Abstract
The NLRP3 inflammasome is a multiprotein complex that regulates the activation of caspase-1 leading to the maturation of the proinflammatory cytokines IL-1β and IL-18 and promoting pyroptosis. Classically, the NLRP3 inflammasome in murine macrophages is activated by the recognition of pathogen-associated molecular patterns and by many structurally unrelated factors. Understanding the precise mechanism of NLRP3 activation by such a wide array of stimuli remains elusive, but several signaling events, including cytosolic efflux and influx of select ions, have been suggested. Accordingly, several studies have indicated a role of anion channels in NLRP3 inflammasome assembly, but their direct involvement has not been shown. Here, we report that the chloride intracellular channel proteins CLIC1 and CLIC4 participate in the regulation of the NLRP3 inflammasome. Confocal microscopy and cell fractionation experiments revealed that upon LPS stimulation of macrophages, CLIC1 and CLIC4 translocated into the nucleus and cellular membrane. In LPS/ATP-stimulated bone marrow-derived macrophages (BMDMs), CLIC1 or CLIC4 siRNA transfection impaired transcription of IL-1β, ASC speck formation, and secretion of mature IL-1β. Collectively, our results demonstrate that CLIC1 and CLIC4 participate both in the priming signal for IL-1β and in NLRP3 activation.
Keywords: chloride channel, inflammasome, lipopolysaccharide (LPS), macrophage, NLRP3, CLIC1, CLIC4, IL-1β
Introduction
Interleukin-1 (IL-1β) is a central proinflammatory cytokine that plays a critical role in the innate immunity and inflammation (1–3). Induction of IL-1β is tightly regulated by the inflammasome, a multiprotein complex that upon recognition of a large number of inflammatory factors mediates activation of caspase-1, which processes pro-IL-1β and IL-18 into their mature active forms and also causes a type of cell death called pyroptosis (4, 5). Several inflammasome complexes have been described to date; however, the NLRP3 inflammasome is of special interest because of its ability to recognize a wide range of noxious substances. Importantly, dysregulation of the NLRP3 inflammasome is recognized as the common feature of a wide spectrum of chronic inflammatory and metabolic diseases, such as gout, rheumatoid arthritis, osteoarthritis, Alzheimer's disease, Parkinson's disease, type-2 diabetes, and obesity (6–14).
The classical activation of the NLRP3 inflammasome in murine macrophages requires two distinct signals. Signal 1, also called priming, involves the recognition of pathogen-associated molecular patterns (PAMPs), such as the Gram-negative bacterial product LPS, by Toll-like receptor 4 (TLR4), which leads to the increased expression of pro-IL1β and NLRP3 (15). The second signal is triggered by the recognition of a myriad of structurally unrelated factors including endogenous markers of cellular and metabolic distress such as monosodium urate (MSU), adenosine triphosphate (ATP), amyloid-β, islet amyloid polypeptide (IAPP), exogenous agents such as asbestos, silica or alum, and obesity-related factors such as ceramide or fatty acids (16–19). All of these factors lead to the activation and assembly of the inflammasome complex. Understanding of the precise mechanism of NLRP3 activation by such a wide array of stimuli remains elusive, but to date several signaling events have been suggested to induce NLRP3 activation. These include cytosolic efflux of K+ (20), Ca2+ influx, lysosome rupture, cathepsin B activation, and mitochondrial reactive oxygen species generation (21). Increasing amounts of evidence suggest that the NLRP3 inflammasome activity is sensitive to changes in ion concentrations and in particular to K+ efflux. Two classical stimuli are known to cause K+ efflux, namely bacterial toxins, such as nigericin, by forming pores in the plasma membrane, and extracellular ATP by activating the ionotropic P2X7 receptor (15). More importantly, increasing the potassium concentration in the media, which inhibits K+ efflux, can potently inhibit NLRP3 activation by many different stimuli.
Ionic fluctuations result in changes in intracellular pH, membrane potential, and cell volume, which must be tightly regulated to maintain cell function and homeostasis. A remarkable feature of LPS-ATP-stimulated macrophages is cell-swelling (22), a response that is controlled by the coordinated action of K+ and Cl−, and the activity of multiple ion channels, a process known as regulatory volume decrease (RVD). A study by Compan et al. (23) demonstrated that hypotonic-induced cell swelling promotes NLRP3 activation through a pathway that involves translocation of the TRP7 and TRPV2 cationic channels into the plasma membrane where they modulate Ca2+ and Mg2+ fluctuations and promote TAK1 phosphorylation. Based on the fact that all inflammasome activators induce changes in cell volume, the authors proposed that mechanosensitive ion channels may be general regulators of inflammasome activation. In accordance with this hypothesis, application of hypertonic solutions blocked the release of IL-1β in response to all NLRP3 activators in vitro and reduced inflammasome activity in a rat model of brain inflammation (23). Furthermore, several studies indicated a role of anion channels in the modulation of the NLRP3 inflammasome based on the inhibitory effects of extracellular NaCl and the general chloride inhibitors 5-nitro-2-(3-phenylpropyl-amino) benzoic acid (NPPB)2 and 4,4′-diisothiocyanatostilbene 2,2′-disulfonic acid (DIDS) (22–25).
Chloride intracellular channel proteins (CLICs 1–6) are a family of evolutionary conserved proteins whose structure belongs to glutathione S-transferase (GST)-fold superfamily. In response to increased cytoplasmic oxidation or pH changes, cytoplasmic soluble CLIC proteins insert into cellular membranes where they act as anion channels that counteract cationic fluctuations. CLIC proteins participate in a number of biological functions, including regulation of membrane potential, cell volume regulation, cell proliferation, and apoptosis (26–29). Upon phagocytosis, CLIC1 has been shown to translocate into the phagosomal membrane where it colocalizes with NADH oxidase and modulates phagosomal acidification. CLIC1−/− macrophages displayed impaired phagosomal proteolytic capacity and reduced reactive oxygen species production (30). CLIC4 is an NF-κB and IRF-3 early response gene in LPS-stimulated macrophages (31). CLIC4 knock-out macrophages exhibit dysregulation of multiple inflammatory mediators during the early response to LPS (32, 33). In response to LPS, CLIC4 is S-nitrosylated and translocates into the nucleus where it functions to enhance TGF-β signaling (33). In addition, compounds highly related to the NLRP3 inhibitor MCC950 have been shown to interact with the chloride intracellular channel CLIC1 (34, 35).
These studies prompted us to investigate the role of CLIC1 and CLIC4 channels during the activation of the NLRP3 inflammasome. We provide evidence for a role for CLIC1 and CLIC4 in both the induction of pro-IL1β and the activation of NLRP3, identifying CLICs as key regulators of the IL-1β system.
