Abstract
During early fetal development, paracrine Hedgehog (HH) ligands secreted from the foregut epithelium activate Gli transcription factors in the surrounding mesenchyme to coordinate formation of the respiratory system, digestive track and the cardiovascular network. Although disruptions to this process can lead to devastating congenital defects, the underlying mechanisms and downstream targets, are poorly understood. We show that the zinc finger transcription factor Osr1 is a novel HH target as Osr1 expression in the foregut mesenchyme depends on HH signaling and the effector of HH pathway Gli3 binds to a conserved genomic loci near Osr1 promoter region. Molecular analysis of mouse germline Osr1 mutants reveals multiple functions of Osr1 during foregut development. Osr1 mutants exhibit fewer lung progenitors in the ventral foregut. Osr is then required for the proper branching of the primary lung buds, with mutants exhibiting miss-located lung lobes. Finally, Osr1 is essential for proper mesenchymal differentiation including pulmonary arteries, esophageal and tracheal smooth muscle as well as tracheal cartilage rings. Tissue specific conditional knockouts in combination with lineage tracing indicate that Osr1 is required cell autonomously in the foregut mesenchyme. We conclude that Osr1 is a novel downstream target of HH pathway, required for lung specification, branching morphogenesis and foregut mesenchymal differentiation.
Keywords: Odd-skipped, Hedgehog, Lung, Trachea, Esophagus, Specification, Lobulation, Mesenchyme
1. Introduction
The formation of the respiratory system, upper digestive track and cardiovascular network requires the coordinated development of the foregut epithelium and the surrounding mesenchyme so that these organ systems are properly integrated for breathing and feeding. The hedgehog (HH) signaling pathway has emerged as a critical mediator of the paracrine signals between the epithelium and mesenchyme to regulate multiple steps in foregut organogenesis (McCulley et al. 2015; Swarr and Morrisey 2015). HH pathway mutations in humans and animal models result in severe congenital birth defects in the foregut including tracheoesophageal stenosis, lung hypoplasia with disrupted lobulation, tracheal-bronchial cartilage malformations (Kugler et al. 2015) and aberrant pulmonary arteries (Peng et al. 2013). Although the HH pathway is clearly a critical regulator of foregut development, its downstream target genes that control organogenesis are poorly understood.
During fetal development HH ligands secreted from the foregut epithelium (Shh and Ihh) cause Gli2 and Gli3 (Gli2/3) transcription factors to be proteolyticaly activated in the surrounding mesenchyme to stimulate HH-target gene expression and regulate multiple aspects of foregut organogenesis. At embryonic day (E) 9.5, HH ligands signal from the epithelium to the surrounding mesenchyme to support the expression of Wnt and Bmp ligands, which in turn induce the specification of the respiratory lineage, marked by the expression of homeobox gene Nkx2-1, in the ventral foregut endoderm (Rankin et al. 2016). After specification, Shh is enriched at the distal tip of the lung epithelial to regulate lobulation and further branching morphogenesis (Swarr and Morrisey 2015). Mesenchymal differentiation also depends on Shh signaling. Various HH and Gli mutants demonstrate the necessity of this pathway in tracheoesophageal differentiation including proper formation of the C-shaped cartilage rings in the ventral-lateral side of trachea (Motoyama et al. 1998; Park et al. 2010). Moreover, epithelial HH signaling is critical to support smooth muscle differentiation in the more distal GI tract (Mao et al. 2010). HH is also required for the development of common cardiopulmonary progenitor cells that contribute to the outflow tract and pulmonary arteries that connect the heart and lungs (Peng et al. 2013).
The tissue specific target genes that HH/Gli regulate to control foregut organogenesis are not well understood. The mesenchymal Fork head transcription factor Foxf1 is one of the well-characterized targets of Shh in small intestine (Madison et al. 2009) and in the second heart field (Hoffmann et al. 2014). However the Foxf1 mutant only partially recapitulates the foregut phenotypes of Shh and Gli2/3 mutants. For example, Foxf1 germline mutant mice die about E8.5 due to extra-embryonic defects (Mahlapuu et al. 2001) while the Foxf1 heterozygous show lung lobe fusion and medial lobe malformation (Lim et al. 2002). Thus it’s unclear if and to what extent Foxf1 might mediate HH function during specification of the respiratory lineage. Endothelial specific Foxf1 mutants display similar distal vasculature defects as in the Shh mutants (Li et al. 2004; Ren et al. 2014), but formation of the pulmonary arteries is not examined. Recent studies in early lung and heart development implicated the zinc finger transcription factor Odd Skipped Related 1 (Osr1) as a potential Gli target (Xie et al. 2012; Rankin et al. 2016). Moreover studies in Xenopus show that Osr1 and Osr2 regulate lung-inducing signals such as Wnt2, Wnt2b and Bmp4 in the splanchnic mesoderm (Rankin et al. 2012), further supporting the idea that Osr factors might mediate some of the effects of HH in the developing respiratory system. Osr1−/− germ line null mutants exhibit heart, kidney and tongue malformations (Wang et al. 2005; Liu et al. 2013), and die between E12.5–E16.5 due to cardiac defects. We have also noticed that these embryos display lung tissue hypoplasia based on histology, but the respiratory and digestive systems have not been examined in detail.
In this study, we tested the hypothesis that Osr1 regulates foregut organogenesis downstream of HH/Gli signaling. We show that Osr1 expression in the splanchnic mesoderm surrounding the foregut is dependent on Gli2 and Gli3 activity. We show that Osr1 plays crucial roles at multiple stages of foregut development. First Osr1 supports robust respiratory specification by promoting expression of lung-inducing mesenchymal signals and then Osr1 directs precise lobe formation at the correct locations. Lastly, Osr1 is required for proper development of the main pulmonary arteries and differentiation of both the tracheal cartilage rings and esophageal smooth muscle. Our data established Osr1 as a novel mediator of HH signaling during foregut organogenesis, which may inform the developmental basis of congenital birth defects.
2. Materials and methods
2.1 Animals
Animal husbandry was performed in accordance with protocols approved by the Cincinnati Children’s Hospital Medical Center IACUC. Gli2+/− (Or Gli2lzki) (Bai and Joyner 2001), Gli3+/− (Or Gli3Xt-J) (Johnson 1967), Osr1+/− (Wang et al. 2005), Osr1f/f (Lan et al. 2011), Foxa2creER (Park et al. 2008), Dermo1cre (Sosic et al. 2003), Osr1creER (Mugford et al. 2008), Gli1lacZ (Bai et al. 2002), ROSAmT/mG (Muzumdar et al. 2007) reporter mice have been described previously. All mice were kept in a mixed background. Timed matings were performed and the date when a plug was detected was noted as embryonic day 0.5. Control and mutant littermate embryos were age-matched by somite numbers.
For Osr1creER lineage tracing, tamoxifen was intraperitoneally injected at E7.5 and E8.5 at 2mg/40g body weight dosage and embryos were collected at E12.5. For Foxa2creER conditional deletion, oral gavage of tamoxifen of 0.12mg/g body weight was performed at E6.75.
2.2 Analysis of public ChIP data
Gli3-flag ChIP data were downloaded from GEO database (accession number GSE44756) (Hoffmann et al. 2014) and reanalyzed as follows. The fastq DNA sequence files were quality control assessed and raw reads were trimmed using FastQC and Trimmomatic. Reads were mapped to the mouse genome assembly mm10 using Bowtie2 (Langmead 2012) at default thresholds. Duplicate and multi-mapped reads were removed using picard and samtools respectively and ChIP-seq peaks were called using MACS2 (Zhang et al. 2008) using mentioned thresholds [--bw 200 --mfold 2 200 -p 0.05 -g 2e9 --call-summits] (Hoffmann et al. 2014). Mapped reads were converted into BigWig format for visualization purposes. We used Homer’s (Heinz et al. 2010) annotatePeaks.pl for obtaining genes close to the peaks. Transcription factor binding motif analysis was performed in the CIS-BP website (Weirauch et al. 2014) (http://cisbp.ccbr.utoronto.ca, last time accessed 2/28/2017).
2.3 Histology, immunostaining and in situ hybridization assays
Mouse embryos were harvested at specific developmental stages and fixed in 4% paraformaldehyde in PBS at 4°C overnight and then washed with PBS. H&E staining was performed according to standard protocol after paraffin embedding. For all others, cryosections were used. After fixation, tissue were embedded in OCT and sectioned at 10μm thickness. For mouse in situ hybridization, DIG-labeled probes were generated using linearized full-length mouse cDNA plasmids. Immunofluorescent (IF) staining or in situ hybridization on sections as well as whole mount IF staining were performed as described previously (Rankin et al. 2016). Primary antibodies used are listed in Table s1. For whole foregut IF followed with quantification, 3D confocal images were captured on a Nikon A1Rsi inverted confocal microscope. The numbers of phospho-histone H3+ (pHH3), Foxa2+ and Nkx2-1+ cells in dissected foregut (n>3 control and n=3 Osr1 mutant embryos) were quantified using IMARIS software. For lobulation quantification, following IF staining with dissected lungs, the three dimensional distance were quantified (n>3 control and n=3 Osr1 mutant embryos) using IMARIS software. The center of branching points and distal point of trachea bifurcation were used to determine the distances. If located anterior to trachea bifurcation, the distance was given as minus. Pairwise student T-tests between control and mutants were used to determine statistical significance *p<0.05.
