm. Binding to the Lo domains was observed in the FITC filter channel, as indicated by the presence of strong fluorescence signal in corresponding areas that emitted little or no signal in the Texas-Red channel as shown in Fig. 6.
Figure 6.
Targeting binding of OG-DHPE-containing NLP1 to a lipid multibilayer (MBL) on mica. The MBL was made using a lipid composition consisting of 50% DOPC, 30% DPPC, 2% DPIDA, 18% cholesterol. The scale bar represents a length of 50 µm.
In Fig. 7, a lipid composition of 50% DOPC, 20% DPPC, 12% DPIDA and 18% cholesterol was used. In comparison to Fig. 6, this higher DPIDA content corresponds to a much higher number of binding sites in the Lo domains. In Fig. 7A, induced mixing is observed once NLP1 is bound. The mixing is indicated by the Lo domains’ eventual disappearance over the course of 10 minutes. Reversibility can easily be assessed using MBLs since they remain associated with the surface during exchange of buffers. Hence, 2 mM EDTA was added to the sample to remove DPIDA-bound Cu2+. Within 1 minute, the reappearance of Lo domains was observed, as illustrated in Fig. 7B. Over the course of at least 15 minutes, the domains continued to coalesce and grow. This phenomena was also observed for NLP3. No significant trace of Oregon Green-DHPE in the MBL was present (See Supporting Information), which strongly suggests that NLPs prepared with DOPC and MSPs do not spontaneously fuse to lipid bilayers or exchange lipids over typical experimental time scales. Instead they behaved as stable molecular assemblies.
Figure 7.
(A) Time lapse of NLP1 induced lipid mixing in multibilayer (MBL) on mica. (B) The removal of bound NLP1 using 2 mM EDTA and reappearance of Lo domains over time. The lipid composition used was 50% DOPC, 20% DPPC, 12% DPIDA, and 18% cholesterol. The scale bar represents a length of 50µm.
DISCUSSION
Reversibility, Size, and Packing Effects in Steric Pressure-induced Mixing
Histidine tagged green fluorescent protein and differently-sized discoidal nanolipoprotein particles (NLPs) were all observed to bind to the Lo region of phase separated DOPC/DPPC/cholesterol GUV bilayers through a metal chelate interaction when DPIDA was incorporated. At sufficiently high concentrations of DPIDA, induced mixing of phases was observed for GUVs incubated with GFP, NLP1, and NLP3. Moreover, phase-separation was quickly restored in a lipid multibilayer platform by removal of bound proteins using EDTA. This is the first report of reversibility in crowding-induced lipid phase mixing, demonstrating the usefulness of the MBL-IDA platform and its potential for developing reusable biosensors that exploit changes in mixing behavior.
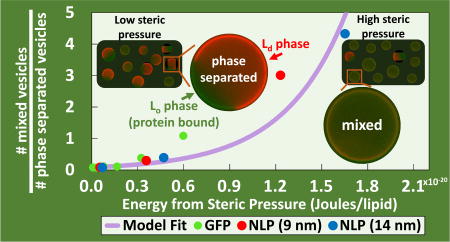
In GUV experiments, NLPs were found to be more effective at inducing mixing than GFP due to their significantly larger size. This trend in size dependent crowding behavior was previously observed with proteins of varying size but has never been observed using large molecular assemblies such as NLPs.24 This trend inherently makes sense, as larger particles generally occupy more space per binding site, which results in larger particles exhibiting larger steric pressure at a given concentration. It would have been expected for NLP3 of diameter approximately 14 nm to generate significantly more steric pressure than NLP1 of diameter approximately 9 nm, assuming flat orientations with circular projected areas for both particles. However, NLP3 and NLP1 were very similar in crowding behavior. For crowding efficacy, the ratio of the projected area to the number of bound histidine-tags is most important; the projected area and bound histidine-tags are directly coupled when calculating surface coverage (η) (See Supporting Information). As long as this ratio remains constant, it is expected that different particles (i.e. NLP1 and NLP3) will behave almost identically, despite a difference in size. Therefore, similar steric pressures can be obtained in flat orientations if both histidine tags (i.e. divalent) on NLP3 bind to surface IDA groups while only one histidine tag (monovalent) on NLP1 binds to surface IDA groups. Alternatively, similar steric pressures can be obtained if both bind via one histidine tag but assume different orientations that lead to the same projected area, an upright orientation of projected rectangular shape of NLP3 and a flat orientation of projected circular shape for NLP1. In order to distinguish the most likely scenario for the orientation and valency of NLP1 and NLP3, we turn to the limiting surface coverage (ηmax) which is related to the shape of the projected area of the crowding agent.