Results
NaCl and KCl inhibited ATP-induced IL-1β release
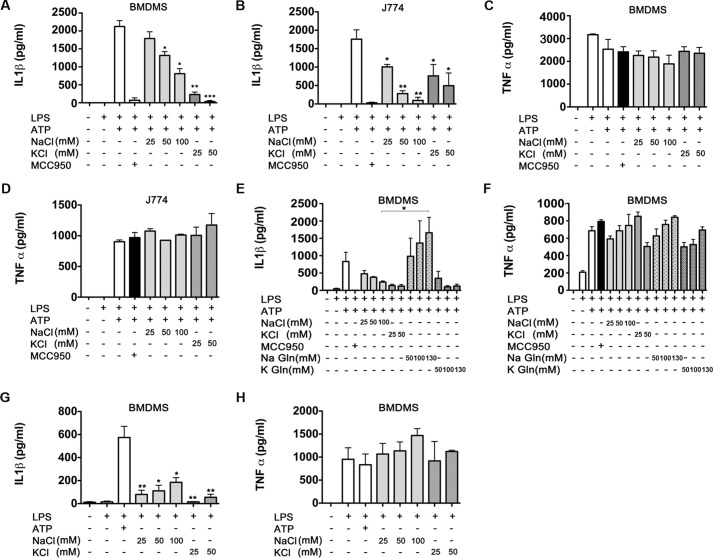
K+ efflux is a common denominator in NLRP3 activation and subsequent IL-1β secretion, a mechanism that can be blocked by increasing the extracellular K+ (21, 36). However, an increase of extracellular Cl− also inhibits IL-1β release (22, 37), suggesting that anions or anion channels may play a role in the IL-1 β response. We reexamined the effects of culturing macrophages in a high extracellular KCl or NaCl medium. ATP-induced IL-1β release in LPS-primed macrophages, BMDMs, and J774 cells were exposed to different concentrations of NaCl or KCl for 30 min after LPS priming before ATP stimulation. The addition of different concentrations of KCl into the extracellular media, ranging from 25 to 50 mm, triggered a significant decrease in a dose-dependent manner of secreted IL-1β in both BMDMs and J774 cells (Fig. 1, A and B). The addition of 25 mm or 50 mm KCl resulted in a >50% reduction on the amount of secreted IL-1β. We used the NLRP3 inhibitor MCC950 (34) as a positive control for inhibition. KCl did not have any significant impact on the production of TNF-α in BMDMs or J774 cells (Fig. 1, C and D, respectively). This result confirms that increasing the extracellular concentration of K+, which in turn impedes a net K+ efflux, can block secretion of IL-1β. However, the addition of different concentrations of NaCl, ranging from 50 mm to 100 mm, to the extracellular media also resulted in a significant dose-dependent decrease of IL-1β release in both BMDMs and J774 cells (Fig. 1, A and B). Furthermore, production of TNF-α remained at similar levels regardless of the extracellular NaCl concentrations (Fig. 1, C and D), mimicking the results observed in cells exposed to high KCl conditions.
Figure 1.
Extracellular osmolarity modulated IL-1β secretion. BMDMs (A and C) and J774 cells (B and D) were stimulated with LPS for 3 h and then incubated with 100 nm MCC950 (used as a control) or increasing concentrations of NaCl or KCl (A–D) or NaGln or KGln (E and F). After 30 min of incubation, cells were stimulated with 5 mm ATP for 1 additional hour. Secreted IL-1β (A, B, and E) and TNF-α (C, D, and F) was quantified by ELISA. G and H, BMDMs were stimulated with LPS for 3 h and then incubated overnight with increasing concentrations of NaCl or KCl as indicated. Secreted IL-1β (G) and TNF-α (H) were quantified by ELISA. Data are expressed as pg/106 cells (mean ± S.E., n = 3). *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
To address the effects of the removal of extracellular Cl−, Na+ and K+ were counterbalanced with the impermeant anion gluconate. Enzyme-linked immunosorbent assay (ELISA) analysis demonstrated that extracellular NaGln, ranging from 50 to 130 mm, actually increased the production of ATP-induced IL-1β in a dose-dependent manner relative to BMDMs cultured with a high extracellular NaCl (100 mm) (Fig. 1E). As expected, when BMDMs were treated with equimolar concentrations of KGln, IL-1β secretion was inhibited, as K+ efflux is impeded by the high extracellular K+ concentration (Fig. 1E). However, treatments with NaGln or KGln had no effect on TNF-α production (Fig. 1F). Taken together, these data suggest that the efflux of Cl− and/or other anions could be involved in the maturation of IL-1β.
In the absence of ATP, NaCl has been reported to induce activation of the NLRP3 inflammasome in BMDMs in a K+ efflux-independent manner (38). To verify the dual role of NaCl in NLRP3 activation, BMDMs primed with 100 ng/ml LPS for 3 h were placed in a high NaCl medium (ranging from 25 mm to 100 mm) overnight. We were able to confirm the increased release of IL-1β, although the effect was much more modest than with ATP (Fig. 1G). No significant differences were observed on the production of TNF-α (Fig. 1H). In contrast to NaCl, when BMDMs were treated with 25–50 mm KCl after LPS, no increase in IL-1β secretion was detected (Fig. 1G). Furthermore, the addition of different concentrations of KCl or NaCl did not significantly change TNF-α production (Fig. 1H), again indicating the specificity of the effect on IL-1β-processing and release. The dual effect of NaCl on IL-1β release suggests that differences in treatment timings lead to different molecular mechanisms that either inhibit or induce IL-1β release.
Taken together, these results confirm what was previously reported by Perregaux and Gabel (22) and Ip et al. (38) studies, and this prompted us to further investigate the role of Cl− and anion channels in the maturation of IL-1β.
Chloride inhibitors NPPB and DIDS blocked the ATP-induced IL-1β release
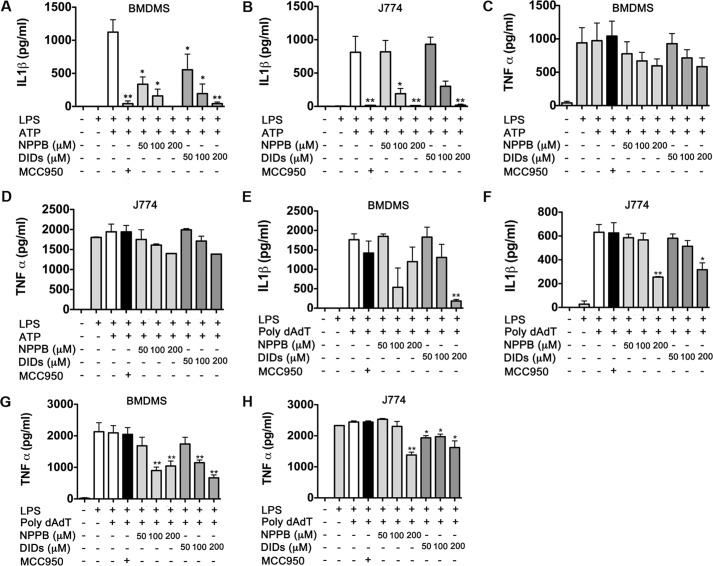
The inhibitory effect of both KCl and NaCl on IL-1β secretion and the promoting effect of NaGln suggested that chloride efflux might mediate this effect. To evaluate this further, we examined two chloride channel inhibitors NPPB and DIDS. Both agents had a strong dose-dependent inhibitory effect on IL-1β secretion induced by ATP (Fig. 2, A and B). A dose of 200 μm NPPB or DIDS prevented IL-1β secretion to the same extent as that observed using the NLRP3 selective inhibitor MCC950. Neither agent had a significant impact on secretion of TNF-α by BMDMs or J774 cells (Fig. 2, C and D, respectively). These results indicated that chloride ion efflux might be required for NLRP3 activation. We next examined whether chloride ions could also impact on IL-1β release triggered by another inflammasome, the double-stranded DNA sensor AIM2. Interestingly, NPPB and DIDS chloride inhibitors only partially inhibited IL-1β release and only at the highest concentration tested (200 μm) (Fig. 2, E and F). However, a slight decrease of TNF-α secretion was also observed on cells treated with 200 μm NPPB or DIDS (Fig. 2G), suggesting that the inhibitory effects observed could be related to toxicity. To ensure that the NPPB and DIDS concentrations used did not cause major toxic effects on the cells, an lactate dehydrogenase assay was performed on the supernatant of each of the treated cells. Based on the amounts of lactate dehydrogenase released into the extracellular media, only a 5% reduction on cell viability was observed on cells treated with 200 μm NPPB or DIDS. No significant reduction on cell viability was observed when using lower concentrations (data not shown).
