Abstract
Twenty-four hours of fasting is known to blunt activation of the human NLRP3 inflammasome. This effect might be mediated by SIRT3 activation, controlling mitochondrial reactive oxygen species. To characterize the molecular underpinnings of this fasting effect, we comparatively analyzed the NLRP3 inflammasome response to nutrient deprivation in wild-type and SIRT3 knock-out mice. Consistent with previous findings for human NLRP3, prolonged fasting blunted the inflammasome in wild-type mice but not in SIRT3 knock-out mice. In SIRT3 knock-out bone marrow–derived macrophages, NLRP3 activation promoted excess cytosolic extrusion of mitochondrial DNA along with increased reactive oxygen species and reduced superoxide dismutase 2 (SOD2) activity. Interestingly, the negative regulatory effect of SIRT3 on NLRP3 was not due to transcriptional control or priming of canonical inflammasome components but, rather, occurred via SIRT3-mediated deacetylation of mitochondrial SOD2, leading to SOD2 activation. We also found that siRNA knockdown of SIRT3 or SOD2 increased NLRP3 supercomplex formation and activation. Moreover, overexpression of wild-type and constitutively active SOD2 similarly blunted inflammasome assembly and activation, effects that were abrogated by acetylation mimic–modified SOD2. Finally, in vivo administration of lipopolysaccharide increased liver injury and the levels of peritoneal macrophage cytokines, including IL-1β, in SIRT3 KO mice. These results support the emerging concept that enhancing mitochondrial resilience against damage-associated molecular patterns may play a pivotal role in preventing inflammation and that the anti-inflammatory effect of fasting-mimetic diets may be mediated, in part, through SIRT3-directed blunting of NLRP3 inflammasome assembly and activation.
Keywords: inflammasome, mitochondria, mtDNA, NLRP3, reactive oxygen species (ROS), sirtuin, superoxide dismutase (SOD)
Introduction
Sterile inflammation linked to obesity is mediated in part by activation of the Nod-like receptor pyrin domain–containing 3 (NLRP3)2 inflammasome (1). The activation of the NLRP3 inflammasome, as a component of the innate immune system, similarly exacerbates obesity-linked diseases, including insulin resistance, diabetes, and asthma (2, 3). Obesity triggers the engagement of toll-like receptors to initiate transcriptional priming of the NLRP3 inflammasome via adipose tissue hypertrophy with macrophage infiltration and cytokine secretion, elevated circulating saturated fatty acids, and/or obesity-linked endotoxemia (4–7). These, in turn, activate NFκB-dependent transcription to up-regulate genes encoding NLRP3 and its canonical cytokines pro–IL-1β and pro–IL-18. This transcriptional induction of NLRP3 inflammasome components is termed priming. Interestingly, mitochondrial reactive oxygen species (ROS) have been found to play a role in inflammasome priming (8, 9).
Subsequent inflammasome activation, as the cornerstone of intracellular surveillance, is initiated in response to either additional pathogen-associated molecular patterns or host-cell derived damage-associated molecular patterns (DAMPs) that promote the assembly and self-oligomerization of canonical inflammasome constituents. Emerging evidence supports that the disruption of mitochondrial integrity with extrusion of mitochondrial content can function as a DAMP to activate the NLRP3 inflammasome. The mechanisms whereby mitochondrial content extrusion into the cytoplasm functions as a DAMP include a role of the mitochondrial membrane cardiolipin via direct interaction with NLRP3 (10), via the release of mitochondrial reactive oxygen species (11), and/or because of the intrinsic composition of hypomethylated CpG motifs of mitochondrial DNA that resemble the immunogenic properties of bacterial CpG DNA motifs (12). In addition, mitochondria are emerging as a structural foundation on which the multiple proteins that constitute the NLRP3 inflammasome can nucleate and assemble (13–16). The NLRP3 complex then promotes caspase-1 activation and cleavage of pro–IL-1β and pro–IL-18 into bioactive cytokines that amplify inflammation (17, 18).
Chronic and intermittent caloric restriction are associated with reduced circulating inflammatory signatures (19, 20), and, more specifically, caloric restriction in obese type 2 diabetes blunts the NLRP3 inflammasome (1). At the same time, caloric restriction has been shown to enhance mitochondrial integrity via the augmentation of mitochondrial quality control programs and the control of mitochondrial ROS levels (21–24). This is mediated in part by activation of sirtuin enzymes that function to enhance mitochondrial function and integrity (25–27). Furthermore, fasting has been employed as a caloric restriction mimetic, and this temporary nutrient deprivation intervention has been found to activate sirtuin-dependent augmentation of mitochondrial functioning (28, 29).
