Abstract
Secretory Immunoglobulin A (sIgA) is a major glycoprotein in milk and plays a key role in mediating immune protection of the gut mucosa. Although it is a highly glycosylated protein, its site-specific glycosylation and associated glycan micro-heterogeneity has still not been fully elucidated. In this study, the site-specific glycosylation of sIgA isolated from human colostrum (n = 3) was analyzed using a combination of LC/MS and LC/MS/MS and in-house software (Glycopeptide Finder). The majority of the glycans found are bi-antennary structures with one or more acidic Neu5Ac residue, however a large fraction belonged to truncated complex structures with terminal GlcNAc. Multiple glycosites were identified with nearly 30 glycan compositions located at seven sites on the secretory component, six compositions at a single site on the J-chain, and 16 compositions at five sites on the IgA heavy (H) chain. Site-specific heterogeneity and relative quantitation of each composition and the extent of occupation at each site was determined using non-specific proteases. Additionally, 54 O-linked glycan compositions located at the IgA1 hinge region (HR) were identified by comparison against a theoretical O-glycopeptide library. This represents the most comprehensive report to date detailing the complexity of glycan micro-heterogeneity with relative quantitation of glycoforms for each glycosylation site on milk sIgA. This strategy further provides a general method for determining site-specific glycosylation in large protein complexes.
Keywords: Glycoproteomics, Glycan microheterogeneity, Mass spectrometry, sIgA, Site-specific glycosylation
INTRODUCTION
Secretory immunoglobulin A (sIgA) is the dominant immunoglobulin in the gut mucosal surface giving the mucosa specialized innate and adaptive defense against ingested pathogens and their toxins 1–3. sIgA is a protein complex consisting of two identical IgA monomers, joined together by a 16-kDa joint chain (J chain) with one potential N-glycosylation site, and the secretory component (SC) with seven potential N-glycosylation sites 4–6. The four polypeptide chains of sIgA are produced by two distinct cell types 6. Plasma cells close to the epithelium produce dimeric IgA with an attached J chain, while epithelial cells express the polymeric immunoglobulin receptor (pIgR) that binds to dimeric IgA. Notably, the SC is derived from the pIgR after cleavage of the trans-membrane tail on the surface of epithelial cells 7.
Glycosylation is one of the most common but also the most structurally complicated post-translational modification (PTM) of proteins. Approximately 70% of human proteins are predicted to be glycosylated 8, 9. sIgA is heavily glycosylated with both N- and/or O-linked oligosaccharides 10, which vary according to the isotype and allotype of the immunoglobulin. Its oligosaccharides are attributed significant roles in immune protection, cell signaling, cell-cell recognition, microbial adhesion and invasion 11–13. sIgA is one of the major antibodies produced in human milk and is essential for the passive immunity of infants against infection 14–16. Specific binding between sialylated glycans of sIgA and pathogens such as S-firmbriated Escherichia coli protects newborns from sepsis and meningitis due to infection 17. The large diversity of N-glycan structures particularly on the SC provides many glycan epitopes that are potent decoys for lectins on bacterial surfaces thereby inhibiting infection by attachment with epithelial surfaces 18–20. sIgA also carries galactose-terminating glycans that protect against toxins including ricin, a particularly toxic galactose-specific lectin 21.
Despite numerous studies establishing the effective protection provided by sIgA, the glycan heterogeneity and glycosite occupancy of this highly glycosylated protein remains incomplete. There have been studies showing the glycosylation of IgA in serum 22, 23, however milk is unique in that the protein also contains a highly glycosylated secretory component 10. The first comprehensive attempt to characterize the glycosylation of sIgA from a commercial sample was reported by Royle et al. 4, whereby polypeptide components were separated using gel-electrophoresis. Glycans were released for the respective gel spots corresponding to different polypeptides and characterized by mass spectrometry. The nature of the method yielded no site-specific information. Furthermore, the glycoforms spread the location of the glycoprotein on the gel, so releasing glycans from the largely-spread gel spot may not necessarily represent the majority of the glycoforms 24. Site-specific N-glycan analysis of sIgA was first performed by Deshpande et al. using in-gel trypsin digestion of a commercial sample 25. The results provided the characterization of several sites with the associated glycan heterogeneity. However, trypsin often produces large peptides, and in this case it could not resolve the glycosylation of 83Asn and 90Asn on the secretory component. Additionally, missed cleavages are common occurrences particularly if the tryptic sites are near sites of glycosylation 26–28. For these reasons, employing only tryptic digestion will often yield incomplete glycan information.
A more complicated aspect is the O-glycosylation analysis of sIgA. The IgA1 hinge region contains up to nine potential O-glycosylation sites clustering in a 19 amino acid peptide sequence 29. A variety of MS analytical approaches have been used to characterize the glycan heterogeneity present in this portion of the immunoglobulin with only moderate success30–33. Additionally, IgA1 O-glycan site localization has been reported combining different protease digestions and ECD/ETD 29, 32, 33. However in these studies sialic acid was removed enzymatically in order to simplify the glycopeptide heterogeneity.
In this paper, we present an extensive and comprehensive glycan analysis of sIgA from commercial pooled human colostrum and isolated from milk colostrum collected from three healthy women. This laboratory has developed methods for site-specific analysis using combinations of specific and non-specific proteolysis 34–40. The method includes an automated annotation software (Glycopeptide Finder) to analyze data from a nanoflow liquid chromatography coupled with quadrupole time-of-flight analyzer (nanoLC-Q-TOF). This analysis yielded both N- and O-glycosylation. Glycopeptide assignments were based on a combination of accurate mass, retention time analysis and tandem MS yielding glycopeptides with scalable lengths but including both non-specific and specific cleavages.
Materials and Methods
Chemicals and Materials
SSL7/Agarose and phosphate-buffered saline (PBS), and TRIzol were purchased from Invitrogen (San Diego, CA). Pronase E, cyanogen bromide (CNBr) activated sepharose 4B (S4B) beads, and pooled human colostrum IgA were purchased from Sigma-Aldrich (St. Louis, MO). PNGase F was purchased from New England Biolab (Ipswich, MA). Sequencing grade modified trypsin and proteoMAX solution were purchased from Promega (Madison, WI). Dialysis tubing (10MWCO) were purchased from Spectrum Lab (Rancho Dominguez, CA). Ammonium acetate and acetic acid were of analytical grade from Merck (Darmstadt, Germany). Graphitized carbon cartridges were purchased from Grace Davison Discovery Sciences (Deerfield, IL).
Human Colostrum Samples
Colostrum samples were collected from three healthy donors enrolled in the UC Davis Lactation Study who gave birth to term infants (> 38 weeks). Colostrum samples were manually collected on day 3–6 postpartum from one breast and transferred into polypropylene Falcon tubes, and frozen immediately in their kitchen freezers (−20 ºC) until weekly sample pick up by the study staff. Samples were transported to the lab on dry ice and stored in −80 ºC until processing.
The University of California Davis Institutional Review Board approved all aspects of the study and informed consent was obtained from all subjects. This trial was registered on clinicaltrials.gov (ID: NCT01817127).
Secretory Immunoglobulin A Extraction
sIgA from donor colostrum was extracted using Staphylococcal Superantigen-Like Protein 7 (SSL 7)/agarose 41. Briefly, whole human colostrum samples (0.5 mL) were centrifuged at 4000 xg, for 30 minutes at 4°C. A CaCl2 solution (pH 4.6) was added to the lower layer after the removal of top layer fat and lipids to a final concentration of 60 mM. The mixture was incubated 1 hour at room temperature (~25°C), and further centrifuged at 6750 xg for 30 minutes. The top layer was extracted as whey proteins. The empty column was packed with 1ml of SSL 7/agorose resin and equilibrated with 1X phosphate buffer saline (running buffer). The whey fractions were loaded onto the columns and the flow-through was collected and reloaded onto the column three times. Upon washing with 10 mL of the running buffer, sIgA bound to SSL 7 was eluted with 5 mL of 0.1 M glycine (pH 2–3) followed by the pH adjustment of the eluate to pH 7.5 by adding a neutralization buffer (1M Tris pH 8). Fractions were collected, dialyzed using a dialysis membrane with a molecular weight cut-off of 10,000 against nano-pure water, concentrated and stored at −20°C. Protein concentration was determined using the Bradford assay and 5 μl aliquots were assayed by SDS-PAGE to confirm protein purity. A 5-μL sample was mixed with 5 μL of laemli buffer, 1 μL of 550 mM dithiothreitol (DTT) at 60 °C for 1 hour prior to incubating with 2 μL of 450 mM iodoacetamide (IAA) for 30 min in the dark. The SDS-PAGE was performed at 140 V for 1 hr, followed by staining with coomasie blue for 1 hr. The gel was then unstained with water overnight.
Trypsin Digestion
A 100μL aliquot of 50 mM NH4HCO3 buffer was added to 10 μg isolated sIgA as described above, followed by reducing with 2 μL of 550 mM DTT for 50 min at 60 °C. A 4 μL solution of 450 mM IAA was then added, and carboxymethylation was performed by incubation for 30 min at room temperature in the dark. Reduced and carboxymethylated sIgA was digested using trypsin 1 μL (1 μg/μL) in 100 μL of 50 mM NH4HCO3 buffer, for 18 hr at 37 °C. The digests were purified on a reverse-phase C18 pipette zip-tip (Agilent Technology, Wilmington, DE, USA). The C18 zip-tip was preconditioned successively with acetonitrile (ACN) (150 μL, 10 times) and water (150 μL, 10 times). The tryptic digest was loaded on to the zip-tip by pipetting 20 times followed by similar 10 washings with nanopure water (150 μL). Tryptic peptides from sIgA were eluted with 0.05% formic acid (FA) in 80% ACN in water (v/v) (200 μL, 20 times extractions), dried down, and reconstituted in 20 μL nanopure water. Standard proteomic analysis of the extracted sIgA was performed to further verify the purity.