Figure 2.
NPPB and DIDS chloride inhibitors impaired secretion of IL-1β. BMDMs (A and C) and J774 cells (B and D) were stimulated with LPS for 3 h and then incubated with 100 nm MCC950 (used as a control) or increasing concentrations of NPPB or DIDS as indicated. After 30 min of incubation, cells were stimulated with 5 mm ATP for 1 additional hour. Secreted IL-1β (A and B) and TNF-α (C and D) was quantified by ELISA. To assess activation of the AIM2 inflammasome, BMDMs (E and G) and J774 cells (F and H) were stimulated with LPS, NPPB, and DIDS as above and then stimulated with a complex of 1 μg of poly(dA:dT) and 3 μl/ml Lipofectamine for 2 h. Data are expressed as pg/106 cells (mean ± S.E., n = 3). *, p ≤ 0.05; **, p ≤ 0.01.
CLIC1 and CLIC4 expression and localization were responsive to LPS stimulation
Because the increase of extracellular osmolarity and the inhibition of chloride channels impaired the release of IL-1β, we sought to determine whether chloride channels could control NLRP3 activation. Compounds highly related to the NLRP3 inhibitor MCC950 have been shown to interact with the chloride intracellular channel CLIC1 (35). We, therefore, examined CLIC proteins here, focusing on CLIC1 and CLIC4, which are highly expressed in macrophages (30, 33).
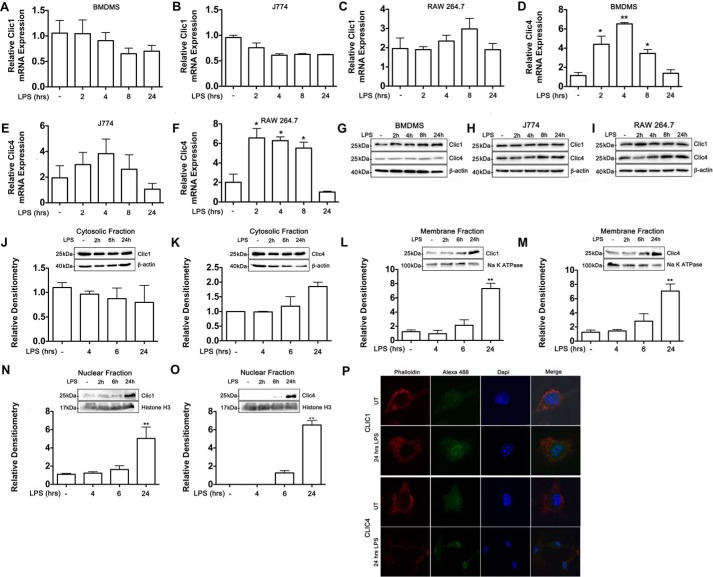
Expression of CLIC1 and CLIC4 mRNA was evaluated by RT-quantitative PCR (qPCR) in BMDMs, J774, and RAW 264.7 cells in a time-course study. CLIC1 mRNA was not significantly changed in LPS-stimulated BMDMs, J774, or RAW 264.7 cells during the time-course studied (Fig. 3, A–C). However, a significant increase of CLIC4 mRNA expression was observed at 2, 4, and 8 h after LPS stimulation in BMDMs and RAW 264.7 (Fig. 3, D and F, respectively). CLIC4 mRNA expression in J774 was not significantly changed after LPS stimulation; however, a trend to increased expression was observed (Fig. 3E). Western blot analysis revealed that CLIC1 protein levels were similar across the different LPS time points, and a slight increase of CLIC4 was observed in J774 and RAW 264.7 cells (Fig. 3, G–I).
Figure 3.
LPS drove subcellular translocation of CLIC1 and CLIC4 proteins. A–I, BMDMs, J774, and RAW 264.7 cells were treated with LPS (100 ng/ml), and RNA and protein were isolated for the indicated time points. RT-qPCR was performed to examine CLIC1 and CLIC4 mRNA levels in BMDMs (A and D), J774 (B and E), and RAW 264.7 (C and F). Expression was normalized to 18S rRNA. -Fold change in expression of the target gene transcript relative to unstimulated cells was determined using the comparative cycle threshold method (2−ΔΔCT). All values shown are the means ± S.E. of three independent experiments. G–I, Western blot analysis of CLIC1 and CLIC4 in whole cell lysates of BMDMs (G), J774 (H), and RAW 264.7 (I). J–O, Western blot and densitometry analysis in the cytoplasmic, membrane, and nuclear fractions of unstimulated or LPS-stimulated RAW 264.7 cells. Equivalent amounts of cytosolic, membrane, or nuclear fraction were subjected to immunoblot against CLIC1 and CLIC4. β-Actin, Na,K-ATPase, and histone H3 antibodies were used as loading controls for the cytosolic, membrane, and nuclear fractions, respectively. Bar graphs were obtained by densitometric analysis and expressed relative to the loading control and normalized to the control (unstimulated cells), except for graph O, which is relative to 6-h LPS, as no protein was detected in unstimulated cells. P, confocal microscopy analysis of unstimulated (UT) or LPS-stimulated BMDMs stained with phalloidin, DAPI, and CLIC1 or CLIC4 antibodies followed by an Alexa-488-conjugated secondary antibody. Merged signals are shown in the right panels. Data are representative of three independent experiments. *, p ≤ 0.05; **, p ≤ 0.01.
The above results confirmed that LPS induces expression of CLIC4, which agrees with He et al. (32). Given that CLIC1 and CLIC4 functionality is redox-dependent and they translocate to different cellular compartments in response to LPS (30, 32, 33, 39), it was important to further characterize the protein abundance and localization changes of these two proteins in response to LPS. Cytosolic, membrane, and nuclear fractions of LPS-stimulated RAW 264.7 cells were analyzed by Western blotting. In unstimulated cells, CLIC1 and CLIC4 proteins were primarily located in the cytosolic fraction (Fig. 3, J and K, respectively) and to a lesser extent in the membrane fraction (Fig. 3. L and M, respectively). CLIC1 is present in the nucleus of unstimulated BMDMs at low levels (Fig. 3N), whereas CLIC4 was undetectable by Western blot analysis (Fig. 3O). However, the expression patterns of CLIC1 and CLIC4 changed dramatically upon LPS stimulation. A marked increase in CLIC1 and CLIC4 levels in the membrane fraction was observed 6 h after LPS stimulation and was especially prominent at 24 h (Fig. 3, L and M, respectively). Interestingly, LPS stimulation also caused increased expression of CLIC1 and CLIC4 in the nuclear compartment. Although nuclear CLIC1 was lowly expressed in unstimulated cells, significant levels of the protein were observed 24 h after LPS (Fig. 3N). Similarly, CLIC4 was undetectable in the nuclear compartment in unstimulated cells, but the presence of the protein increased in a time-dependent manner, being especially prominent 24 h after LPS stimulation (Fig. 3O). Using confocal imaging we observed increased expression of CLIC1 and CLIC4 in BMDMs stimulated with LPS for 24 h relative to unstimulated cells (Fig. 3P). However, translocation into the membrane could not be observed by this technique. It is likely that protein conformational changes produced by membrane insertion may have led to changes in antigen presentation. These data demonstrate that both CLIC1 and CLIC4 are responsive to LPS stimulation, and although CLIC1 expression is not induced, its cellular localization changes in stimulated macrophages.
