Abstract
Metallochaperones are a diverse family of trafficking molecules that provide metal ions to protein targets for use as cofactors. The copper chaperone for superoxide dismutase (Ccs1) activates immature copper-zinc superoxide dismutase (Sod1) by delivering copper and facilitating the oxidation of the Sod1 intramolecular disulfide bond. Here, we present structural, spectroscopic, and cell-based data supporting a novel copper-induced mechanism for Sod1 activation. Ccs1 binding exposes an electropositive cavity and proposed “entry site” for copper ion delivery on immature Sod1. Copper-mediated sulfenylation leads to a sulfenic acid intermediate that eventually resolves to form the Sod1 disulfide bond with concomitant release of copper into the Sod1 active site. Sod1 is the predominant disulfide bond-requiring enzyme in the cytoplasm, and this copper-induced mechanism of disulfide bond formation obviates the need for a thiol/disulfide oxidoreductase in that compartment.
Keywords: chaperone, copper, metalloenzyme, superoxide dismutase (SOD), X-ray crystallography, enzyme activation
Introduction
The copper chaperone for superoxide dismutase (Ccs1)3 activates newly translated copper-zinc superoxide dismutase (Sod1) proteins (1). Sod1 is a critical antioxidant enzyme that detoxifies superoxide, a byproduct of cellular respiration, through redox cycling at its catalytic copper ion (O2˙̄ + 2H+ → H2O2 + O2) (2). Nascent Sod1 is inactive and monomeric (3–7) but is converted to its active homodimeric form through its interaction with Ccs1 (1, 6, 8). Yeast lacking Ccs1 are deficient in Sod1 enzymatic activity unless exogenous copper is added to the culture medium (9–11). Like ccs1Δ yeast, Drosophila melanogaster deficient in Ccs1 lack Sod1 activity and are hypersensitive to oxidative stress (12). Expression of human Sod1 in yeast ccs1Δ cells results in limited Sod1 activity (∼25%) through the ability of a subset of human Sod1 proteins to acquire copper independently of Ccs1 (12, 13).
Ccs1 proteins comprise three domains (D1, D2, and D3), each of which plays a specialized role in Sod1 activation (6, 8, 9). The N-terminal domain (D1) contains an MXCXXC copper-binding motif that is required under copper-limiting in vivo yeast growth conditions and in vitro activation of human Sod1 (6, 9). Ccs1 D1 appears to acquire Cu(I) from the membrane transporter Ctr1 (9, 14, 15). The second domain (D2) possesses significant sequence and structural similarity with Sod1 that permits it to mediate interaction with the immature molecule via residues analogous to those found at the Sod1 homodimeric interface (8, 16–19). Like D2, the C-terminal domain (D3) is essential for Sod1 activation and contains an invariant CXC motif (1, 9). Cells containing Ccs1 proteins lacking the D3 CXC motif possess Sod1 proteins devoid of copper and intrasubunit disulfide bonds (8, 9, 20). However, in vitro studies on Sod1 activation revealed that a Ccs1 variant containing only D1 and D2 was competent to transfer Cu(I) to Sod1 but failed to mediate the disulfide bond formation in Sod1 (8). Disulfide bond formation in Sod1 is dependent on Ccs1 D3 (8).
Multiple studies have sought to delineate the mechanism by which Ccs1 activates Sod1. Experiments in yeast demonstrated that Ccs1 acts on newly synthesized Sod1 to facilitate Cu(I) ion insertion and oxidation of the Sod1 intrasubunit disulfide bond (6, 8, 21, 22). The presence of a disulfide bond in Sod1 is unusual for an enzyme found predominantly in the highly reducing cytosolic environment. The two major cellular compartments in eukaryotes in which disulfide oxidation is commonly observed are the lumen of the endoplasmic reticulum and the mitochondrial intermembrane space, both of which contain thiol/disulfide oxidoreductases (for a review, see Ref. 23). Although the mechanism of disulfide oxidation in nascent cytosolic Sod1 had been elusive until now, it is known that the disulfide bond is essential for full catalytic activity because it anchors the side chain of Arg143 adjacent to the catalytic copper ion where it helps to guide superoxide into the active site (24, 25).
Copper ion delivery and disulfide bond oxidation appear to be linked processes (6, 26) but may occur sequentially (8). In Ccs1-containing yeast, Sod1 mutants unable to bind copper in the active site remain largely disulfide-reduced (27). The addition of Cu–Ccs1 to Zn–Sod1 fails to induce oxidation of the disulfide bond anaerobically, but this process is facile in the presence of oxygen or superoxide (21). A previous yeast Sod1–Ccs1 crystal structure revealed an intermolecular disulfide between Cys57 of Sod1 and Cys229 of Ccs1, suggesting that the intermolecular disulfide may be an intermediate that eventually resolves to the Cys57–Cys146 disulfide via a disulfide exchange reaction (19).
Unlike mammalian Sod1 proteins, yeast Sod1 proteins cannot acquire copper via the Ccs1-independent pathway in normal growth medium (10, 28). The electrostatic loop of yeast Sod1 contains two proline residues that preclude the low levels of Ccs1-independent activation observed in the human and mouse enzymes (13, 29). Replacement of the Pro residues (Pro142 and Pro144) in the yeast enzyme with their cognate residues from mammalian Sod1 proteins permits yeast enzyme activation via a Ccs1-indpendent pathway (13). Likewise, ∼25% of the Sod1 pool is active in ccs1−/− null mice (22). The activation of Sod1 proteins without Pro at these positions in ccs1Δ yeast cells (12, 28) or in ccs1−/− null mice suggests that the copper ion may be provided by labile Cu(I) complexes in the yeast cytosol (e.g. Cu–glutathione) (30, 31). These results also highlight the link between copper acquisition and disulfide bond formation.
Here, we present structural, biochemical, and spectroscopic data that suggest a mechanism of Sod1 activation not described previously. Crystallographic studies on a new Sod1–Ccs1 complex reveal a cavity with positive electrostatic potential exposed by a previously unobserved β-hairpin conformation of Ccs1 D3 that embraces the Sod1 “disulfide loop,” displacing it from the β-barrel. Spectroscopic studies establish that Cu(I) is held at a Cys-Cys-His “entry site” adjacent to the electropositive cavity and Sod1–Ccs1 interface. In vivo studies of yeast Sod1 and its variants strongly suggest that the entry site is functionally relevant and that disulfide formation and copper delivery into the Sod1 active site are linked processes dependent on sulfenylation of Cys146. This new model is consistent with the vast majority of the biochemical data on Ccs1-mediated activation in the literature.
Results
Role(s) of Ccs1 D3 in Sod1 activation
Previous studies have highlighted the importance of the CXC motif in Ccs1 D3 for Sod1 activation (8, 9, 32); however, debate continues on its exact role. Early studies suggested that the D3 CXC motif mediated Cu(I) transfer from D1 to Sod1 (9), but subsequent studies point to a more prominent role in Sod1 disulfide bond formation (6, 8, 21). We attempted to glean additional insights into the role of D3 by structurally characterizing the Sod1–Ccs1 heterocomplex and evaluating residue spacing within the CXC motif.
Previously, Lamb et al. (19) determined the structure of a yeast H48F Sod1–Ccs1 heterotetramer containing two Sod1–Ccs1 heterodimers in the asymmetric unit related by 2-fold noncrystallographic symmetry. Heterotetramer assembly was mediated by reciprocal “swapping” interactions in which a highly conserved Ccs1 D3 237WEER240 α-helical motif bound to grooves on D2 of noncrystallographic symmetry-related heterodimers (supplemental Fig. S1) (19). To probe the importance of the 237WEER240 motif, we created a series of mutants targeting these residues. One variant, E238A/E239A/R240A (237WAAA240) demonstrated ∼5-fold greater yield and enhanced resistance to the trypsinolysis observed at Lys66 in the linker region between D2 and D3 in wild-type Ccs1 (18). H46R/H48Q Sod1 possesses an ablated copper-binding site (27) and forms a highly stable, H46R/H48Q Sod1–E238A/E239A/R240A Ccs1 heterocomplex useful here for structural analysis.
New structure of the Sod1–Ccs1 complex
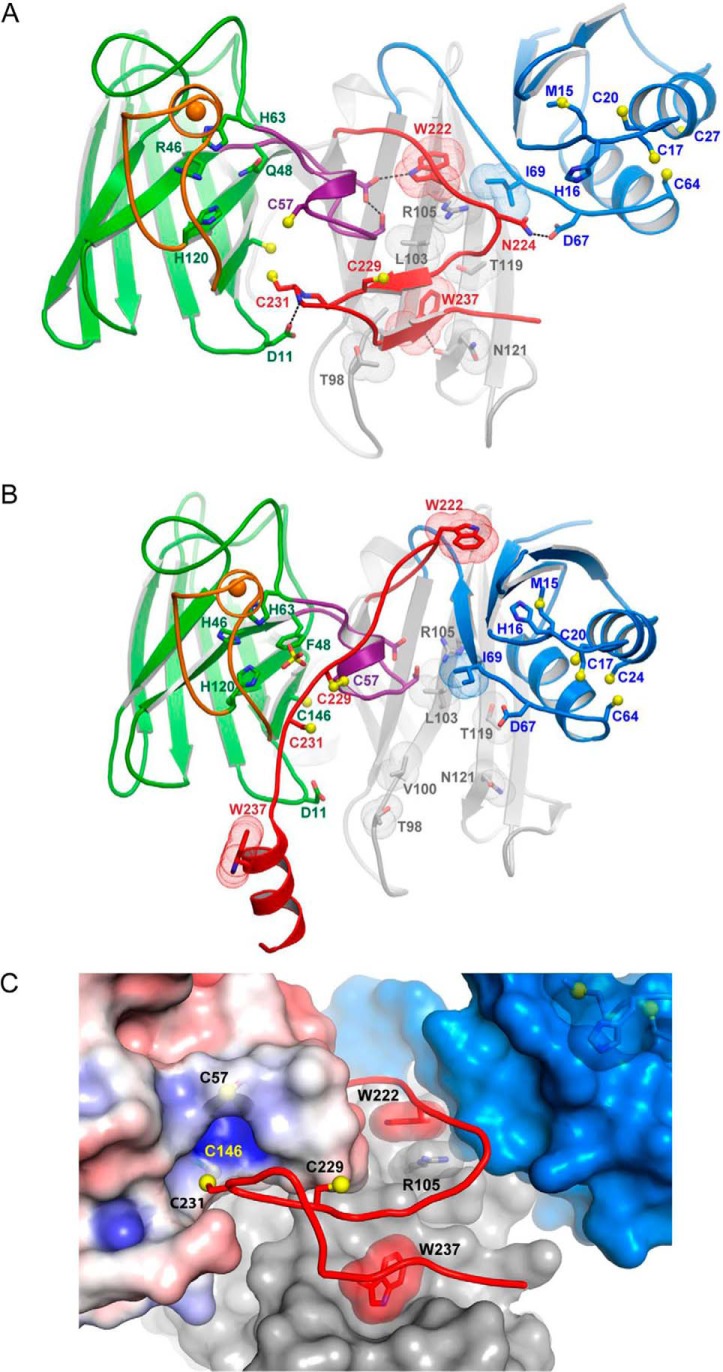
The structure of the H46R/H48Q Sod1–E238A/E239A/R240A Ccs1 complex was determined by molecular replacement and refined in space group P21212 to a resolution of 2.35 Å. Table 1 provides the X-ray diffraction and structure refinement statistics. Although the asymmetric unit contains two Sod1–Ccs1 heterodimers related by noncrystallographic symmetry, the D3 conformation and heterodimeric orientations are dramatically different from the previous yeast H48F Sod1–Ccs1 heterotetramer structure (supplemental Fig. S2) (19). The extended α-helical conformation of Ccs1 D3 is unlikely to exist within the confines of a stable Sod1–Ccs1 heterodimer. Previous data consistently indicate that the Sod1–Ccs1 complex exists as a heterodimer in solution (17, 27), and in contrast to the α-helical and/or disordered conformations of D3 observed in earlier work (18, 19), D3 in the new structure adopts a compact β-hairpin conformation that strengthens the Sod1–Ccs1 interaction (Fig. 1, A and B).