Whole mount in situ hybridization was performed as follows. Briefly, after fixation, tissue was rehydrated and digested with proteinase K. A short fixation was performed after digestion. In situ probes were then incubated with tissue at 70°C overnight. Serial washes were done before blocking and adding anti-DIG alkaline phosphatase. Overnight 4°C incubation and several washes were done before color development using BM purple.
3. Results
3.1 Osr1 expression in the foregut mesenchyme is regulated by HH/Gli signaling
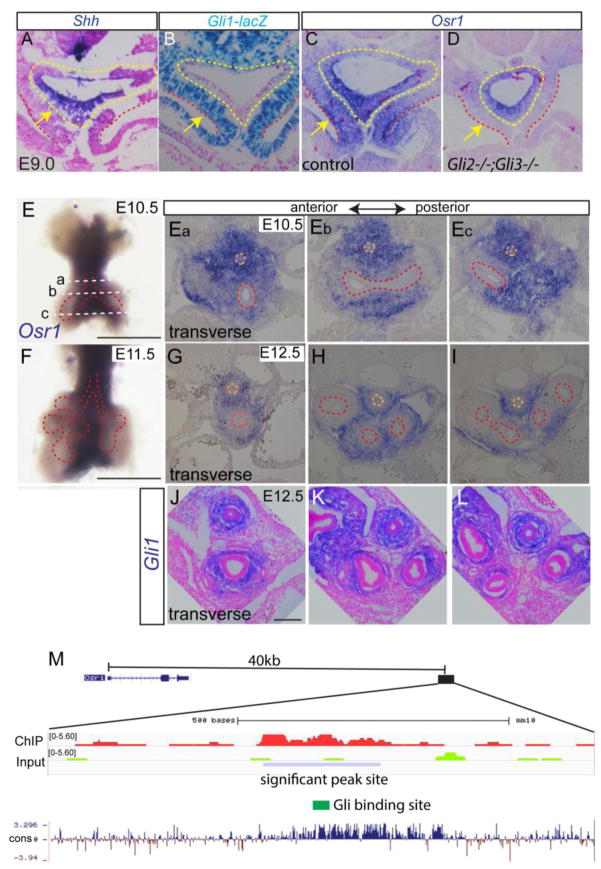
Our recent analysis of the signaling networks controlling respiratory system specification suggested that the zinc finger transcription factor Osr1 is a Gli-target in the splanchnic mesenchyme (Rankin et al. 2016). To directly test this possibility, we first compared the expression pattern of Osr1 to Shh and Gli1. Noteworthy is the fact that Gli1 transcription is directly activated by HH pathway effectors, and thus is a reliable readout of Hh-responsive tissue (Bai et al. 2002). In the E9.0 (12–16 somite numbers) foregut, Shh was restricted to the epithelium (Fig. 1A) whereas expression of a Gli1-lacZ knock-in allele was restricted to the mesenchyme (Fig. 1B), consistent with previous reports (Rankin et al. 2016). In comparison, Osr1 was robustly expressed in the both the presumptive respiratory epithelium and the surrounding splanchnic mesenchyme (Fig. 1C). Though normal Osr1 expression was observed in Gli2 and Gli3 single mutants (data not shown), analysis of Gli2−/−;Gli3−/− double mutants confirmed that Osr1 expression was specifically lost in the HH-responsive mesenchyme, but not in the foregut epithelium (Fig. 1D). This suggested that the mesenchymal but not epithelial Osr1 expression is HH regulated.
Fig. 1.
Osr1 is a Gli2,3 target and displays dynamic expression pattern during foregut development. (A) Shh in situ staining on E9.0 transverse foregut section. Arrow points to foregut epithelial. (B) LacZ staining in foregut using Gli1lacZ reporter mouse. (C–D) Osr1 in situ on transverse sections of E9.0 foregut in control and Gli2−/−;Gli3−/− double mutant. Yellow dotted lines mark endoderm while the region in between yellow and red dotted lines denotes the splanchnic mesoderm. Arrows point to splanchnic mesenchyme. (E) Whole mount in situ of Osr1 in dissected E10.5 foregut. Scale bar: 500 μm. Dotted lines corresponds to the section levels in Fig. Ea–Ec. (Ea–Ec) Osr1 in situ on transverse sections of foregut region at E10.5 from anterior to posterior. (F) Whole mount Osr1 in situ in E11.5 dissected respiratory tissue. Scale bar: 500 μm. (G–I) Osr1 in situ on transverse sections of foregut at E12.5 from anterior to posterior region. Orange dotted line marks esophagus and red dotted line marks respiratory epithelial cells. (J) Gli1 in situ on transverse sections through trachea and main bronchi. Scale bar: 100 μm. (M) Browser shot of previously published ChIP-seq data in second heart field tissue comparing Gli3-flag ChIP to input around Osr1 region together with mammal pairwise conservation. Blue box indicates the significant Gli3-flag ChIP signal located 40kb downstream of Osr1 transcription start site.
In situ hybridization of E10.5–E12.5 foreguts showed that Osr1 is expressed in the medial mesenchyme surrounding the developing trachea, esophagus and main stem bronchi but is down regulated in the respiratory epithelium and the peripheral lung mesenchyme (Fig. 1E–I). This Osr1 expression pattern in the E12.5 respiratory mesenchyme was similar to that of Gli1 in the mesenchyme, suggesting a continuous regulation by HH signaling (1K–1M).
To test the hypothesis that Osr1 transcription is directly regulated by HH/Gli, we re-analyzed the published Chromatin Immuno-precipitation followed with genomic sequencing (ChIP-seq) data of Gli3T-flag transgenic mice (GEO accession number GSE44756) where chromatin binding sites of HH effector Gli3 were identified in E9.5 second heart field tissue (Hoffmann et al. 2014). The Gli3/Osr1-expressing splanchnic mesenchyme partially overlaps with the second heart field at this stage of development. Therefore, we expected that common Gli-binding sites should be detected. We identified one statistically significant Gli3-binding peak (chr12: 9622096–9622310, mm10) located about 40 kb downstream of the Osr1 transcription start site. This genomic region is evolutionarily conserved (Fig. 1M) and transcription factor DNA-binding motif analysis of this peak confirmed the presence of a Gli DNA-binding consensus sequence (GGGACCACCCTG) in the middle of the peak, supporting the hypothesis that Osr1 is a direct target of HH/Gli in the foregut mesenchyme. Together these data indicate that Osr1 is a transcriptional target of HH/Gli signaling in the foregut mesenchyme.
3.2 Osr1 mutants exhibit lung hypoplasia, lobe deformation and defective mesenchymal differentiation
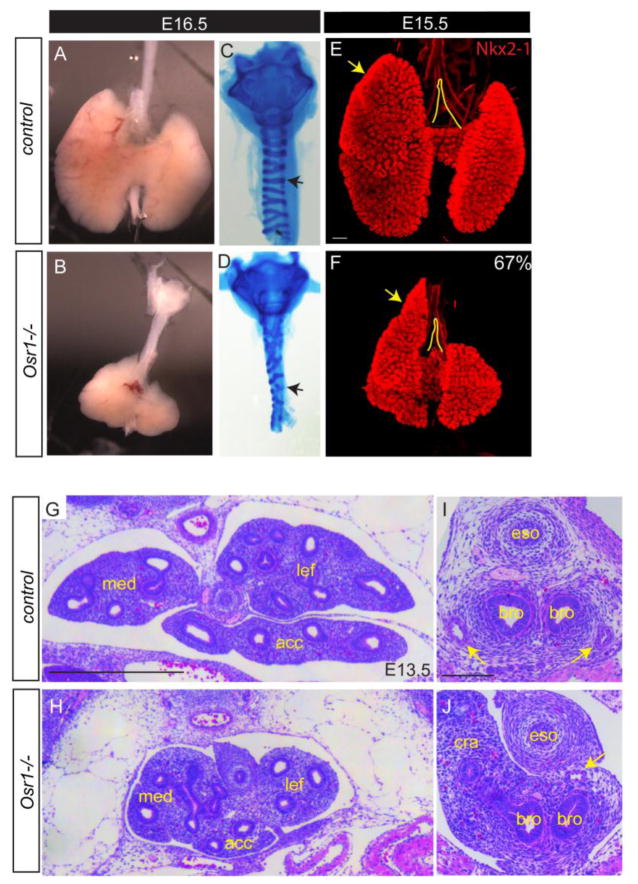
To further test the hypothesis that Osr1 partially mediates HH function, we examined foregut development in Osr1−/− germ line null mutants (Wang et al. 2005). Osr1 mutants die between E12.5–E16.5 due to heart defects, but examination of surviving mutants at revealed much smaller lungs than litermate controls (Fig. 2A,B) despite comparable body size (Fig. s2A). Additionally, about 67% (n= 10/15) of Osr1 mutants display reproducible misshaping of the lung lobes, with the cranial lobe extending more rostrally with a pointed shape as opposed to a rounded shape in the control littermates (Fig. 2E,F). Histological sections at E13.5 confirmed the reduced lung size, though overall the major lung tissue organization appeared relatively normal in mutants (Fig. 2G,H).