Given this understanding, we initially proposed that fasting may blunt the NLRP3 inflammasome relative to the fed state and that this may be mediated in part via the activation of sirtuin deacetylase enzymes. We initially tested this in a group of human volunteers and evaluated inflammasome activation in peripheral blood mononuclear cells and monocytes following a 24-h fast and then again 3 h after a fixed caloric meal. In that study, we found that the NLRP3 inflammasome was blunted in the fasted compared with the refed state. Additionally, indirect data supported the activation of the mitochondrial enriched sirtuin deacetylase (SIRT3) by fasting. Furthermore, this activation was linked to the deacetylation and activation of the canonical SIRT3 substrate superoxide dismutase 2 (SOD2) with a modest reduction in mitochondrial ROS levels. Additionally, probable non-SIRT3-dependent priming effects were mediated by refeeding, although whether this nutrient intervention modulated mitochondrial integrity was not explored (30). To more clearly delineate the fasting effect on the NLRP3 inflammasome and the role of SIRT3 in this biology, we have now undertaken a study of this innate immune inflammatory program, comparing wild-type with SIRT3 knock-out mice.
In this study, we found that prolonged fasting blunted the NLRP3 inflammasome in a SIRT3-dependent manner. The SIRT3 effects modulated the execution of the NLRP3 inflammasome by enhancing mitochondrial resilience to DAMPs and by attenuating mitochondrial ROS levels. SOD2 was identified as an integral component of this anti-inflammatory effect, and acetylation of SOD2 abrogated the anti-inflammatory effects of SIRT3.
Results
Fasting-mediated blunting of the NLRP3 inflammasome is dependent on SIRT3
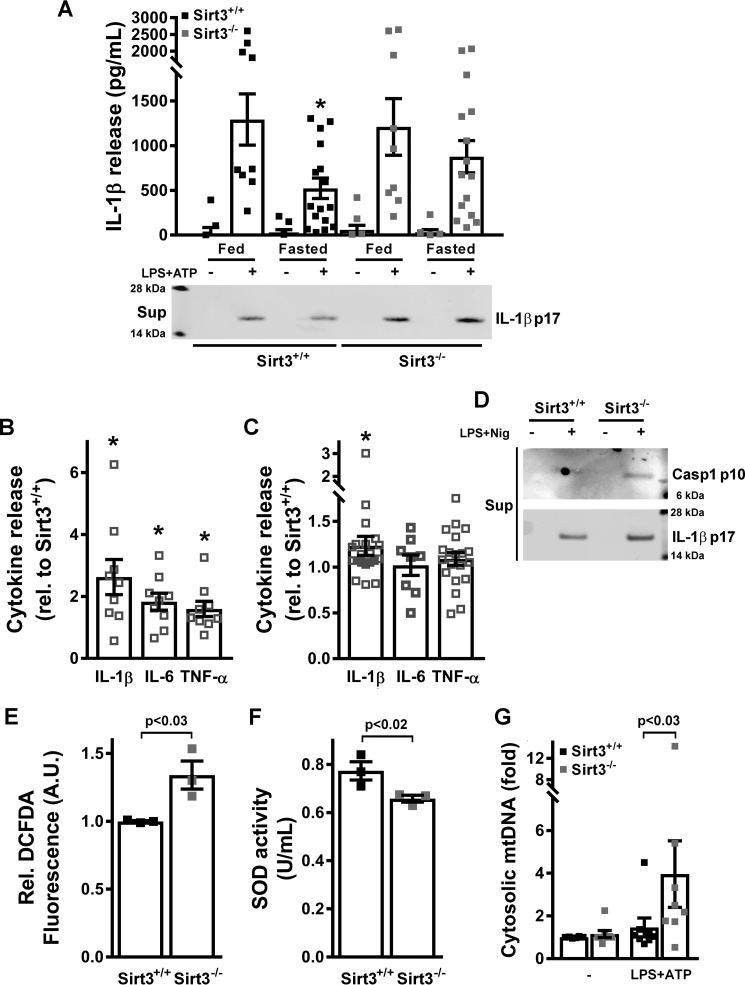
To begin to explore whether fasting blunted the NLRP3 inflammasome, WT and SIRT3 KO mice were fed an ad libitum diet or fasted for 24 or 48 h prior to the extraction of primary peritoneal macrophages. The NLRP3 inflammasome was assayed by measuring IL-1β release following priming with LPS and activation by ATP. No significant difference was found in IL-1β release in fed versus 24-h fasting WT and SIRT3 KO peritoneal macrophages (supplemental Fig. 1A). In contrast, WT macrophages showed robust fasting-mediated suppression of IL-1β secretion (Fig. 1A) following 48 h of food deprivation. This blunting effect of fasting was, however, not evident in SIRT3 KO mice (Fig. 1A). The reduction of IL-1β protein levels in the supernatant was further confirmed by immunoblotting (Fig. 1A, lower panel). To explore a broader array of cytokines released at 48 h comparing the fed versus the fasted state, we measured the release of IL-6 and TNF. Although these downstream cytokines showed higher levels in the fasted SIRT3 KO–derived peritoneal macrophages, the induction was less pronounced than that of the primary NLRP3 inflammasome substrate IL-1β (Fig. 1B). In primary bone marrow–derived macrophages (BMDMs), the relative induction of IL-1β was modestly but still significantly higher in KO cells, whereas the induction of IL-6 and TNF was not different between WT and KO-derived BMDMs (Fig. 1C). Exposure of these cells to LPS and nigericin as a second NLRP3 inflammasome trigger similarly augmented the release of cleaved caspase-1 and IL-1β in the KO cells (Fig. 1D). In parallel, stable knockdown (KD) of SIRT3 in murine J774A.1 macrophages also showed increased caspase-1 cleavage in response to LPS and ATP or nigericin (supplemental Fig. 1B).