Table 1.
Data collection, phasing, and refinement statistics
r.m.s., root mean square.
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 99.1, 184.4, 48.5 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 0.97918 |
| Resolution (Å) | 48.51–2.35 (2.48–2.35)a |
| Rsym | 0.112 (1.125) |
| Rpim | 0.051 (0.480) |
| Mean (I/σI) | 13.8 (2.0) |
| Completeness (%) | 99.9 (100) |
| Redundancy | 7.0 (7.3) |
| Refinement | |
| Resolution (Å) | 46.92–2.35 |
| No. reflections | 37,948 |
| Rwork/Rfree | 0.186/0.235 |
| No. atoms | |
| Protein | 5,734 |
| Ligand | 3 Zn2+ |
| Solvent | 120 |
| B-factors | |
| Protein | 57.4 |
| Ligand | 73.0 |
| Solvent | 42.9 |
| r.m.s deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.142 |
| Ramachandran statistics | |
| Favored, allowed, outliers (%) | 98.7, 1.3, 0.0 |
| PDB 5U9M | |
a Values in parentheses are for the highest-resolution shell.
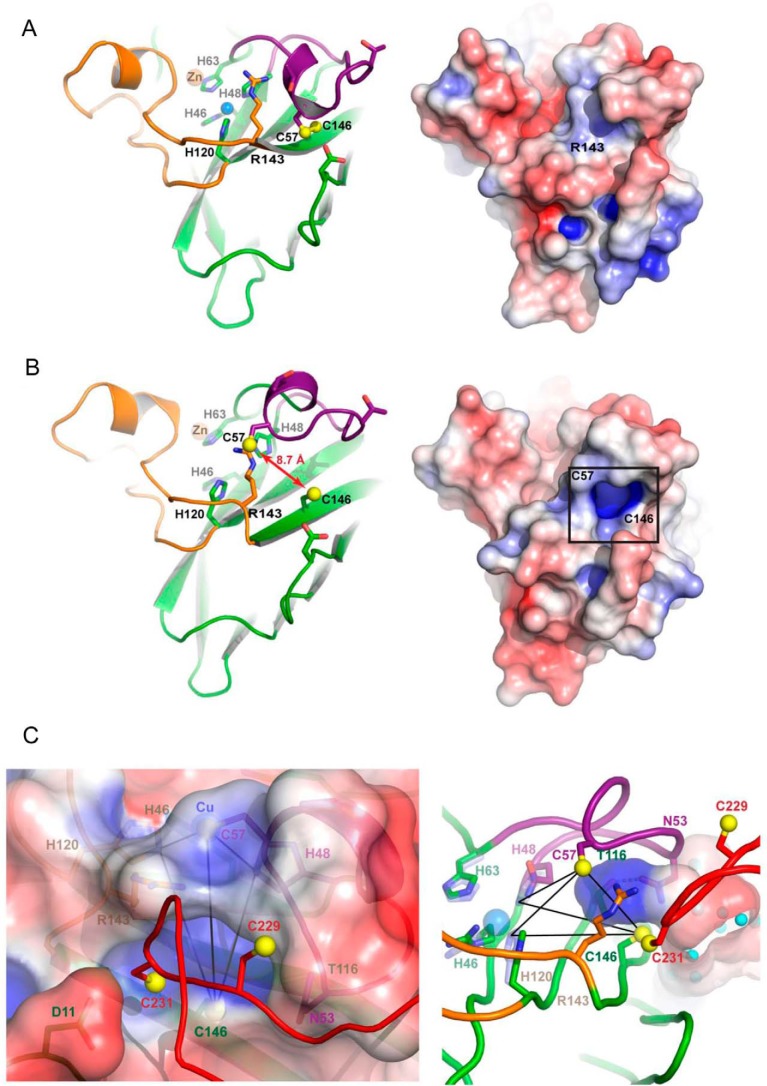
Figure 1.
Structures of Sod1–Ccs1 complexes. In all panels, the Sod1 β-barrel is shown in green with the electrostatic loop orange and disulfide loop purple. Ccs1 D1 is blue, D2 is gray, and D3 is red. The Sod1 zinc ions are shown as orange spheres. A, the heterodimeric complex between human H46R/H48Q Sod1 and yeast E238A/E239A/R240A Ccs1 determined in the present work (Protein Data Bank code 5U9M). The reduced disulfide bond of H46R/H48Q Sod1 between Cys57 and Cys146 permits the displacement of the disulfide loop from the Sod1 β-barrel where it interacts extensively with residues from Ccs1 D2 and D3. The β-hairpin portion of D3 begins just downstream of Asn224 with Cys231 residing on the loop between the two short β-strands. B, the heterodimeric complex between yeast H48F Sod1 and yeast Ccs1 (Protein Data Bank code 1JK9 (19)). C, intercalation of the D3 β-hairpin between the Sod1 β-barrel and disulfide loop. Sod1 is shown as an electrostatic surface contoured at ±4kT. The reduced Cys57–Cys146 Sod1 disulfide bond exposes an electropositive “hole” near Cys146. Domains 1 (blue) and 2 (gray) of Ccs1 are shown as surfaces. The stabilizing positioning of the conserved Ccs1 tryptophan residues Trp222 and Trp237 from D3 are highlighted.
Multiple interactions with residues coming from Ccs1 D1 and D2 stabilize the β-hairpin conformation of Ccs1 D3 (Fig. 1C). The indole ring of Trp222 stacks with the side chain of Arg105 from D2, immobilizing the residues immediately N-terminal to the β-hairpin (Fig. 1, A and C). The buried location of Trp222 is dramatically different from the solvent-exposed environment observed in a previous Sod1–Ccs1 complex structure (Fig. 1B). The invariant 237WEER240 tryptophan fits nicely into a hydrophobic pocket on D2 formed by residues Ile83, Val100, and Leu103 and seemingly acts as a “latch” stabilizing the D3 β-hairpin (Fig. 1, A and C). Ccs1 D3 highlights the major structural differences between the Sod1–Ccs1 complex presented here and the previous structure from Rosenzweig and co-workers (19). The two differing forms of Sod1 used in the compared structures are determined to be nearly identical in their conformation with an overall root mean square deviation of 0.55 over 781 atoms (supplemental Fig. S3C). The other side of the major dimeric interface (Ccs1 D2) aligns with an even lower root mean square deviation of 0.48 over 839 atoms (supplemental Fig. S3C).
Previous studies in solution reported that Ccs1 D3 is required not only for Sod1 activation but also for robust Sod1–Ccs1 interaction (8, 33). The Sod1–Ccs1 complex structure presented here provides a clear molecular basis for the latter observation. Fig. 1C highlights the interface between the Sod1 disulfide loop region and the Ccs1 D3 β-hairpin as a stereoimage of the representative αA electron density with coefficients 2mFo − DFc contoured at 1.2 σ superimposed on the refined model. The large number of interactions between Sod1 and Ccs1 (supplemental Table S1) can occur only when the Sod1 disulfide bond is reduced, consistent with previous observations of stable Sod1–Ccs1 binding only under reducing conditions (27). These interactions, together with the unanticipated intercalation of the D3 CXC motif cysteine (C231) between the disulfide loop and β-barrel of Sod1, create a cysteine-rich “notch-into-groove” complementary surface (supplemental Fig. S3).
Spacing of D3 CXC residues is critical for Sod1 activation
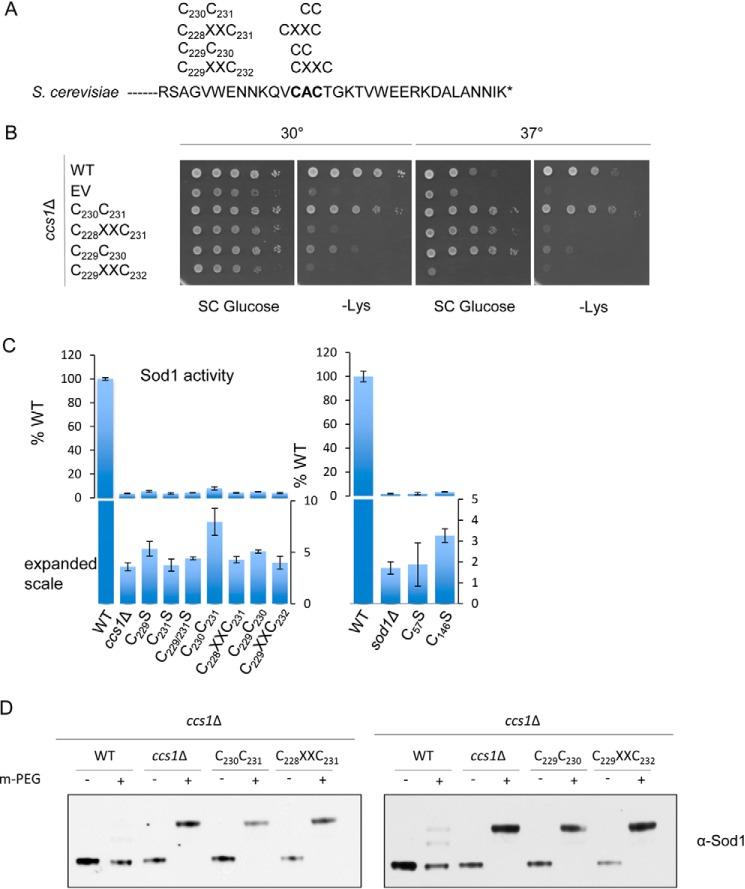
By altering the spacing of the Ccs1 D3 CXC cysteines, we can begin to differentiate the roles of copper delivery and disulfide bond formation catalyzed by Ccs1. Yeast mutants lacking mature (i.e. metal-bound and disulfide-oxidized) Sod1 are lysine auxotrophs and exhibit impaired growth in −Lys medium. Growth of the CXC variant yeast strains was largely compromised (Fig. 2, A and B, and supplemental Fig. S4), and lysates from these cells exhibited nominal Sod1 activity (Fig. 2C). Because only 2% of endogenous Sod1 activity is required for cell growth suppressing the lysine auxotrophy (34), the impaired growth of most Ccs1 CXC variants suggests that Sod1 activation is compromised by greater than 98%. However, cells expressing the Cys230-Cys231 Ccs1 variant exhibited significant growth in −Lys medium at both 30 and 37 °C, and lysates exhibited upward of 7% Sod1 activity (Fig. 2, B and C). With a dithiol configuration, only the Cys230-Cys231 Ccs1 variant shows any significant Sod1 activation. These results suggest that two thiolates are necessary in Ccs1 D3 for Sod1 activation and that a strong positional effect is observed for both. The Cys230-Cys231 Ccs1 variant, which cannot form an intramolecular D3 disulfide, ostensibly rules out prior models implicating Ccs1 D3-mediated shuffling of a preformed disulfide bond to Sod1 during activation (8, 21, 35).