CLIC1 and CLIC4 deficiency impaired pro-IL1β and ASC speck formation in classical activated macrophages
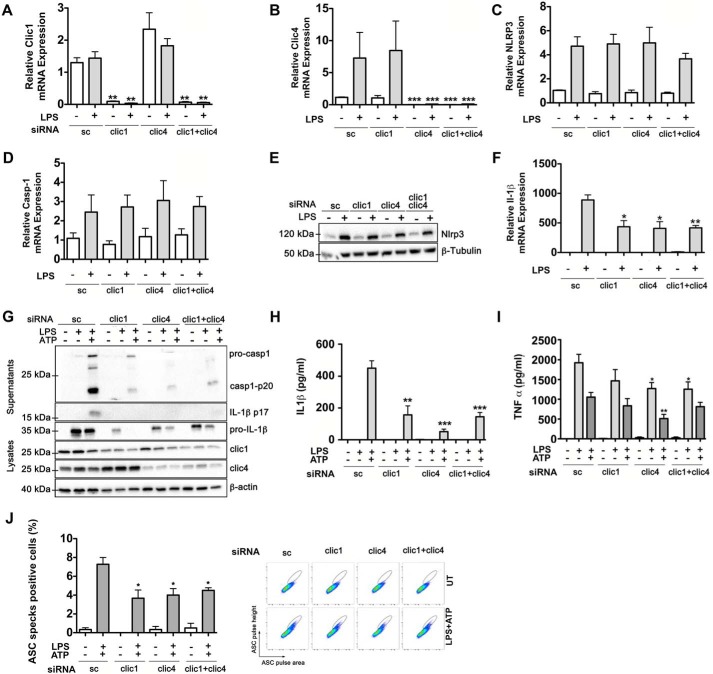
We next tested the role of CLIC1 and CLIC4 by knocking them down using siRNA oligo nucleotides. CLIC1 and CLIC4 knockdown significantly inhibited the expression of CLIC1 and CLIC4 mRNA (Fig. 4, A and B, respectively) as well as protein expression (Fig. 4G). There was no significant difference on the mRNA expression of NLRP3 or caspase-1 in the CLIC1- and CLIC4-transfected cells compared with the scrambled control (Fig. 4, C and D). No difference was observed in NLRP3 protein expression (Fig. 4E). However, knockdown of CLIC1 and/or CLIC4 resulted in a significant reduction of pro-IL-1β mRNA expression after LPS treatment (Fig. 4F), indicating that both proteins modulate the LPS priming step in macrophages.
Figure 4.
Inhibition of CLIC1 and CLIC4 impaired transcription of IL-1β and ASC speck formation. BMDMs were transiently transfected with CLIC1 and/or CLIC4 or a scramble siRNA as indicated. 36 h after transfection cells were LPS- and ATP-stimulated, and supernatants and adherent cells were harvested for RT-qPCR, Western blot, and ELISA analysis. Shown are RT-qPCR of CLIC1 (A), CLIC4 (B), NLRP3 (C), caspase-1 (D), and Il-1β (F). Expression normalized to 18S rRNA. -Fold change in expression of the target gene transcript relative to unstimulated cells was determined using the comparative cycle threshold method (2−ΔΔCT). E and G, Western-blotting analysis of NLRP3 in whole cell lysates ran in a 10% polyacrylamide gel (E) and pro-casp1, IL-1β p17 in supernatants and pro-IL1b, CLIC1, and CLIC4 in whole cell lysates (G) as indicated. H and I, secretion of Il-1β (H) and TNF-α (I) was quantified by ELISA. J, analysis of ASC speck formation in BMDMs transiently transfected with scrambled (sc), CLIC1, and/or CLIC4 siRNAs after LPS and ATP stimulation was evaluated by FACS analysis. Cells were fixed, permeabilized, and incubated with an antibody to ASC and an Alexa Fluor 488®-conjugated secondary antibody. The bar graph represents the percentage of ASC speck- positive cells from three independent experiments. Dot plots show the percentage of ASC speck formation.
Next, we assessed the impact of CLIC1 and CLIC4 inhibition in LPS- and ATP-stimulated samples by Western-blotting and ELISA assays. Impairment of LPS-induced pro-IL-1β protein was observed in CLIC1 and/or CLIC4 siRNA-transfected samples compared with the scramble control (Fig. 4G), which is consistent with the reduction of pro-IL-1β mRNA. However, no further effect was observed in samples when both genes were simultaneously ablated. Analysis of cell supernatants revealed that secretion of mature IL-1β p17 was completely abolished, and importantly, cleaved caspase-1 p20 was greatly reduced (Fig. 4G). We also confirmed the significant reduction of secreted IL-1β in CLIC1- and/or CLIC4-ablated samples by ELISA (Fig. 4H). Again, no further effect was observed when both CLIC1 and CLIC4 were ablated relative to each on their own. Secretion of TNF-α protein in the same samples revealed no differences between the CLIC1 siRNA and the scrambled siRNA-transfected samples (Fig. 4I). However, individual and combined knockdown of CLIC1 and CLIC4 produced a significant slight decrease in TNF-α secretion (Fig. 4I). Reduction of TNF-α in CLIC4-inhibited samples is consistent with previous findings (32) and further supports the role of CLIC4 in the modulation of proinflammatory cytokines. Overall, these results suggest that both CLIC1 and CLIC4 are required for the induction of IL-1β. However, whereas CLIC4 is also involved in the regulation of TNF-α, CLIC1 is not.
ASC speck is a distinguishing feature of inflammasome activation (40). To further characterize the role of CLIC1 and CLIC4 in NLRP3 activation, we assessed whether CLIC1 and CLIC4 knockdown could also modulate ASC speck formation. This led to an overall 50% reduction of ASC speck formation (Fig. 4J).
Discussion
In the classical model of NLRP3 activation, NLRP3 activators such as ATP are known to induce major ion fluxes, such as K+ efflux or Ca2+ influx, with K+ efflux being clearly required for NLRP3 activation (20, 43). A role for K+ efflux in IL-1β secretion was in fact reported some years before the discovery of NLRP3 (36). However, release of IL-1β from LPS-stimulated human and murine macrophages is compromised when anion transport processes are disrupted (44). Therefore, changes to the intracellular ionic environment and thereby anion transporters appear to be a necessary component required for the post-transcriptional maturation of pro-IL1β. A role of chloride in the activation of the NLRP3 inflammasome has also been indicated from the inhibitory effect of high extracellular NaCl and the chloride inhibitors NPPB and DIDS (23, 24, 44). In addition, the use hypertonic solutions has been widely used in the treatment of acute conditions, such as ischemic stroke and intracerebral hemorrhage (45, 46), and has also shown to have beneficial effects in the treatment of patients with septic shock (47–49). Contrary to these studies, Ip and Medzhitov (38) reported high extracellular osmolarity as proinflammatory.
Equally, however, from our study, preconditioning of high extracellular NaCl of LPS-stimulated macrophages acts to limit inflammation by inhibiting the post-transcriptional activation of IL-1β. The effect inhibitory effect of NaCl on IL-1β was found to be lower than that of KCl, indicating that the coordinated action of K+ and Cl− is needed for IL-1β release. In fact, it is likely that the inhibitory effect of high extracellular KCl is more potent because both K+ and Cl− efflux are blocked. Moreover, when Cl− efflux is blocked with Cl− inhibitors such as NPPB and DIDS, IL-1β secretion is strongly inhibited. Importantly, the inhibitory effect of NPPB on IL-1β secretion has no effect on K+ efflux (23). Overall, our results provide evidence of Cl− required for NLPR3-mediated IL-1β release, with no role in AIM2 activation or TNF-α production.
We also confirmed that high extracellular NaCl promotes secretion of IL-1β in the absence of NLRP3 activators. Therefore, Cl− efflux seems to have a dual role; that is, inhibition of efflux in the absence of signal 2 drivers will activate NLRP3, whereas in the presence of such drivers it will be inhibitory. The latter situation is likely to dominate in vivo.