Figure 1.
SIRT3 blunts the NLRP3 inflammasome. A, IL-1β release into the supernatant of cultured peritoneal macrophages obtained from fed or 48-h–fasted SIRT3+/+ or SIRT3−/− mice left untreated or primed with LPS and stimulated with 5 mm ATP was assessed by ELISA (top panel) and immunoblot analysis (bottom panel). Error bars represent mean ± S.E. (n = 9–17). *, p < 0.05. Sup, supernatant. B and C, ratio of release of IL-1β, IL-6, and TNF comparing SIRT3−/− with SIRT3+/+ 48-h–fasted mouse peritoneal macrophages primed with LPS and stimulated with 5 mm ATP (B) or SIRT3−/− with SIRT3+/+ BMDMs primed with LPS and stimulated with 3 mm ATP (C). Cytokines were measured by ELISA. Error bars represent mean ± S.E. (n = 9–21). *, p < 0.05. D, immunoblot of release of active caspase-1 (p10 subunit) and mature IL-1β (p17 subunit) into the supernatants of cultured BMDMs left untreated or primed with LPS and stimulated with nigericin. E, ROS levels in LPS-treated BMDMs. Error bars represent mean ± S.E. (n = 3). A.U., arbitrary units. F, SOD2 activity in LPS-treated BMDM mitochondria. Error bars represent mean ± S.E. (n = 3). G, measurement of mitochondrial genomic DNA extrusion in SIRT3+/+ and SIRT3−/− BMDMs left untreated or primed with LPS and stimulated with 3 mm ATP. Error bars represent mean ± S.E. (n = 8).
Inflammasome activation disrupted mitochondrial fidelity in the absence of SIRT3
Given the quality control functions of SIRT3 (25), we then explored aspects of known mitochondrial orchestrated events in the execution of the NLRP3 inflammasome (31). In response to NLRP3 priming, the SIRT3 KO BMDMs showed evidence of increased ROS levels, as measured by DCFDA and MitoSOX fluorescence (Fig. 1E and supplemental Fig. 1C) and a reduction in SOD2 activity (Fig. 1F). This excessive ROS response to NLRP3 priming was reproduced in SIRT3 KD J774A.1 cells (supplemental Fig. 1D). Interestingly, and as observed previously in THP-1 cells (30), the mitochondrion-targeted superoxide dismutase mimetic MitoTEMPO blunted differences in IL-1β release when comparing wild-type and KO BMDMs (supplemental Fig. 1E), supporting that increased ROS levels were involved in the exaggerated activation of this program in SIRT3-deficient cells.
To further explore this SIRT3–ROS link, we employed different doses of ATP and studied the capacity of MitoTEMPO to block the NLRP3 inflammasome. Interestingly, although IL-1β release driven by 1 mm ATP was fully blocked by the ROS scavenger, this scavenging effect was completely abrogated using 5 mm ATP (supplemental Fig. 1F). This showed that different NLRP3 activation conditions differentially relied on mitochondrial ROS as an activator, as shown previously (32). In parallel, the differential activation of the inflammasome in SIRT3 KO versus WT BMDMs followed the same ATP dose dependence (supplemental Fig. 1G), with no difference at all between the genotypes at 5 mm ATP (where MitoTEMPO does not have an inhibitory effect, and, thus, ROS are not involved in NLRP3 activation) and maximal difference at 1 mm ATP (where the ROS scavenger fully blocks the inflammasome, and, thus, ROS are critical for NLRP3 activation). Together, these data further support that increased ROS levels orchestrate the exaggerated NLRP3 inflammasome activation in the SIRT3 KO BMDMs.
Interestingly, the release of mitochondrial DNA into the cytosol following genetic disruption of mitochondrial integrity (heterozygous knockdown of transcription factor A of mitochondria (TFAM)) has been found previously to engage the cytosolic DNA–sensing cyclic GAMP synthetase (cGAS) to promulgate inflammation through interferon signaling (33). We explored whether this pathway was induced following the genetic depletion of SIRT3. However, cyclic GAMP levels were below HPLC and MS detection levels in WT and SIRT3 KO BMDMs in the basal state or in response to LPS or LPS and ATP, and the phosphorylation of downstream interferon regulatory factor 3 (IRF3) was similarly not induced (data not shown). Despite this, and in parallel with the TFAM knockdown study, we found that the increased mitochondrial DNA extrusion in SIRT3 KD J774A.1 cells probably primed the antiviral innate response; these cells showed higher induction of interferon-β transcript levels and increased phosphorylation of IRF3 after transfection with polycytidylic acid (poly(I:C)) (supplemental Fig. 2A).