Pronase E Digestion and Glycopeptide Cleanup
Pronase E was covalently coupled to CNBr activated sepharose beads via coupling chemistry as reported previously 34, 36, 38. A 100-μg sample was dissolved in 100 μL of 50 mM NH4HCO3 buffer followed by reduction with 2 μL of 550 mM DTT for 50 min at 60 °C. A 4-μL solution of 450 mM IAA was added, and carboxymethylation was performed for 50 min at room temperature in the dark. Reduced and carboxymethylated samples were added to the pronase beads and incubated at 37 ºC for 18 h with gentle agitation. The glycopeptide digest was desalted and enriched via solid phase extraction (SPE) using graphitized carbon cartridges. After loading the glycopeptide on preconditioned 150-mg bed volume graphitized carbon cartridges (Grace Davison Discovery Sciences, Deerfield, IL, USA), the cartridges were washed with 6-mL nanopure water. A clean mixture of glycopeptides were eluted with 6 mL of 0.05% Trifluoroacetic acid (TFA) in 40% ACN in water (v/v) and 6 mL of 0.05% TFA in 80% ACN in water (v/v). Collected fractions were dried down and then reconstituted in 20 μL of nanopure water. A 2-μL volume was used for MS analysis.
Determination of O-glycosylation by PNGase F followed by Trypsin Digestion
A 10-μg sample of sIgA dissolved in 50 μL NH4HCO3 buffer solution was reduced by DTT (1 μL, 550 mM) and carboxymethylated by adding IAA (2 μL, 450 mM). The solution was digested by the addition of PNGase F (1 μL) for 16 hours at 37 °C to remove the N-glycans. Trypsin digestion was added to the solution, which was incubated at 37 °C for 18 hours for digestion. Glycopeptides in the digest were purified via hydrophilic affinity separation according to a method described previously 42. Briefly, O-glycopeptides were enriched with a 15 μL packed volume of sepharose CL4B in 1 mL of an organic solvent of 1-butanol/ethanol/H2O (5:1:1, v/v). Followed by shaking for 45 min, the gel was washed twice with the same organic solvent prior to incubating with an aqueous solvent of ethanol/H2O (1:1, v/v) for 30 min. The solution phase was collected after centrifugation and dried prior to reconstitution with 20 μL nanopure water.
LC/MS analysis of glycopeptides
A nano-HPLC-Chip Q-TOF instrument using the Agilent 1200 series microwell-plate autosampler (maintained at 6 ºC by the thermostat), capillary pump, nano pump, HPLC-Chip interface, and the Agilent 6520 Q-TOF MS (Agilent Technologies, Inc., Santa Clara, CA) was used in this study. The chip is equipped with a 40 nL enrichment column and a 43 × 0.075 mm ID analytical column. A graphitized carbon stationary phase was used for pronase produced glycopeptides, while C18 stationary phase was used for the tryptic peptides. Both mobile phases consists of 0.1% formic acid in 3% ACN in water (v/v) and 0.1% formic acid in 90% ACN in water (v/v) as solvent A and B, respectively. The gradient was performed on the analytical column to separate the pronase glycopeptides with a flow rate at 0.4 μL/min. The samples were eluted in 45 min with the following gradient: 0% B (0.00–2.50 min); 0 to 16% B (2.50–20.00 min); 16 to 44% B (20.00–30.00 min); 44 to 100% B (30.00–35.00 min) and 100% B (35.00–45.00 min). The tryptic glycopeptides/peptides were eluted in 87 min with the following gradient: 1% to 8% B (0.00–5.00 min); 8% to 26.5% B (5.00 min–48.00 min); 26.5% to 73% B (48.00 min–75 min); 73% to 99% (75.00 min–77.00 min); 99%B (77.00 min–87.00 min).
The Agilent 6520 Q-TOF MS was operated in the positive ion mode for MS and MS/MS analysis. The recorded mass ranges were m/z 500–3,000 for MS-only and m/z 50–3,000 for MS/MS. Acquisition rates were 0.63 spectrum/s for both MS and MS/MS. All mass spectra were internally calibrated using the G1969-85000 ESI tuning mix (Agilent Technologies, Inc., Santa Clara, CA), with reference masses at m/z 922.010, and 1,521.971 in the positive ion mode. The parameters used were as follows: drying gas (N2) temperature 325°C, drying gas flow rate 3 L/min, capillary voltage 1800 V, fragmentor voltage 150 V, skimmer voltage 65 V, and capillary cunnrent 0.08 μA, respectively. For MS/MS mode, the collision energies for each compound were calculated as follows:
Data Processing
The MS/MS data of the tryptic peptides were analyzed using X! Tandem (www.thegpm.org). X! Tandem was set up to search the Swissprot human complete proteome database. X! Tandem was searched with a fragment ion mass tolerance of 80 ppm and a parent ion tolerance of 100 ppm. Iodoacetamide derivative of cysteine was specified in X! Tandem as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan were specified in X! Tandem as variable modifications.
Data analyses for N-glycopeptides were performed with the MassHunter Qualitative Analysis software ver. B.03.01 (Agilent Technologies, Inc., Santa Clara, CA). Deconvolution was performed in the software with the grouping peak spacing tolerant of 0.0025 m/z plus 7 ppm and isotope mode was set for peptides. The mass list of the glycopeptide precursor ions and the associated tandem spectra were analyzed via an in-house software (Glycopeptide Finder)39 set to a 5% false-discovery rate. All N-glycopeptide assignments were made based on the mass of each potential glycopeptide, the amino acid sequence(s) of the protein(s), and a specific tolerance level (< 20 ppm). The presence of carbohydrate-specific oxonium fragment ions was used as diagnostic ions to sort the product tandem spectra.
For the O-glycopeptide identification, the tryptic digest after PNGase F treatment was analyzed in both MS and MS/MS mode. A theoretical O-glycopeptide library was created using the tryptic peptide from the HR (HYTNPSQDVTVPCPVPSTPPTPSPSTPPTPSPSCCHPR) with glycan compositions within a specified range (HexNAc 0–6, Hex 0–6, Sialic Acid 0–12) based on previous results25, 30, 33 and 0 to 3 carbamidomethyl groups. The resulting library is composed by 780 theoretical HR O-glycopeptides ranging in mass between 3964.818 Da and 9819.821 Da being the smaller difference between compounds larger than 1 Da. The library included empirical formulae in order to compare isotopic patterns. MS data was analyzed with MassHunter Qualitative Analysis software ver. B.03.01 (Agilent Technologies, Inc., Santa Clara, CA). Compound ion signals with different charge states were grouped using the Molecular Feature algorithm (MFE) and compared against the O-glycopeptide library with an error tolerance of 30 ppm. Only ions with a signal-to-noise ratio above 3 were considered. Isotopic distributions were manually checked. The extracted compound chromatograms (ECCs) for each O-glycopeptide were obtained and their corresponding retention times compared.
Results and Discussion
Glycan-heterogeneity of sIgA from Human Colostrum
Colostrum sIgA was extracted from three individual donors. Isolation of sIgA was performed as outlined in the methods section. The purity of sIgA was assessed by SDS-PAGE with four main bands corresponding to the SC, H chain and light chain of IgA, and J chain as shown in Figure S1. Standard proteomic analysis was performed on the isolated sIgA to further confirm the purity of the protein. The results are listed in Table S1 and indicate that the isolated sIgA is relatively pure with only very minor protein contaminants.
The extracted compound chromatograms (ECCs) produced by the molecular feature extractor (MFE) demonstrate similar glycopeptide profiles as well as abundances for the three colostrum sIgA samples (Figure S2). A list of glycopeptides from the pronase-digested sIgA isolated from one representative colostrum sample is shown in Table 1. The ECCs produced by the MFE yielded intensities that were used for obtaining relative abundances. Abundances for all peptides and glycoforms representing a specific site were added 37.