After addition of NLPs to GUV samples, the percentage of vesicles containing Lo domains did not decrease to zero with increasing DPIDA, but reached plateaus, with each plateau steadily decreasing as cholesterol mol% increased. It was not until 30 mol% cholesterol, when the free energy of mixing approaches zero, that GUVs displaying domains reached a plateau close to zero (1%). This plateau behavior could be attributed to a limiting surface coverage associated with bound particles. As shown in equation 8, the population distribution exhibits an exponential dependence. In order to reach 0% domains (i.e. Nmixed/Nunmixed approaches ∞), the ΔFp term would have to diverge towards infinity. This would require η (Eqn. 6 and 7), the fractional surface coverage of particles, to approach unity. However, there is a maximal packing limit associated with certain shapes. We found that these maximal limits (ηmax) occurred around 0.92 for NLP1 and 0.94 for NLP3, corresponding closely to hexagonally packed circles where ηmax is 0.9069.39 Therefore, both NLP1 and NLP3 likely have a flat orientation of circular projected area relative to the bilayer rather than an upright orientation of rectangular projected area in which a value of η=1 is attainable.36 The similarity in mixing behavior despite the large difference in projected circular areas indicates that NLP1 and NLP3 bind the surface via monovalent and divalent interactions respectively. This difference in valency could be related to structural differences in MSP1 and MSP3. For example, the N-terminus of MSP1 includes a flexible, loosely-membrane-associated, region40 where the histidine tag is located, that may form a copper-histidine complex with histidines on a neighboring MSP1, rather than with surface IDA lipids. This flexible region has been truncated in the case of MSP3 - the histidine tag is located in the membrane-associated region of the N-terminus.
Free Energy of Mixing in DOPC/DPPC/cholesterol Ternary Mixtures
The free energy of mixing (ΔFmix) determines whether or not a multi-component system will phase separate or undergo mixing. Spontaneous mixing will occur at values of ΔFmix < 0. We have proposed and used a methodology for experimentally determining ΔFmix in lipid bilayers by thermodynamic analysis of a statistical distribution of steric pressure-induced mixing. Free energies of mixing for DOPC/(DPPC+DPIDA)/cholesterol bilayers were determined at various lipid compositions through targeting GFP and NLP to the Lo phase. Publications providing numerical values for ΔFmix in lipid bilayers are limited. By utilizing temperature integration of heat capacity measurements in phase-separated mixtures through transition points, values for energy of mixing have been estimated to be on the order of 4.8 mJ/m2.24, 41–42 Assuming the average area of a lipid head group as 60 Å2,43 this energy of mixing converts to roughly 3×10−21 J/lipid. Interaction energies between individual lipids can also be used for order of magnitude approximations of mixing energy. These interaction energies can fluctuate on the order of −300 to +300 cal/mol for phosphocholine lipids in the presence of cholesterol.25, 44–45 By treating a bilayer as a two-dimensional, hexagonally-packed lattice,25, 46 an order of magnitude calculation can be utilized to demonstrate that the energy of mixing can range anywhere between 0 to 3×10−20 J/lipid (0 to 3600 cal/mol). Theoretical ΔFmix values can also be determined by use of Flory-Huggins solution theory and treating the Lo and Ld phases as two separate fluids mixing in a lattice space (see Supporting Information). All of these methods give a range of values for energy of mixing that are consistent with our experimentally determined values of ΔFmix, which vary from 5.0×10−23 to 1.3×10−20 J/lipid. In addition, the observation that that all statistical distributions for GFP, NLP1, and NLP3 collapse to the same regressed line for a particular cholesterol content demonstrates the validity of our statistical model for steric-pressure induced mixing.