SIRT3-deficient BMDMs showed no evidence of exaggerated priming of the NLRP3 inflammasome program
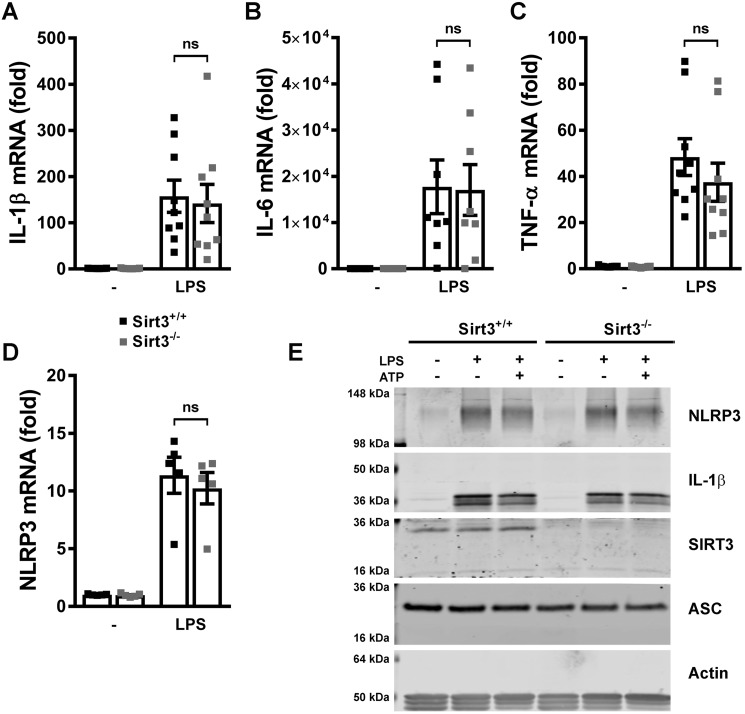
Previous data showed that mitochondrial ROS initiated NLRP3 inflammasome priming (9). We therefore assessed whether the absence of SIRT3 with concurrent reduced dismutase and increased ROS levels was sufficient to selectively evoke NLRP3 inflammasome priming. Interestingly, in response to LPS administration priming was evident to a similar extent in WT and SIRT3 KO BMDMs, as measured by the induction of transcripts encoding for IL-1β, IL-6, TNF, and NLRP3 itself (Fig. 2, A–D). In parallel, the steady-state protein levels of pro IL-1β and NLRP3 were also not differentially induced by LPS and/or by administration of ATP (Fig. 2E). This observation was recapitulated in control and SIRT3 KD J774A.1 cells (supplemental Fig. 2B). Interestingly, SOD2 acetylation was significantly more prominent in SIRT3 KD J774A.1 cells following LPS and ATP compared with control cells (supplemental Fig. 2B).
Figure 2.
SIRT3-deficient BMDMs show no evidence of increased priming by LPS. A–D, quantitative RT-PCR analysis of Il1b (A), Il6 (B), Tnf (C), and Nlrp3 (D) transcripts in SIRT3+/+ and SIRT3−/− BMDMs left untreated or primed with LPS. Error bars represent mean ± S.E. (n = 5–9). ns, not significant. E, immunoblot analysis of steady-state protein levels of inflammasome components in SIRT3+/+ and SIRT3−/− BMDMs left untreated or primed with LPS and left unstimulated or stimulated with 3 mm ATP.
SOD2 mutagenesis studies
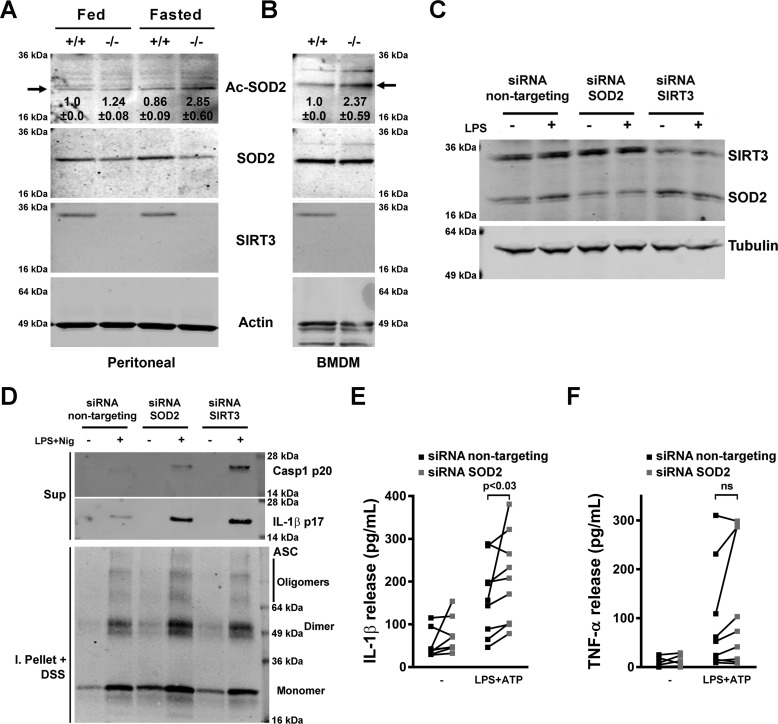
In our human study, fasting reduced monocyte SOD2 acetylation in parallel with blunting of the inflammasome (30). To assess whether this regulation was operational during murine fasting, we assessed the levels of SOD2 acetylation in primary peritoneal macrophages. The relative levels of SOD2 acetylation after 48 h of fasting were higher in SIRT3 KO compared with WT peritoneal macrophages (Fig. 3A), and this SOD2 acetylation difference was also evident in SIRT3 KO BMDMs (Fig. 3B). To directly investigate the effects of SIRT3 and the activation of SOD2 on inflammasome function, we genetically modulated SOD2 levels in THP-1 macrophages. siRNA-targeted KD of SOD2 resulted in an ≈50% reduction in SOD2 levels at baseline and in response to LPS priming (Fig. 3C). In response to NLRP3 inflammasome priming and activation by LPS and nigericin, both SIRT3 and SOD2 KD THP-1 macrophages exhibited a significantly greater release of cleaved caspase-1 and IL-1β in parallel with evidence of increased dimerization and oligomerization of ASC (Fig. 3D). IL-1β but not TNF levels were similarly induced in response to LPS and ATP in response to SOD2 KD (Fig. 3, E and F).