Table 1.
List of N-glycopeptides from isolated sIgA from collected milk sample
| RT (min) | M/Z | Mass (Da) | PPM error | Protein | Sequence | Site | Hex | HexNAc | Fuc | Neu5Ac | Intensity |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 33.869 | 1187.807 | 3560.4003 | 5 | IGA2 | HVKHYTNPSQ | 92 | 5 | 4 | 1 | 2 | 68197072 |
| 13.293 | 832.324 | 2493.9477 | 8 | IGA2 | YTNPS | 92 | 5 | 4 | 0 | 1 | 464028 |
| 26.164 | 878.6541 | 2632.9404 | 20 | IGA2 | YTNPSQDV | 92 | 5 | 3 | 0 | 1 | 775421 |
| 26.196 | 946.348 | 2836.0191 | 20 | IGA2 | YTNPSQDV | 92 | 5 | 4 | 0 | 1 | 2130024 |
| 29.815 | 1043.384 | 3127.1324 | 13 | IGA2 | YTNPSQDV | 92 | 5 | 4 | 0 | 2 | 3291937 |
| 23.32 | 977.0724 | 2928.1911 | 9 | IGJ | ENISDPTSPL | 71 | 5 | 3 | 1 | 1 | 1381861 |
| 26.32 | 1141.784 | 3422.3268 | 3 | IGJ | ENISDPTSPL | 71 | 5 | 4 | 1 | 2 | 1661523 |
| 21.692 | 953.0611 | 2856.1589 | 5 | IGJ | NISDPTSPL | 71 | 5 | 4 | 0 | 1 | 860383 |
| 22.502 | 1001.749 | 3002.2246 | 7 | IGJ | NISDPTSPL | 71 | 5 | 4 | 1 | 1 | 329314 |
| 25.502 | 1050.09 | 3147.2477 | 2 | IGJ | NISDPTSPL | 71 | 5 | 4 | 0 | 2 | 1405679 |
| 17.758 | 900.6658 | 2698.9762 | 6 | SC | NDT | 135 | 5 | 4 | 1 | 2 | 777521 |
| 24.384 | 1147.841 | 3440.5043 | 0 | SC | VSLEVSQGPGLLNDTK | 135 | 6 | 4 | 0 | 0 | 1123577 |
| 24.136 | 1215.534 | 3643.5889 | 0 | SC | VSLEVSQGPGLLNDTK | 135 | 6 | 5 | 0 | 0 | 4305616 |
| 20.277 | 952.4099 | 3805.6105 | 7 | SC | VSLEVSQGPGLLNDTK | 135 | 7 | 5 | 0 | 0 | 253514 |
| 23.925 | 952.4168 | 3805.6334 | 1 | SC | VSLEVSQGPGLLNDTK | 135 | 7 | 5 | 0 | 0 | 6387259 |
| 23.798 | 962.6741 | 3846.6648 | 0 | SC | VSLEVSQGPGLLNDTK | 135 | 6 | 6 | 0 | 0 | 4180562 |
| 19.877 | 1003.179 | 4008.685 | 8 | SC | VSLEVSQGPGLLNDTK | 135 | 7 | 6 | 0 | 0 | 174310 |
| 23.585 | 1003.189 | 4008.7254 | 1 | SC | VSLEVSQGPGLLNDTK | 135 | 7 | 6 | 0 | 0 | 7677994 |
| 20.071 | 847.3634 | 3385.4165 | 7 | SC | NYTGRIRLD | 186 | 6 | 5 | 0 | 1 | 1881945 |
| 19.575 | 898.1307 | 3588.496 | 6 | SC | NYTGRIRLD | 186 | 6 | 6 | 0 | 1 | 1677348 |
| 16.882 | 850.3206 | 2547.9384 | 6 | SC | NPN/PGNG | 186/421 | 5 | 4 | 0 | 2 | 1504821 |
| 18.096 | 815.9875 | 2444.9406 | 3 | SC | VPGN(PGNV) | 469 | 5 | 4 | 1 | 1 | 508112 |
| 20.296 | 913.0201 | 2736.0374 | 3 | SC | VPGN(PGNV) | 469 | 5 | 4 | 1 | 2 | 1157752 |
| 24.476 | 1020.688 | 4078.721 | 13 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 5 | 0 | 0 | 6116531 |
| 26.152 | 1052.947 | 4207.7536 | 15 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 5 | 5 | 0 | 1 | 3595040 |
| 24.225 | 1071.457 | 4281.7958 | 14 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 6 | 0 | 0 | 6857053 |
| 26.003 | 1093.458 | 4369.8037 | 15 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 5 | 0 | 1 | 8980128 |
| 25.819 | 1103.718 | 4410.835 | 14 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 5 | 6 | 0 | 1 | 4215505 |
| 24.654 | 1139.731 | 4554.8928 | 10 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 6 | 0 | 1 | 3272015 |
| 25.691 | 1144.232 | 4572.8955 | 12 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 6 | 0 | 1 | 9554195 |
| 26.363 | 1161.731 | 4642.8965 | 13 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 5 | 0 | 2 | 4439261 |
| 25.359 | 1184.75 | 4734.9699 | 7 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 7 | 6 | 0 | 1 | 3199461 |
| 26.062 | 1212.498 | 4845.959 | 16 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 6 | 0 | 2 | 1359938 |
| 24.086 | 1212.501 | 4845.9763 | 12 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 6 | 0 | 2 | 597825 |
| 27.84 | 1217.005 | 4863.9906 | 11 | SC | LTNFPENGTFVVNIAQLSQ | 90 | 6 | 6 | 0 | 2 | 6714054 |
| 25.776 | 984.6771 | 3934.6744 | 9 | SC | EGYVSSKYAGRANLT | 83 | 5 | 6 | 0 | 1 | 4837727 |
| 25.585 | 1025.191 | 4096.7328 | 10 | SC | EGYVSSKYAGRANLT | 83 | 6 | 6 | 0 | 1 | 9760395 |
| 22.825 | 1025.194 | 4096.7449 | 13 | SC | EGYVSSKYAGRANLT | 83 | 6 | 6 | 0 | 1 | 990066 |
| 25.424 | 1035.447 | 4137.7591 | 10 | SC | EGYVSSKYAGRANLT | 83 | 5 | 7 | 0 | 1 | 5184296 |
| 22.162 | 1075.941 | 4299.7436 | 5 | SC | EGYVSSKYAGRANLT | 83 | 6 | 7 | 0 | 1 | 924140 |
| 25.21 | 1075.962 | 4299.8158 | 11 | SC | EGYVSSKYAGRANLT | 83 | 6 | 7 | 0 | 1 | 12223087 |
| 28.002 | 1097.964 | 4387.8253 | 9 | SC | EGYVSSKYAGRANLT | 83 | 6 | 6 | 0 | 2 | 6682655 |
| 24.758 | 1097.964 | 4387.8259 | 9 | SC | EGYVSSKYAGRANLT | 83 | 6 | 6 | 0 | 2 | 1611696 |
| 27.447 | 1148.735 | 4590.9081 | 9 | SC | EGYVSSKYAGRANLT | 83 | 6 | 7 | 0 | 2 | 8800010 |
| 9.058 | 710.7651 | 1419.5156 | 1 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 5 | 2 | 0 | 0 | 266033 |
| 8.906 | 617.9116 | 1850.7109 | 3 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 3 | 5 | 1 | 0 | 548567 |
| 9.366 | 671.9295 | 2012.7634 | 3 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 4 | 5 | 1 | 0 | 361962 |
| 9.703 | 720.275 | 2157.8008 | 3 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 4 | 5 | 0 | 1 | 598056 |
| 10.14 | 774.292 | 2319.8541 | 3 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 5 | 5 | 0 | 1 | 343641 |
| 15.706 | 822.9688 | 2465.8852 | 8 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 5 | 5 | 1 | 1 | 199925 |
| 12.099 | 822.9805 | 2465.9174 | 5 | SC//IGA1//IGA2 | AN | 83//144//131/205 | 5 | 5 | 1 | 1 | 261020 |
| 9.281 | 961.37 | 1920.725 | 2 | SC//IGA1/IGA2 | ENGT//SEAN | 90//144/131 | 3 | 5 | 0 | 0 | 493651 |
| 10.416 | 656.2664 | 1965.7749 | 20/14//4 | SC//IGA1/IGA2 | ENG/WN//NVS | 90/499//340/327 | 3 | 5 | 1 | 0 | 556088 |
| 10.699 | 710.2852 | 2127.8318 | 20/15//5 | SC//IGA1/IGA2 | ENG/WN//NVS | 90/499//340/327 | 4 | 5 | 1 | 0 | 1079828 |
| 13.368 | 807.3168 | 2418.9288 | 20/15//5 | SC//IGA1/IGA2 | ENG/WN//NVS | 90/499//340/327 | 4 | 5 | 1 | 1 | 640041 |
| 13.258 | 861.3339 | 2580.9775 | 17/11//3 | SC//IGA1/IGA2 | ENG/WN//NVS | 90/499//340/327 | 5 | 5 | 1 | 1 | 715784 |
| 16.21 | 958.3694 | 2872.0847 | 19/14//7 | SC//IGA1/IGA2 | ENG/WN//NVS | 90/499//340/327 | 5 | 5 | 1 | 2 | 610683 |
| 11.434 | 724.7811 | 1447.5476 | 1 | IGA1//IGA2//SC | NV | 340//47/327//469 | 5 | 2 | 0 | 0 | 254065 |
| 9.941 | 805.8059 | 1609.5973 | 0 | IGA1//IGA2//SC | NV | 340//47/327//469 | 6 | 2 | 0 | 0 | 153825 |
| 10.484 | 681.271 | 2040.7925 | 2 | IGA1//IGA2//SC | NV | 340//47/327//469 | 4 | 5 | 1 | 0 | 582738 |
| 8.54 | 917.8562 | 1833.6979 | 5//14//14 | IGA1//IGA2//SC//IGJ | EAN//NVT//NIS | 144//47/131//469//71 | 3 | 5 | 0 | 0 | 325261 |
| 19.811 | 921.6835 | 2762.0284 | 22//4 | IGA1/IGA2//SC | LLGSEAN//EEPGNGT | 131/144//421 | 5 | 4 | 1 | 1 | 586605 |
| 19.407 | 970.3717 | 2908.0924 | 19//6 | IGA1/IGA2//SC | LLGSEAN//EEPGNGT | 131/144//421 | 5 | 4 | 2 | 1 | 306195 |
| 22.092 | 1018.719 | 3053.1319 | 17//6 | IGA1/IGA2//SC | LLGSEAN//EEPGNGT | 131/144//421 | 5 | 4 | 1 | 2 | 1448849 |
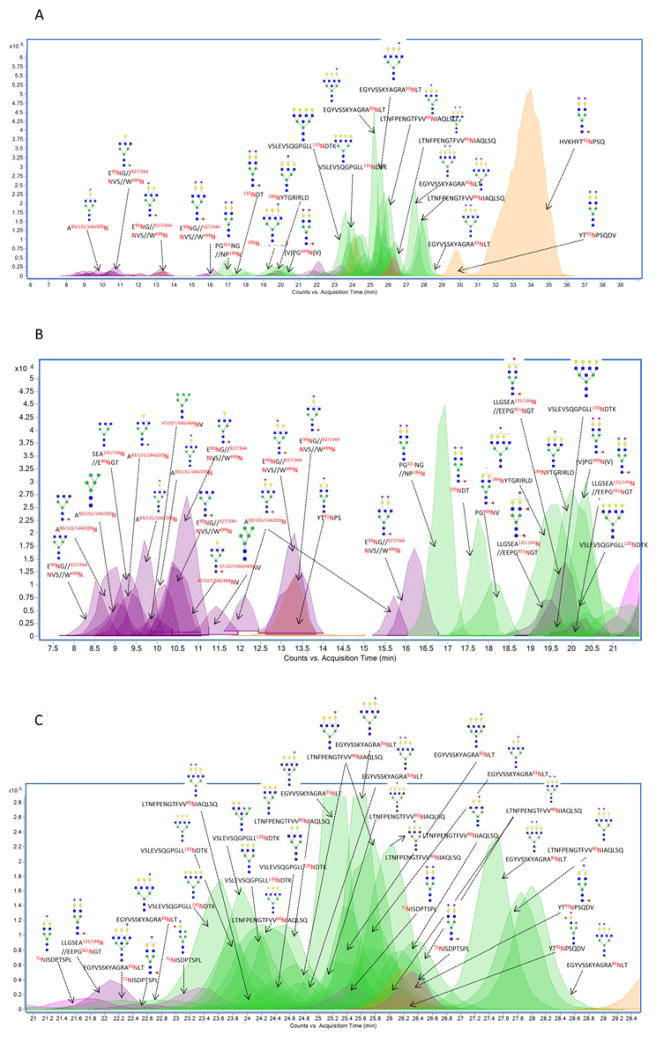
The elution order of glycopeptides on PGC follows loosely a set of empirical rules43, 44. Smaller peptides elute before larger ones with the same glycoforms. For example, the most abundant complex-type glycan, Hex5HexNAc4Fuc1NeuAc2 attached to the peptide HVKHYT92NPSQ from IgA2, eluted late at 33.9 min, while the same glycan composition on the peptide 135NDT from the SC region eluted much earlier at 17.8 min. Meanwhile, increasing the number of sialic acid residues increases the elution time. For example, the glycopeptide 71NISDPTSPL+Hex5HexNAc4NeuAc1 (21.7 min) with one sialic acid eluted earlier than the glycopeptide 71NISDPTSPL+Hex5HexNAc4NeuAc2 (25.5 min) with two sialic acids. In general, sialylated species elute later than neutral species 43, 44.