The decreasing values for ΔFmix as cholesterol content is increased (at a constant DOPC:[DPPC+DPIDA] ratio of 3:2) is consistent with a changing lipid composition approaching a critical point. Beyond the critical point, phase separation no longer occurs,4 thus ΔFmix should approach zero at the critical point. Although the values for ΔFmix obtained by regressing Equations 8 and 9 are in general agreement, the errors associated with values derived from Equation 9 are substantially larger. This was due to the fact that ΔFmix on a J/vesicle basis can be orders of magnitude larger than kT. Observable populations for GUVs (i.e. Nmix/Nunmix) are on the order of 0.01 – 100. Since Equation 9 requires the natural log of these values, kTln(Nmix/Nunmix) is on the order of 10−20 J/vesicle, a very small energy difference on a per vesicle basis. Therefore, the purpose of utilizing this method was to provide an additional method for regressing ΔFmix.
Phase and Area in Steric Pressure-induced Mixing
GFP, NLP1, and NLP3 were observed to bind to the Ld region of phase separated DOPC/DPPC/cholesterol GUVs when DOIDA was incorporated. The binding-induced mixing observed at a higher DOIDA concentration was in complete qualitative agreement with the corresponding DPIDA results, discussed above, verifying that NLPs possessed a greater crowding efficacy than GFP attributed to their significantly larger size. Similarly, NLP1 was as effective at inducing mixing as NLP3, which may be attributed to a difference in binding valency (monovalent vs. divalent) while maintaining circular packing. To our knowledge, this is the first report of lipid mixing induced by targeted binding and crowding in the Ld phase.
Interestingly, GUV samples containing DOIDA or DPIDA exhibited similar crowding induced mixing statistics even though Ld regions occupy significantly more surface area than Lo regions. Since there is more Ld phase present, the Ld phase would undergo an overall smaller area change after mixing than the Lo phase. Using Equation 4, it can be seen that a smaller area change (dAc) should result in a smaller ΔFp. Thus Ld phase targeting would have been expected to result in reduced crowding efficacy for a given composition. However, the Ld phase is also capable of containing more bound particles since it possessed larger area. The number of bound particles (N) is directly proportional to ΔFp, as shown in Equation 7. Therefore, a larger number of particles contributes to a larger ΔFp. That these terms negate one another rationalizes the observed phenomena of Lo and Ld targeting resulting in nearly identical crowding efficacies.
CONCLUSIONS
We examined the dissolution of membrane domains through the phase-selective binding of large molecular assemblies, nanolipoprotein particles (NLPs), and a small protein, GFP, to test a statistical thermodynamic model for the crowding pressure-induced mixing of lipid phases. The validity of the model was confirmed by finding that all of the statistical mixing distributions, in terms of crowding energies, for GUVs bound by GFP, NLP1, and NLP3 collapse to the same regressed line for each tested cholesterol content. In addition, free energies of mixing, with increasing cholesterol content, determined by fitting our statistical mixing data to the model, are in agreement with limited experimental and theoretical literature values. There appears to be a maximum packing density of membrane-bound NLPs consistent with circular packing and NLP1 and NLP3 binding the bilayer via monovalent and divalent interactions respectively. Using fluorescence microscopy, our results indicated that histidine-tagged GFP and NLPs bound to targeted regions of GUVs at low iminodiacetic (IDA)-lipid concentrations, and induced phase mixing when targeting either Lo (DPIDA) or Ld (DOIDA) phase at sufficiently high IDA-lipid concentrations due to steric crowding. To our knowledge, this is the first report of lipid mixing induced by targeted binding and crowding in the Ld phase. In a supported lipid multibilayer (MBL) with Lo targeting, NLPs exhibited the same qualitative behavior as observed in GUVs. In addition, we observed the demixing of lipid phases after removal of bound NLPs with EDTA, the first report of reversibility in this type of system. Future work in this area could investigate mixing thermodynamics at different membrane compositions in GUV and MBL systems, or applications of this system towards the development of biomaterials for purposes of biosensors and high-density arrays.
Acknowledgments
MLL, SHR, and WFZ acknowledge partial support from the National Science Foundation under award number DMR-1500275. WFZ was partially supported by Grant Number T32-GM008799 from NIGMS-NIH. SHR also acknowledges partial support derived from his Blacutt-Underwood Endowed Chair funds. DYS was supported by the US Department of Energy, Office of Basic Energy Sciences, Division of Materials Science and Engineering. Sandia National Laboratories is a multi-program laboratory managed and operated by Sandia Corporation, a wholly owned subsidiary of Lockheed Martin Corporation, for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL85000.