Figure 3.
SOD2 blunts the NLRP3 inflammasome. A, immunoblot analysis of relative SOD2 protein acetylation in peritoneal macrophages obtained from fed or 48-h–fasted SIRT3+/+ or SIRT3−/− mice. B, immunoblot analysis of relative SOD2 protein acetylation in cultured BMDMs obtained from SIRT3+/+ or SIRT3−/− mice. Inset numbers show the average relative intensity in two independent experiments ± S.D., and the arrow shows the acetylated Lys-68 residue on SOD2. C, immunoblot analysis of cell lysates from untreated or LPS-treated human THP-1 macrophages transfected with control siRNA or siRNA targeting SOD2 or SIRT3. D, immunoblot analysis of release of active caspase-1 (Casp1, p20 subunit) and mature IL-1β (p17) into supernatants (Sup) from human THP-1 macrophages transfected with control siRNA or siRNA targeting SOD2 or SIRT3, left untreated or primed with LPS and stimulated with nigericin (Nig), and lysates of the same cells solubilized with Triton X-100–containing buffer, followed by cross-linkage of insoluble pellets with DSS (I. Pellet + DSS) to capture ASC dimers and oligomers. E and F, ELISA of IL-1β (E) and TNF (F) release into supernatants of human THP-1 macrophages transfected with control siRNA or siRNA targeting SOD2, left untreated or primed with LPS and stimulated with 5 mm ATP. Error bars represent mean ± S.E. (n = 9). ns, not significant.
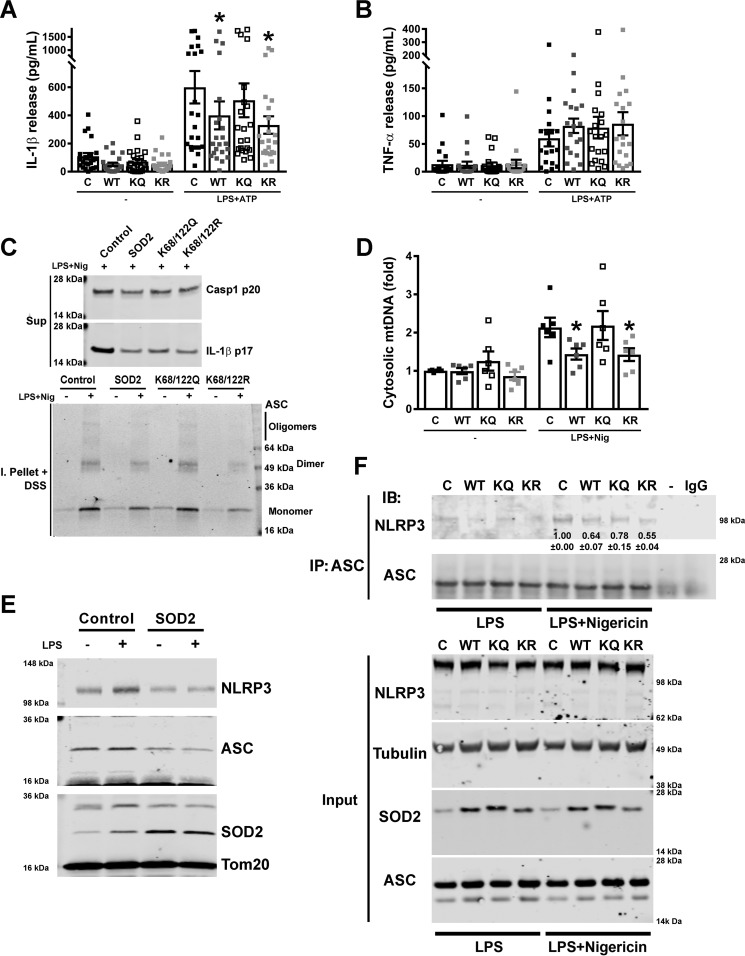
Given that the modulation of canonical lysine residues on SOD2 has been found previously to alter its enzyme activity, we employed this genetic approach to validate the role of SOD2 activity in mediating the SIRT3 effect on the NLRP3 inflammasome. We explored the effect of overexpression of WT, lysine deacetylation mimetic (K68R/K122R), and lysine acetylation mimetic (K68Q/K122Q) SOD2 constructs on the NLRP3 inflammasome. Consistent with our prior data, the WT and K68R/K122R constructs blunted IL-1β levels, but not TNF levels, in response to LPS and ATP (Fig. 4, A and B). In contrast, the lysine acetylation mimetic SOD2 showed higher cytokine release to an extent similar to overexpression of an empty vector (Fig. 4, A and B). In parallel, exposure of these stable overexpressing THP-1 cells to LPS and nigericin showed that the WT and lysine deacetylation mimetic SOD2 similarly reduced caspase-1 cleavage, IL-1β release, and ASC multimerization (Fig. 4C). Furthermore, overexpression of the WT and the deacetylation mimetic SOD2, but not the acetylation mimetic, blunted LPS- and nigericin-linked mitochondrial DNA extrusion into the cytosol in THP-1 cells (Fig. 4D), and, as with the primary BMDMs, cyclic GAMP levels were not detectable, and IRF3 was not phosphorylated in the basal state or in response to NLRP3 inflammasome priming or activation (data not shown). In keeping with the data from the primary cells, modest SOD2 induction blunted inflammasome activation, whereas high levels of SOD2 activity had a broader effect, blunting both priming (pro–IL-1β levels) and NLRP3 activation (supplemental Fig. 3A).