Figure 2.
The spacing of the D3 CXC Cys residues is critical for Sod1 activation. A, mutations were made in the CXC motif of Ccs1 to alter the spacing to either a CC or a CXXC motif without disrupting downstream residues. B, viability tests were performed by plating cells on synthetic complete medium with or without lysine at 30 or 37 °C. C, Sod1 activity for various Ccs1 mutants was quantified; error bars represent S.D. D, the status of the disulfide bond was visualized by lysing cells in the presence of a PEG-maleimide alkylating agent that selectively reacts with free thiols.
To assess the redox status of the Sod1 disulfide bond in these studies, thiol-specific alkylation experiments were performed. Modification of cysteines with polyethylene glycol-modified N-ethylmaleimide (m-PEG) selectively adds 2 kDa per free thiol. Sod1 present in WT cells is resistant to m-PEG modification, whereas Sod1 in ccs1Δ cells is readily modified (Fig. 2D). Likewise, Sod1 modification is similar for cells harboring Ccs1 CXC spacer mutants (Fig. 2D), suggesting that the bulk of cellular Sod1 in these strains exists in the disulfide-reduced inactive state. However, there is likely a small fraction of oxidized Sod1 in the cells expressing Ccs1 Cys230-Cys231 that cannot be visualized by alkylation, which gives rise to the cell viability seen on plate assays.
Stalled Sod1–Ccs1 complexes
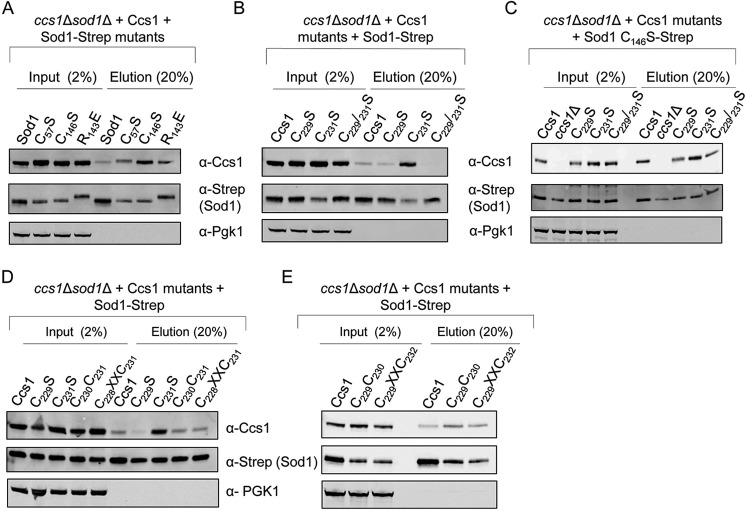
Previous studies have noted and our current Sod1–Ccs1 structure deciphers that immature forms of Sod1 are the primary targets of Ccs1 (Fig. 1C) (17, 27). Fig. 3 shows the results from Strep-Sod1–Ccs1 pulldowns from yeast where Ccs1 does not significantly co-purify with WT Strep-Sod1 (Fig. 3A). Co-purification of stalled Sod1–Ccs1 complexes was enhanced in cells expressing the Sod1 variant C146S and to a lesser degree with C57S or the catalytically inactive R143E mutant (Fig. 3A). The Ccs1 D3 CXC variant C231S, but not C229S, enhanced co-purification relative to WT Ccs1 (Fig. 3B). Enhancement persists when the Sod1 variant C146S is mixed with Ccs1 D3 CXC variants (Fig. 3C). However, D3 CXC spacing mutants with Cys231 still in place did not affect complex stalling (Fig. 3, D and E). The positional requirement of Cys231, but not Cys229, posits deviating roles during the Sod1 activation process that may be explained by their relative arrangements in our Sod1–Ccs1 complex (Fig. 1, A and C). The structure shows that Cys231 is near the proposed entry site as Cys229 points away and does not share a proximate relationship with the other proposed entry site residues.
Figure 3.
Stalled Sod1–Ccs1 complexes. A, lysates from sod1Δ cells transformed with Sod1-Strep mutants were subject to affinity purification using Strep-Tactin resin where 2% (20 μg) of total input and 20% of total elution were loaded and analyzed by Western blotting. B, Strep affinity purification from ccs1Δ cells harboring Ccs1 CXC motif mutants. C, Strep affinity purification from ccs1Δ sod1Δ cells harboring Sod1 C146S-Strep and co-transformed with vectors encoding various Ccs1 CXC mutants. D and E, Strep affinity purification from lysates containing Ccs1 CXC spacing mutants coexpressing Sod1-Strep.
Sod1 disulfide formation secures copper delivery
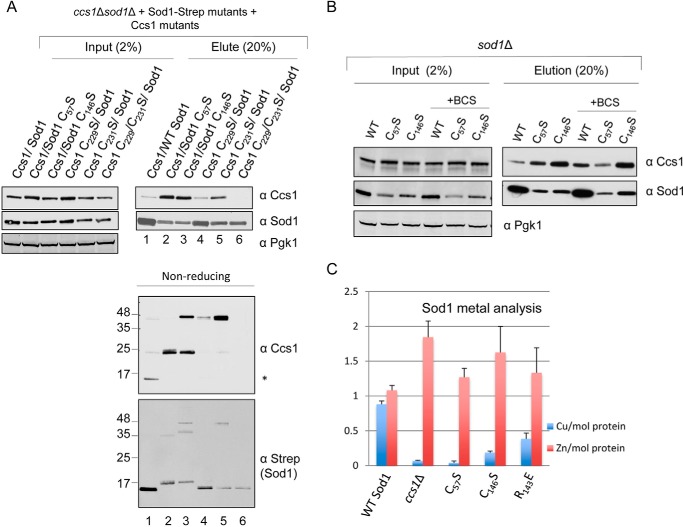
Next, we further examined the link between disulfide bond formation and metal delivery to Sod1. Various affinity-purified heterocomplexes were assessed by non-reducing SDS-PAGE for the presence of an intermolecular disulfide bond. A ∼45-kDa molecular mass band containing both Sod1 and Ccs1 was not only observed in samples when C146S Sod1 was utilized but also when C229S or C231S Ccs1 was present (Fig. 4A). Without both Ccs1 D3 CXC cysteines present, the process guiding formation of the Sod1 Cys57–Cys146 intrasubunit disulfide bond likely becomes trapped as an intermolecular disulfide intermediate. Interestingly, although a high mass Sod1–Ccs1 complex was not visualized with Sod1 C57S, significant levels of Ccs1 were co-purified with this Sod1 mutant (compare Fig. 4A with Fig. 3A). However, when we purify Sod1 after the addition of 20 mm N-ethylmaleimide (NEM) to the lysate, a clear cross-linked Sod1–Ccs1 adduct is seen with the C57S Sod1 sample (Fig. S5). The intermolecular cross-link between C57S Sod1 and Ccs1 in the absence of NEM is susceptible to reduction or resolution via disulfide exchange reactions. In contrast, the intermolecular disulfide linking C146S Sod1 and Ccs1 is more resistant to reduction. The addition of NEM resulted in all of the single mutants forming the high molecular weight complex; nonetheless, at least one of the D3 CXC cysteines is necessary (supplemental Fig. S5). Interestingly, a Ccs1 D3 variant lacking both Cys residues was attenuated in its interaction with Sod1, consistent with an intermolecular disulfide in the stalled Sod1–Ccs1 interaction. To rule out that the stalled heterocomplexes are bridged by Cu(I) coordination, the Cu(I) chelator bathocuproine disulfonic acid (BCS) was added to the reactions, and stalled complexes persisted even in the presence of excess BCS (Fig. 4B).
Figure 4.
Sod1 can form an intermolecular disulfide with Ccs1. A, cell lysates from ccs1Δ sod1Δ cells co-transformed with WT or Ccs1 CXC mutants and WT Sod1-Strep or Sod1-Strep mutants were subjected to affinity purification where 2% (20 μg) of total input and 20% of total elution were loaded and analyzed by Western blotting. Reducing SDS-polyacrylamide (top) as well as non-reducing SDS-polyacrylamide (bottom) gels were run and blotted for Sod1 or Ccs1. B, WT Sod1-Strep or mutants were purified in the presence or absence of 500 μm BCS and analyzed for Ccs1 interaction. C, Sod1-Strep (WT) or various Sod1 mutations were affinity-purified, and metal ratios were determined by ICP-OES; error bars represent S.D.
To gauge whether Sod1 in the Ccs1 mutant cells contained bound metal ions, N-terminally Strep-tagged Sod1 purified from cells with WT and mutant Ccs1 variants were probed for metals by ICP-OES (Fig. 4C). WT Sod1 contained ∼1 molar eq of both copper and zinc, whereas Sod1 purified from ccs1Δ cells contained essentially no copper but ∼2 molar eq of zinc. Sod1 purified from cells containing Ccs1 D3 variants or Sod1 with a C57S substitution was largely devoid of copper. In contrast, the C146S Sod1 was isolated with appreciable levels (∼0.3 molar eq) of copper relative to ccs1Δ cells. The difference in the copper content of C57S versus C146S Sod1 may suggest that these two cysteines play differing roles in copper loading and/or retention. These in vivo studies support the previous in vitro studies showing the importance of the Sod1 cysteines in copper acquisition (36). Taken together, the variant forms of Sod1 or Ccs1 that hinder efficient disulfide bond formation also hinder the ability of Sod1 to receive its full complement of copper.