Footnotes
SUPPORTING INFORMATION
The Supporting Information is available free of charge on the ACS Publications website.
References
- 1.Lingwood D, Simons K. Lipid Rafts as a Membrane-Organizing Principle. Science (Washington, DC, US.) 2010;327:46–50. doi: 10.1126/science.1174621. [DOI] [PubMed] [Google Scholar]
- 2.Juhasz J, Sharom FJ, Davis JH. Quantitative Characterization of Coexisting Phases in Dopc/Dppc/Cholesterol Mixtures: Comparing Confocal Fluorescence Microscopy and Deuterium Nuclear Magnetic Resonance. Biochim. Biophys. Acta. 2009;1788:2541–2552. doi: 10.1016/j.bbamem.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Marsh D. Cholesterol-Induced Fluid Membrane Domains: A Compendium of Lipid-Raft Ternary Phase Diagrams. Biochim. Biophys. Acta. 2009;1788:2114–2123. doi: 10.1016/j.bbamem.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Veatch SL, Keller SL. Seeing Spots: Complex Phase Behavior in Simple Membranes. Biochim. Biophys. Acta. 2005;1746:172–185. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E. Effect of Chain Length and Unsaturation on Elasticity of Lipid Bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Juhasz J, Davis JH, Sharom FJ. Fluorescent Probe Partitioning in Guvs of Binary Phospholipid Mixtures: Implications for Interpreting Phase Behavior. Biochim. Biophys. Acta. 2010;1818:19–26. doi: 10.1016/j.bbamem.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Momin N, Lee S, Gadok A, Busch D, Bachand G, Hayden C, Stachowiak J, Sasaki DY. Designing Lipids for Selective Partitioning into Liquid Ordered Membrane Domains. Soft Matter. 2015;11:3241–3250. doi: 10.1039/c4sm02856b. [DOI] [PubMed] [Google Scholar]
- 8.Stelzle M, Weissmuller G, Sackmann E. On the Application of Supported Bilayers as Receptive Layers for Biosensors with Electrical Detection. J. Phys. Chem. 1993;97:2974–2981. [Google Scholar]
- 9.Bally M, Bailey K, Sugihara K, Grieshaber D, Voros J, Stadler B. Liposome and Lipid Bilayer Arrays Towards Biosensing Applicaitons. Small. 2010;6:2481–2497. doi: 10.1002/smll.201000644. [DOI] [PubMed] [Google Scholar]
- 10.Cornell BA, Breach-Maksvytis VLB, King LG, Osman PDJ, Raguse B, Weieczorek L, Pace RJ. A Biosensor That Uses Ion Channel Switches. Nature (London, UK.) 1997;387:580–582. doi: 10.1038/42432. [DOI] [PubMed] [Google Scholar]
- 11.Karlsson M, Sott K, Cans AS, Karlsson A, Karlsson R, Orwar O. Micropipette-Assisted Formation of Microscopic Networks for Unilamellar Lipid Bilayer Nanotubes and Containers. Langmuir. 2001;17:6754–6758. [Google Scholar]
- 12.Ogunyankin MO, Huber DL, Sasaki DY, Longo ML. Nanoscale Patterning of Membrane-Bound Proteins Formed through Curvature-Induced Partitioning of Phase-Specific Receptor Lipids. Langmuir. 2013;29:6109–15. doi: 10.1021/la401011d. [DOI] [PubMed] [Google Scholar]
- 13.Seitz M, Wong JY, Park CK, Alcantar NA, Israelachvili J. Formation of Tethered Supported Bilayers Via Membrane-Inserting Lipids. Thin Solid Films. 1998;327:767–771. [Google Scholar]
- 14.Svedhem S, Pfeiffer I, Larsson C, Wingren C, Borrebaeck C, Hook F. Patterns of DNA-Labeled and Scfv-Antibody-Carrying Lipid Vesicles Directed by Material-Specific Immobilization of DNA and Supported Lipid Bilayer Formation on Au/Sio2 Template. ChemBioChem. 2003;4:339–343. doi: 10.1002/cbic.200390055. [DOI] [PubMed] [Google Scholar]