Figure 4.
SOD2 inhibition of the NLRP3 inflammasome is acetylation-level dependent and modulates NLRP3 localization to mitochondria and its interaction with ASC. A and B, ELISA of IL-1β (A) and TNF (B) release into supernatants of human THP-1 macrophages stably transduced with control empty vector or with WT or mutant SOD2 vectors, left untreated or primed with LPS and stimulated with 5 mm ATP. Error bars represent mean ± S.E. (n = 23). *, p < 0.05. C, immunoblot analysis of release of active caspase-1 (Casp1, p20 subunit) and mature IL-1β (p17) to supernatants (Sup) from human THP-1 macrophages stably transduced with control empty vector or with wild-type or mutant SOD2 vectors, primed with LPS and stimulated with nigericin (Nig), and lysates of the same cells solubilized with Triton X-100–containing buffer, followed by cross-linkage of insoluble pellets with DSS (I. Pellet + DSS) to capture ASC dimers and oligomers. D, measurement of mitochondrial genomic DNA extrusion in human THP-1 macrophages stably transduced with control empty vector or with wild-type or mutant SOD2 vectors, left untreated or primed with LPS and stimulated with nigericin. Error bars represent mean ± S.E. (n = 6). *, p < 0.05. E, representative immunoblot analysis for NLRP3 and ASC on mitochondrial fractions from untreated or LPS-treated human THP-1 macrophages stably transduced with control empty vector or with SOD2. F, representative immunoprecipitation (IP) and immunoblot (IB) analysis of the interaction between NLRP3 and ASC in human THP-1 macrophages stably transduced with control empty vector or with wild-type or mutant SOD2 vectors, primed with LPS and left untreated or stimulated with nigericin. Inset numbers show the relative intensity of the bands compared with the control treated with LPS and nigericin ± S.E. (n = 3). The asterisks show significant (p < 0.05) reduction in WT and KR lanes compared with the control and KQ lanes. − indicates immunoprecipitation of a control sample primed with LPS and stimulated with nigericin but without an antibody, and IgG indicates immunoprecipitation of the same sample with an unspecific antibody. Input indicates immunoblot analysis of the same samples before immunoprecipitation. C, control vector; WT, wild-type SOD2 vector; KQ, mutant K68Q/K122Q SOD2 vector; KR, mutant K68R/K122R SOD2 vector.
SOD2 effects on NLRP3 assembly and localization
Emerging data show that the assembly of the NLRP3 multiprotein complex on mitochondria enables subsequent NLRP3 activation (13). We therefore explored the localization of NLRP3 and the apoptosis-associated speck-like protein containing a CARD (ASC) adaptor protein in response to SOD2 overexpression. We found that overexpression of SOD2 reduced colocalization of NLRP3 and ASC with mitochondria (Fig. 4E). The relative purity of the mitochondrial fractions for these colocalization assays is shown in supplemental Fig. 3B. We then explored the role of SOD2 in modulating inflammasome assembly by assessing the interactions between NLRP3 and ASC using ASC immunoprecipitation with analysis of NLRP3 levels. In response to LPS and nigericin, NLRP3 more avidly bound to ASC in control THP-1 cells compared with cells overexpressing WT SOD2 (Fig. 4F; replicates are shown in supplemental Fig. 3C). In parallel, K68Q/K122Q (lysine acetylation mimetic)–overexpressing cells showed modestly higher interaction between NLRP3 and ASC compared with overexpression of K68R/K122R cells (lysine deacetylation mimetic) (Fig. 4F and supplemental Fig. 3C).