Cysteine residues from Sod1 and/or Ccs1 form a Cu(I) entry site
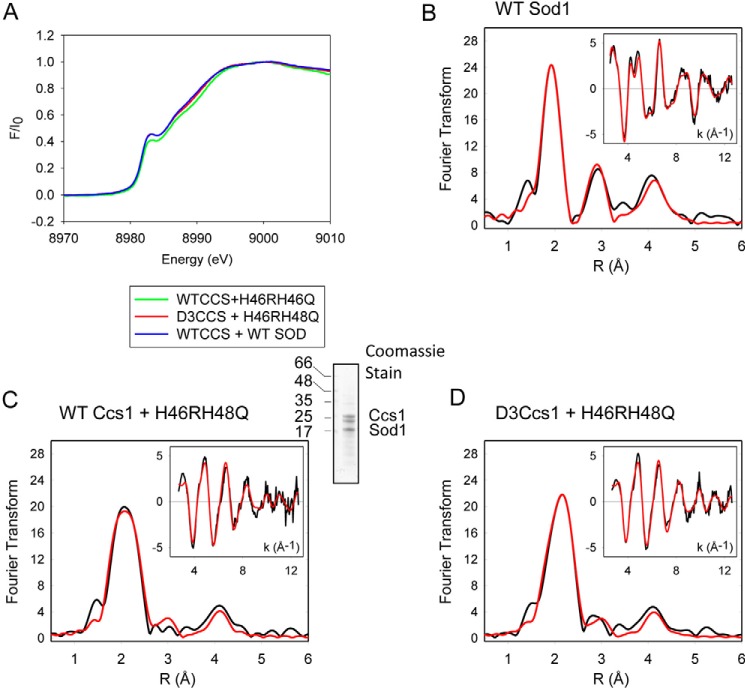
Next, we wanted to address the position and coordination of copper during delivery to Sod1. WT and variant forms of Strep-Sod1 were purified from yeast cells containing either WT or mutant Ccs1. Again, the mutant Sod1 samples, but not the WT, co-purify with Ccs1. X-ray absorption spectroscopy was then utilized to probe copper coordination. Absorption edge features can distinguish Cu(I) from Cu(II) and digonal from trigonal complexes by comparison with extensive data from Cu(I) and Cu(II) model complexes (37). Fig. 5A shows the X-ray absorption spectroscopy CuK edge for WT and mutant Sod1. The absorption edge exhibited a feature at ∼8983 eV consistent with a 1s → 4p transition of the copper ion. For all three samples, the energy and intensity of this near-edge feature relative to the absorption continuum are characteristic of trigonal Cu(I) complexes (37).
Figure 5.
Cysteine residues from Sod1 and/or Ccs1 form a novel Cu(I) entry site. Sod1 was purified from yeast lysates containing the indicated Sod1 and Ccs1 mutations. X-ray absorption spectroscopy was used to determine the copper coordination environment of Sod1. A, the CuK edge of WT samples along with WT Ccs1 + Sod1 H46R/H48Q and Ccs1 C17S/C20S/C27S/C64S/C159S + Sod1 H46R/H48Q. B, EXAFS region for WT samples. C, EXAFS region for sample containing WT Ccs1 + Sod1 H46R/H48Q. D, EXAFS region for sample containing C17S/C20S/C27S/C64S/C159S + Sod1 H46R/H48Q.
Fig. 5B shows the copper K-edge extended X-ray absorption fine structure (EXAFS) spectra and Fourier transform of the EXAFS for WT Sod1. The Fourier transform consists of a major peak at 1.96 Å and two outer shell scatter peaks at ∼3 and ∼4 Å. Simulations with different ligand types reveal a best fit for the 1.96-Å peak with three nitrogen/oxygen ligands, which agrees well with the known three-histidyl ligand coordination of Cu(I) ion in the Sod1 active site (38). The outer shell scatter peaks correspond to backscattering effects of the imidazole ring carbons. Analyses of the H46R/H48Q Sod1 mutant, which cannot bind copper at the active site, was carried out after purification from yeast containing WT Ccs1 or the mutant Ccs1 containing only the D3 CXC cysteines (Fig. 5, C and D, respectively). The copper K-edge feature in both samples was indistinguishable from WT Sod1, suggestive of trigonal Cu(I) coordination (Fig. 5A). In contrast to WT Sod1, the EXAFS analyses of the two variant samples reveal broader first shell peaks that best fit to mixed sulfur and nitrogen/oxygen coordination of the Cu(I) ion. Cu(I) binding to Ccs1 consists of only thiolate ligands, so the candidate nitrogen ligand is likely from Sod1. The EXAFS Fourier transform of the mutant Sod1 purified with WT Ccs1 is best fit with a model containing two sulfur ligands at 2.21 Å (2.22 Å for Cys variant Ccs1) and a single nitrogen/oxygen at 1.92 Å (1.93 Å for Cys variant Ccs1) (Fig. 5, C and D). The Cu(I) site thiolate ligands are not fully distinguished but are consistent with a model in which Cu(I) is coordinated at an entry site generated by the interaction between immature Sod1 and Ccs1.
Ccs1 exposes an electropositive cavity on immature Sod1
The reduced disulfide bond in Sod1 permits the intercalation of the Ccs1 D3 β-hairpin between the disulfide loop and β-barrel, which exposes an electropositive cavity on the Sod1 surface inaccessible in the disulfide-oxidized/mature enzyme (Fig. 6, A and B). The positive surface potential arises primarily through the amide nitrogen atoms of Ala60, Thr116, and Leu117. The cavity is immediately adjacent to the Sγ atom of Cys231 coming from the Ccs1 D3 CXC motif and Sod1 residues Cys57 and Cys146 that become oxidized to form the disulfide bond. Replacement of the glutamine residue at position 48 in this structure with the normal active site histidine suggests a structural equivalence between His48 and His120 such that either could provide a ligand in a putative Cys-Cys-His Cu(I) entry site (Fig. 6C and supplemental Fig. S6). Sod1 disulfide bond formation likely drives concomitant copper delivery to the active site as mutants in which the disulfide bond is not formed are defective in copper acquisition/retention (Fig. 4c). These structural details provide relevance for our hypothesis that an electropositive cavity and proximal copper ion entry site exposed on Ccs1-bound Sod1 link copper ion delivery and disulfide oxidation.
Figure 6.
Electropositive cavity and copper ion entry site on immature Sod1. A, WT Sod1 (colored as in Figs. 1 and 2) shown in its disulfide-oxidized mature conformation (left). The electrostatic surface is contoured at ±4kT (right). B, the disulfide reduced, copper-free Sod1 molecule from the current Sod1–Ccs1 complex with residues 46 and 48 exchanged back to histidine for demonstrative purposes. The box in the right panel highlights an electropositive cavity on Sod1 made accessible through the intercalation of the Ccs1 D3 β-hairpin. C, the Cu(I) entry site on immature Sod1 (solid black lines) as shown here contains two cysteine sulfhydryl groups (Cys57 and Cys146 from Sod1) and one histidyl side chain from His48 or His120. The left panel shows a semitransparent electrostatic surface covering the entry site from the view of the Ccs1 D3 β-hairpin. The right panel is rotated back and counterclockwise (both ∼90°) from the left panel, and the electropositive cavity surface is shown to emphasize its depth and positioning with regard to the entry site ligands.
Copper-dependent sulfenylation at the Sod1 entry site
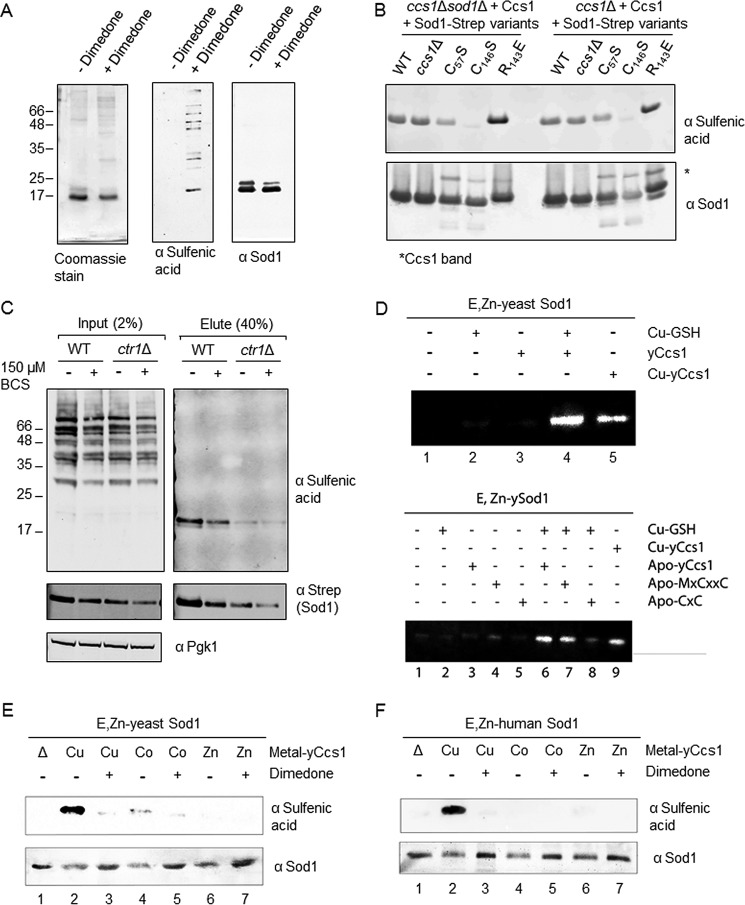
The dismutation of superoxide anions by Sod1 yields both molecular oxygen and hydrogen peroxide. Thiols are particularly prone to modification by reactive oxygen species in the cell, and sulfenic acid is one product produced upon oxidation by H2O2 (39, 40). The propensity of cysteine residues to sulfenylation is influenced by their nucleophilicity, accessibility to H2O2, and the protein microenvironment (41, 42). The formation of sulfenic acid on glutathione peroxidase 3 is a critical precursor for the subsequent disulfide transfer to the transcriptional activator Yap1 under oxidative stress (43, 44). Could the Cu(I) entry site Sod1 intermediate with its electropositive hole mediate cysteine sulfenylation?
Purified Sod1 treated with dimedone, which specifically traps sulfenic acid modifications, was analyzed by Western blotting using a dimedone adduct-specific antibody. A gel band visualized by this technique comigrated with a band imaged with an Sod1 antibody (Fig. 7A). Notably, sulfenylation was observed on mutant Sod1 proteins purified from yeast also containing untagged WT Sod1, so the modification did not arise from a buildup of cellular reactive oxygen species. The sulfenylated Sod1 intermediate is only observed in samples purified without the addition of a reductant. In the presence of reductant, no sulfenylated Sod1 is seen. The sulfenylated adduct is detectable in all Sod1 mutants except for C146S, advocating a preferred site for adduct formation (Fig. 7B). In mature Sod1, Arg143 is important for electrostatic guidance of superoxide toward the active site. During the maturation process, the role of Arg143 is not well understood. Interestingly, sulfenylation is more prevalent in the R143E variant. Architectural effects of this mutation on the entry site may enhance the lifetime of the adduct, although addition of a negative charge at this site may simply help direct superoxide toward the vicinal electropositive cavity and entry site.
Figure 7.