Figure 1A shows the overlaid ECCs of glycopeptides from isolated sIgA with the major peaks annotated. The most abundant peak was determined to be HVKHYT92NPSQ+Hex5HexNAc4Fuc1NeuAc2 from the conserved site (92Asn) of IgA2 (34 min). Glycosylation on this site is unusual for the reasons discussed below. More IgA2 glycopeptides were observed associated with complex sialylated or high mannose type glycans (peaks in yellow). Glycopeptides corresponding to SC (peaks in green) were observed mostly with complex type glycans varying from biantennary to tetraantennary with a varying number of Neu5Ac residues eluting mostly from 16–21 minutes, and 23–29 min. The J chain glycopeptides (peaks in pink) were among the less intense peaks and originated from the single site on the J chain. All the glycopeptides from the J chain were observed to contain at least one Neu5Ac. The glycopeptides eluting between 7 min to 29 min illustrate the large complexity and the dynamic range of the glycosylation (Figures 1B and 1C).
Figure 1.
Extracted Compound chromatogram (ECC) and associated structural assignments of glycopeptides from pronase digested sIgA isolated from an individual donor. (A) ECC of major glycopeptides eluted between 6 and 38 min; (B) ECC of all glycopeptides eluted between 7 and 21 min; (C) ECC of all glycopeptides eluted between 21 to 29 min. Glycosites corresponding to the four polypeptides was denoted in different colors. Green Circles, yellow circles, blue squares, red triangles, and purple diamonds represent mannose, galactose, GlcNAc, fucose and Neu5Ac, respectively.
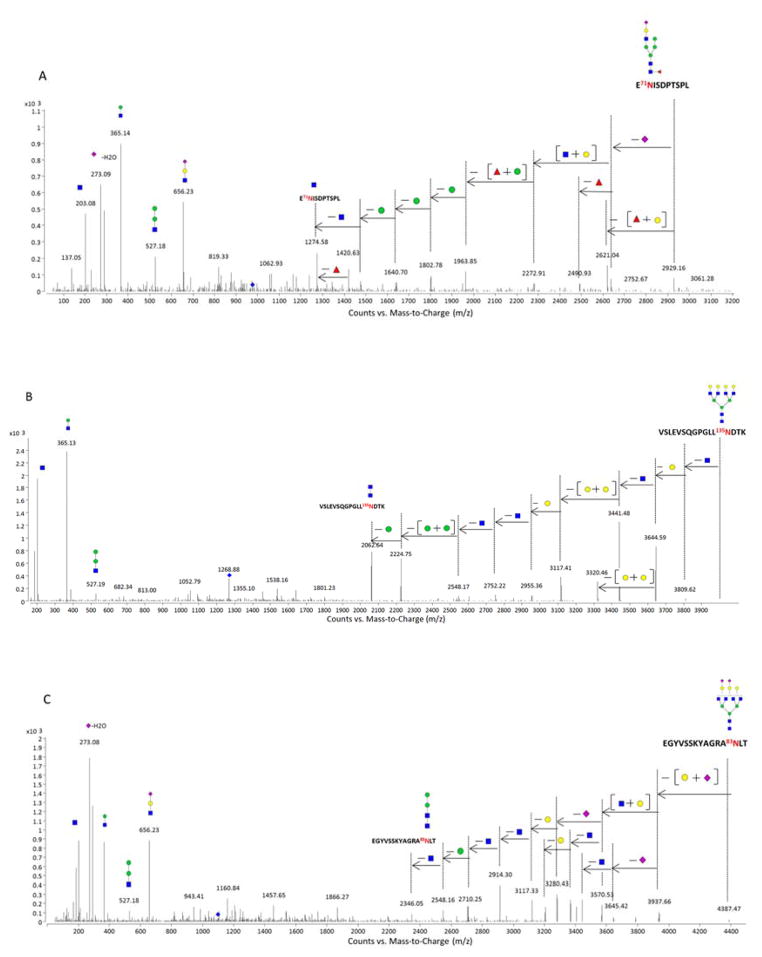
To obtain structural information from the peaks in Figure 1, tandem MS spectra were performed (shown in Figure S3). We have previously reported characteristic fragmentation of glycopeptides. Glycan fragmentation yielding B-type ions is commonly observed in the collision induced dissociation (CID) spectra of glycopeptides 45. A typical tandem MS spectrum is shown for the glycopeptide E71NISDPTSPL+Hex5HexNAc3Fuc1NeuAc1 (Figure 2A). The tandem mass spectrum shows fragment products consistent with hybrid-type glycan structures. The fragments include a series of Hex losses (162 Da) prior to the loss of core HexNAc (203 Da). Similarly, Y-type ions corresponding to the initial neutral loss of (Hex+Fucose) are also associated with hybrid structures. Core fucosylation was confirmed via the loss of fucose (146 Da) to yield the Y1 ion of (E71NISDPTSPL+HexNAc1). Figure 2B is representative of deconvoluted MS/MS spectrum for a SC glycopeptide containing a neutral complex type tetraantennary N-glycan (VSLEVSQGPGLL135NDTK+Hex7HexNAc6). In this study, higher abundances of tetraantennary structures were detected from sIgA compared with earlier work 4. Figure 2C depicts the CID data with the loss of the two Neu5Ac (291 Da) residues to yield the extent of sialylation. With sialylation, B-type ions with neutral masses of 273 Da, 291 Da, 656 Da, and 818 Da were observed corresponding to Neu5Ac–H2O, Neu5Ac, Neu5Ac+Hex+HexNAc, and Neu5Ac+Hex2+HexNAc. Due to the labile nature of sialic acids and the fact that they generally reside in the terminal positions, losses of Neu5Ac and Neu5Ac+Hex+HexNAc from the precursor peaks are common.
Figure 2.
Deconvoluted MS/MS spectra of three N-linked glycopeptides from sIgA. (A) MS/MS data for a N-linked peptide from J chain; (B) MS/MS data for a neutral N-linked peptide from SC; (C) MS/MS data for an acidic N-linked peptide from SC. Green Circles, yellow circles, blue squares, red triangles, and purple diamonds represent mannose, galactose, GlcNAc, fucose and Neu5Ac, respectively.
In several of the large glycopeptides, the loss of H2O, specifically from Neu5Ac, was observed. For example, after deconvolution, glycopeptides with neutral masses 4554.9 Da and 4572.9 Da, differing by 18 Da, were assigned as LTNFPE90NGTFVVNIAQLSQ+Hex6HexNAc6NeuAc2–H2O and LTNFPE90NGTFVVNIAQLSQ+Hex6HexNAc6NeuAc2. Figure S4 represents deconvoluted MS/MS spectra for these two glycopeptides. Neutral loss of Neu5Ac (291 Da) was observed followed by the initial loss of one Hex (Figure S4A). However, Figure S4B reveals a loss of 273 Da corresponding to (Neu5Ac – H2O) as the first loss. Y-type ions with the intact peptide, LTNFPE90NGTFVVNIAQLSQ associated with the trimannosyl core are present in both spectra indicating the same peptide were present in the two glycopeptides.
Glycan-heterogeneity of pooled human colostrum sIgA
The same experiments were performed on pooled human colostrum sIgA obtained commercially. A list of glycopeptides from the pronase-digested sIgA is shown in Table S2. A much lower diversity of peptides and glycan structures were observed with the commercial pooled sIgA as compared to the isolated sIgA from individual donors. The glycopeptide HVKHYT92NPSQ+Hex5HexNAc4Fuc1NeuAc2 from sIgA H chain remained the most abundant species. However, 186Asn was not found with glycosylation in the pooled sIgA. In general, sIgA extracted from the colostrum from individuals contained greater glycan diversity than those obtained from the commercial sIgA, including more triantennary and tetraantennary glycan structures at multiple sites particularly in the SC.
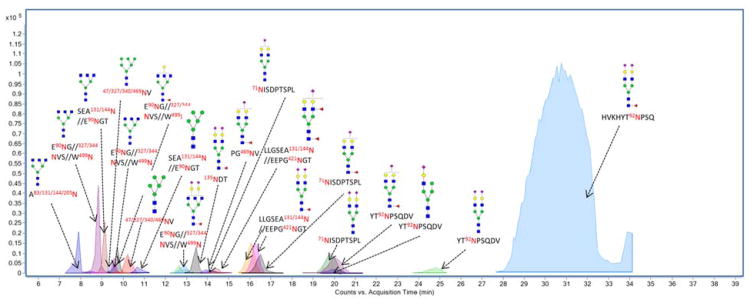
Figure 3 shows the overlaid ECCs of the glycopeptides from the commercial sIgA sample with the major peaks annotated. The majority of the observed glycopeptides contained Neu5Ac residue(s) with complex-type bi-antennary N-glycans. Included in this group are several truncated structures with terminal GlcNAc. Although less abundant, other glycan types were observed, including high mannose and hybrid. Over 80% of the glycopeptide abundances belonged to structures with core fucosylation, which were verified by tandem MS (see below). In general, essentially all singly fucosylated glycans were core fucosylated. Only one glycan was observed with two fucose residues.
Figure 3.
Extracted compound chromatogram (ECC) of glycopeptides from pronase digested commercial pooled human colostrum IgA with corresponding structural assignments. Different colors represent various sites. Green Circles, yellow circles, blue squares, red triangles, and purple diamonds represent mannose, galactose, GlcNAc, fucose and Neu5Ac, respectively.
Figure S5 and S6 show the deconvoluted MS/MS spectra of several N-glycopeptides representing different sites on the pooled sIgA. Figure S5A and S5B are representative of two deconvoluted MS/MS spectra of the glycopeptides from J chain and SC, respectively. The fragment denoted as peptide+HexNAc was observed and highly informative for identifying the two glycopeptides. Figure S6A shows the fragmentation of one of the most abundant glycopeptide (HVKHYT92NPSQ+Hex5HexNAc4Fuc1NeuAc2). This peptide is highly unusual because it does not conform to the N-X-S sequon 25, where X cannot be proline. However, we have a number of confirming evidences for the glycosylation of this site. The tandem MS shows the progression of saccharide loss to the peptide+GlcNAc core (1412.65 Da), which gives indication as to the mass of the peptide. The GP Finder software does not provide other structures corresponding to this mass that were consistent with the tandem MS. Additionally, tandem MS of other glycopeptides representing the same site was also obtained. For example, tandem MS spectra representative for glycopeptide YT92NPSQDV+Hex5HexNAc4NeuAc1 and glycopeptide YT92NPSQDV+ Hex5HexNAc4NeuAc2 are shown in Figure S6B and S6C, which provides the glycan heterogeneity of this site. The fact that glycan heterogeneity was observed at this site with various peptide backbone as shown in Table 1 is a confirmation that this site is glycosylated. There are also reports in literature that support glycosylation on this site. In a report by Picariello et al., they found tryptic peptides from this site is potentially modified and glycosylated 46. A 1Da mass shift from the peptide after PNGase F release is indicative of a putative N-glycan site.