Fig. 2.
Osr1 mutants display lung hypoplasia, dysmorphyc lobulation and mesenchymal differentiation defects. (A–B) Bright field view of dissected esophagus, trachea and lung. (C–D) Alcian blue staining of dissected trachea. Arrows show broken and disorganized cartilage rings in Osr1 mutant compared to control. (E–F) Whole mount IF of Nkx2-1 in wildtype and mutant lungs. Arrows point to the anterior extension of the cranial lobe relative to the trachea bifurcation. Scale bar: 200 μm. (G–J) H&E staining of transverse sections lung (G–H, scale bar: 500 μm) and main bronchi (I–J, scale bar: 100 μm) regions. Arrows indicate pulmonary arteries. Med, medial lobe; lef, left lobe; acc, accessory lobe; eso, esophagus; bro, bronchus; cra, cranial lobe.
Similar to the HH mutants, mesenchymal differentiation defects were also identified in Osr1 mutants. We examined E16.5 dissected tracheas using alcian blue staining, which revealed reduced and disrupted cartilage rings in the Osr1 mutants (Fig. 2C,D), similar to what has been described in Shh mutants (Park et al. 2010), though not as severe. Furthermore, histological analysis of the main stem bronchi showed pulmonary artery defects, including loss of the right artery and mislocation of the left artery in between the esophagus and bronchi instead of ventral-lateral to the bronchi (n=5/6) (Fig. 2I,J).
3.3 Osr1−/− mutants specify fewer respiratory progenitors
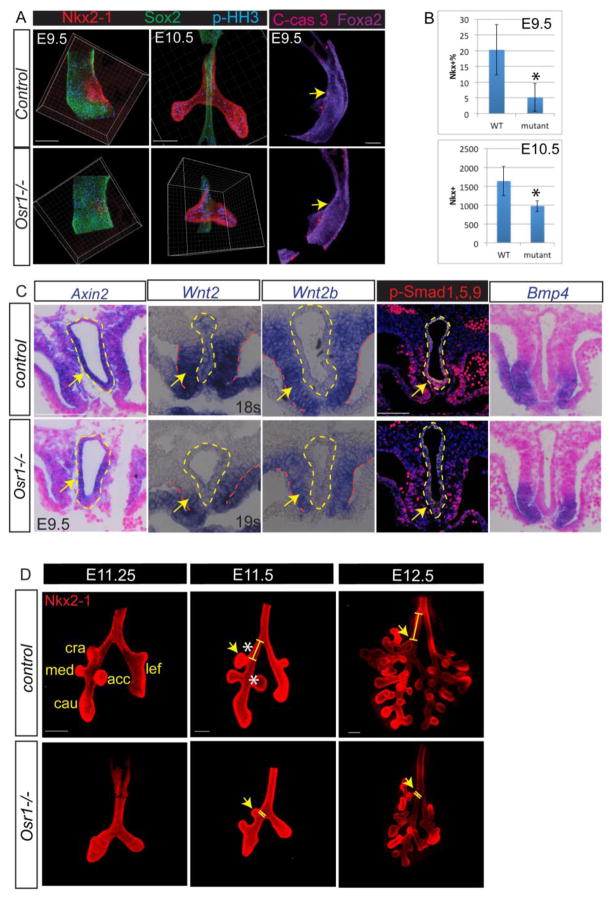
To better understand why the lungs are smaller and dysmorphic in the Osr1 mutants we sought to identify the earliest defects. Since Osr1 is implicated in specification of respiratory progenitors in Xenopus (Rankin et al. 2012), we examined the expression of Nkx2-1, the earliest marker of respiratory progenitors at E9.5. Whole mount immunostaining of Nkx2-1 and Sox2 (a marker of foregut/esophageal epithelium) in dissected foreguts revealed dramatically fewer Nkx2-1+ respiratory progenitors in Osr1 mutants compared with somite-matched litermate controls (Fig. 3A). Quantitative image analysis with Imaris software confirmed a four-fold reduction in the percentage of Nkx2-1+ progenitor cells (normalized to the total number of foregut epithelial cells from the pharynx to the liver bud) at E9.5. The smaller respiratory domain in Osr1 mutants persisted at E10.5. Analysis of cell mitosis by phospho-Histone H3 (pHH3) staining and apoptosis as marked by cleaved Caspase-3 showed no difference between mutants and litermate controls (Fig. 3A,B). There results indicate that the overall lung hypoplasia in Osr1 mutants is due, at least in part, to fewer progenitors being specified early in development.
Fig. 3.
Osr1 is required for sufficient respiratory specification and accurate lobe formation. (A) Whole mount immunostaining of foregut with respiratory marker (Nkx2-1), esophageal marker (Sox2), proliferation (p-HH3) and apoptosis (C-caspase 3) markers in dissected foreguts. (B) Quantification of respiratory specification marked by Nkx2-1+ cells normalized by the entire foregut epithelial cells in between pharynx and liver at E9.5 and the total number of Nkx2-1+ cells at E10.5. (C) In situ staining of Axin2, Wnt2, 2b and Bmp4 and IF stainging of p-Smad 1,5,9 in wildtypes and Osr1 mutants. (D) Whole mount IF with dissected lungs. Yellow lines indicate the distance between cranial lobe and trachea bifurcation. Cra, cranial lobe; med, medial lobe; cau, caudal lobe; lef, left lobe; acc, accessory lobe. Asterisks indicate the distances from cranial lobe to tracheal bifurcation and from accessory lobe to medial lobe are both significantly changed. Arrows indicate cranial lobe locations. Scale bars: 100 μm.
In Xenopus embryos, Osr1 and Osr2 are redundantly required for specification of the respiratory lineage (Rankin et al. 2012). To test this redundancy in mice, we examined Osr2 expression in control and Osr1 mutants. We found that unlike Xenopus, Osr2 was not expressed in wildtype foregut at E9.5 (Fig. s1B). Even in the absence of Osr1, Osr2 was not up-regulated (Fig. s1C). Consistently, Osr1−/−;Osr2−/− double mutants were indistinguishable from Osr1−/− single mutants as judged by Nkx2-1 expression at E9.5 and lung bud morphology at E10.5 (Fig. s1D), indicating that Osr2 is not involved in lung specification in mice.
Next, we sought to determine why Osr1 mutants had fewer respiratory progenitors. We have previously shown that HH/Gli signal is required for the expression of lung-inducing factors Wnt2, Wnt2b and Bmp4 in the splanchnic mesenchyme. Therefore, we assessed the expression and activity of these growth factors. We found a modest reduction in Wnt2 and Wnt2b expression along with reduced levels of the Wnt-target gene Axin2 (Fig. 3C). On the other hand Bmp4 in the splanchnic mesenchyme appeared normal in Osr1−/− mutants, although p-Smad 1,5,9 staining in the foregut epithelium appeared reduced (Fig. 3C). Given that the foregut is smaller in the Osr1 mutants than in somite-matched controls, we also compared Wnt2 expression in a Osr1 mutant with a four-somites younger control, so that the overall foregut morphology is more comparable (Fig. s3A). Wnt2 expression does not differ in this comparison, consistent with the observation that respiratory specification is delayed but not completely abolished. Taken together, these data show that Osr1 is required for robust specification of respiratory progenitors, which might be explained in part by reduced Wnt and Bmp signals.