Copper-dependent sulfenylation at the Sod1 entry site and a role for reduced GSH. A, WT Sod1-Strep purified from yeast cells and treated with DMSO vehicle or dimedone at a final concentration of 20 mm. Coomassie staining of the purified Sod1 samples is in the left panel, and a Western blot using a cysteine sulfenic antibody or Sod1 antibody is in the right panels. B, Purified Sod1 with various mutations was treated with dimedone and visualized by Western blot analysis. C, WT cells or ctr1Δ cells were transformed with a Sod1-Strep-encoding plasmid. The cells were grown in the absence or presence of 150 μm BCS and subjected to Strep-Tactin affinity purification where 2% (20 μg) of total input and 40% of total elution were loaded and analyzed by Western blotting. D, in vitro activation of yeast Sod1. Zn–Sod1 (copper-free, disulfide-reduced) was mixed with either Cu–GSH, Cu–Ccs1, or apo-Ccs1 aerobically in 50 mm Tris, pH 7.6, 100 mm NaCl and loaded on the gel for an in-gel Sod1 enzymatic activity. Top panel, lane 1, Zn–Sod1; lane 2, Zn–Sod1 + Cu–GSH; lane 3, Zn–Sod1 + apo-Ccs1; lane 4, Zn–Sod1 + Cu–GSH + apo-Ccs1; lane 5, Zn–Sod1 + Cu–Ccs1. Bottom panel, the same Sod1 activation assay as the top panel but including the Ccs1 D1 MXCXXC (lanes 4 and 7) and D3 CXC (lanes 5 and 8) mutants. E, copper-mediated sulfenylation of yeast Sod1. Apo-Sod1 was preloaded with 1 molar eq of Zn(II) and mixed with yeast Ccs1 either preloaded with 1 molar eq of Cu(I), Co(II), or Zn(II). After mixing, the samples were incubated with 20 mm dimedone prior to loading on the gel. The top panel is the visualization of the dimedone-sulfenylated Sod1 adduct using the anti-sulfenic acid antibody, and the bottom panel shows the level of Sod1 protein as visualized with an anti-Sod1 antibody. F, copper-mediated sulfenylation of human Sod1. Apo-human Sod1 was preloaded with 1 molar eq of Zn(II) and incubated with yeast Ccs1 either preloaded with 1 molar eq of Cu(I), Co(II), or Zn(II). The conditions and two panels are as described in E.
Sod1 sulfenylation is attenuated in yeast cells that are rendered copper-deficient by inclusion of the copper chelator BCS in the growth medium or selective knock-out of the influx copper transporter Ctr1 (Fig. 7C). This strongly suggests that sulfenylation at Sod1 Cys146 is a copper-mediated reaction. Additionally, Sod1 isolated from ccs1Δ cells is sulfenylated, implying that the primary role of Ccs1 may not be in copper delivery. Sulfenylation at this site likely promotes Cys57–Cys146 disulfide oxidation in Sod1. We cannot be certain whether the sulfenylation of Cys146 or the subsequent disulfide bond formation triggers release of copper from its entry site ligands into the nearby Sod1 active site (see Fig. 5C).
Sod1 activation under copper-replete conditions
For yeast Ccs1 (but not necessarily mammalian), D1 is required only under copper-limiting conditions in vivo (9, 10, 32). The conditional necessity of D1 in yeast Ccs1 suggests that the cellular environment may alter the mechanism of copper delivery. During normal cellular conditions, copper ions are present in the cytosol as small molecule complexes with reduced glutathione (GSH) or metallothionein scavengers (45). However, only mammalian cells are known to utilize Cu(I)–GSH as an alternate route for Sod1 activation (13). Fig. 7D shows the results for an in vitro yeast Sod1 activation assay incorporating Cu(I)–GSH. The addition of Cu(I)–GSH to E,Zn–ySod1 (where E indicates an empty copper-binding site) results in only minimal Sod1 activation (lane 2). When E,Zn–ySod1 is mixed with apo-Ccs1 and Cu(I)–GSH, abundant Sod1 activity is detected (lane 4) that is similar to the activity of apo-Sod1 mixed with Cu(I)–Ccs1 (lane 5). In addition, an apo-Ccs1 D1 MXCXXC variant (C17A/C20A) can still promote copper delivery by Cu–GSH to E,Zn–ySod1 (Fig. 7D, lower panel, lane 7), but the D3 CXC variant (C229A/C231A) fails to do so (lane 8). These results strongly support a facilitatory role for Ccs1 in the copper delivery process of reduced glutathione without copper coordination by Ccs1 D1. Relatedly, the addition of dimedone to in vitro Sod1 activation assays precludes complete Sod1 activation, likely through inhibition of disulfide exchange at the entry site and the associated copper delivery (supplemental Fig. S7). These data suggest that when copper is readily available Ccs1 is not required to directly coordinate copper by D1 and may simply expose the electropositive cavity and Cu(I) entry site at the Sod1–Ccs1 interface to allow entry of Cu(I) from either Ccs1 or a labile source such as glutathione and then facilitate disulfide formation as described above.
Entry site-specific cysteine sulfenylation examined by MS
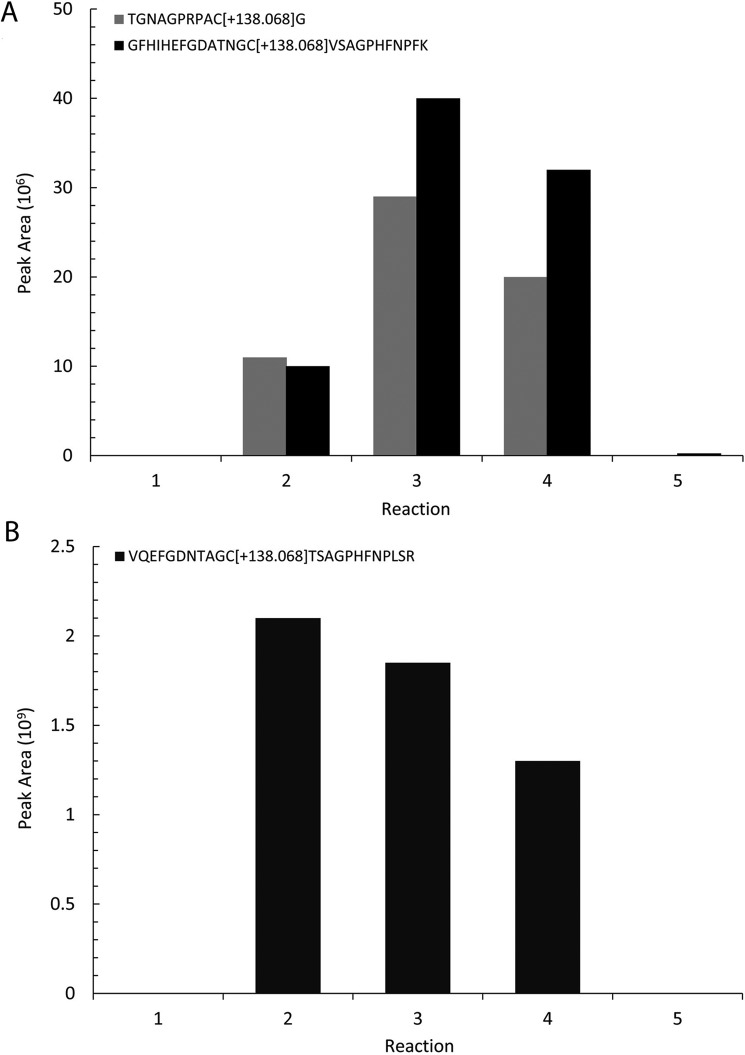
Sulfenylation reactions were analyzed by mass spectrometry to identify peptides with dimedone adducts. Briefly, reactions were digested by trypsin, fractionated online using reversed-phase chromatography, and analyzed by tandem mass spectrometry. Dimedone-modified peptides were identified by a characteristic mass shift of +138.068080 on the modified cysteine residue. Entry site cysteine residues on Sod1 (Cys57 and Cys146) were both modified in the yeast Sod1 sample. For human Sod1, the Cys57 site was modified. No cysteines outside of the entry site (e.g. Cys6 and Cys111) on either form of Sod1 were modified, showing specificity for the additions. Annotated MS2 spectra for each modified peptide (supplemental Fig. S8) as well as the relative abundance of each peptide across the set of reactions is shown in Fig. 8, A and B.
Figure 8.
Relative abundance of dimedone-modified Sod1 peptides calculated from the extracted ion chromatogram for each peptide across the five reactions. Reaction 1, 5 μm apo-Sod1, BCS buffer (50 mm Tris, pH 7.6, 100 mm NaCl, 0.5 mm tris(2-carboxyethyl)phosphine, 200 μm BCS). Reaction 2, 5 μm apo-Sod1, 5 μm H2O2, 5 mm dimedone, 20 μm Zn2SO4, 25 μm Cu(I)–yCcs1, BCS buffer. Reaction 3, 5 μm apo-Sod1, 25 μm H2O2, 5 mm dimedone, 20 μm Zn2SO4, 25 μm Cu(I)–yCcs1, BCS buffer. Reaction 4, 5 μm apo-Sod1, 5 mm dimedone, 20 μm Zn2SO4, 25 μm Cu(I)–yCcs1, BCS buffer. Reaction 5, 5 μm apo-Sod1, 20 μm Zn2SO4, 25 μm Cu(I)–yCcs1, BCS buffer. In A, 1–5 indicates yeast Sod1 reactions, and in B, 1–5 indicates human Sod1 reactions.
Discussion
Based upon the data presented here, Ccs1-mediated Sod1 maturation appears to proceed via a copper-mediated sulfenylation intermediate that resolves to form the stable Sod1 disulfide bond. Our new Sod1–Ccs1 complex structure demonstrates that upon binding to immature Sod1 interactions between Ccs1 and the disulfide loop of Sod1 orient the D3 β-hairpin toward Sod1. The indole side chain of Trp237 fits into a hydrophobic pocket on Ccs1 D2 securing the D3 β-hairpin. The intercalation of Cys231, together with hydrogen bonding interactions between the Asp52 side chain of Sod1 and the indole nitrogen of Trp222, pulls the disulfide loop away from the Sod1 β-barrel and exposes a copper ion entry site and adjacent electropositive cavity. A Cu(I) ion coming from Ccs1 D1 or a labile Cu(I) pool can now be coordinated at the entry site, and either superoxide or H2O2 is attracted toward the electropositive cavity harboring Sod1 Cys146. If superoxide anion is attracted, copper ion oxidation (e.g. Cu(I) → Cu(II)) reduces the superoxide to hydrogen peroxide. This newly generated H2O2 molecule promotes sulfenylation of a cysteine, likely Sod1 Cys146, although sulfenylation could initially occur on a Ccs1 D3 cysteine. Alternatively, attraction of H2O2 to the Cu(I) entry site could directly sulfenylate the cysteine with no change in the copper oxidation state. Sulfenylation subsequently mediates disulfide bond formation through disulfide exchange reactions involving the Ccs1 D3 cysteines. This reaction likely drives the entry site copper ion to the active site. Sulfenylation of Sod1 is not dependent on Ccs1 (Fig. 7B), but the resolution to a disulfide bond is dependent on Ccs1. The driving force for copper migration to the tetrahistidine environment of the mature catalytic site may be either sulfenylation of Cys146 or disulfide bond formation. Sod1 disulfide oxidation and copper ion release into the active site expel the Ccs1 D3 β-hairpin as the formation of the disulfide bond closes the electropositive cavity and terminates interaction between Sod1 and Ccs1.