Assignment of the glycopeptide structures using the software GP Finder was performed with 95% confidence based on decoy data. Some assignments were complicated by the large number of potential sites for sIgA. One complication is when the peptide mass was indistinguishable between different glycopeptides from different sites. For example, the glycopeptide with the peptide sequence E90NGT from the SC and SEA131/144N from IgA1/IgA2 have nearly identical masses corresponding to 419.165 Da. Tandem MS could not resolve the peptides because the peptides were short and did not yield suitable sequence data. Other problems arose when the peptides were even shorter. For example peptides such as NV and AN were observed as products and corresponded to four different glycosylation sites. It is noted that these glycopeptide abundances would affect the determination of site occupancy. However, these short peptides were minor products with very low abundances (<5%).
Compared with previous glycan analyses of sIgA 4, 23, 25, 46, our method revealed a more comprehensive glycan micro-heterogeneity from each site. We find glycans with heterogeneity for all potential N-sites including seven for the SC, two for IgA1, five for IgA2, and one for the J chain. The most comprehensive analysis before this report obtained five sites from the SC, two sites for IgA1, two IgA2 and one for the J chain 25. A potentially controversial assignment is that for 92Asn on IgA2, because it goes against the putative sequon. We find this site to be glycosylated, consistent with an earlier proposal but not found in a subsequent analysis 25.
Semi-quantitative Site-specific Glycosylation
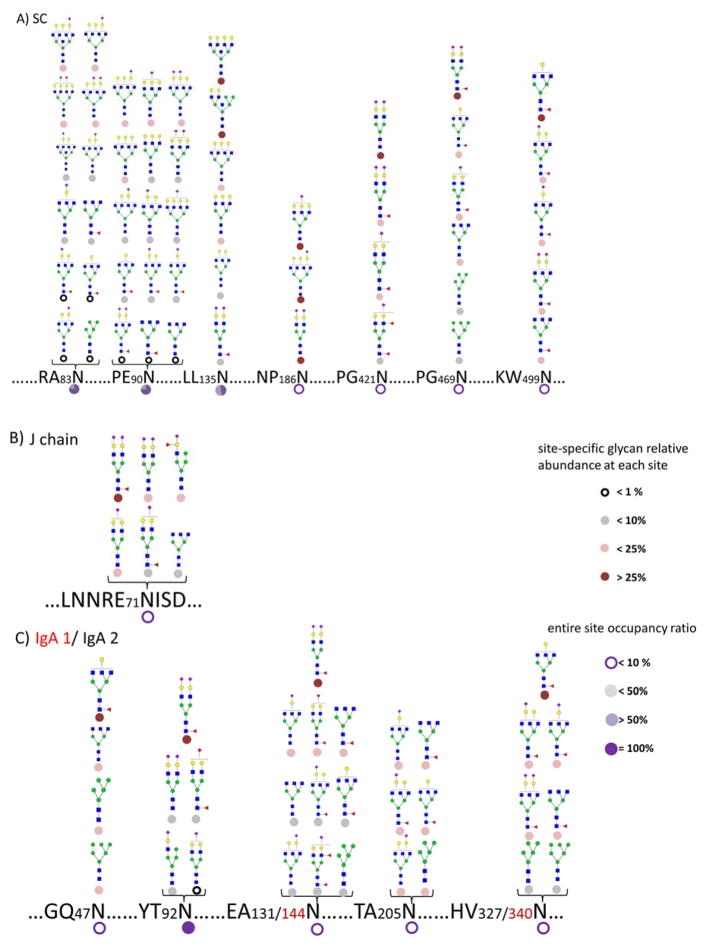
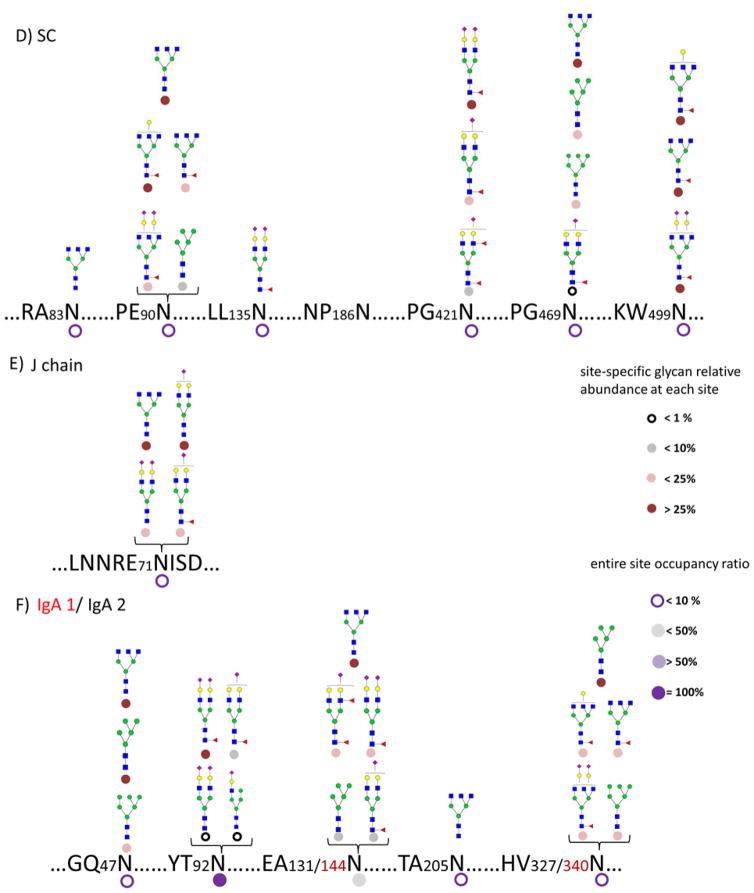
A summary of detailed and comprehensive glycan site heterogeneity of the sIgA isolated from a single human colostrum sample is depicted in Figure 4A–C. The result of the pooled commercial sample is shown in Figure 4D–F. The sIgA glycopeptide profiles were found to be similar between the fresh human colostrum samples from three donors. However, these samples were more highly glycosylated than the pooled commercial sample. For example, commercial sIgA was not found glycosylated in 186N of the SC while fresh colostrum was glycosylated. A greater site-heterogeneity was observed with most of the sites in the fresh colostrum sIgA. The result is not unexpected as commercial enrichment, chemical processing, and the storage may degrade glycans.
Figure 4.
(A–C) Glycosite occupancy and heterogeneity in pronase digested sIgA isolated from an individual donor, SC, J chain, and IgA1/IgA2, respectively. (D–F) Glycosite occupancy and heterogeneity in pooled commercial sIgA, SC, J chain, and IgA1/IgA2, respectively. Circle shading under each site is representative of site occupancy ratio to the most occupied site in 92Asn from IgA2. Circle shading under each glycan is representative of the relative glycan abundance at each site.
Quantitative information was obtained based on the abundances of the glycopeptides. Two types of quantitative information are provided in the figures. The small circular symbol under the glycan structure designates the abundance of that glycan relative to all the glycans at that glycosite. The circular symbol under the amino acid represents the occupancy of the site. To obtain these values, the MS abundances for all glycoforms associated with a site were added. The abundances were then combined for each glycoform and normalized to the total site abundance for the relative glycan abundance at each site. The sum for each site represents the occupancy while the sum for each glycan represents the relative abundance of the glycoform. The occupancy is further normalized to the most occupied site (100%). Quantitation of glycan site occupancy remains a difficult task. We propose a semi-quantitative method using ion abundances but, for the moment, neglecting issues such as differences in ionization efficiency of different peptides and different glycoforms as well as the dispersion of ion abundances across different peptides due to the nature of the nonspecific proteases. Nonetheless, a previous study by Hua et al. reported the quantitative accuracy and reproducibility of non-specific glycosylation analysis and revealed the efficiency of this method 47. In this way, we find that the most abundant site is 92Asn of the IgA2 in the protein complex.
Figure 4A illustrates the site heterogeneity of the SC. The most abundant glycans at 135Asn correspond to the tetra-antennary compositions. The glycans on the SC illustrate general diversity of the structures. The majority of structures were tri- and tetraantennary complex-type glycans further decorated with Neu5Ac and core fucosylation, in accordance with previous reports 4, 25. A small amount of high mannose (Man5 and Man6) and neutral complex glycan structures were observed. Glycosylation was found to be most abundant at 90Asn among the seven sites on SC, followed by 83Asn and 135Asn.
The characterization of the site heterogeneity on the J chain is summarized in Figure 4B. Krugman et al. 48 previously reported that the substitution of the Asn with Ala in J chain prevents IgA dimer formation, indicating the significance of glycosylation to the tertiary structure. Also, the glycans associated with J chain are essential for binding and trans-locating pIgR across epithelial cells 49. However, the results obtained here show a single site (71Asn) with less than 20% occupancy corresponding to mainly sialylated complex type glycans. The results are consistent with previous work but it calls to question contribution of this glycosite given the relatively low occupancy.
The H chain of IgA is significantly more glycosylated than the J chain and the SC. Figure 4C details the abundances of site-specific IgA1 H chain and IgA2 H chain glycans. Because the amino acid sequence near sites 144Asn for IgA1 are identical to those of 131Asn for IgA2, and similarly for 340Asn of IgA1 and 327Asn of IgA2, the glycopeptide assignments could not be more precise and were therefore combined for both IgA1 and IgA2. The analysis shows that the majority of N-glycans on the H chains are sialylated and/or terminated by galactose (Gal) residues, in agreement with previous studies of serum IgA 22, 50. The results are therefore consistent with the formation of milk sIgA, which originates from B cells. While 131/144Asn and 327/340Asn sites were observed as being slightly glycosylated, the 92Asn from IgA2 was characterized as being mostly glycosylated mainly due to the most abundant glycopeptide (HVKHYT92NPSQ+Hex5HexNAc4Fuc1NeuAc2). Moreover, bi-antennary complex type glycans were observed at 92Asn associated with one or two sialic acids. Core fucosylation was detected, as discussed previously from the MS/MS data, mainly due to the observation of the loss of fucose to yield an informative Y1 ion.