- 15.Groves JT, Wulfing C, Boxer SG. Electrical Manipulation of Glycan Phohatidyl Inositol Tethered Proteins in Planar Supported Bilayers. Biophys. J. 1996;71:2716–2723. doi: 10.1016/S0006-3495(96)79462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darst SA, Ahlers M, Meller PH, Kubalek EW, Blankenburg R, Ribi HO, Ringsdorf H, Kornberg RD. 2-Dimensional Crystals of Strepavidin on Biotinylated Lipid Bilayers and Their Interactions with Biotinylated Macromolecules. Biophys. J. 1991;59:387–396. doi: 10.1016/S0006-3495(91)82232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nye JA, Groves JT. Kinetic Control of Histidine-Tagged Protein Surface Density on Supported Lipid Bilayers. Langmuir. 2008;24:4145–4149. doi: 10.1021/la703788h. [DOI] [PubMed] [Google Scholar]
- 18.Bornhorst JA, Falke JJ. Purifcation of Proteins Using Polyhistidine Affinity Tags. Methods Enzymol. 2000;326:245. doi: 10.1016/s0076-6879(00)26058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng K, Pack DW, Sasaki DY, Arnold FH. Engineering Protein-Lipid Interactions: Targeting of Histidine-Tagged Proteins to Metal-Chelating Lipid Monolayers. Langmuir. 1995;11:4048–4055. [Google Scholar]
- 20.Hayden CC, Hwang JS, Abate EA, Kent MS, Sasaki DY. Directed Formation of Lipid Membrane Microdomains as High Affinity Sites for His-Tagged Proteins. J. Am. Chem. Soc. 2009;131:8728–8729. doi: 10.1021/ja901157c. [DOI] [PubMed] [Google Scholar]
- 21.Stachowiak JC, CC H, Sanchez MAA, Wang J, Bunker BC, Voigt JA, Sasaki DY. Targeting Proteins to Liquid-Ordered Domains in Lipid Membranes. Langmuir. 2010;27:1457–1462. doi: 10.1021/la1041458. [DOI] [PubMed] [Google Scholar]
- 22.Stachowiak JC, Hayden CC, Sasaki DY. Steric Confinement of Proteins on Lipid Membranes Can Drive Curvature and Tubulation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7781–7786. doi: 10.1073/pnas.0913306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, Sasaki DY, Sherman MB, Geissler PL, Fletcher DA, Hayden CC. Membrane Bending by Protein-Protein Crowding. Nat. Cell Biol. 2012;14:944–949. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 24.Scheve CS, Gonzales PA, Momin N, Stachowiak JC. Steric Pressure between Membrane-Bound Proteins Opposes Lipid Phase Separation. J. Am. Chem. Soc. 2013;135:1185–1188. doi: 10.1021/ja3099867. [DOI] [PubMed] [Google Scholar]
- 25.Almeida PF. Thermodynamics of Lipid Interactions in Complex Bilayers. Biochim. Biophys. Acta. 2009;1788:72–85. doi: 10.1016/j.bbamem.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Chromy BA, Arroyo E, Blanchette CD, Bench G, Benner H, Cappuccio JA, Coleman MA, Hoeprich PD. Different Apolipoproteins Impact Nanolipoprotein Particle Formation. J. Am. Chem. Soc. 2007;129:14348–14354. doi: 10.1021/ja074753y. [DOI] [PubMed] [Google Scholar]
- 27.Bricarello DA, Smilowitz JT, Zivkovic AM, German JB, Parikh AN. Reconstituted Lipoprotein: A Versatile Class of Biologically-Inspired Nanostructures. ACS Nano. 2011;5:42–57. doi: 10.1021/nn103098m. [DOI] [PubMed] [Google Scholar]
- 28.Bayburt TH, Grinkova YV, Sligar SG. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. ACS Nano. 2002;2:853–856. [Google Scholar]
- 29.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed Self-Assembly of Monodisperse Phospholipid Bilayer Nanodiscs with Controlled Size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 30.Zeno WF, Hilt SL, Risbud SH, Voss JC, Longo ML. Spectroscopic Characterization of Structural Changes in Membrane Scaffold Proteins Entrapped within Mesoporous Silica Gel Monoliths. ACS Appl. Mater. Interfaces. 