3.4 Osr1 is required for proper establishment of the primary lung lobes
In addition to having smaller lungs at E13.5–E16.5 (Fig. 1), the Osr1 mutants exhibited disrupted lung lobe morphology. The stereotypical lobe pattern is established at E11.5 when three additional branches form on the primary right lung bud to generate total of five lobes in mice. Examination of early branching morphogenesis revealed delayed primary lobulation and misplaced stereotypical locations in the Osr1 mutants (Fig. 3D). The primary lung bud branch that gives rise to the cranial lobe is normally located distal to the tracheal bifurcation where the trachea bifurcates into the two main stem bronchi. However, in 67% of Osr1 mutants, the cranial lobe was anteriorly shifted to about the same level as the tracheal bifurcation (Fig. 3D). Quantification using confocal images of wholemount staining of E11.5 fetal lungs revealed that the cranial lobe was located at 169 ±20 μm distal to the tracheal bifurcation in controls, but was on average 25 ± 43 μm proximal to the bifurcation in the Osr1 mutants. This phenotype is similar to a human condition known as tracheal bronchus, or “pig bronchus”, where a bronchus emerges from the trachea in an abnormally more rostral position. This condition is often non-symptomatic but, it can occur with tracheal stenosis, which significantly complicates the surgical repair (Morita et al. 2016).
Although the cranial lobe is the most severely disrupted, several other lobes were also mis-positioned in Osr1 mutants. For example, while the medial lobe is relatively less affected, the length of the left lobe as well as the secondary branches on the left lobe is much shorter and delayed. Also, the accessory lobe is significantly shifted distally relative to the medial lobe (control: 18±13μm and mutant: 81±27μm, n>3, p<0.05). Interestingly, the subsequent branching morphogenesis after the establishment of the initial five lobes appears similar between controls and mutants, consistent with Osr1 expression pattern in the proximal large airways but absent from distal lung where most of branching morphogenesis occurs. This suggests that the overall lung misshaping might derive from the earlier events when the primary lobes were established.
During branching morphogenesis, the lungs also establish the proximal and distal domains marked by Sox2 and Sox9 expression respectively. Immunostaining analysis with these markers at E16.5 demonstrated that the overall proximal-distal patterning was normal. However consistent with the mislocalization of the primary cranial lobe, the distal Sox9+ domain does appear to have a slight medial and anterior shift even as early as E10.5 and the cells at the tracheal bifurcation level adopt a Sox9+ fate instead of Sox2+ at E11.5 (Fig. s2B).
The disrupted lung bud morphogenesis in the Osr1 mutants is reminiscent of what is seen in some Hh pathway mutants. Shh−/− mutants (Li et al. 2004) and HH feedback inhibitor Hhip mutants (Chuang et al. 2003) fail to establish lobes. Gli2 mutants (Motoyama et al. 1998) only form one lobe on the right. Gli3 mutants have narrowed caudal and left lobes (Grindley et al. 1997). Epithelial Shh is well-known to regulate branching morphogenesis through a signaling network including Fgf10, Bmp and Wnt signals in the distal lung bud tips (Swarr and Morrisey 2015). To test the possibility that Osr1 might participate in this signaling network, we examined Shh, Wnt2, Bmp4 and Fgf10 expression in control and Osr1 mutant lungs. However we found little if any difference in expression between mutants and controls (Fig. s3A–C) suggesting that Osr1 regulates lobulation independently of these known pathways, or acts downstream of them.
3.5 Osr1 is required for mesenchymal differentiation and proper formation of the pulmonary arteries
HH signaling is known to be required for several aspects of mesenchymal differentiation including formation of the pulmonary arteries, tracheal cartilage, and digestive tract smooth muscle. Osr1 mutants appeared to exhibit similar defects (Fig. 1), prompting us to investigate in more detail.
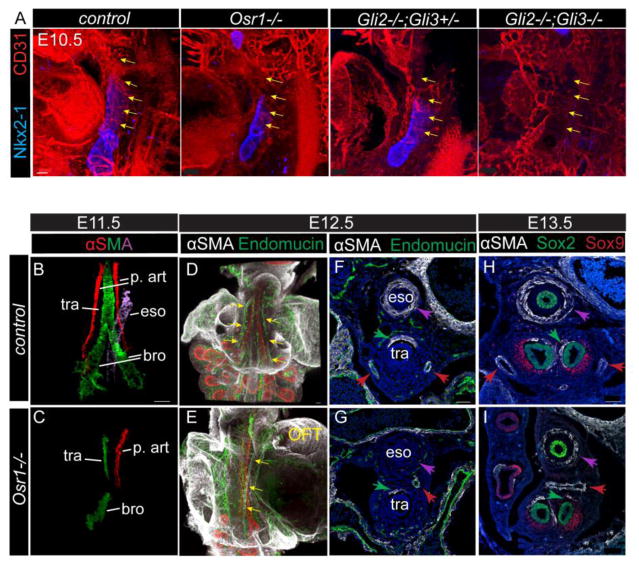
First we characterized the earliest vascular defect by examining the initial vascular plexus between the cardiac outflow tract and fetal lungs at E10.5, which is thought to give rise to pulmonary arteries later. The formation of this pulmonary vasculature from cardiopulmonary progenitors is known to be regulated by Shh signaling (Peng et al. 2013). Immunostaining for CD31, an endothelial marker, demonstrated that Osr1 mutants had reduced blood vessels in the distal region of the trachea where the pulmonary artery connects to the lungs; a phenotype that was similar to that observed in the Gli2−/−;Gli3+/− mutants (Fig. 4A). In the more severe case where all alleles of Gli2/3 were mutated, the connecting plexus appeared completely absent between the outflow tract and the presumptive lung region (Fig. 4A). This is consistent with the previously reported disrupted connections between the heart and lungs in Shh−/− mutants and the mesenchymal conditional knockout of the HH receptor Smoothened (Peng et al. 2013). This suggests that the Shh-Osr1 axis is crucial for the proper connection between the heart and the lungs.
Fig. 4.
Mesenchymal development including pulmonary arteries and smooth muscle differentiation in foregut is dependent on Osr1. (A) Whole mount IF staining in E10.5 embryos. Yellow arrows mark the vascular plexus connecting the out flow tract to the lungs. Scale bar: 50 μm. (B–C) αSMA whole mount staining of dissected foregut. Epithelial staining (not shown) is used to guide the identification of different smooth muscle cell structures. Different regions of the staining are isolated and pseudo-colored using Imaris. (D–E) Whole mount staining of the dissected foregut with the heart attached. Arrows point to the pulmonary arteries. Scale bars: 100 μm. (F–I) IF on transverse sections at the trachea level (F–G) and the main bronchi level (H–I). Purple arrows indicate esophagus SMC, green arrows indicate tracheal SMC, while red arrows indicate pulmonary arteries SMC. OFT, out flow tract; p. art, pulmonary artery; tra, trachea; eso, esophagus; bro, bronchi. Scale bars: 50 μm.
After E11.5, pulmonary arteries can be clearly identified as endothelial vessels connecting the outflow tract and the lungs. Those vessels are lined by endothelial cells marked by endomucin, surrounded by smooth muscle cells marked by α-smooth muscle actin (αSMA). Our initial histological analysis indicated that pulmonary arteries were misplaced or absent in Osr1 mutants (Fig. 1). To examine this in more detail we dissected the trachea, esophagus and lungs with associated vasculature and mesenchyme from E11.5 embryos and performed whole mount immunostaining for αSMA and epithelial marker Foxa2. To better visualize the structures, the different αSMA+ structures were pseudo-colored differently for different regions (smooth muscle associated with pulmonary arteries in red; esophagus purple, with trachea and bronchi green) (Fig. 4B,C). We found that Osr1 mutants consistently (n=5/6) had only the left pulmonary artery that was mislocated dorsally to the trachea from its normal location on either side of the ventral trachea (Fig. 4B,C; Vid. s1,2), in agreement with the histology analysis (Fig1). By E12.5–13.5 the mutant left artery appears to extend into the right side crossing between the esophagus and the main bronchi as evidenced in both whole mount immunostaining (Fig. 4D,E; Vid. S3,4) and transverse sections at E12.5 (Fig. s5A) and E13.5 (Fig. 4H,I). This phenotype is similar to a “pulmonary artery sling”, where the left artery branches from the right artery instead of the pulmonary trunk and crosses between esophagus and trachea. This is an uncommon congenital pediatric condition identified in humans, usually associated with respiratory distress and requiring surgical intervention (Tretter et al. 2016). The etiology of such lesion is completely unknown and the similarity of Osr1 mutant phenotype offers an interesting model to explore this further. Pertinently, the disruption to the vasculature was restricted to the main vessels as the endothelial plexus in the distal lung region at E12.5 was indistinguishable between controls and mutants (Fig. s4A). Additionally the expression of Gli target Foxf1 is largely normal in Osr1 mutants between E9.5 and E12.5, suggesting its independence from cross regulation of Osr1 (Fig. s4A).
SMA staining also revealed reduced smooth muscle differentiation on the dorsal trachea, the main bronchi and around the esophagus in the Osr1−/− mutants (Fig. 4B,C). In addition, the remnant tracheal smooth muscle mislocated to the right instead of on the dorsal side (Fig. 4F,G). Meanwhile smooth muscle were expanded around the right bronchus but were reduced around the left bronchus (Fig. 4H,I). Finally there was a dramatic reduction of αSMA+ smooth muscle around the esophagus of E11.5–13.5 Osr1 mutants (Fig. 4B,C,F–I), although the esophageal epithelial tube is still present and the surrounding mesenchymal cells are in similar numbers.