Overall, from the colostrum of individual donors, sialylated glycans were found to be the most abundant (82%), from which mono-sialylation (35%) and bi-sialylation (47%) were both present. Fucosylation (36%) were observed mostly as core fucosylation.
In the commercial sIgA, the most abundant glycopeptide was HVKHYT92NPSQ+Hex5HexNAc4Fuc1NeuAc2 with 92Asn being the most occupied site. The absence of branched complex type N-glycans, in particular from the SC, was notable possibly due to the processing methods of the commercial sample (Figure 4D–4F).
O-glycosylation Analysis
We have previously determined N- and O-glycosylation simultaneously for other glycoproteins,24, 36, 37, 47 however the same comprehensive analysis described above failed to detect O-glycopeptides in the hinge region of IgA. The presence of peptides and N-glycopeptides in the IgA hydrolysate complicate the detection of the O-glycopeptides. In order to observe the O-glycopeptides, we removed the N-glycans from IgA, digest the protein with trypsin, and enriched the O-glycopeptides using hydrophilic affinity methods.
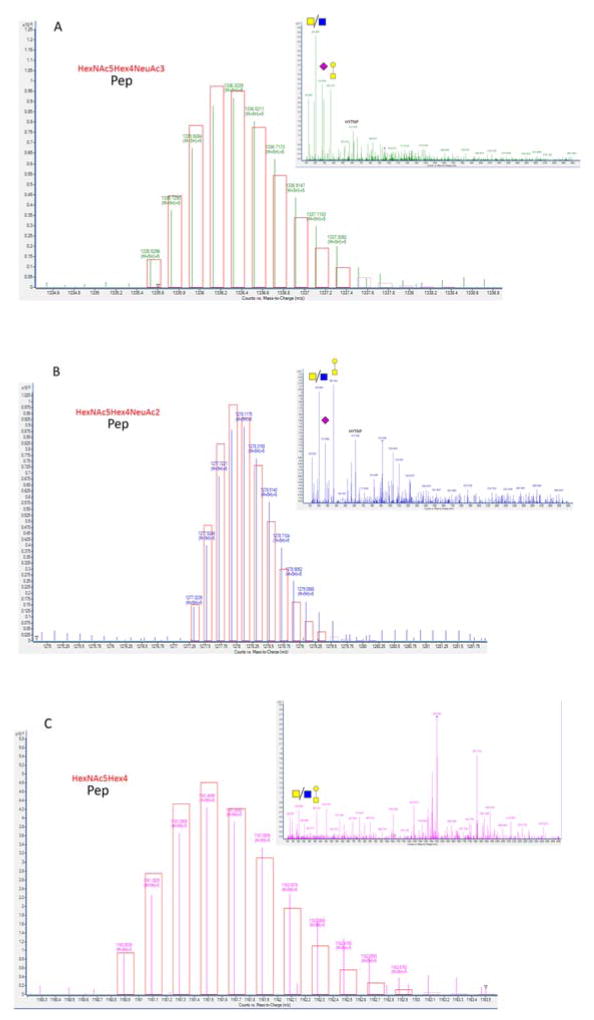
The enriched O-glycopeptide fraction was analyzed in both MS and MS/MS mode. A glycopeptide library was employed to identify the glycopeptides using accurate mass and the theoretical isotopic distribution. This approach revealed 54 compounds compiled in Table 2. Complete cysteine alkylation was observed for all hinge region O-glycopeptides. The identified compounds eluted in a narrow interval (between minute 22 and 28) in agreement with the elution behavior in C18 of a family of glycopeptides sharing the same peptide moiety and multiple glycan compositions. Tandem-MS of the O-glycopeptides (Figure 5A–5C) revealed characteristic oxonium ions and the presence of peptide backbone fragment ion b11 (621.79 m/z and 612.79 m/z) corresponding to the sequence HYTNPSQDVTV, which unambiguously identifies the compounds as HR glycopeptides. Due to the overlap of the glycopeptide peaks and their low abundances, tandem-MS spectra were obtained only for the more abundant species. However, O-glycopeptide assignments were confirmed by comparing the elution order. The elution order follows a certain pattern as illustrated in Figure 6 for the O-glycopeptide series containing five N-acetylgalactosamine, three galactose and different number of sialic acid residues.
Table 2.
List of sIgA HR O-glycopeptides
| RT (min) | Base Peak (Da) | Z | Error (ppm) | Formula | Hex | HexNAc | Neu5Ac | Relative intensity (%) |
|---|---|---|---|---|---|---|---|---|
| 22.35 | 1441.7946 | 5 | −3.5 | C294 H459 N59 O143 S3 | 6 | 6 | 3 | 0.07 |
| 22.12 | 1383.5586 | 5 | −15.9 | C283 H442 N58 O135 S3 | 6 | 6 | 2 | 0.07 |
| 21.93 | 1325.1411 | 5 | −15.4 | C272 H425 N57 O127 S3 | 6 | 6 | 1 | 0.04 |
| 23.12 | 1467.5875 | 5 | −14.1 | C299 H466 N60 O146 S3 | 6 | 5 | 4 | 0.15 |
| 22.55 | 1409.3685 | 5 | −14.6 | C288 H449 N59 O138 S3 | 6 | 5 | 3 | 0.45 |
| 22.28 | 1351.1494 | 5 | −15.3 | C277 H432 N58 O130 S3 | 6 | 5 | 2 | 0.57 |
| 22.04 | 1292.9313 | 5 | −15.2 | C266 H415 N57 O122 S3 | 6 | 5 | 1 | 0.32 |
| 21.76 | 1234.7120 | 5 | −16.1 | C255 H398 N56 O114 S3 | 6 | 5 | 0 | 0.08 |
| 23.40 | 1435.1755 | 5 | −15.4 | C293 H456 N60 O141 S3 | 6 | 4 | 4 | 0.38 |
| 22.83 | 1376.9572 | 5 | −15.5 | C282 H439 N59 O133 S3 | 6 | 4 | 3 | 1.64 |
| 22.49 | 1318.7403 | 5 | −14.6 | C271 H422 N58 O125 S3 | 6 | 4 | 2 | 1.35 |
| 22.25 | 1260.1283 | 5 | −9.6 | C260 H405 N57 O117 S3 | 6 | 4 | 1 | 0.31 |
| 21.95 | 1502.3845 | 4 | −10.2 | C249 H388 N56 O109 S3 | 6 | 4 | 0 | 0.14 |
| 23.59 | 1344.5323 | 5 | −26.5 | C276 H429 N59 O128 S3 | 6 | 3 | 3 | 0.94 |
| 23.20 | 1286.3315 | 5 | −13.5 | C265 H412 N58 O120 S3 | 6 | 3 | 2 | 1.15 |
| 22.81 | 1227.9189 | 5 | −8.9 | C254 H395 N57 O112 S3 | 6 | 3 | 1 | 0.70 |
| 22.43 | 1169.7008 | 5 | −8.5 | C243 H378 N56 O104 S3 | 6 | 3 | 0 | 0.34 |
| 24.71 | 1485.1758 | 5 | −24.0 | C302 H470 N60 O149 S3 | 5 | 5 | 5 | 0.59 |
| 23.72 | 1427.1532 | 5 | −27.4 | C291 H453 N59 O141 S3 | 5 | 5 | 4 | 1.87 |
| 23.19 | 1368.7517 | 5 | −15.7 | C280 H436 N58 O133 S3 | 5 | 5 | 3 | 2.39 |
| 22.78 | 1310.5358 | 5 | −14.0 | C269 H419 N57 O125 S3 | 5 | 5 | 2 | 2.21 |
| 22.39 | 1252.3178 | 5 | −13.8 | C258 H402 N56 O117 S3 | 5 | 5 | 1 | 1.56 |
| 22.10 | 1193.9069 | 5 | −7.6 | C247 H385 N55 O109 S3 | 5 | 5 | 0 | 0.57 |
| 24.56 | 1452.7646 | 5 | −24.9 | C296 H460 N60 O144 S3 | 5 | 4 | 5 | 0.10 |
| 24.18 | 1394.5549 | 5 | −19.2 | C285 H443 N59 O136 S3 | 5 | 4 | 4 | 6.72 |
| 23.69 | 1336.3333 | 5 | −21.9 | C274 H426 N58 O128 S3 | 5 | 4 | 3 | 9.20 |
| 23.13 | 1278.1190 | 5 | −19.2 | C263 H409 N57 O120 S3 | 5 | 4 | 2 | 7.93 |
| 22.65 | 1219.7091 | 5 | −12.6 | C252 H392 N56 O112 S3 | 5 | 4 | 1 | 5.30 |
| 22.29 | 1161.4932 | 5 | −10.5 | C241 H375 N55 O104 S3 | 5 | 4 | 0 | 1.85 |
| 24.41 | 1303.9263 | 5 | −19.8 | C268 H416 N58 O123 S3 | 5 | 3 | 3 | 3.78 |
| 23.93 | 1245.5141 | 5 | −15.2 | C257 H399 N57 O115 S3 | 5 | 3 | 2 | 4.08 |
| 23.27 | 1187.3040 | 5 | −8.4 | C246 H382 N56 O107 S3 | 5 | 3 | 1 | 2.48 |
| 22.76 | 1129.0857 | 5 | −8.1 | C235 H365 N55 O99 S3 | 5 | 3 | 0 | 0.68 |
| 24.79 | 1213.1057 | 5 | −13.8 | C251 H389 N57 O110 S3 | 5 | 2 | 2 | 0.33 |
| 24.10 | 1154.8932 | 5 | −8.8 | C240 H372 N56 O102 S3 | 5 | 2 | 1 | 0.37 |
| 25.42 | 1412.1573 | 5 | −19.6 | C288 H447 N59 O139 S3 | 4 | 4 | 5 | 0.27 |
| 25.14 | 1353.9268 | 5 | −28.9 | C277 H430 N58 O131 S3 | 4 | 4 | 4 | 6.64 |
| 24.74 | 1295.7254 | 5 | −16.5 | C266 H413 N57 O123 S3 | 4 | 4 | 3 | 6.67 |
| 24.14 | 1237.5065 | 5 | −17.1 | C255 H396 N56 O115 S3 | 4 | 4 | 2 | 5.48 |
| 23.46 | 1179.0949 | 5 | −11.6 | C244 H379 N55 O107 S3 | 4 | 4 | 1 | 3.20 |
| 22.87 | 1120.8728 | 5 | −15.0 | C233 H362 N54 O99 S3 | 4 | 4 | 0 | 0.96 |
| 25.65 | 1321.5394 | 5 | −12.0 | C271 H420 N58 O126 S3 | 4 | 3 | 4 | 0.31 |
| 25.54 | 1263.3206 | 5 | −12.4 | C260 H403 N57 O118 S3 | 4 | 3 | 3 | 4.12 |
| 25.13 | 1204.8921 | 5 | −20.7 | C249 H386 N56 O110 S3 | 4 | 3 | 2 | 3.67 |
| 24.34 | 1146.6773 | 5 | −18.1 | C238 H369 N55 O102 S3 | 4 | 3 | 1 | 1.63 |
| 23.56 | 1088.4603 | 5 | −17.2 | C227 H352 N54 O94 S3 | 4 | 3 | 0 | 0.50 |
| 26.05 | 977.2487 | 6 | −8.2 | C243 H376 N56 O105 S3 | 4 | 2 | 2 | 0.16 |
| 24.69 | 928.7259 | 6 | −16.0 | C232 H359 N55 O97 S3 | 4 | 2 | 1 | 0.06 |
| 25.75 | 1222.5168 | 5 | −2.9 | C252 H390 N56 O113 S3 | 3 | 3 | 3 | 2.60 |
| 25.16 | 1164.2785 | 5 | −19.5 | C241 H373 N55 O105 S3 | 3 | 3 | 2 | 1.62 |
| 24.18 | 1106.2704 | 5 | −10.7 | C230 H356 N54 O97 S3 | 3 | 3 | 1 | 0.24 |
| 26.16 | 1131.8795 | 5 | −9.9 | C235 H363 N55 O100 S3 | 3 | 2 | 2 | 1.09 |
| 28.19 | 1199.5245 | 4 | −8.6 | C202 H310 N52 O77 S3 | 1 | 1 | 1 | 0.06 |
| 28.80 | 1035.4722 | 4 | −5.6 | C177 H270 N50 O59 S3 | 0 | 0 | 0 | 0.02 |
Figure 5.