2015;7:8640–8649. doi: 10.1021/acsami.5b00898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cappusccio J, Blanchette CD, Sulchek TA, Arroyo ES, Kralj J, Hinz A, Kuhn EA, Chromy A, Segelke BW, Rothschild K, Fletcher JE, Katzen F, Peterson T, Kudlicki WA, Bench G, Hoeprich PD, Coleman MA. Cell-Free Co-Expression of Functional Membrane Proteins and Apolipoprotein, Forming Soluble Nanolipoprotein Particles. Mol. Cell. Proteomics. 2008;7:2246–2253. doi: 10.1074/mcp.M800191-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bayburt TH, Grinkova YV, Sligar SG. Assembly of Single Bacteriorhodopsin Trimers in Bilayer Nanodics. Arch. Biochem. Biophys. 2006;450:215–222. doi: 10.1016/j.abb.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 33.Israelachvili JN. Intermolecular and Surface Forces: Revised Third Edition. Academic press; 2011. [Google Scholar]
- 34.Carnahan NF, Starling KE. Equation of State for Nonattracting Rigid Spheres. J Chem Phys. 1969;51:635–636. [Google Scholar]
- 35.Tian A, Johnson C, Wang W, Baumgart T. Line Tension at Fluid Membrane Domain Boundaries Measured by Micropipette Aspiration. Phys. Rev. Lett. 2007;98:208102. doi: 10.1103/PhysRevLett.98.208102. [DOI] [PubMed] [Google Scholar]
- 36.Lee SB, Torquato S. Monte Carlo Study of Correlated Continuum Percolation: Universality and Percolation Thresholds. Phys. Rev. A: At., Mol, Opt. Phys. 1990;41:5338. doi: 10.1103/physreva.41.5338. [DOI] [PubMed] [Google Scholar]
- 37.Pack DW, Chen G, Maloney KM, Chen C-T, Arnold FH. A Metal-Chelating Lipid for 2d Protein Crystallization Via Coordination of Surface Histidines. J. Am. Chem. Soc. 1997;119:2479–2487. [Google Scholar]
- 38.Zeno WF, Hilt SL, Aravagiri KA, Risbud SH, Voss JC, Parikh AN, Longo ML. Analysis of Lipid Phase Behavior and Protein Conformational Changes in Nanolipoprotein Particles Upon Entrapment in Sol–Gel-Derived Silica. Langmuir. 2014;30:9780–9788. doi: 10.1021/la5025058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldberg M. The Packing of Equal Circles in a Square. Math. Mag. 1970;43:24–30. [Google Scholar]
- 40.Morgan CR, Hebling CM, Rand KD, Stafford DW, Jorgenson JW, Engen JR. Conformational Transitions in the Membrane Scaffold Protein of Phospholipid Bilayer Nanodiscs. Mol. Cell. Proteomics. 2011;10:M111–010876. doi: 10.1074/mcp.M111.010876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hac AE, Seeger HM, Fidorra M, Heimburg T. Diffusion in Two-Component Lipid Membranes—a Fluorescence Correlation Spectroscopy and Monte Carlo Simulation Study. Biophys. J. 2005;88:317–333. doi: 10.1529/biophysj.104.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ivanova VP, Heimburg T. Histogram Method to Obtain Heat Capacities in Lipid Monolayers, Curved Bilayers, and Membranes Containing Peptides. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2001;63:041914. doi: 10.1103/PhysRevE.63.041914. [DOI] [PubMed] [Google Scholar]
- 43.White SH, King GI. Molecular Packing and Area Compressibility of Lipid Bilayers. Proc. Natl. Acad. Sci. 1985;82:6532–6536. doi: 10.1073/pnas.82.19.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugár IP, Thompson TE, Biltonen RL. Monte Carlo Simulation of Two-Component Bilayers: Dmpc/Dspc Mixtures. Biophys. J. 1999;76:2099–2110. doi: 10.1016/S0006-3495(99)77366-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugár IP, Biltonen RL, Mitchard N. Monte Carlo Simulations of Membranes: Phase Transition of Small Unilamellar Dipalmitoylphosphatidylcholine Vesicles. Methods Enzymol. 1994;240:569–593. doi: 10.1016/s0076-6879(94)40064-4. [DOI] [PubMed] [Google Scholar]
- 46.Hill TL. An Introduction to Statistical Thermodynamics. Courier Corporation; 2012. [Google Scholar]