Sox9 is both a marker of chondrocytes and also an important transcription factor supporting normal cartilage development. During tracheal mesenchymal differentiation, Sox9 and αSMA expression occupy the ventral and dorsal side, respectively, in a complementary pattern. Subsequently the longitudinal strip of Sox9 domain remodels into horizontal C-shaped rings (Yi et al. 2009). In the Osr1 mutant, the ventral tracheal Sox9 expression is still quite comparable to controls at E12.5 (Fig. s5A), suggesting that the cartilage ring phenotype might derive from the later remodeling of Sox9+ domains. HH, Wnt and Fgf10 signaling pathways have been shown to regulate mesenchymal patterning (Park et al. 2010; Sala et al. 2011; Snowball et al. 2015), but they appear to be largely comparable between Osr1 mutants and controls (Fig. s5B). Taken together Osr1 is likely to function downstream of those signaling pathways and function within the mesenchyme cell-autonomously to regulate patterning.
In summary, we showed that Osr1 mutants partially pheno-copy HH mutants in pulmonary artery, cartilage and smooth muscle formation, further supporting that Osr1 mediates HH function during foregut development.
3.6 Osr1 is required cell autonomously in the mesenchyme
Since Osr1 is expressed in both epithelial and mesenchymal cells in the foregut at E9 but only the mesenchymal Osr1 is dependent on HH signaling, we next performed tissue-specific conditional deletion of Osr1 using floxed Osr1f/f mice to identify the tissue in which Osr1 function is required. When combined with ubiquitously expressed EIIa-Cre, the Osr1f/f embryos recapitulated the germ line null mutant phenotypes, indicating that Osr1f allele can be efficiently inactivated by Cre (Lan et al. 2011). We crossed Dermo1Cre; Osr1+/− and Foxa2creER; Osr1+/− male mice to Osr1f/f females, respectively, to generate embryos with Osr1 inactivation specifically in mesenchymal or epithelial cells.
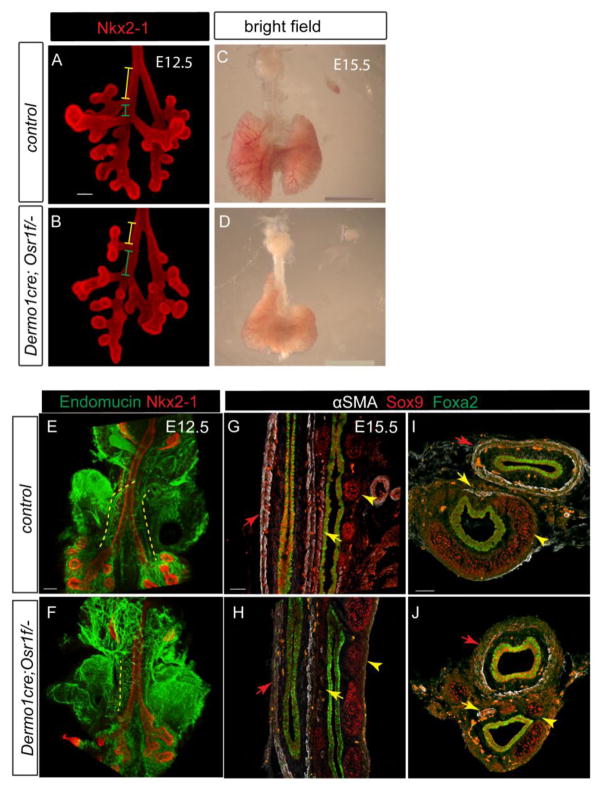
Epithelial-specific Osr1-deleted embryos showed the respiratory domains of similar size and shape as in control littermates at E10.5 (Fig. s6B), indicating that the epithelial Osr1 is dispensable in supporting lung development. In contrast, the mesenchymal-specific Osr1-deleted embryos recapitulated the germ line null phenotype. In the Dermo1Cre;Osr1f/− mutants, the cranial lobe shifted anteriorly at E12.5 (Fig. 5A,B) and hypoplastic lungs with anteriorly extended right lobes were observed at E15.5 (Fig. 5C,D). Moreover, the mesenchymal specific mutants also had only one pulmonary artery with reduced expression of endothelial marker, reduced esophageal and tracheal smooth muscle and disorganized Sox9+ cartilage domains (Fig. 5E–J). Similar to the germline knockout, the proximal-distal patterning and distal vasculature were not affected in the mesenchymal specific knockout (Fig. s6A). The less severe phenotype in the mesenchymal specific knockout is likely due to incomplete deletion of Osr1. These data support the conclusion that the Osr1 is predominantly required in the foregut mesenchyme to regulate foregut development.
Fig. 5.
Osr1 is required in the mesenchyme to support foregut development. (AD) Confirmation of loss of Osr1 in the mesenchyme. Exon specific in situ detecting either exon1 which is not floxed, and exon2 which is within the floxed region. (E–F) In the mesenchymal conditional knockout of Osr1, cranial lobe anterior shifting is evident at E12.5 (the relative distance ratio of yellow to green line), though to a lesser extent compared to the germline Osr1 mutant. Scale bars: 100 μm. (G–H) At E15.5, the mesenchymal conditional knockout lung demonstrates very similar hypoplasia and misshaping as the germline mutant. Scale bar: 2 mm. (I–J) Whole mount IF shows compromised pulmonary artery (dotted line) development in mesenchymal Osr1 knockout. Scale bar: 100 μm. (K–N) Longitudinal (K,L) and transverse (M–N) sections demonstrate failure of two layers of smooth muscle (red arrows) formation surrounding esophagus without Osr1 in mesenchyme, sparse trachea smooth muscle (yellow arrows) formation as well as disorganized tracheal cartilage (yellow arrowheads). Scale bars: 50 μm.
3.7 Osr1 expressing multi-potent cells gave rise to smooth muscle and chondrocytes in the foregut
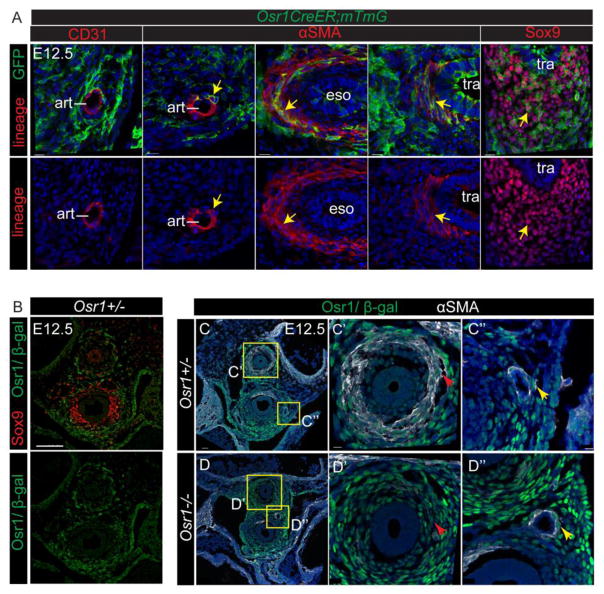
To further narrow down which of the mesenchymal defects were cell autonomous, we first determined which cell populations were derived from Osr1 expressing cells by lineage tracing experiment. Osr1creER were crossed with RosamTmG reporter mice. When the mTmG locus is recombined upon CreER activation with tamoxifen, the CreER expressing cells are permanently labeled with membrane bound GFP expression. Tamoxifen was administered to dams at E7.5–E8.5 to label Osr1 expressing cells and foreguts were analyzed at E12.5. We found GFP+ cells which co-expressed markers of the smooth muscle around the arteries, esophagus and trachea as well as in the Sox9+ tracheal chondrocytes, but not arterial endothelial cells (Fig. 6A). This demonstrated that Osr1 expression marks a multi-potential cell population at E7.5–E8.5 in the foregut, indicating that the mesenchymal tissue defects in the Osr1 mutants were likely to be cell-autonomous.
Fig. 6.
Osr1 expressing progenitors contribute to multiple mesenchymal lineages. (A) GFP lineage labeling is co-stained with multiple mesenchymal lineage markers at E12.5. The upper panel shows co-staining while the lower panel shows the lineage markers alone. Arrowheads indicate co-stainings. Art, artery; eso, esophagus; tra, trachea. Scale bars: 15 μm. (B) β-gal co-staining with Sox9 on transverse sections. Scale bar: 100 μm. (C–D) β-gal co-staining with αSMA on transverse sections. Red arrows indicate esophageal smooth muscle, while yellow arrows indicate pulmonary artery. Scale bar: 50 μm. Scale bars in inserts: 10 μm.