We focused on the CLIC family of chloride channels as possible participants here. Both CLIC1 and CLIC4 are highly expressed in macrophages, and several lines of evidence suggest that oxidative activation of CLIC proteins promotes their integration into membranes where they act as anion channels that remove the excess of negative charges. Because reactive oxygen species has been implicated in NLRP3 activation, we wondered if CLICs would be involved. Second, and more importantly, we reported previously that the sulfonyl urea MCC950 is a potent and selective inhibitor of NLRP3 (34). The related compound CRID2 has been shown to interact with CLIC1 (35), and so we addressed whether CLIC1 or CLIC4 might be required for NLRP3 activation. We found that in response to LPS, CLIC4, but not CLIC1, was slightly increased in its expression, confirming what previously reported by He et al. (32). However, profound changes in protein localization were observed after LPS stimulation for both CLIC1 and CLIC 4. LPS induced the translocation of both CLIC1 and CLIC4 to the plasma membrane, the effect on CLIC1 evident from 6 h. Confocal imagining confirmed the increased expression or nuclear translocation of both nuclear CLIC1 and CLIC4. However, we were not able to detect translocation into the cellular membrane confocally. This could be due to technical limitations probably related to antigen masking. It has been shown that S-nitrosylation induces nuclear translocation of CLIC4 (50) and nuclear-targeted CLIC4 overexpression inhibits IL-1β expression (33). Several studies have also demonstrated that NO acts as a suppressor of NLRP3 activation (51). We speculate that this effect might involve CLIC4 nuclear translocation.
Importantly, however, we have found that in response to LPS and ATP stimulation, transcription and secretion of IL-1β is impaired after knockdown of CLIC1 and/or CLIC4, whereas no changes in the transcription of NLRP3 of caspase-1 were detected. Interestingly, although the knockdown of CLIC4 diminished production of TNF-α, no change was detected when CLIC1 was knocked down, indicating that although both proteins are required for the transcription of IL-1β, they can specifically modulate different cytokines. Based on our results, showing that CLIC1 is present in the nucleus at an earlier time point than CLIC4, we hypothesize that the orchestrated function of this two proteins is required to modulate the inflammatory response.
More importantly, this study demonstrates for the first time that both CLIC1 and CLIC4 are not only involved in the priming step of LPS-induced IL-1β but also modulate signal 2 of NLRP3 activation. Knockdown of CLIC1 and/or CLIC4 in LPS and ATP-stimulated BMDMs produced a marked decrease in the production of ASC specks, which is a mark of NLRP3 activation.
Because high extracellular NaCl, Cl− inhibitors NPPB and DIDS, and the knockdown of both CLIC1 and CLIC4 using siRNAs produced impaired IL-1β production, the question arises as to whether this is due to the CLIC-mediated chloride currents. However, inhibition of the CLIC-mediated Cl− currents is technically challenging. To the best of our knowledge no specific inhibitors for the CLIC proteins are known to date. The IAA-94 down-regulates Cl− current; however, it is not very specific and has only been tested in planar lipid bilayers (41, 42). Others reported that IAA-94 treatment in CLIC1+/+ macrophages raised phagosomal pH, (where CLIC1 acts a pH modulator), whereas this had no effect on phagosomal pH of CLIC1−/− macrophages (30). The authors assumed that IAA-94 blocked CLIC1-mediated currents but did not measure this. Therefore, whether the Cl− channel activity of CLICs is requested for their role here remains to be investigated.
Based on these results, we speculate that translocation of CLIC1 and CLIC4 proteins to the plasma membrane and the nucleus could be part of the LPS priming of NLRP3. The CLIC proteins might be required to compensate the efflux of K+ associated with NLRP3 activation, whereas nuclear CLIC1 and CLIC4 may be involved in the transcriptional regulation of IL-1β and other cytokines. Overall, our data indicate that both CLIC1 and CLIC4 are regulators of NLRP3 and IL-1β and suggest them as potential therapeutic targets for limiting inflammation.
Experimental procedures
Materials
LPS used in in vitro studies was from Escherichia coli, serotype EH100 (RA) (TLRgradeTM), Enzo Life Sciences/Alexis, Lörrach, Germany. Chloride inhibitors 4,4′-diisothiocyanatostilbene-2,2′-disulfonic acid disodium salt hydrate (D3514), and 5-nitro-2-(3-phenylpropylamino)benzoic acid (N4779), and adenosine 5′-triphosphate disodium salt hydrate (A2383) were purchased from Sigma. The indanyloxyacetic acid IAA-94 was purchased from Santa Cruz Biotechnology. Poly(deoxyadenylic-deoxythymidylic) acid sodium salt poly(dA:dT) (#tlrl-patn, tlrl-patn-1) was purchased from InvivoGen, San Diego, CA.
Cell culture and treatments
The murine macrophage-like cell line RAW 264.7 was purchased from the American Type Culture Collection and maintained in DMEM+ GlutaMAXTM-I medium (Gibco, ThermoFisher Scientific, Waltham, MA) supplemented with 10% fetal bovine serum and 2 mm penicillin/streptomycin. The murine macrophage-like cell line J774 was kindly provided by Kingston Mills (Trinity College Dublin) and was originally from the American Type Culture Collection, Manassas, VA. J774 cells were grown in DMEM+ GlutaMAXTM-I supplemented with 10% fetal bovine serum and 2 mm penicillin/streptomycin. Bone marrow cells from C57BL/6, CLIC1 −/−, and 129/svj mice were differentiated for 7 days in DMEM+ GlutaMAXTM-I containing 10% FCS, 2 mm penicillin/streptomycin, and 20% L929 cell media. On day 6, cells were resuspended in DMEM+ GlutaMAXTM-I containing 10% FCS, 2 mm penicillin/streptomycin, and 10% L929 cell media then counted and seeded at 106/ml for treatment. BMDMs were cultured in DMEM medium supplemented with 20% macrophage-colony stimulating factor, 10% FBS, and 2 mm penicillin/streptomycin.
LPS treatments and Inflammasome activation assays
RAW 264.7, J774, and primary BMDM cells were plated at 106 cells/ml in 6-well plates and stimulated with 100 ng/ml LPS from E. coli, serotype 0111:B4 (Enzo Life Sciences) for the designated time points. For experiments involving assays caspase-1 activation and IL-1β release, cells were serum-starved for 30 min before LPS priming (10 ng/ml) for 3 h. The culture media was then removed and replaced for fresh media containing 5 mm ATP for 1 h. To test the effects of extracellular Cl−, different concentrations ranging from 50 mm to 200 mm NaCl, 50 mm to 100 mm KCl, or 50 mm to 130 mm NaGln or KGln were added into the media for 30 min and followed by the addition of 5 mm ATP. Similarly, for experiments using the chloride inhibitors, 50–100 μm NPPB or DIDS or 30–150 μm IAA94 was added into the media for 30 min and followed by the addition of 5 mm ATP for 1 h. Supernatants were assayed for IL-1β and TNFα by ELISA and also Western blotting for secreted Il-1β. The remaining adherent cells were collected and further analyzed by real-time qPCR and Western blotting.
Immunofluorescence confocal microscopy
BMDMs were plated on 12-mm diameter coverslips at a density of 0.3 × 106 cells. The following day cells were LPS-stimulated (1 μg of LPS for 24 h) or unstimulated, fixed with 2% paraformaldehyde for 30 min, and permeabilized with 0.2% Triton-X-100 in 1% BSA-PBS for 20 min at room temperature. Cells were then stained with antibodies against CLIC1 (356.1), CLIC4 (45.42) (Santa Cruz Biotechnology, Santa Cruz, CA) (1:100) at 4 °C overnight. After washing 3 times with phosphate-buffered saline (PBS), cells were stained with Alexa Fluor 488 goat antimouse IgG (H+L) secondary antibody (Invitrogen) (1:1000) for 1 h at room temperature. For phalloidin staining, cells were incubated with 1 μg/ml phalloidin-TRITC (Sigma) for 30 min at room temperature. Coverslips were then rinsed 3 times with PBS and mounted on microscope slides with Duolink® in situ mounting medium with DAPI (Sigma). Stained coverslips were imaged on a Leica SP8 confocal microscope using a 40× oil-objective lens. A number of images were taken for each treatment.