MS spectra of three tryptic O-glycopeptides from sIgA and the corresponding isotopic patterns and tandem spectra. (A) O-glycopeptide with glycan composition HexNAc5Hex4Neu5Ac3; (B) O-glycopeptide with glycan composition HexNAc5Hex4Neu5Ac2; (C) O-glycopeptide with glycan composition HexNAc5Hex4. Red boxes are representative of the predicted isotopic pattern.
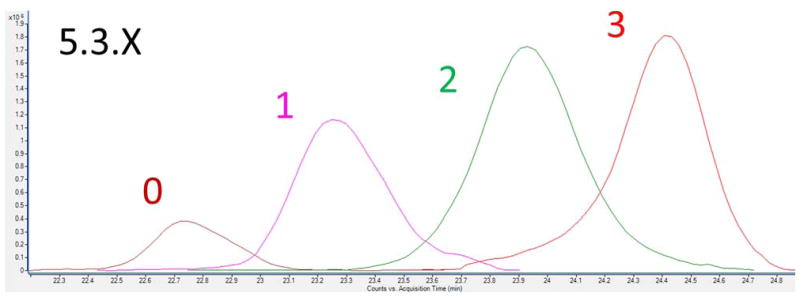
Figure 6.
Extracted compound chromatograms (ECCs) of four sIgA HR O-glycopeptides containing same number of N-acetylgalactosamine and galactose residues and 0–3 sialic acid monosaccharides.
In every case, the addition of sialic acid residues increases the retention time of the corresponding O-glycopeptides (Figure S7). Similarly, the elution time order can be compared with the number of hexoses or hexosamines residues. The addition of these monosaccharides decreased gradually the retention time (Figure S8). The combined comparison of the elution orders confirmed the compound assignments. Isomeric o-glycopeptides were found for 5 different compositions when examining the extracted compounds chromatograms (Figure S7). The results are in agreement with those published previously 25.
CONCLUSION
The results demonstrate the detection and characterization of a complicated large glycoprotein complex sIgA containing multiple polypeptides and numerous sites of glycosylation. N-glycosylation analysis of sIgA from human colostrum is performed at the glycopeptide level using pronase digestion and subsequent LC-MS and LC-MS/MS analysis. Extensive and comprehensive site-specific glycosylation analysis was obtained, which will aid our understanding of the complex biological roles of sIgA. O-glycosylation was identified via trypsin digestion with 54 most likely glycan compositions present on this multiply O-glycosylated peptide. This study demonstrates the wealth of quantitative site-occupancy information on protein complexes that include with multiple glycosites.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Research reported in this publication was supported by the National Institute of Child Health and Human Development, National Institute of General Medicine, National Center of Complimentary and Alternative Medicine of the National Institutes of Health under award number R01HD061923, R01GM049077, and 1U24DK097154.
Abbreviations
- sIgA
secretory immunoglobulin A
- pIgR
polymeric immunoglobulin receptor
- HPLC
high-performance liquid chromatography
- MFE
molecular feature extractor
- Q-TOF
quadrupole time-of-flight
- CID
collision-induced dissociation
- SPE
solid phase extraction
- Hex
hexose
- HexNAc
N-acetylhexosamine
- Neu5Ac
neuraminic acid (sialic acid)
- Fuc
fucose
- GCC
graphitized carbon cartridge
- ACN
acetonitrile
- HR
hinge region
- ECC
Extracted compound chromatogram
References
- 1.Renegar K, Small P., Jr Passive immunization: systemic and mucosal. Mucosal Immunology. 1994;3:841–851. [Google Scholar]
- 2.Russell M, Kilian M, Lamm M. Biological activities of IgA. Mucosal immunology. 1999:225. [Google Scholar]
- 3.Phalipon A, Cardona A, Kraehenbuhl JP, Edelman L, Sansonetti PJ, Corthésy B. Secretory component: a new role in secretory IgA-mediated immune exclusion in vivo. Immunity. 2002;17(1):107–115. doi: 10.1016/s1074-7613(02)00341-2. [DOI] [PubMed] [Google Scholar]
- 4.Royle L, Roos A, Harvey DJ, Wormald MR, Van Gijlswijk-Janssen D, Redwan ERM, Wilson IA, Daha MR, Dwek RA, Rudd PM. Secretory IgA N-and O-glycans provide a link between the innate and adaptive immune systems. Journal of Biological Chemistry. 2003;278(22):20140–20153. doi: 10.1074/jbc.M301436200. [DOI] [PubMed] [Google Scholar]
- 5.Johansen FE, Braathen R, Brandtzaeg P. The J chain is essential for polymeric Ig receptor-mediated epithelial transport of IgA. The Journal of Immunology. 2001;167(9):5185–5192. doi: 10.4049/jimmunol.167.9.5185. [DOI] [PubMed] [Google Scholar]
- 6.Mostov KE. Transepithelial transport of immunoglobulins. Annual review of immunology. 1994;12(1):63–84. doi: 10.1146/annurev.iy.12.040194.000431. [DOI] [PubMed] [Google Scholar]
- 7.Underdown BJ, Mestecky J. Mucosal immunoglobulins. Handbook of mucosal immunology. 1994:79. [Google Scholar]
- 8.An HJ, Peavy TR, Hedrick JL, Lebrilla CB. Determination of N-glycosylation sites and site heterogeneity in glycoproteins. Analytical chemistry. 2003;75(20):5628–5637. doi: 10.1021/ac034414x. [DOI] [PubMed] [Google Scholar]
- 9.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochimica et Biophysica Acta (BBA)-General Subjects. 1999;1473(1):4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 10.Zauner G, Selman MH, Bondt A, Rombouts Y, Blank D, Deelder AM, Wuhrer M. Glycoproteomic analysis of antibodies. Molecular & Cellular Proteomics. 2013;12(4):856–865. doi: 10.1074/mcp.R112.026005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takahashi M, Kuroki Y, Ohtsubo K, Taniguchi N. Core fucose and bisecting GlcNAc, the direct modifiers of the N-glycan core: their functions and target proteins. Carbohydrate research. 2009;344(12):1387–1390. doi: 10.1016/j.carres.2009.04.031. [DOI] [PubMed] [Google Scholar]
- 12.Marth JD, Grewal PK. Mammalian glycosylation in immunity. Nature Reviews Immunology. 2008;8(11):874–887. doi: 10.1038/nri2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Sato Y, Kariya Y, Isaji T, Taniguchi N, Fukuda T. A Mutual Regulation between Cell–Cell Adhesion and N-Glycosylation: Implication of the Bisecting GlcNAc for Biological Functions. Journal of proteome research. 2008;8(2):431–435. doi: 10.1021/pr800674g. [DOI] [PubMed] [Google Scholar]
- 14.Newburg DS, Ruiz-Palacios GM, Morrow AL. Human milk glycans protect infants against enteric pathogens. Annu Rev Nutr. 2005;25:37–58. doi: 10.1146/annurev.nutr.25.050304.092553. [DOI] [PubMed] [Google Scholar]
- 15.Hanson LA. Breastfeeding provides passive and likely long-lasting active immunity. Annals of Allergy, Asthma & Immunology. 1998;81(6):523–537. doi: 10.1016/S1081-1206(10)62704-4. [DOI] [PubMed] [Google Scholar]
- 16.Peterson R, Cheah WY, Grinyer J, Packer N. Glycoconjugates in human milk: Protecting infants from disease. Glycobiology. 2013;23(12):1425–1438. doi: 10.1093/glycob/cwt072. [DOI] [PubMed] [Google Scholar]
- 17.Schroten H, Stapper C, Plogmann R, Köhler H, Hacker J, Hanisch FG. Fab-independent antiadhesion effects of secretory immunoglobulin A on S-fimbriated Escherichia coli are mediated by sialyloligosaccharides. Infection and immunity. 1998;66(8):3971–3973. doi: 10.1128/iai.66.8.3971-3973.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, Endo T, Edén CS. Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infection and immunity. 1990;58(9):3073–3077. doi: 10.1128/iai.58.9.3073-3077.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Falk P, Roth KA, Boren T, Westblom TU, Gordon JI, Normark S. An in vitro adherence assay reveals that Helicobacter pylori exhibits cell lineage-specific tropism in the human gastric epithelium. Proceedings of the National Academy of Sciences. 1993;90(5):2035–2039. doi: 10.1073/pnas.90.5.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262(5141):1892–1895. doi: 10.1126/science.8018146. [DOI] [PubMed] [Google Scholar]
- 21.Mantis NJ, Farrant SA, Mehta S. Oligosaccharide side chains on human secretory IgA serve as receptors for ricin. The Journal of Immunology. 2004;172(11):6838–6845. doi: 10.4049/jimmunol.172.11.6838. [DOI] [PubMed] [Google Scholar]
- 22.Tanaka A, Iwase H, Hiki Y, Kokubo T, Ishii-Karakasa I, Toma K, Kobayashi Y, Hotta K. Evidence for a site-specific fucosylation of N-linked oligosaccharide of immunoglobulin A1 from normal human serum. Glycoconjugate journal. 1998;15(10):995–1000. doi: 10.1023/a:1006989910120. [DOI] [PubMed] [Google Scholar]