Furthermore, β-gal immuno-staining in the Osr1lacZ/+ heterozygous knockin embryos revealed LacZ-expressing mesenchymal cells around the esophagus, trachea, bronchi and pulmonary arteries at E12.5 (Fig. 6B, C), consistent with the lineage tracing (Fig. 6A) and Osr1 in situ data (Fig. 1). Interestingly, Osr1LacZ expression was very low in the Sox9-expressing chondrocytes, even though the lineage tracing indicate that this tissue is derived from Osr1-expressing cells (Fig. 6A). This suggests that Osr1 is down regulated as the proximal mesenchyme around the trachea differentiated into chondrocyte (Fig. 6B,C,D). Examination of LacZ expression in the Osr1lacZ/lacZ mutant embryos (Fig. 6C) revealed comparable β-gal expressing mesenchymal tissue around the esophagus and pulmonary arteries of both homozygous mutants and heterozygous controls, suggesting that the Osr1 mutant cells did not die or fail to proliferate, but rather did not differentiate into αSMA expressing smooth muscle.
Together these data indicate that Osr1 is cell-autonomously required in the foregut mesenchymal cells for proper development of the esophagus, airway and pulmonary artery smooth muscle as well as trachea cartilage.
4. Discussion
In this study we demonstrated that the zinc finger transcription factor Osr1 is a transcriptional target of HH signaling in the mouse embryonic foregut mesenchyme. We showed that Osr1 expression is dependent on HH activity and the HH effectors Gli3 binds to a potential enhancer region near the Osr1 genomic loci. Analysis of Osr1 mutant embryos revealed that Osr1 function can account for some of the key roles of HH in foregut development including respiratory specification, lobulation and pulmonary artery development (Fig. 7).
Fig. 7.
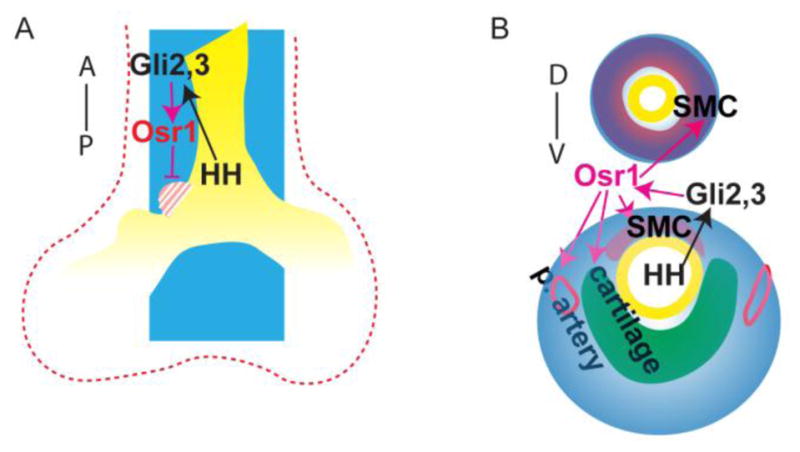
In conclusion, Osr1 functions downstream of HH pathway and regulate multiple aspects of foregut development. (A) during lobulation, Osr1, which expression is restricted in the medial mesenchyme, prevents medial/anterior shifting of cranial lobe; (B) during trachea and esophagus mesenchyme differentiation, Osr1 is expressed in the peripheral mesenchyme surrounding trachea and esophagus, supporting esophagus and trachea smooth muscle differentiation, trachea cartilage ring formation and pulmonary arteries development. Blue spaces indicate Osr1 expression domains; red dotted line outlines the splanchnic mesenchyme. Yellow marks the endoderm; red spaces mark esophageal smooth muscle in the dorsal and tracheal smooth muscle in the ventral; green space marks the cartilage; red circles show pulmonary artery smooth muscle. D, dorsal; V, ventral; A, anterior; P, posterior.
We recently demonstrated that HH ligands are essential, in both mice and frogs, to activate expression of lung-inducing signals Wnt2/2b and Bmp in the splanchnic mesoderm (Rankin et al. 2016), but it was unclear from these studies whether these were direct or indirect HH/Gli targets. In the present study we found that Osr1−/− embryos have fewer lung progenitors, possible due in part to reduced lung-inducing Wnt signals. However our data suggest that Osr1 alone cannot account for Wnt2/2b and Bmp4 transcription and it is likely they that are activated by HH/Gli or other HH-dependent factors. Such factors include Tbx transcription factors, supported by the evidence that Tbx5 is reduced without Gli2/3 (Rankin et al. 2016) and Tbx4/5 regulate Wnt2/2b in the fetal lung (Arora et al. 2012). Interestingly, those HH targets might cross-regulate each other as well, to fine-tune the transcriptional network. For example, Osr1 has been suggested to be a downstream target of Tbx5 in the second heart field (Xie et al. 2012). It remains to be determined if and how Tbx5 cooperates with the HH pathway to regulate Osr1 during foregut development.
Our analysis indicates that Osr1 is required for the proper location of the primary branching of the lung lobes. Based on the medial expression pattern of Osr1, one possibility was that Osr1 needs to be down regulated in regions of the growing lung buds where branching occurs. However, preliminary experiments when we expressed an inducible Osr1 using a ubiquitous Cre driver to ectopically express Osr1 in all cells including the branching regions, we did not observe obvious changes in branching (data not shown). Nevertheless, the fact that Osr1 has a defect in primary lobulation but not branching morphogenesis as such makes Osr1 mutants a good model for further studies into the distinct molecular mechanisms that control lobulation versus branching.
Our data indicate that Osr1 is a potential effector of HH pathway to support pulmonary artery development. Osr1 mutants have reduced vasculature connection between the heart and the lungs, similar to HH mutants. Meanwhile the known HH target Foxf1 mutants appear only recapitulate the distal lung vasculature plexus defects of HH pathway (Li et al. 2004; Ren et al. 2014). Interestingly, HH activity is required in mesenchymal but not in endothelial or smooth muscle cells during pulmonary artery formation (Peng et al. 2013). Given that Osr1 is expressed in arterial smooth muscle cells as well as the surrounding mesenchymal cells, it remains to be determined the more lineage specific functions of Osr1 using particular lineage restricted Cre lines. Various congenital pulmonary artery defects are identified in humans, including the previously mentioned “pulmonary artery sling” and pulmonary atresia, where one or two arteries are lacking (Lofland 2009). The similar phenotype of Osr1 mouse mutant suggests Osr1 as a potential risk allele for such a distinct lesion. Moreover an OSR1 gene variant is associated with high blood pressure in a GWAS study (Kato et al. 2015). Our finding here that Osr1 is expressed in smooth muscle cells surrounding pulmonary arteries provides some clue for this association.
The absence or malformation of cartilage rings is a pathological feature of tracheomalacia patients where the trachea walls collapse during expiration due to lack of support and rigidity provided by the cartilage (Kugler and Stanzel 2014). The HH pathway is required to support tracheal cartilage differentiation (Miller et al. 2004), and the disorganization of cartilage rings observed in Osr1 mutants suggested that Osr1 functions downstream of HH signaling to regulate such events. Little is known how the cartilage rings remodeling occurs and the observation here suggests that either Osr1 is required in progenitor cells before Sox9 differentiation to prime those cells for proper remodeling later, or that Osr1 is re-expressed in subpopulations of such cells to downregulate Sox9 to segregate the domain into rings.
Despite the functional importance of esophageal muscle layers for food intake and transport to stomach (Krauss et al. 2016), very little is known about their development, especially the early events that establish the initial two layers of smooth muscle. We show here that the Osr1 is required for smooth muscle differentiation, likely in a cell autonomous way. HH signaling is required in stomach and intestine SMC differentiation (Mao et al. 2010). The HH target Foxf1 is also required in smooth muscle cells to support such differentiation as smooth muscle cell conditional Foxf1 mutants display thinner esophageal smooth muscle (Hoggatt et al. 2013), which is apparently less severe than in the HH mutants. Considering the almost absent esophageal smooth muscle in the Osr1 mutants, we propose that the HH-Foxf1/Osr1 regulatory element is required for esophageal SMC development.
In summary, our study suggests Osr1 coordinates development of the mammalian respiratory system, digestive track and the cardiovascular network downstream of HH signaling. So far OSR1 has been associated with congenital kidney and heart defects in both animal models (Wang et al. 2005) and in human patients (Zhang et al. 2016). Our results suggest Osr1 is a potential gene associated with human respiratory and digestive diseases as well, serving as another screening candidate and potential treatment target in humans with congenital respiratory and digestive defects.