Reverse transfection of siRNAs
BMDMs cells were plated in 6-well plates at 1 × 106/well 24 h before transfection. 25 nm siRNA CLIC1, siRNA CLIC4, or siRNA negative control (Life Technologies, #s100126, #s78122, #AM4636) were transfected using 5 μl of Lipofectamine® siRNA Max (Life Technologies) according to the manufacturer's recommendations. 36 h after transfection cells were treated with LPS and ATP as described above to examine inflammasome activation.
Immunostaining cells for ASC for flow cytometry analysis
After transfection of BMDMs with siRNA CLIC1 and/or CLIC4 or scrambled control, cells were harvested and transferred to FACS tubes. Cells were fixed with ice-cold 100% ethanol for 15 min at room temperature, pelleted at 600 × g for 10 min, resuspended in ASC speck buffer (PBS, 0.1% sodium azide, 0.1% BSA, 1.5% FBS), and incubated for 20 min at room temperature. Cells were stained with anti-ASC (N15) antibody (Santa Cruz Biotechnology) (1:1500) and Alexa 488 goat anti-rabbit IgG (1:1500). Samples were processed on a FACSCanto II (BD Biosciences) and analyzed with FlowJo. Samples were gated to exclude debris (forward light scatter (FSC-area versus side scatter-area), and then any cell doublets were excluded using FSC-area versus FSC-width analysis. Cells were gated and used for further analysis of inflammasome activation state by either pulse width to pulse area profile (W:A) or high pulse height to area (H:A) analysis.
Enzyme-linked immunosorbent assay
Supernatants were analyzed for murine IL-1β and murine TNFα using ELISA kits according to the manufacturer's instructions (R&D Systems). The optical density values were measured using a microplate reader set to 450 nm.
Preparation of whole cell lysates for Western blot analysis
Total protein was isolated from cells using a radioimmunoprecipitation assay (RIPA) lysis buffer (Sigma). Cell pellets were washed with PBS, solubilized in radioimmunoprecipitation assay containing protease and phosphatase inhibitors for 30 min, and centrifuged at 14,000 × g for 15 min. Protein concentration was measured using the PierceTM BCA assay (ThermoFisher Scientific). 2× Laemmli sample buffer was added to the samples and then boiled for 5 min at 99 °C.
Preparation of secreted proteins for Western-blotting analysis
At the termination of experiments, supernatants were collected and transferred to microcentrifuge tubes. The protein content of the supernatants was concentrated using StrataClean TM resin (Agilent Genomics, Santa Clara, CA). 1 μl of resin was added per 100 μl of supernatant, and the mixture was vortexed for 1 min and then centrifuged at 40 × g for 1 min. The supernatant was aspirated, and 5× Laemmli sample buffer was added to the remaining resin. Samples were boiled for 5 min at 99 °C.
Subcellular fractionation of proteins for Western-blotting analysis
RAW 264.7 cells grown to confluence in T175 flasks and treated with 100 ng/ml LPS for 2, 4, 6, and 24 h. Cells were collected, resuspended in PBS, and centrifuged at 1000 × g for 5 min. Cytosolic, membrane, and nuclear protein fractions were isolated using the Qproteome Cell Compartment Kit (Qiagen, West Sussex, UK) according to the manufacturer's recommendations.
Western-blotting analysis
Protein lysates (∼ 20 μg of total protein) were analyzed by SDS-PAGE (10% or 12%), electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA), and then incubated overnight (4 °C) with antibodies for CLIC1 (356.1) and CLIC4 (45.42) (Santa Cruz Biotechnology; sc-81873 and sc-135739, respectively), IL-1β (R&D Systems; AF-401-NA), anti-caspase-1 (p20) (AdipopGen, San Diego, CA; AG-20B-0042), sodium-potassium ATPase (EP1845Y) (Abcam, Cambridge, UK), and histone H3 and β-tubulin, both from Cell Signaling Technology; #4499 and #2146, respectively. Immunoblots were labeled with secondary HRP-conjugated antibodies (goat anti-mouse or-rabbit) for 1 h, Jackson ImmunoResearch Laboratories), and developed by LumiGLO® Chemiluminescent substrate (Cell Signaling Technology; #7003) using the ChemiDocTM Imaging Systems (Bio-Rad).
RNA extractions
Total RNA was extracted using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions. RNA quality and quantity were measured using the Nanodrop Spectrophotometer (Thermoscientific, Loughborough, UK).
Stem-loop reverse transcription and real-time quantitative PCR
cDNA synthesis was performed using the High-Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA). Reverse transcription reactions were carried out using 500 ng of total RNA in a final volume of 20 μl of reaction mix. Cycling conditions were as follows: 10 min at 25 °C, 120 min at 37 °C, 5 min at 85 °C, and indefinitely at 4 °C. All reactions were performed on a Veriti® Thermal Cycle (Applied Biosystems). Real-time qPCRs were carried out on the 7900HT Fast Real-Time PCR System (Applied Biosystems) using specific Taqman gene expression or gene-specific SYBR primers. Primers were designed using NCBI/Primer-BLAS software and purchased from Eurofins MWG Operon (Eurofins Genomics, Ebersberg, Germany). Sequences used to analyze gene expression include CLIC4 forward (5′-AACTCA AGA CCA GAG GCT AAT G-3′) and reverse (5′-TGA TGT CCT CCA TGC TGT TC-3′), IL-1β forward (5′-GGA AGC AGC CCT TCA TCT TT-3′) and reverse (5′-TGG CAA CTG TTC CTG AAC TC-3′), caspase-1 forward (5′-CCA GGC AAG CCA AAT CTT TAT C-3′) and reverse (5′-TCA GCT GAT GGA GCT GAT TG-3′), NLRP3 forward (5′-CCT CTT GGT GAC CTC ATG TAA T-3′) and reverse (5′-AGA AAG ATA GCG ATG ATG ATA-3′), and 18S rRNA forward (GTA ACC CGT TGA ACC CCA TT-3′) and reverse (5′-CCA TCC AAT CGG TAG TAG GG-3′). 18S rRNA was used as an endogenous control, and CT (threshold cycle) values were normalized to levels of 18S rRNA expression. Individual qPCRs were carried out on the 7900HT Fast Realtime System (Applied Biosystems). A relative -fold change in expression of the target gene transcript was determined using the comparative cycle threshold method (2−ΔΔCT). Results were averaged from three independent experiments.
Statistical analysis
An unpaired Student's t test was used to determine statistical significance at p < 0.05 in all functional assays.
Author contributions
R.-D. F. designed the study and wrote the paper. R. C. C. performed the preliminary experiments on CLIC1−/− BMDMs. J. K. analyzed the experiments shown in Fig. 4J. S. B. provided the CLIC1−/− mice used in preliminary experiments. L. A. J. O. coordinated the study. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by grants from the European Research Council (268155_MicroInnate; to L. A. J. O.). The authors declare that they have no conflicts of interest with the contents of this article.
- NPPB
- 5-nitro-2-(3-phenylpropyl-amino) benzoic acid
- DIDS
- 4,4′-diisothiocyanatostilbene 2,2′-disulfonic acid
- IAA-94
- indanyloxyacetic acid
- CILC
- chloride intracellular channel protein
- BMDMs
- bone marrow-derived macrophages
- qPCR
- quantitative PCR
- TRITC
- tetramethylrhodamine isothiocyanate.