- 23.Gomes MM, Wall SB, Takahashi K, Novak J, Renfrow MB, Herr AB. Analysis of IgA1 N-Glycosylation and Its Contribution to FcαRI Binding†. Biochemistry. 2008;47(43):11285–11299. doi: 10.1021/bi801185b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nwosu CC, Huang J, Aldredge DL, Strum JS, Hua S, Seipert RR, Lebrilla CB. In-gel nonspecific proteolysis for elucidating glycoproteins: a method for targeted protein-specific glycosylation analysis in complex protein mixtures. Analytical chemistry. 2012;85(2):956–963. doi: 10.1021/ac302574f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deshpande N, Jensen PH, Packer NH, Kolarich D. GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. Journal of proteome research. 2010;9(2):1063–1075. doi: 10.1021/pr900956x. [DOI] [PubMed] [Google Scholar]
- 26.LOO TW, CLARKE DM. The human multidrug resistance P-glycoprotein is inactive when its maturation is inhibited: potential for a role in cancer chemotherapy. The FASEB journal. 1999;13(13):1724–1732. doi: 10.1096/fasebj.13.13.1724. [DOI] [PubMed] [Google Scholar]
- 27.Cauchi MR, Henchal E, Wright PJ. The sensitivity of cell-associated dengue virus proteins to trypsin and the detection of trypsin-resistant fragments of the nonstructural glycoprotein NS1. Virology. 1991;180(2):659–667. doi: 10.1016/0042-6822(91)90079-q. [DOI] [PubMed] [Google Scholar]
- 28.Bezouška K, Sklenář J, Novák P, Halada P, Havlíček V, Kraus M, Tichá M, Jonáková V. Determination of the complete covalent structure of the major glycoform of DQH sperm surface protein, a novel trypsin-resistant boar seminal plasma O-glycoprotein related to pB1 protein. Protein science. 1999;8(7):1551–1556. doi: 10.1110/ps.8.7.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahashi K, Smith AD, Poulsen K, Kilian M, Julian BA, Mestecky J, Novak J, Renfrow MB. Naturally Occurring Structural Isomers in Serum IgA1 O-Glycosylation. Journal of Proteome Research. 2011;11(2):692–702. doi: 10.1021/pr200608q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada Y, Dell A, Haslam SM, Tissot B, Canis K, Azadi P, Bäckström M, Costello CE, Hansson GC, Hiki Y. Comparison of Methods for Profiling O-Glycosylation HUMAN PROTEOME ORGANISATION HUMAN DISEASE GLYCOMICS/PROTEOME INITIATIVE MULTI-INSTITUTIONAL STUDY OF IgA1. Molecular & Cellular Proteomics. 2010;9(4):719–727. doi: 10.1074/mcp.M900450-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Renfrow MB, Mackay CL, Chalmers MJ, Julian BA, Mestecky J, Kilian M, Poulsen K, Emmett MR, Marshall AG, Novak J. Analysis of O-glycan heterogeneity in IgA1 myeloma proteins by Fourier transform ion cyclotron resonance mass spectrometry: implications for IgA nephropathy. Analytical and bioanalytical chemistry. 2007;389(5):1397–1407. doi: 10.1007/s00216-007-1500-z. [DOI] [PubMed] [Google Scholar]
- 32.Renfrow MB, Cooper HJ, Tomana M, Kulhavy R, Hiki Y, Toma K, Emmett MR, Mestecky J, Marshall AG, Novak J. Determination of aberrant O-glycosylation in the IgA1 hinge region by electron capture dissociation fourier transform-ion cyclotron resonance mass spectrometry. Journal of Biological Chemistry. 2005;280(19):19136–19145. doi: 10.1074/jbc.M411368200. [DOI] [PubMed] [Google Scholar]
- 33.Takahashi K, Wall SB, Suzuki H, Smith AD, Hall S, Poulsen K, Kilian M, Mobley JA, Julian BA, Mestecky J, Novak J, Renfrow MB. Clustered O-Glycans of IgA1: DEFINING MACRO- AND MICROHETEROGENEITY BY USE OF ELECTRON CAPTURE/TRANSFER DISSOCIATION. Molecular & Cellular Proteomics. 2010;9(11):2545–2557. doi: 10.1074/mcp.M110.001834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seipert RR, Dodds ED, Lebrilla CB. Exploiting differential dissociation chemistries of O-linked glycopeptide ions for the localization of mucin-type protein glycosylation. Journal of proteome research. 2008;8(2):493–501. doi: 10.1021/pr8007072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seipert RR, Dodds ED, Clowers BH, Beecroft SM, German JB, Lebrilla CB. Factors that influence fragmentation behavior of N-linked glycopeptide ions. Analytical chemistry. 2008;80(10):3684–3692. doi: 10.1021/ac800067y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German BJ, Lebrilla CB. Simultaneous and extensive site-specific N-and O-glycosylation analysis in protein mixtures. Journal of proteome research. 2011;10(5):2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hua S, Nwosu CC, Strum JS, Seipert RR, An HJ, Zivkovic AM, German JB, Lebrilla CB. Site-specific protein glycosylation analysis with glycan isomer differentiation. Analytical and bioanalytical chemistry. 2012;403(5):1291–1302. doi: 10.1007/s00216-011-5109-x. [DOI] [PubMed] [Google Scholar]
- 38.Clowers BH, Dodds ED, Seipert RR, Lebrilla CB. Site determination of protein glycosylation based on digestion with immobilized nonspecific proteases and Fourier transform ion cyclotron resonance mass spectrometry. Journal of proteome research. 2007;6(10):4032–4040. doi: 10.1021/pr070317z. [DOI] [PubMed] [Google Scholar]
- 39.Strum JS, Nwosu CC, Hua S, Kronewitter SR, Seipert RR, Bachelor RJ, An HJ, Lebrilla CB. Automated Assignments of N-and O-Site Specific Glycosylation with Extensive Glycan Heterogeneity of Glycoprotein Mixtures. Analytical chemistry. 2013;85(12):5666–5675. doi: 10.1021/ac4006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hong Q, Lebrilla CB, Miyamoto S, Ruhaak LR. Absolute quantitation of immunoglobulin G and its glycoforms using multiple reaction monitoring. Analytical chemistry. 2013;85(18):8585–8593. doi: 10.1021/ac4009995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langley R, Wines B, Willoughby N, Basu I, Proft T, Fraser JD. The staphylococcal superantigen-like protein 7 binds IgA and complement C5 and inhibits IgA-FcαRI binding and serum killing of bacteria. The Journal of Immunology. 2005;174(5):2926–2933. doi: 10.4049/jimmunol.174.5.2926. [DOI] [PubMed] [Google Scholar]
- 42.Wada Y, Tajiri M, Yoshida S. Hydrophilic affinity isolation and MALDI multiple-stage tandem mass spectrometry of glycopeptides for glycoproteomics. Analytical chemistry. 2004;76(22):6560–6565. doi: 10.1021/ac049062o. [DOI] [PubMed] [Google Scholar]
- 43.Ninonuevo M, An H, Yin H, Killeen K, Grimm R, Ward R, German B, Lebrilla C. Nanoliquid chromatography-mass spectrometry of oligosaccharides employing graphitized carbon chromatography on microchip with a high-accuracy mass analyzer. Electrophoresis. 2005;26(19):3641–3649. doi: 10.1002/elps.200500246. [DOI] [PubMed] [Google Scholar]
- 44.Chu CS, Niñonuevo MR, Clowers BH, Perkins PD, An HJ, Yin H, Killeen K, Miyamoto S, Grimm R, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9(7):1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nwosu CC, Seipert RR, Strum JS, Hua SS, An HJ, Zivkovic AM, German BJ, Lebrilla CB. Simultaneous and Extensive Site-specific N- and O-Glycosylation Analysis in Protein Mixtures. Journal of Proteome Research. 2011;10(5):2612–2624. doi: 10.1021/pr2001429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picariello G, Ferranti P, Mamone G, Roepstorff P, Addeo F. Identification of N-linked glycoproteins in human milk by hydrophilic interaction liquid chromatography and mass spectrometry. Proteomics. 2008;8(18):3833–3847. doi: 10.1002/pmic.200701057. [DOI] [PubMed] [Google Scholar]
- 47.Hua S, Hu CY, Kim BJ, Totten SM, Oh MJ, Yun N, Nwosu CC, Yoo JS, Lebrilla CB, An HJ. Glyco-Analytical Multispecific Proteolysis (Glyco-AMP): A Simple Method for Detailed and Quantitative Glycoproteomic Characterization. Journal of Proteome Research. 2013;12(10):4414–4423. doi: 10.1021/pr400442y. [DOI] [PubMed] [Google Scholar]
- 48.Krugmann S, Pleass R, Atkin J, Woof J. Mutagenesis of J chain residues critical for IgA dimer assembly. Biochemical Society transactions. 1997;25(2):323S. doi: 10.1042/bst025323s. [DOI] [PubMed] [Google Scholar]
- 49.Vaerman JP, Langendries A, Giffroy D, Brandtzaeg P, Kobayashi K. Lack of SC/pIgR-mediated epithelial transport of a human polymeric IgA devoid of J chain: in vitro and in vivo studies. Immunology. 1998;95(1):90. doi: 10.1046/j.1365-2567.1998.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattu TS, Pleass RJ, Willis AC, Kilian M, Wormald MR, Lellouch AC, Rudd PM, Woof JM, Dwek RA. The glycosylation and structure of human serum IgA1, Fab, and Fc regions and the role of N-glycosylation on Fcα receptor interactions. Journal of Biological Chemistry. 1998;273(4):2260–2272. doi: 10.1074/jbc.273.4.2260. [DOI] [PubMed] [Google Scholar]