Supplementary Material
Fig. s1. Osr2 is dispensable during respiratory specification in mouse. (A–C) Osr2 in situ staining on transverse sections of kidney (A) as a positive control and in foreguts (B,C). Yellow dotted lines mark endoderm. (D) IF staining on transverse sections at E9.5 and whole mount at E10.5. Scale bar in the upper panel: 50 μm. Scale bar in the lower panel: 200 μm.
Table S1. Antibodies used in immunofluorescence staining.
3D rotational view of whole mount staining of αSMA with dissected E11.5 control foregut. Different regions of αSMA were isolated and pseudo-colored. Red marks the smooth muscle associated with the pulmonary arteries; green marks the smooth muscle associated with trachea; purple marks the smooth muscle associated with esophagus.
Overall body size and Proximal-distal patterning is independent on Osr1. (A) Bright field view of E12.5 embryos. Scale bar: 2mm. (B) IF staining on the transverse sections in foregut, whole mount of dissected lungs and frontal sections of lung. Scale bar in the first row: 50 μm; second row: 100 μm; third row: 200 μm; fourth row: 150 μm.
Osr1 functions downstream of the branching signals Wnt2, Shh, Bmp4 and Fgf10. (A) In situ of Wnt2 on transverse sections of foregut at E9.5 and dissected lungs during E10.5–E12.5; in situ of Shh on transverse sections of foregut at E9.5 and E10.5 as well as dissected lungs during E11.5–E12.5. (B) pSmad1 IF staining on transverse foregut at E10.5 and Bmp4 whole mount in situ with dissected foregut from E10.5 to E12.5. (C) Whole mount in situ of Fgf10 on dissected foregut from E10.5–E12.5. Scale bars in whole mount view: 500 μm; transverse view: 100 μm.
Foxf1 expression and distal vascular plexus development is comparable between Osr1 mutants and controls. (A) IF staining on transverse sections and whole mount dissected lungs. Scale bars in whole mount view: 200 μm; transverse view: 100 μm.
Sox9 patterning as well as HH, Wnt and Fgf10 signals are comparable without Osr1 at E12.5. (A) IF staining on transverse sections from anterior trachea region to posterior lung region. Yellow arrows point to the mesenchymal Sox9 expression while the red arrows point to pulmonary arteries. Scale bar: 50 μm. (B) In situ on transverse sections at the level of trachea and main bronchi. Scale bar: 100 μm.
Mesenchymal Osr1 expressiong is dispensible for proximal-distal epithelial pattering and epithelial Osr1 is dispensible during foregut development. (A) IF staining on frontal and longitudinal sections of lungs. (C) Whole mount staining on embryos. Scale bars: 100 μm.
3D rotational view of whole mount staining of αSMA with dissected E11.5 mutant foregut. Different regions of αSMA were isolated and pseudo-colored. Red marks the smooth muscle associated with the pulmonary arteries; green marks the smooth muscle associated with trachea; purple marks the smooth muscle associated with esophagus.
Slice views of E12.5 dissected foregut with heart attached from control embryos. Endomucin is in green, αSMA is in purple and Nkx2-1 is in red.
Slice views of E12.5 dissected foregut with heart attached from Osr1 mutant embryos. Endomucin is in green, αSMA is in purple and Nkx2-1 is in red.
Highlights.
Zinc finger transcription factor Osr1 is a Hedgehog target in the foregut mesenchyme.
Osr1 promotes respiratory specification through modulating mesenchymal signals.
Osr1 is required for proper positioning of primary lung buds.
Osr1 expression marks multi-potential foregut mesenchymal progenitors.
Osr1 is required in the foregut mesenchyme for development of pulmonary arteries, smooth muscle and cartilage rings.
Acknowledgments
Funding
This work was supported by HL114898 to AMZ.
We thank Confocal Imaging Core for helping with imaging and analysis. We also thank Dr. Debora Sinner, Dr. Susan Wert, Dr. Anne Karina Perl, Dr. Samantha Brugmann, all Dr. Aaron Zorn and Dr. James Wells lab members for continuous support and critical feedbacks.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arora R, Metzger RJ, Papaioannou VE. Multiple roles and interactions of Tbx4 and Tbx5 in development of the respiratory system. PLoS genetics. 2012;8:e1002866. doi: 10.1371/journal.pgen.1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai CB, Auerbach W, Lee JS, Stephen D, Joyner AL. Gli2, but not Gli1, is required for initial Shh signaling and ectopic activation of the Shh pathway. Development. 2002;129:4753–4761. doi: 10.1242/dev.129.20.4753. [DOI] [PubMed] [Google Scholar]
- Bai CB, Joyner AL. Gli1 can rescue the in vivo function of Gli2. Development. 2001;128:5161–5172. doi: 10.1242/dev.128.24.5161. [DOI] [PubMed] [Google Scholar]
- Chuang PT, Kawcak T, McMahon AP. Feedback control of mammalian Hedgehog signaling by the Hedgehog-binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes & development. 2003;17:342–347. doi: 10.1101/gad.1026303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley JC, Bellusci S, Perkins D, Hogan BL. Evidence for the involvement of the Gli gene family in embryonic mouse lung development. Developmental biology. 1997;188:337–348. doi: 10.1006/dbio.1997.8644. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, Vokes SA, McMahon AP, Kalinichenko VV, Moskowitz IP. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS genetics. 2014;10:e1004604. doi: 10.1371/journal.pgen.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggatt AM, Kim JR, Ustiyan V, Ren X, Kalin TV, Kalinichenko VV, Herring BP. The transcription factor Foxf1 binds to serum response factor and myocardin to regulate gene transcription in visceral smooth muscle cells. The Journal of biological chemistry. 2013;288:28477–28487. doi: 10.1074/jbc.M113.478974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DR. Extra-toes: anew mutant gene causing multiple abnormalities in the mouse. Journal of embryology and experimental morphology. 1967;17:543–581. [PubMed] [Google Scholar]
- Kato N, Loh M, Takeuchi F, Verweij N, Wang X, Zhang W, Kelly TN, Saleheen D, Lehne B, Mateo Leach I, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47:1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauss RS, Chihara D, Romer AI. Embracing change: striated-for-smooth muscle replacement in esophagus development. Skeletal muscle. 2016;6:27. doi: 10.1186/s13395-016-0099-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugler C, Stanzel F. Tracheomalacia. Thoracic surgery clinics. 2014;24:51–58. doi: 10.1016/j.thorsurg.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Kugler MC, Joyner AL, Loomis CA, Munger JS. Sonic hedgehog signaling in the lung. From development to disease. American journal of respiratory cell and molecular biology. 2015;52:1–13. doi: 10.1165/rcmb.2014-0132TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y, Liu H, Ovitt CE, Jiang R. Generation of Osr1 conditional mutant mice. Genesis. 2011;49:419–422. doi: 10.1002/dvg.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead S. 2012 AHCA/NCAL National Quality Award: commitment, achievement, excellence. Provider. 2012;38:40–42. 44, 47–48. [PubMed] [Google Scholar]
- Li Y, Zhang H, Choi SC, Litingtung Y, Chiang C. Sonic hedgehog signaling regulates Gli3 processing, mesenchymal proliferation, and differentiation during mouse lung organogenesis. Developmental biology. 2004;270:214–231. doi: 10.1016/j.ydbio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Lim L, Kalinichenko VV, Whitsett JA, Costa RH. Fusion of lung lobes and vessels in mouse embryos heterozygous for the forkhead box f1 targeted allele. American journal of physiology Lung cellular and molecular physiology. 2002;282:L1012–1022. doi: 10.1152/ajplung.00371.2001. [DOI] [PubMed] [Google Scholar]
- Liu H, Lan Y, Xu J, Chang CF, Brugmann SA, Jiang R. Odd-skipped related-1 controls neural crest chondrogenesis during tongue development. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18555–18560. doi: 10.1073/pnas.1306495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lofland GK. An overview of pulmonary atresia, ventricular septal defect, and multiple aorta pulmonary collateral arteries. Progress in Pediatric Cardiology. 2009;26:65–70. doi: 10.1053/j.pcsu.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Madison BB, McKenna LB, Dolson D, Epstein DJ, Kaestner KH. FoxF1 and FoxL1 link hedgehog signaling and the control of epithelial proliferation in the developing stomach and intestine. The Journal of biological chemistry. 2009;284:5936–5944. doi: 10.1074/jbc.M808103200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlapuu M, Ormestad M, Enerback S, Carlsson P. The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development. 2001;128:155–166. doi: 10.1242/dev.128.2.155. [DOI] [PubMed] [Google Scholar]