References
- 1. van der Meer J. W., Barza M., Wolff S. M., and Dinarello C. A. (1988) A low dose of recombinant interleukin 1 protects granulocytopenic mice from lethal gram-negative infection. Proc. Natl. Acad. Sci. U.S.A. 85, 1620–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li P., Allen H., Banerjee S., Franklin S., Herzog L., Johnston C., McDowell J., Paskind M., Rodman L., and Salfeld J. (1995) Mice deficient in IL-1β-converting enzyme are defective in production of mature IL-1β and resistant to endotoxic shock. Cell 80, 401–411 [DOI] [PubMed] [Google Scholar]
- 3. Dinarello C. A. (1996) Biologic basis for interleukin-1 in disease. Blood 87, 2095–2147 [PubMed] [Google Scholar]
- 4. Bergsbaken T., Fink S. L., and Cookson B. T. (2009) Pyroptosis: host cell death and inflammation. Nat. Rev. Microbiol. 7, 99–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brough D., and Rothwell N. J. (2007) Caspase-1-dependent processing of pro-interleukin-1β is cytosolic and precedes cell death. J. Cell Sci. 120, 772–781 [DOI] [PubMed] [Google Scholar]
- 6. Denoble A. E., Huffman K. M., Stabler T. V., Kelly S. J., Hershfield M. S., McDaniel G. E., Coleman R. E., and Kraus V. B. (2011) Uric acid is a danger signal of increasing risk for osteoarthritis through inflammasome activation. Proc. Natl. Acad. Sci. U.S.A. 108, 2088–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gustot A., Gallea J. I., Sarroukh R., Celej M. S., Ruysschaert J. M., and Raussens V. (2015) Amyloid fibrils are the molecular trigger of inflammation in Parkinson's disease. Biochem. J. 471, 323–333 [DOI] [PubMed] [Google Scholar]
- 8. Heneka M. T., Kummer M. P., Stutz A., Delekate A., Schwartz S., Vieira-Saecker A., Griep A., Axt D., Remus A., Tzeng T. C., Gelpi E., Halle A., Korte M., Latz E., and Golenbock D. T. (2013) NLRP3 is activated in Alzheimer's disease and contributes to pathology in APP/PS1 mice. Nature 493, 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee H. M., Kim J. J., Kim H. J., Shong M., Ku B. J., and Jo E. K. (2013) Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes 62, 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Legrand-Poels S., Esser N., L'homme L., Scheen A., Paquot N., and Piette J. (2014) Free fatty acids as modulators of the NLRP3 inflammasome in obesity/type 2 diabetes. Biochem. Pharmacol. 92, 131–141 [DOI] [PubMed] [Google Scholar]
- 11. Martinon F., Pétrilli V., Mayor A., Tardivel A., and Tschopp J. (2006) Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 440, 237–241 [DOI] [PubMed] [Google Scholar]
- 12. Meng D. M., Zhou Y. J., Wang L., Ren W., Cui L. L., Han L., Qu Z. H., Li C. G., and Zhao J. J. (2013) Polymorphisms in the NLRP3 gene and risk of primary gouty arthritis. Mol. Med. Rep. 7, 1761–1766 [DOI] [PubMed] [Google Scholar]
- 13. Vandanmagsar B., Youm Y. H., Ravussin A., Galgani J. E., Stadler K., Mynatt R. L., Ravussin E., Stephens J. M., and Dixit V. D. (2011) The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 17, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Westwell-Roper C., Nackiewicz D., Dan M., and Ehses J. A. (2014) Toll-like receptors and NLRP3 as central regulators of pancreatic islet inflammation in type 2 diabetes. Immunol. Cell Biol. 92, 314–323 [DOI] [PubMed] [Google Scholar]
- 15. Mariathasan S., Weiss D. S., Newton K., McBride J., O'Rourke K., Roose-Girma M., Lee W. P., Weinrauch Y., Monack D. M., and Dixit V. M. (2006) Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 [DOI] [PubMed] [Google Scholar]
- 16. Duncan J. A., Bergstralh D. T., Wang Y., Willingham S. B., Ye Z., Zimmermann A. G., and Ting J. P. (2007) Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. U.S.A. 104, 8041–8046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Masters S. L., Dunne A., Subramanian S. L., Hull R. L., Tannahill G. M., Sharp F. A., Becker C., Franchi L., Yoshihara E., Chen Z., Mullooly N., Mielke L. A., Harris J., Coll R. C., Mills K. H., et al. (2010) Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 11, 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peeters P. M., Perkins T. N., Wouters E. F., Mossman B. T., and Reynaert N. L. (2013) Silica induces NLRP3 inflammasome activation in human lung epithelial cells. Part. Fibre Toxicol. 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salminen A., Ojala J., Suuronen T., Kaarniranta K., and Kauppinen A. (2008) Amyloid-β oligomers set fire to inflammasomes and induce Alzheimer's pathology. J. Cell. Mol. Med. 12, 2255–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Muñoz-Planillo R., Kuffa P., Martínez-Colón G., Smith B. L., Rajendiran T. M., and Núñez G. (2013) K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity 38, 1142–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., and Tschopp J. (2007) Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ 14, 1583–1589 [DOI] [PubMed] [Google Scholar]
- 22. Perregaux D. G., Laliberte R. E., and Gabel C. A. (1996) Human monocyte interleukin-1β posttranslational processing. Evidence of a volume-regulated response. J. Biol. Chem. 271, 29830–29838 [DOI] [PubMed] [Google Scholar]
- 23. Compan V., Baroja-Mazo A., López-Castejón G., Gomez A. I., Martínez C. M., Angosto D., Montero M. T., Herranz A. S., Bazán E., Reimers D., Mulero V., and Pelegrín P. (2012) Cell volume regulation modulates NLRP3 inflammasome activation. Immunity 37, 487–500 [DOI] [PubMed] [Google Scholar]
- 24. Daniels M. J., Rivers-Auty J., Schilling T., Spencer N. G., Watremez W., Fasolino V., Booth S. J., White C. S., Baldwin A. G., Freeman S., Wong R., Latta C., Yu S., Jackson J., Fischer N., Koziel V., Pillot T., et al. (2016) Fenamate NSAIDs inhibit the NLRP3 inflammasome and protect against Alzheimer's disease in rodent models. Nat. Commun. 7, 12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marty V., Médina C., Combe C., Parnet P., and Amédée T. (2005) ATP binding cassette transporter ABC1 is required for the release of interleukin-1β by P2X7-stimulated and lipopolysaccharide-primed mouse Schwann cells. Glia 49, 511–519 [DOI] [PubMed] [Google Scholar]
- 26. Littler D. R., Harrop S. J., Fairlie W. D., Brown L. J., Pankhurst G. J., Pankhurst S., DeMaere M. Z., Campbell T. J., Bauskin A. R., Tonini R., Mazzanti M., Breit S. N., and Curmi P. M. (2004) The intracellular chloride ion channel protein CLIC1 undergoes a redox-controlled structural transition. J. Biol. Chem. 279, 9298–9305 [DOI] [PubMed] [Google Scholar]
- 27. Singh H., and Ashley R. H. (2006) Redox regulation of CLIC1 by cysteine residues associated with the putative channel pore. Biophys. J. 90, 1628–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cross M., Fernandes M., Dirr H., and Fanucchi S. (2015) Glutamate 85 and glutamate 228 contribute to the pH-response of the soluble form of chloride intracellular channel 1. Mol. Cell. Biochem. 398, 83–93 [DOI] [PubMed] [Google Scholar]