The Sod1–Ccs1 interaction during the maturation process is likely short-lived and is terminated by successful intrasubunit Cys57–Cys146 disulfide bond formation in Sod1. However, this transient binding event can be trapped as an intermolecular disulfide-linked intermediate in cells containing either C146S Sod1 or C231S Ccs1; however, substitution of both Ccs1 D3 CXC cysteines attenuates trapping as expected. Enhanced trapping of the Sod1–Ccs1 interaction observed with Sod1 catalytic site His mutants blocks copper migration to the active site. This may lead to resolution of the Cys146 sulfenic acid by a Ccs1-derived thiol, which may in turn generate intermolecular cross-links of the type observed by Lamb et al. (19), and our present results show that at least one Ccs1-derived thiol is required for formation of the cross-link. Our data support a model where Ccs1-mediated disulfide exchange reactions between cysteine residues at the Cu(I) entry site drive Sod1 maturation to completion.
Clear evidence for a Cu(I) entry site on immature Sod1 bound by Ccs1 is provided by X-ray absorption and related techniques. Cells harboring a Sod1 H46R/H48Q variant unable to bind copper at the active site coordinate Cu(I) using a Cys-Cys-His pseudotrigonal planar geometry. Consistent with this geometry and ligand set, the new heterocomplex structure reveals that the Sγ atoms of Cys231 and Cys57 and the position of copper in the activated enzyme are co-planar with liganding nitrogen atoms from His48 and His120 equidistant and normal to this plane (Fig. 6C). Thus, the entry site may be fluxional, sampling some or all of the four possible thiolate ligands. A Cu(I) resting state observed in the WT EXAFS analysis is consistent with in vivo NMR analysis performed on the maturation steps of Sod1 (6, 7). In a separate in vitro analysis, transfer of copper from Cu(I)-Ccs1 to Sod1 in the presence of oxygen showed a stable Cu(II)–Sod1 (26). The difference between these two experiments may be that H2O2-driven sulfenylation may predominate in vivo, yielding a Cu(I)–Sod1 complex. This is not unduly surprising because it is likely that the concentration of free H2O2 vastly exceeds that of free superoxide in the cytosol. However, we emphasize that both pathways are chemically plausible, and it may be that both are operative, perhaps under different cellular conditions.
A readily accessible Cu(I) entry site requires direct delivery and “handoff” by a copper ion carrier. Cu–GSH complexes have shown the ability to activate human Sod1 in the absence of Ccs1, a process not replicable for yeast Sod1 due to proline residues just upstream of Cys146 (13). Here, we show evidence that Cu–GSH can deliver copper to yeast Sod1 but requires interaction with metal-free Ccs1. Under normal cellular conditions, the major role of Ccs1 may be to pull the disulfide loop away from the Sod1 β-barrel, thereby exposing the electropositive cavity and entry site on immature Sod1.
The copper dependence of the sulfenylation reaction may arise from two effects. If superoxide is the substrate, peroxide is generated through a copper redox reaction. The copper dependence of a H2O2-mediated sulfenylation reaction may occur, orienting the thiols at the reaction site or enhancing the cysteine nucleophilicity. Finally, we note that coordination may increase the fidelity of the sulfenylation reaction at the critical thiol (Cys146) via a specific interaction with the Cu(I), which may electronically activate this thiol for reactivity.
In conclusion, our biochemical studies and Sod1–Ccs1 complex structure suggest a novel mechanism for Sod1 maturation. Copper delivery to an exposed entry site on immature Sod1 leads to a copper-mediated sulfenylation intermediate. Copper donation may occur either through Ccs1 D1 or via a small labile Cu(I) complex. In the later case, Ccs1 may still facilitate entry site copper binding through its stabilization of a nascent conformation of Sod1. Ccs1 directs Sod1 disulfide formation through sulfenylation of Cys146, which enters disulfide exchange reactions concluding in a stable Sod1 Cys57–Cys146 disulfide, copper ion placement in the active site, and termination of the interaction with the now mature Sod1. The maturation path is a concerted process requiring a specific conformation of thiolates in the entry complex accompanied by coupled sulfenylation, copper ion migration, and resolution to disulfide with the driving force dependent on all three molecular events. In this context, the Cu(I) entry site and electropositive cavity are integral to normal Sod1 function. Our data and ensuing mechanism also require that Sod1 maturation is initiated by oxidative chemistry as reported previously by O'Halloran and co-workers (21). This latter conclusion is significant because it implies that Sod1 metallation and activation are regulated by the presence of its substrate such that its precious copper cargo is only loaded when the cell or organelle experiences oxidative stress.
Experimental procedures
Materials
Monobasic and dibasic potassium phosphate, acetonitrile (Optima grade), formic acid, sodium hydroxide, yeast extract, peptone, dextrose (glucose), EDTA, sodium chloride, and sodium acetate were obtained from Fischer Scientific. Ammonium sulfate was purchased from US Biological. Polyethylene glycol 1000 came from Fluka. Tris was purchased from Research Products International. Primers came from Invitrogen. Agarose glycerol, DEAE-Sephadex, DTT, and PMSF were obtained from Sigma. Pfu DNA polymerase and deoxyribonucleotides were purchased from Stratagene. Glass beads were obtained from Biospec. Crystallization screening kits and crystal growth trays were purchased from Hampton Research. Phenyl-Sepharose and Sephadex G-75 came from GE Healthcare. All solutions were prepared using deionized water passed through a Millipore ultrapurification system.
Sod1 cloning, expression, and purification
DNA fragments encoding the human H46R/H48Q Sod1 double mutant were amplified by polymerase chain reaction (PCR) and ligated into the YEP351-hSOD plasmid where expression of the Sod1 protein is directed under the control of its own promoter. The protein was expressed, purified, and characterized as described previously (27) with the addition of a DEAE-Sephadex chromatography step between the hydrophobic interaction chromatography and gel filtration column steps. The metal content of purified Sod1 and Ccs1 proteins was determined using inductively coupled plasma mass spectrometry at the Chemical Analysis Laboratory at the University of Georgia.
Ccs1 cloning, expression, and purification
DNA fragments encoding yCcs1 were generated by PCR from plasmids generously supplied by J. S. Valentine (UCLA). Ccs1 constructs were cloned into a pkA6H vector, which contains an inducible lacZ promoter, an N-terminal His6 tag, and a tobacco etch virus (TEV) cleavage site. Residues 237WEER240 in D3 were substituted to 237WAAA240 via QuikChange mutagenesis, and these substitutions dramatically enhanced the overall yield of recombinant Ccs1 (∼5-fold) while simultaneously obviating the instability and proteolysis observed in wild-type Ccs1 otherwise identically prepared. Yeast Ccs1 was expressed in Escherichia coli BL21(DE3). Transformed cells were grown in LB medium at 37 °C to an A600 nm of 0.6–0.8. After induction with 0.1 mm isopropyl 1-thio-β-d-galactopyranoside, the cells were transferred to 30 °C for an additional 4 h before being harvested. Ccs1 was purified using a HisTrap HP Ni2+ affinity column purchased from GE Healthcare. After purification, the hexa-His tag was removed by digestion overnight with TEV protease produced in-house and engineered to contain its own non-cleavable hexa-His tag. After digestion, the cleaved His tag and TEV protease were removed from the Ccs1 sample by a final pass through the nickel column. This procedure leaves a two-residue (Gly-His) extension on the Ccs1 N terminus.
Crystallization, structure determination, and refinement
The H46R/H48Q Sod1–E238A/E239A/R239 Ccs1 complex was prepared in solution as described previously (27). All crystals were grown at room temperature using the hanging drop vapor diffusion method. Purified Sod1–Ccs1 complex at 20 mg/ml in 50 mm MES, pH 6.5, 150 mm sodium chloride, 10 mm tris(2-carboxyethyl)phosphine, 50 μm ZnSO4 was mixed with an equal volume of reservoir solution (100 mm Bis-Tris, pH 6.5, 200 mm ammonium sulfate, 25% (w/v) PEG 3350). Rectangular crystals grew in space group P21212 within 2 weeks at room temperature. Suitable specimens were soaked in a cryoprotectant consisting of reservoir solution with 10% (v/v) ethylene glycol before flash cooling by plunging into liquid nitrogen. X-ray diffraction data were measured to a resolution of 2.5 Å at the Argonne National Laboratory Northeastern Collaborative Access Team (NE-CAT) beamline 24-ID-E, which is equipped with a microfocus MD-E microdiffractometer. All diffraction data were processed using the HKL2000 program suite (48). The structure was determined by molecular replacement with the program MOLREP (49) using the structures of human S134N Sod1 (Protein Data Bank code 1OZU (50)) and yCcs1 D2 (Protein Data Bank code 1JK9 (19)) as the search models. The Sod1 component was positioned in the unit cell first, and then yeast Ccs1 D2 followed by rigid body refinement using the program PHENIX (51). The model was refined iteratively in PHENIX without noncrystallographic symmetry restraints, invoking simulated annealing in the early rounds of refinement and alternating with manual model adjustment into σA-weighted electron density map using the program Coot (52). No stereochemical restraints were applied to the metal-ligand distances or bond angles. All structural figures were created using the PyMOL Molecular Graphics System, version 1.8, Schrödinger, LLC.
Yeast growth, strains, and plasmids
Saccharomyces cerevisiae strains were grown in SC-glucose (dextrose) (SCD) medium at 30 °C unless otherwise noted. BY4741 ccs1Δ::KanMX knock-out was generously provided by Dr. Val Culotta. The sod1Δ mutant in BY4741 as well as in the ccs1Δ::KanMX background was generated with a URA3 disruption. Dr. Culotta generously provided both pRS313-CCS1 and YEP351-ySOD1 plasmid for in vivo and EXAFS experiments. N-terminal Strep tag (WSHPQFEK) with a two-Gly linker and all subsequent mutations in ySOD1 were generated by Phusion mutagenesis PCR.
Analysis of Sod1 disulfide redox state and activity measurements
ccs1Δ cells were transformed with CCS1-containing plasmids harboring specific spacing mutations. 50-ml cultures were harvested at an A600 nm of ∼1. Pellets were resuspended in 8 m urea, 3 mm EDTA, 0.5% Triton X-100, 50 mm Tris, pH 8.0, and split into two aliquots containing or lacking PEG-maleimide (molecular weight, 2000) purchased from Nanocs. The cells were lysed by bead beating and centrifuged at 12,000 × g for 10 min. The supernatant was placed at 37 °C for 1 h, and ∼30 μg of lysate was analyzed by Western blotting. Sod1 activity was measured using the SOD assay kit purchased from Sigma.