- Mao J, Kim BM, Rajurkar M, Shivdasani RA, McMahon AP. Hedgehog signaling controls mesenchymal growth in the developing mammalian digestive tract. Development. 2010;137:1721–1729. doi: 10.1242/dev.044586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulley D, Wienhold M, Sun X. The pulmonary mesenchyme directs lung development. Current opinion in genetics & development. 2015;32:98–105. doi: 10.1016/j.gde.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LA, Wert SE, Clark JC, Xu Y, Perl AK, Whitsett JA. Role of Sonic hedgehog in patterning of tracheal-bronchial cartilage and the peripheral lung. Developmental dynamics: an official publication of the American Association of Anatomists. 2004;231:57–71. doi: 10.1002/dvdy.20105. [DOI] [PubMed] [Google Scholar]
- Morita K, Yokoi A, Fukuzawa H, Hisamatsu C, Endo K, Okata Y, Tamaki A, Mishima Y, Oshima Y, Maeda K. Surgical intervention strategies for congenital tracheal stenosis associated with a tracheal bronchus based on the location of stenosis. Pediatric surgery international. 2016 doi: 10.1007/s00383-016-3928-8. [DOI] [PubMed] [Google Scholar]
- Motoyama J, Liu J, Mo R, Ding Q, Post M, Hui CC. Essential function of Gli2 and Gli3 in the formation of lung, trachea and oesophagus. Nat Genet. 1998;20:54–57. doi: 10.1038/1711. [DOI] [PubMed] [Google Scholar]
- Mugford JW, Sipila P, McMahon JA, McMahon AP. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Developmental biology. 2008;324:88–98. doi: 10.1016/j.ydbio.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Park EJ, Sun X, Nichol P, Saijoh Y, Martin JF, Moon AM. System for tamoxifen-inducible expression of cre-recombinase from the Foxa2 locus in mice. Developmental dynamics: an official publication of the American Association of Anatomists. 2008;237:447–453. doi: 10.1002/dvdy.21415. [DOI] [PubMed] [Google Scholar]
- Park J, Zhang JJ, Moro A, Kushida M, Wegner M, Kim PC. Regulation of Sox9 by Sonic Hedgehog (Shh) is essential for patterning and formation of tracheal cartilage. Developmental dynamics: an official publication of the American Association of Anatomists. 2010;239:514–526. doi: 10.1002/dvdy.22192. [DOI] [PubMed] [Google Scholar]
- Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, Morrisey EE. Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature. 2013;500:589–592. doi: 10.1038/nature12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Gallas AL, Neto A, Gomez-Skarmeta JL, Zorn AM. Suppression of Bmp4 signaling by the zinc-finger repressors Osr1 and Osr2 is required for Wnt/beta-catenin-mediated lung specification in Xenopus. Development. 2012;139:3010–3020. doi: 10.1242/dev.078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Han L, McCracken KW, Kenny AP, Anglin CT, Grigg EA, Crawford CM, Wells JM, Shannon JM, Zorn AM. A Retinoic Acid-Hedgehog Cascade Coordinates Mesoderm-Inducing Signals and Endoderm Competence during Lung Specification. Cell reports. 2016 doi: 10.1016/j.celrep.2016.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Ustiyan V, Pradhan A, Cai Y, Havrilak JA, Bolte CS, Shannon JM, Kalin TV, Kalinichenko VV. FOXF1 transcription factor is required for formation of embryonic vasculature by regulating VEGF signaling in endothelial cells. Circulation research. 2014;115:709–720. doi: 10.1161/CIRCRESAHA.115.304382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala FG, Del Moral PM, Tiozzo C, Alam DA, Warburton D, Grikscheit T, Veltmaat JM, Bellusci S. FGF10 controls the patterning of the tracheal cartilage rings via Shh. Development. 2011;138:273–282. doi: 10.1242/dev.051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowball J, Ambalavanan M, Whitsett J, Sinner D. Endodermal Wnt signaling is required for tracheal cartilage formation. Developmental biology. 2015;405:56–70. doi: 10.1016/j.ydbio.2015.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosic D, Richardson JA, Yu K, Ornitz DM, Olson EN. Twist regulates cytokine gene expression through a negative feedback loop that represses NF-kappaB activity. Cell. 2003;112:169–180. doi: 10.1016/s0092-8674(03)00002-3. [DOI] [PubMed] [Google Scholar]
- Swarr DT, Morrisey EE. Lung endoderm morphogenesis: gasping for form and function. Annual review of cell and developmental biology. 2015;31:553–573. doi: 10.1146/annurev-cellbio-100814-125249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter JT, Tretter EM, Rafii DY, Anderson RH, Bhatla P. Fetal Diagnosis of Abnormal Origin of the Left Pulmonary Artery. Echocardiography. 2016;33:1258–1261. doi: 10.1111/echo.13239. [DOI] [PubMed] [Google Scholar]
- Wang Q, Lan Y, Cho ES, Maltby KM, Jiang R. Odd-skipped related 1 (Odd 1) is an essential regulator of heart and urogenital development. Developmental biology. 2005;288:582–594. doi: 10.1016/j.ydbio.2005.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirauch MT, Yang A, Albu M, Cote AG, Montenegro-Montero A, Drewe P, Najafabadi HS, Lambert SA, Mann I, Cook K, et al. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014;158:1431–1443. doi: 10.1016/j.cell.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Developmental cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi L, Domyan ET, Lewandoski M, Sun X. Fibroblast growth factor 9 signaling inhibits airway smooth muscle differentiation in mouse lung. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:123–137. doi: 10.1002/dvdy.21831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang KK, Xiang M, Zhou L, Liu J, Curry N, Heine Suner D, Garcia-Pavia P, Zhang X, Wang Q, Xie L. Gene network and familial analyses uncover a gene network involving Tbx5/Osr1/Pcsk6 interaction in the second heart field for atrial septation. Human molecular genetics. 2016;25:1140–1151. doi: 10.1093/hmg/ddv636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. s1. Osr2 is dispensable during respiratory specification in mouse. (A–C) Osr2 in situ staining on transverse sections of kidney (A) as a positive control and in foreguts (B,C). Yellow dotted lines mark endoderm. (D) IF staining on transverse sections at E9.5 and whole mount at E10.5. Scale bar in the upper panel: 50 μm. Scale bar in the lower panel: 200 μm.
Table S1. Antibodies used in immunofluorescence staining.
3D rotational view of whole mount staining of αSMA with dissected E11.5 control foregut. Different regions of αSMA were isolated and pseudo-colored. Red marks the smooth muscle associated with the pulmonary arteries; green marks the smooth muscle associated with trachea; purple marks the smooth muscle associated with esophagus.
Overall body size and Proximal-distal patterning is independent on Osr1. (A) Bright field view of E12.5 embryos. Scale bar: 2mm. (B) IF staining on the transverse sections in foregut, whole mount of dissected lungs and frontal sections of lung. Scale bar in the first row: 50 μm; second row: 100 μm; third row: 200 μm; fourth row: 150 μm.
Osr1 functions downstream of the branching signals Wnt2, Shh, Bmp4 and Fgf10. (A) In situ of Wnt2 on transverse sections of foregut at E9.5 and dissected lungs during E10.5–E12.5; in situ of Shh on transverse sections of foregut at E9.5 and E10.5 as well as dissected lungs during E11.5–E12.5. (B) pSmad1 IF staining on transverse foregut at E10.5 and Bmp4 whole mount in situ with dissected foregut from E10.5 to E12.5. (C) Whole mount in situ of Fgf10 on dissected foregut from E10.5–E12.5. Scale bars in whole mount view: 500 μm; transverse view: 100 μm.
Foxf1 expression and distal vascular plexus development is comparable between Osr1 mutants and controls. (A) IF staining on transverse sections and whole mount dissected lungs. Scale bars in whole mount view: 200 μm; transverse view: 100 μm.
Sox9 patterning as well as HH, Wnt and Fgf10 signals are comparable without Osr1 at E12.5. (A) IF staining on transverse sections from anterior trachea region to posterior lung region. Yellow arrows point to the mesenchymal Sox9 expression while the red arrows point to pulmonary arteries. Scale bar: 50 μm. (B) In situ on transverse sections at the level of trachea and main bronchi. Scale bar: 100 μm.
Mesenchymal Osr1 expressiong is dispensible for proximal-distal epithelial pattering and epithelial Osr1 is dispensible during foregut development. (A) IF staining on frontal and longitudinal sections of lungs. (C) Whole mount staining on embryos. Scale bars: 100 μm.
3D rotational view of whole mount staining of αSMA with dissected E11.5 mutant foregut. Different regions of αSMA were isolated and pseudo-colored. Red marks the smooth muscle associated with the pulmonary arteries; green marks the smooth muscle associated with trachea; purple marks the smooth muscle associated with esophagus.
Slice views of E12.5 dissected foregut with heart attached from control embryos. Endomucin is in green, αSMA is in purple and Nkx2-1 is in red.
Slice views of E12.5 dissected foregut with heart attached from Osr1 mutant embryos. Endomucin is in green, αSMA is in purple and Nkx2-1 is in red.