The role of SIRT3 in vivo
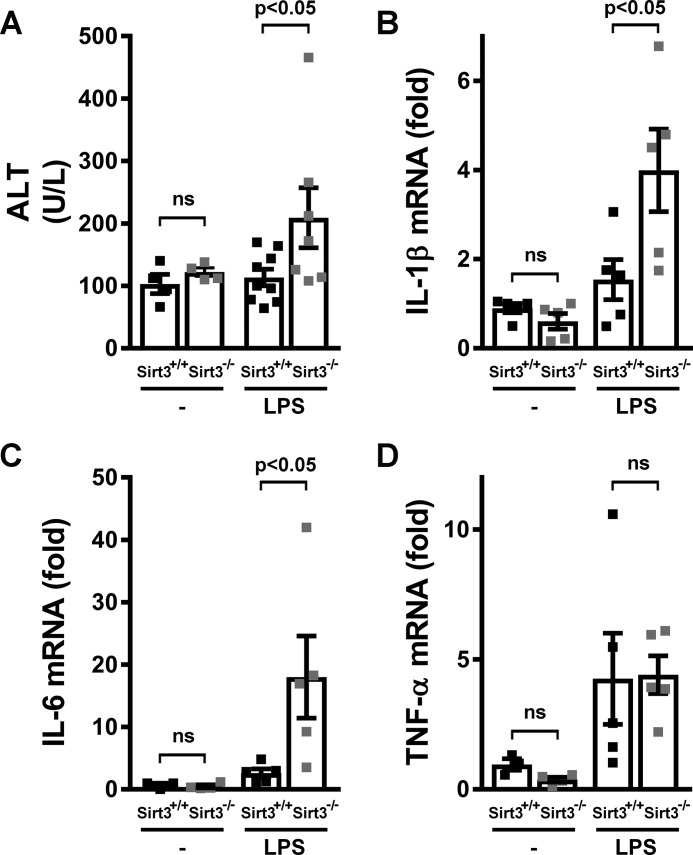
Finally, the effect of fasting on the NLRP3 inflammasome in vivo was compared in SIRT3 wild-type and KO mice. Following a 48-h fast, i.p. administration of LPS resulted in significant elevation of the circulating liver enzyme alanine aminotransferase (ALT) (Fig. 5A) in SIRT3 KO mice, suggesting inflammation-induced liver injury, with a similarly increased induction of IL-1β and IL-6 transcript levels in cells of the peritoneal cavity (Fig. 5, B and C). In contrast, peritoneal TNF transcript levels were not significantly different between the genotypes in response to LPS (Fig. 5D). In parallel, there was no significant difference in ALT levels and IL-1β or IL-6 transcript expression between the presence or absence of SIRT3 in the ad libitum–fed state (supplemental Fig. 4, A–C).
Figure 5.
SIRT3 inhibited inflammation in an in vivo model. A—D, 48-h–fasted SIRT3+/+ or SIRT3−/− mice were injected intraperitoneally with PBS (−) or 1 mg/kg LPS. Two hours later, ALT (A) levels were measured in serum, and the transcript levels of genes encoding IL-1β (B), IL-6 (C), and TNF were quantified (D) from the cells mobilized to the peritoneal cavity. Error bars represent mean ± S.E. (A, n = 7–9; B—D, n = 5).
Discussion
The beneficial immune-modulatory effects of intermittent fasting are increasingly being recognized, although the mechanisms orchestrating these effects are less well understood. As a component of this, a role of SIRT3 in blunting of the NLRP3 inflammasome has recently been shown. This is consistent with the known activation of SIRT3 by fasting, the role of SIRT3 in augmenting mitochondrial function, and the role of disrupted mitochondrial integrity in NLRP3 inflammasome activation. In this study, we have tk;4demonstrated that the canonical SIRT3 target SOD2 blunts inflammasome activation by reducing mitochondrial reactive oxygen species levels, by sustaining mitochondrial integrity, and by preventing the assembly and activation of the NLRP3 inflammasome complex. Moreover, we show that this SIRT3–SOD2–mediated effect is independent of NLRP3 inflammasome priming and is in part due to the fasting-mediated, posttranslational control of SOD2 acetylation and activity.
Intermittent fasting and caloric restriction have been found to elicit numerous beneficial effects, including weight loss, improved insulin sensitivity, and a reduction in inflammation, as assessed by the measurement of C-reactive protein (19, 20, 34) and a reduction of exacerbation in inflammation-linked diseases such as asthma (35). Although, to date, the mechanisms by which caloric deprivation modifies immune function have not been well established, SIRT3 has recently been implicated in suppressing the NLRP3 inflammasome (30). SIRT3 is activated by fasting and caloric restriction (28, 29, 36), and a common consequence of its activation in ameliorating pathological effects is reduced ROS levels (37).
The contributions of ROS signaling in priming and activation of the NLRP3 inflammasome have been actively explored (31). Conflicting findings have been reported, with studies showing that ROS signaling was only necessary for NLRP3 inflammasome priming (8, 9), whereas others showed that oxidized mitochondrial DNA, as a consequence of excess ROS, can function as a DAMP and directly bind to and activate the NLRP3 inflammasome (16). This latter finding suggests that excess ROS can distinctly activate the second signal in cells where priming is already evident. Here we show that the absence of SIRT3 did not affect LPS-induced transactivation (priming) of a canonical NLRP3 regulatory transcript and/or protein levels. Conversely, the lack of SIRT3 resulted in the amplification of NLRP3 inflammasome activation in part via mitochondrial ROS signaling. At the same time, the possibility that additional targets of SIRT3 activity, e.g. by maintaining mitochondrial integrity, contribute to excessive execution of the inflammasome in the absence of SIRT3 has not been excluded.