- 29. Peter B., Polyansky A. A., Fanucchi S., and Dirr H. W. (2014) A Lys-Trp cation-pi interaction mediates the dimerization and function of the chloride intracellular channel protein 1 transmembrane domain. Biochemistry 53, 57–67 [DOI] [PubMed] [Google Scholar]
- 30. Jiang L., Salao K., Li H., Rybicka J. M., Yates R. M., Luo X. W., Shi X. X., Kuffner T., Tsai V. W., Husaini Y., Wu L., Brown D. A., Grewal T., Brown L. J., Curmi P. M., and Breit S. N. (2012) Intracellular chloride channel protein CLIC1 regulates macrophage function through modulation of phagosomal acidification. J. Cell Sci. 125, 5479–54888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ogawa S., Lozach J., Benner C., Pascual G., Tangirala R. K., Westin S., Hoffmann A., Subramaniam S., David M., Rosenfeld M. G., and Glass C. K. (2005) Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 122, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He G., Ma Y., Chou S. Y., Li H., Yang C., Chuang J. Z., Sung C. H., and Ding A. (2011) Role of CLIC4 in the host innate responses to bacterial lipopolysaccharide. Eur. J. Immunol. 41, 1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Malik M., Jividen K., Padmakumar V. C., Cataisson C., Li L., Lee J., Howard O. M., and Yuspa S. H. (2012) Inducible NOS-induced chloride intracellular channel 4 (CLIC4) nuclear translocation regulates macrophage deactivation. Proc. Natl. Acad. Sci. U.S.A. 109, 6130–6135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coll R. C., Robertson A. A., Chae J. J., Higgins S. C., Muñoz-Planillo R., Inserra M. C., Vetter I., Dungan L. S., Monks B. G., Stutz A., Croker D. E., Butler M. S., Haneklaus M., Sutton C. E., Núñez G., et al. (2015) A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 21, 248–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laliberte R. E., Perregaux D. G., Hoth L. R., Rosner P. J., Jordan C. K., Peese K. M., Eggler J. F., Dombroski M. A., Geoghegan K. F., and Gabel C. A. (2003) Glutathione S-transferase omega 1–1 is a target of cytokine release inhibitory drugs and may be responsible for their effect on interleukin-1β posttranslational processing. J. Biol. Chem. 278, 16567–16578 [DOI] [PubMed] [Google Scholar]
- 36. Perregaux D., and Gabel C. A. (1994) Interleukin-1β maturation and release in response to ATP and nigericin: evidence that potassium depletion mediated by these agents is a necessary and common feature of their activity. J. Biol. Chem. 269, 15195–15203 [PubMed] [Google Scholar]
- 37. Verhoef P. A., Kertesy S. B., Lundberg K., Kahlenberg J. M., and Dubyak G. R. (2005) Inhibitory effects of chloride on the activation of caspase-1, IL-1β secretion, and cytolysis by the P2X7 receptor. J. Immunol. 175, 7623–7634 [DOI] [PubMed] [Google Scholar]
- 38. Ip W. K., and Medzhitov R. (2015) Macrophages monitor tissue osmolarity and induce inflammatory response through NLRP3 and NLRC4 inflammasome activation. Nat. Commun. 6, 6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Averaimo S., Milton R. H., Duchen M. R., and Mazzanti M. (2010) Chloride intracellular channel 1 (CLIC1): sensor and effector during oxidative stress. FEBS Lett. 584, 2076–2084 [DOI] [PubMed] [Google Scholar]
- 40. Stutz A., Horvath G. L., Monks B. G., and Latz E. (2013) ASC speck formation as a readout for inflammasome activation. Methods Mol. Biol. 1040, 91–101 [DOI] [PubMed] [Google Scholar]
- 41. Tulk B. M., Kapadia S., and Edwards J. C. (2002) CLIC1 inserts from the aqueous phase into phospholipid membranes, where it functions as an anion channel. Am. J. Physiol. Cell Physiol. 282, C1103–C1112 [DOI] [PubMed] [Google Scholar]
- 42. Warton K., Tonini R., Fairlie W. D., Matthews J. M., Valenzuela S. M., Qiu M. R., Wu W. M., Pankhurst S., Bauskin A. R., Harrop S. J., Campbell T. J., Curmi P. M., Breit S. N., and Mazzanti M. (2002) Recombinant CLIC1 (NCC27) assembles in lipid bilayers via a pH-dependent two-state process to form chloride ion channels with identical characteristics to those observed in Chinese hamster ovary cells expressing CLIC1. J. Biol. Chem. 277, 26003–26011 [DOI] [PubMed] [Google Scholar]
- 43. Lee G. S., Subramanian N., Kim A. I., Aksentijevich I., Goldbach-Mansky R., Sacks D. B., Germain R. N., Kastner D. L, and Chae J. J. (2012) The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 492, 123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laliberte R., Perregaux D., Svensson L., Pazoles C. J., and Gabel C. A. (1994) Tenidap modulates cytoplasmic pH and inhibits anion transport in vitro. II. Inhibition of IL-1β production from ATP-treated monocytes and macrophages. J. Immunol. 153, 2168–2179 [PubMed] [Google Scholar]
- 45. Al-Rawi P. G., Tseng M. Y., Richards H. K., Nortje J., Timofeev I., Matta B. F., Hutchinson P. J., and Kirkpatrick P. J. (2010) Hypertonic saline in patients with poor-grade subarachnoid hemorrhage improves cerebral blood flow, brain tissue oxygen, and pH. Stroke 41, 122–128 [DOI] [PubMed] [Google Scholar]
- 46. Zeng W. X., Han Y. L., Zhu G. F., Huang L. Q., Deng Y. Y., Wang Q. S., Jiang W. Q., Wen M. Y., Han Q. P., Xie D., and Zeng H. K. (2017) Hypertonic saline attenuates expression of Notch signaling and proinflammatory mediators in activated microglia in experimentally induced cerebral ischemia and hypoxic BV-2 microglia. BMC Neurosci. 18, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petroni R. C., Biselli P. J., de Lima T. M., Theobaldo M. C., Caldini E. T., Pimentel R. N., Barbeiro H. V., Kubo S. A., Velasco I. T., Soriano F. G. (2015) Hypertonic saline (NaCl 7.5%) reduces LPS-induced acute lung injury in rats. Inflammation 38, 2026–2035 [DOI] [PubMed] [Google Scholar]
- 48. Rizoli S. B., Kapus A., Fan J., Li Y. H., Marshall J. C., and Rotstein O. D. (1998) Immunomodulatory effects of hypertonic resuscitation on the development of lung inflammation following hemorrhagic shock. J. Immunol. 161, 6288–6296 [PubMed] [Google Scholar]
- 49. Wang Y. L., Lam K. K., Cheng P. Y., Kung C. W., Chen S. Y., Chao C. C., Hwang H. R., Chung M. T., and Lee Y. M. (2013) The cardioprotective effect of hypertonic saline is associated with inhibitory effect on macrophage migration inhibitory factor in sepsis. Biomed. Res. Int. 2013, 201614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Malik M., Shukla A., Amin P., Niedelman W., Lee J., Jividen K., Phang J. M., Ding J., Suh K. S., Curmi P. M., and Yuspa S. H. (2010) S-Nitrosylation regulates nuclear translocation of chloride intracellular channel protein CLIC4. J. Biol. Chem. 285, 23818–23828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mao K., Chen S., Chen M., Ma Y., Wang Y., Huang B., He Z., Zeng Y., Hu Y., Sun S., Li J., Wu X., Wang X., Strober W., et al. (2013) Nitric oxide suppresses NLRP3 inflammasome activation and protects against LPS-induced septic shock. Cell Res, 23, 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]