Sod1 purification
For Strep tag affinity purification, ccs1Δ sod1Δ cells were co-transformed with vectors encoding Sod1-Strep and Ccs1 variants and grown to an A600 nm of ∼1 in SCD medium. Cells were resuspended in 50 mm NaH2PO4, 300 mm NaCl, pH 8.0, with Roche Applied Science cOmplete protease inhibitor mixture tablets. Cells were lysed by bead beating and centrifuged at 12,000 × g for 10 min. One milligram of supernatant was diluted to 500 μl with buffer, and a 50-μl bead volume of Strep-Tactin Superflow Plus (Qiagen) was added and incubated for either 4 h or overnight. For non-reducing alkylation experiments, 20 mm NEM was added to overnight incubations of lysates. To test whether copper was involved in heterodimer complex, 500 μm BCS was added to lysates in overnight incubations. Eluates were then analyzed by Western blot analysis. For purification of Sod1 H46R/H48Q, cells were lysed using a French pressure cell press and centrifuged at 10,000 × g to clarify lysates. Ammonium sulfate was added to lysates at 4 °C while stirring to a final concentration of 60%. Upon reaching 60% saturation, an equal volume of methanol was added to the solution and stirred vigorously for 10 min. The supernatant was centrifuged at 10,000 × g for 30 min, and the supernatant was dialyzed overnight into 50 mm phosphate buffer, 150 mm NaCl, pH 7.0. Following dialysis, the eluate was loaded onto a Superdex 75 FPLC column, and the major peak containing Sod1 was dialyzed into 20 mm Tris, pH 8.0.
Sod1 metal analysis
Strep-tagged Sod1 constructs were grown in SCD medium to an A600 nm of 1.5 and harvested. Cells were lysed using a French pressure cell press and centrifuged at 50,000 × g for 1 h at 4 °C. The protein was purified using Strep-Tactin Superflow Plus resin. Eluted proteins were dialyzed overnight in 20 mm Tris, pH 8.0, and analyzed by ICP-OES for metal content.
Sod1 sulfenylation
Sod1 was purified as described above. 5,5-Dimethyl-1,3-cyclohexanedione (dimedone; Sigma-Aldrich) was added to a final concentration of 20 mm to the eluate or lysates. The protein was incubated at 37 °C for 1 h and analyzed by Western blot analysis using an anti-cysteine sulfenic antibody (EMD Millipore) that reacts with cysteine sulfenic acids derivatized with dimedone. For experiments to test the role of copper in sulfenic acid formation, cells were grown overnight in low copper/iron medium to an OD of ∼1. Cells were back-diluted to an OD of ∼1 in low copper/iron medium plus or minus 150 μm BCS and harvested at an OD of ∼1.
X-ray absorption data collection and analysis
Copper K-edge (8.9-keV) EXAFS and X-ray absorption near-edge structure data were measured at the Stanford Synchrotron Radiation Lightsource operating at 3 GeV with currents between 300 and 500 mA and in continuous top-up mode. Samples were measured on beamline 7-3 using a Si(220) monochromator and an upstream rhodium-coated mirror with 13-keV energy cutoff to reject harmonics. Data were collected in fluorescence mode using a Canberra 30-element germanium array detector with maximum count rates per array element set to less than 120 kHz. A Z-1 nickel (NiO) filter and Soller slit assembly inserted in front of the detector were used to reduce elastic scattering relative to the CuKα fluorescence. Four scans of a buffer blank were averaged and subtracted from the averaged data for each protein sample to remove the NiKβ fluorescence and produce a flat pre-edge baseline. Samples were measured as aqueous glasses in 20% ethylene glycol at 10 K. Data reduction and background subtractions were performed using the program modules of EXAFSPAK (53). Spectral simulation was carried out using EXCURVE, version 9.2 (54, 55) as described previously (56–58). The best fit of the simulations of the EXAFS data supported a mixed-shell coordination model consisting of imidazole from histidine and sulfur from cysteine coordination where appropriate. The threshold energy, E0, was chosen at 8985 eV, and refinement of structural parameters included distances (R), coordination numbers (N), Debye-Waller factors (2σ2), and multiple scattering contributions from outer-shell atoms of imidazole rings.
Mass spectrometry data collection and analysis
Sample digestion and data acquisition were performed as described in Caslavka Zempel et al. (59). Modified peptides were identified by the MSGF+ search algorithm using a differential modification search that considered a mass shift of +138.068080 on cysteine residues and performed against a yeast or human database depending on the proteins of interest (60). The search results were combined and analyzed using Percolator and filtered at a spectrum-level q-value of 0.01 (61). Dimedone site localization and site probability were determined using the PTMRS algorithm and filtered at 95% confidence level or higher (62). The spectrum for each peptide was manually annotated using the SeeMS spectrum viewer (46). Skyline software was used to evaluate the presence of each dimedone-modified peptide across reactions and plot the intensity-based bar plot (47).
Author contributions
M. M. F., S. D. B., A. B. T., H. J. K., and D. D. W. conducted the research presented. M. M. F., N. J. B., D. D. W., P. J. H., J. A. W., and D. R. W. contributed to data analysis and interpretation. M. M. F., N. J. B., D. D. W., P. J. H., and D. R. W. drafted and edited the manuscript. D. D. W., N. J. B., J. A. W., and D. R. W. contributed ideas for experimentation. All authors approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Val Culotta for generously providing numerous reagents. Support for Northeastern Collaborative Access Team beamline 24-ID-E is provided by National Institutes of Health Grant P41 GM103403 and United States Department of Energy Grant DE-AC02-06CH11357. The X-Ray Crystallography Core Laboratory at the University of Texas Health Science Center at San Antonio is supported in part by the Office of the Vice President for Research and by San Antonio Cancer Institute Grant P30 CA054174. Use of the Stanford Synchrotron Radiation Lightsource, Stanford Linear Accelerator Center National Accelerator Laboratory, is supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The Stanford Synchrotron Radiation Lightsource Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including Grant P41GM103393).
This work was supported in part by National Institutes of Health Grants R01 GM110755, R01 GM054803, R01 NS39112, R01 GM112763, and R01 GM120252 (to D. R. W., N. J. B., P. J. H., J. A. W., and D. D. W., respectively); United States Department of Veterans Affairs Merit Review Award I01 BX002580 (to P. J. H.); and Robert A. Welch Foundation Grant AQ-1399 (to P. J. H.). The authors declare that they have no conflicts of interest with the contents of this article. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs, the United States government, or the National Institutes of Health.
This article contains supplemental Figs. S1–S8 and Table S1.
The atomic coordinates and structure factors (code 5U9M) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- Ccs1
- the copper chaperone for superoxide dismutase
- Sod1
- copper-zinc superoxide dismutase
- m-PEG
- polyethylene glycol-modified N-ethylmaleimide
- NEM
- N-ethylmaleimide
- BCS
- bathocuproine disulfonic acid
- ICP-OES
- inductively coupled plasma optical emission spectrometry
- EXAFS
- extended X-ray absorption fine structure
- ySod1
- yeast Sod1
- yCcs1
- yeast Ccs1
- TEV
- tobacco etch virus
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- SC
- synthetic complete.
References
- 1. Culotta V. C., Klomp L. W., Strain J., Casareno R. L., Krems B., and Gitlin J. D. (1997) The copper chaperone for superoxide dismutase. J. Biol. Chem. 272, 23469–23472 [DOI] [PubMed] [Google Scholar]
- 2. Fridovich I. (1989) Superoxide dismutases. An adaptation to a paramagnetic gas. J. Biol. Chem. 264, 7761–7764 [PubMed] [Google Scholar]
- 3. Arnesano F., Banci L., Bertini I., Martinelli M., Furukawa Y., and O'Halloran T. V. (2004) The unusually stable quaternary structure of human Cu,Zn-superoxide dismutase 1 is controlled by both metal occupancy and disulfide status. J. Biol. Chem. 279, 47998–48003 [DOI] [PubMed] [Google Scholar]
- 4. Doucette P. A., Whitson L. J., Cao X., Schirf V., Demeler B., Valentine J. S., Hansen J. C., and Hart P. J. (2004) Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J. Biol. Chem. 279, 54558–54566 [DOI] [PubMed] [Google Scholar]
- 5. Lindberg M. J., Normark J., Holmgren A., and Oliveberg M. (2004) Folding of human superoxide dismutase: disulfide reduction prevents dimerization and produces marginally stable monomers. Proc. Natl. Acad. Sci. U.S.A. 101, 15893–15898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banci L., Barbieri L., Bertini I., Luchinat E., Secci E., Zhao Y., and Aricescu A. R. (2013) Atomic-resolution monitoring of protein maturation in live human cells by NMR. Nat. Chem. Biol. 9, 297–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Luchinat E., Barbieri L., Rubino J. T., Kozyreva T., Cantini F., and Banci L. (2014) In-cell NMR reveals potential precursor of toxic species from SOD1 fALS mutants. Nat. Commun. 5, 5502. [DOI] [PubMed] [Google Scholar]
- 8. Banci L., Bertini I., Cantini F., Kozyreva T., Massagni C., Palumaa P., Rubino J. T., and Zovo K. (2012) Human superoxide dismutase 1 (hSOD1) maturation through interaction with human copper chaperone for SOD1 (hCCS). Proc. Natl. Acad. Sci. U.S.A. 109, 13555–13560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmidt P. J., Rae T. D., Pufahl R. A., Hamma T., Strain J., O'Halloran T. V., and Culotta V. C. (1999) Multiple protein domains contribute to the action of the copper chaperone for superoxide dismutase. J. Biol. Chem. 274, 23719–23725 [DOI] [PubMed] [Google Scholar]
- 10. Rae T. D., Schmidt P. J., Pufahl R. A., Culotta V. C., and O'Halloran T. V. (1999) Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science 284, 805–808 [DOI] [PubMed] [Google Scholar]
- 11. Sea K. W., Sheng Y., Lelie H. L., Kane Barnese L., Durazo A., Valentine J. S., and Gralla E. B. (2013) Yeast copper-zinc superoxide dismutase can be activated in the absence of its copper chaperone. J. Biol. Inorg. Chem. 18, 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirby K., Jensen L. T., Binnington J., Hilliker A. J., Ulloa J., Culotta V. C., and Phillips J. P. (2008) Instability of superoxide dismutase 1 of Drosophila in mutants deficient for its cognate copper chaperone. J. Biol. Chem. 283, 35393–35401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carroll M. C., Girouard J. B., Ulloa J. L., Subramaniam J. R., Wong P. C., Valentine J. S., and Culotta V. C. (2004) Mechanisms for activating Cu- and Zn-containing superoxide dismutase in the absence of the CCS Cu chaperone. Proc. Natl. Acad. Sci. U.S.A. 101, 5964–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dancis A., Haile D., Yuan D. S., and Klausner R. D. (1994) The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269, 25660–25667 [PubMed] [Google Scholar]
- 15. Pope C. R., De Feo C. J., and Unger V. M. (2013) Cellular distribution of copper to superoxide dismutase involves scaffolding by membranes. Proc. Natl. Acad. Sci. U.S.A. 110, 20491–20496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamb A. L., Wernimont A. K., Pufahl R. A., O'Halloran T. V., and Rosenzweig A. C. (2000) Crystal structure of the second domain of the human copper chaperone for superoxide dismutase. Biochemistry 39, 1589–1595 [DOI] [PubMed] [Google Scholar]
- 17. Lamb A. L., Torres A. S., O'Halloran T. V., and Rosenzweig A. C. (2000) Heterodimer formation between superoxide dismutase and its copper chaperone. Biochemistry 39, 14720–14727 [DOI] [PubMed] [Google Scholar]
- 18. Lamb A. L., Wernimont A. K., Pufahl R. A., Culotta V. C., O'Halloran T. V., and Rosenzweig A. C. (1999) Crystal structure of the copper chaperone for superoxide dismutase. Nat. Struct. Biol. 6, 724–729 [DOI] [PubMed] [Google Scholar]
- 19. Lamb A. L., Torres A. S., O'Halloran T. V., and Rosenzweig A. C. (2001) Heterodimeric structure of superoxide dismutase in complex with its metallochaperone. Nat. Struct. Biol. 8, 751–755 [DOI] [PubMed] [Google Scholar]
- 20. Proescher J. B., Son M., Elliott J. L., and Culotta V. C. (2008) Biological effects of CCS in the absence of SOD1 enzyme activation: implications for disease in a mouse model for ALS. Hum. Mol. Genet. 17, 1728–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furukawa Y., Torres A. S., and O'Halloran T. V. (2004) Oxygen-induced maturation of SOD1: a key role for disulfide formation by the copper chaperone CCS. EMBO J. 23, 2872–2881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wong P. C., Waggoner D., Subramaniam J. R., Tessarollo L., Bartnikas T. B., Culotta V. C., Price D. L., Rothstein J., and Gitlin J. D. (2000) Copper chaperone for superoxide dismutase is essential to activate mammalian Cu/Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 97, 2886–2891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Mirabal H. R., and Winther J. R. (2008) Redox characteristics of the eukaryotic cytosol. Biochim. Biophys. Acta 1783, 629–640 [DOI] [PubMed] [Google Scholar]
- 24. Ferraroni M., Rypniewski W., Wilson K. S., Viezzoli M. S., Banci L., Bertini I., and Mangani S. (1999) The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited. J. Mol. Biol. 288, 413–426 [DOI] [PubMed] [Google Scholar]
- 25. Polticelli F., Battistoni A., O'Neill P., Rotilio G., and Desideri A. (1998) Role of the electrostatic loop charged residues in Cu,Zn superoxide dismutase. Protein Sci. 7, 2354–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brown N. M., Torres A. S., Doan P. E., and O'Halloran T. V. (2004) Oxygen and the copper chaperone CCS regulate posttranslational activation of Cu,Zn superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 101, 5518–5523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Winkler D. D., Schuermann J. P., Cao X., Holloway S. P., Borchelt D. R., Carroll M. C., Proescher J. B., Culotta V. C., and Hart P. J. (2009) Structural and biophysical properties of the pathogenic SOD1 variant H46R/H48Q. Biochemistry 48, 3436–3447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jensen L. T., and Culotta V. C. (2005) Activation of CuZn superoxide dismutases from Caenorhabditis elegans does not require the copper chaperone CCS. J. Biol. Chem. 280, 41373–41379 [DOI] [PubMed] [Google Scholar]
- 29. Leitch J. M., Jensen L. T., Bouldin S. D., Outten C. E., Hart P. J., and Culotta V. C. (2009) Activation of Cu,Zn-superoxide dismutase in the absence of oxygen and the copper chaperone CCS. J. Biol. Chem. 284, 21863–21871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ciriolo M. R., Desideri A., Paci M., and Rotilio G. (1990) Reconstitution of Cu,Zn-superoxide dismutase by the Cu(I)·glutathione complex. J. Biol. Chem. 265, 11030–11034 [PubMed] [Google Scholar]
- 31. Ascone I., Longo A., Dexpert H., Ciriolo M. R., Rotilio G., and Desideri A. (1993) An X-ray absorption study of the reconstitution process of bovine Cu,Zn superoxide dismutase by Cu(I)-glutathione complex. FEBS Lett. 322, 165–167 [DOI] [PubMed] [Google Scholar]
- 32. Caruano-Yzermans A. L., Bartnikas T. B., and Gitlin J. D. (2006) Mechanisms of the copper-dependent turnover of the copper chaperone for superoxide dismutase. J. Biol. Chem. 281, 13581–13587 [DOI] [PubMed] [Google Scholar]
- 33. Schmidt P. J., Kunst C., and Culotta V. C. (2000) Copper activation of superoxide dismutase 1 (SOD1) in vivo. Role for protein-protein interactions with the copper chaperone for SOD1. J. Biol. Chem. 275, 33771–33776 [DOI] [PubMed] [Google Scholar]
- 34. Corson L. B., Strain J. J., Culotta V. C., and Cleveland D. W. (1998) Chaperone-facilitated copper binding is a property common to several classes of familial amyotrophic lateral sclerosis-linked superoxide dismutase mutants. Proc. Natl. Acad. Sci. U.S.A. 95, 6361–6366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rae T. D., Torres A. S., Pufahl R. A., and O'Halloran T. V. (2001) Mechanism of Cu,Zn-superoxide dismutase activation by the human metallochaperone hCCS. J. Biol. Chem. 276, 5166–5176 [DOI] [PubMed] [Google Scholar]
- 36. Banci L., Cantini F., Kozyreva T., and Rubino J. T. (2013) Mechanistic aspects of hSOD1 maturation from the solution structure of Cu(I)-loaded hCCS domain 1 and analysis of disulfide-free hSOD1 mutants. Chembiochem 14, 1839–1844 [DOI] [PubMed] [Google Scholar]
- 37. Pickering I. J., George G. N., Dameron C. T., Kurz B., Winge D. R., and Dance I. G. (1993) X-ray absorption spectroscopy of cuprous-thiolate clusters in proteins and model systems. J. Am. Chem. Soc. 115, 9498–9505 [Google Scholar]
- 38. Richardson J., Thomas K. A., Rubin B. H., and Richardson D. C. (1975) Crystal structure of bovine Cu,Zn superoxide dismutase at 3 Å resolution: chain tracing and metal ligands. Proc. Natl. Acad. Sci. U.S.A. 72, 1349–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Klomsiri C., Nelson K. J., Bechtold E., Soito L., Johnson L. C., Lowther W. T., Ryu S. E., King S. B., Furdui C. M., and Poole L. B. (2010) Use of dimedone-based chemical probes for sulfenic acid detection evaluation of conditions affecting probe incorporation into redox-sensitive proteins. Methods Enzymol. 473, 77–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Leonard S. E., Reddie K. G., and Carroll K. S. (2009) Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 4, 783–799 [DOI] [PubMed] [Google Scholar]
- 41. Ferrer-Sueta G., Manta B., Botti H., Radi R., Trujillo M., and Denicola A. (2011) Factors affecting protein thiol reactivity and specificity in peroxide reduction. Chem. Res. Toxicol. 24, 434–450 [DOI] [PubMed] [Google Scholar]
- 42. Lo Conte M., and Carroll K. S. (2013) The redox biochemistry of protein sulfenylation and sulfinylation. J. Biol. Chem. 288, 26480–26488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Paulsen C. E., and Carroll K. S. (2009) Chemical dissection of an essential redox switch in yeast. Chem. Biol. 16, 217–225 [DOI] [PubMed] [Google Scholar]
- 44. Delaunay A., Pflieger D., Barrault M. B., Vinh J., and Toledano M. B. (2002) A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell 111, 471–481 [DOI] [PubMed] [Google Scholar]
- 45. Luk E., Jensen L. T., and Culotta V. C. (2003) The many highways for intracellular trafficking of metals. J. Biol. Inorg. Chem. 8, 803–809 [DOI] [PubMed] [Google Scholar]
- 46. Kessner D., Chambers M., Burke R., Agus D., and Mallick P. (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacLean B., Tomazela D. M., Shulman N., Chambers M., Finney G. L., Frewen B., Kern R., Tabb D. L., Liebler D. C., and MacCoss M. J. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 49. Vagin A., and Teplyakov A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 50. Elam J. S., Taylor A. B., Strange R., Antonyuk S., Doucette P. A., Rodriguez J. A., Hasnain S. S., Hayward L. J., Valentine J. S., Yeates T. O., and Hart P. J. (2003) Amyloid-like filaments and water-filled nanotubes formed by SOD1 mutant proteins linked to familial ALS. Nat. Struct. Biol. 10, 461–467 [DOI] [PubMed] [Google Scholar]
- 51. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 52. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 53. George G. N. (1995) EXAFSPAK, Stanford Radiation Laboratory, Menlo Park, CA [Google Scholar]
- 54. Binsted N., and Hasnain S. S. (1996) State-of-the-art analysis of whole X-ray absorption spectra. J. Synchrotron Radiat. 3, 185–196 [DOI] [PubMed] [Google Scholar]
- 55. Gurman S. J. (1995) Interpretation of EXAFS data. J. Synchrotron Radiat. 2, 56–63 [DOI] [PubMed] [Google Scholar]
- 56. Eisses J. F., Stasser J. P., Ralle M., Kaplan J. H., and Blackburn N. J. (2000) Domains I and III of the human copper chaperone for superoxide dismutase interact via a cysteine-bridged dicopper(I) cluster. Biochemistry 39, 7337–7342 [DOI] [PubMed] [Google Scholar]
- 57. Bauman A. T., Broers B. A., Kline C. D., and Blackburn N. J. (2011) A copper-methionine interaction controls the pH-dependent activation of peptidylglycine monooxygenase. Biochemistry 50, 10819–10828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Blackburn N. J., Hasnain S. S., Binsted N., Diakun G. P., Garner C. D., and Knowles P. F. (1984) An extended-X-ray-absorption-fine-structure study of bovine erythrocyte superoxide dismutase in aqueous solution. Direct evidence for three-co-ordinate Cu(I) in reduced enzyme. Biochem. J. 219, 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Caslavka Zempel K. E., Vashisht A. A., Barshop W. D., Wohlschlegel J. A., and Clarke S. G. (2016) Determining the mitochondrial methyl proteome in Saccharomyces cerevisiae using heavy methyl SILAC. J. Proteome Res. 15, 4436–4451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim S., and Pevzner P. A. (2014) MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Käll L., Canterbury J. D., Weston J., Noble W. S., and MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 62. Taus T., Köcher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.