The integrity of mitochondrial membranes is a component of healthy mitochondria. Disruption of this integrity enables mitochondrial genomic content and cardiolipin to leak into the cytoplasm. As discussed, both the extrusion of mitochondrial DNA and cardiolipin have been shown to function as DAMPs to drive inflammation (10, 38). The concept that disrupted mitochondrial integrity regulates the inflammasome is also evident in studies where disrupted mitophagy with diminished mitochondrial clearance played a role in activating the NLRP3 inflammasome (39). The role of SIRT3 in the maintenance of these additional aspects of mitochondrial integrity has not been well studied, although it is indirectly indicated in the modulation of mitochondrial autophagy (40–42). Although we did not explore mitophagy in this study, we did show that inflammasome activation mediated extrusion of mitochondrial DNA into the cytoplasm and that this was exaggerated and resulted in excess mitochondrial ROS in the absence of SIRT3.
Interestingly, the extrusion of mtDNA has been found previously to engage the cGAS to promote STING–IRF3–dependent signaling to activate IFN-β following the heterozygous depletion of TFAM (33) (where STING is stimulator of interferon genes). Under those conditions, the innate disruption of mitochondrial integrity by disruption of TFAM results in a basal leak of mtDNA. Whether this persistent mtDNA leak preferentially activates cGAS versus the more acute mitochondrial injury in response to activation of the NLRP3 inflammasome warrants further study. In addition, recent evidence supports that activation of caspases during apoptosis or with inflammasome activation had an inhibitory effect on IFN-β via direct cleavage of cGAS (43, 44). These counterregulatory controls may support the lack of cGAS activation in response to inflammasome activation. On the other hand, the fact that poly(I:C) exacerbates IRF3 signaling following SIRT3 KD suggests that SIRT3 may play a broader role in the control of innate immune pathway activation. These potential pathways will be explored in future studies. At the same time, although we have not extensively explored the function of mitochondria as a platform for NLRP3 assembly, we found that activation of SOD2 blunted oligomerization of the ASC adaptor protein as a component of the pyroptosome and that SOD2 activation reduced the co-localization of NLRP3 and ASC with mitochondria following inflammasome activation.
The composite of work to date clearly defines SIRT3 as a mitochondrial fidelity protein, with a component of this effect, because of the deacetylation of SOD2, resulting in the activation of SOD2 to dismutate superoxide within the mitochondrial matrix (25, 45). One proposed mechanism of action is that that the direct deacetylation of canonical lysine residues within the catalytic domain (Lys-68 and Lys-122) of SOD2 resulted in enhanced avidity between the negatively charged superoxide anion and the positively charged deacetylated lysine residue (46). A second proposed mechanism was that deacetylation of Lys-68 played an important role in maintaining SOD2 tetramer stability to optimize SOD2 scavenging activity (47). In this study, we showed that constitutively active SOD2, via the substitution of arginine residues at Lys-68 and Lys-122 to mimic deacetylation, blunted inflammasome assembly and activation and that the substitution of glutamine, to mimic acetylation at the same residues, had the opposite effect. These data strongly support that SOD2 is an important SIRT3 substrate in mediating the inflammasome-moderating effect from mitochondrially generated superoxide.
Another consequence of prolonged fasting is the generation of ketone bodies. The relevance of this metabolic response to fasting is that the ketone body β-hydroxybutyrate (BHB), but not acetoacetate, has been shown previously to inhibit the NLRP3 inflammasome (48). Interestingly, in our study, we confirmed the findings of others (49) that the levels of BHB were elevated to a similar level at 24 and 48 h of fasting (data not shown). Despite this, the effect of fasting on blunting of the inflammasome was much more pronounced following a 48-h fast. This distinction between our data and the effects of BHB is also compatible with the finding that BHB-conferred inflammasome inhibition did not appear to be dependent on the inhibition of ROS (48). Taken together, these data suggest that the inflammasome-inhibitory effects of SIRT3 are, at least in part, independent of BHB levels. This ketone-independent, fasting-induced inflammasome blunting was also shown in our clinical study, where only 15% of subjects had fasting induction of serum ketone levels despite an almost uniform blunting of the inflammasome following a 24-h fast in human volunteers (30).
A limitation of this study is that the genetic manipulation of SOD2 may overwhelm or ameliorate additional effects of SIRT3 in the regulation of the inflammasome. Multiple additional SIRT3 substrates modulate mitochondrial quality control and homeostasis (25), and it is possible that effects may be masked by genetic induction or depletion of SOD2. Thus, further characterization of the SIRT3 effects on mitochondrial biology and their modulation of NLRP3 inflammasome susceptibility is required.
In conclusion, emerging evidence strongly supports that SIRT3 has a ameliorating effect on the activation of the NLRP3 inflammasome in both humans (30) and mice (50) and that this effect appears to function in part through the role of SIRT3 in controlling reactive oxygen species levels. In this study, we showed that the effect of SIRT3 regulates the execution rather than priming of the NLRP3 inflammasome and demonstrated that SIRT3-mediated activation of SOD2 was necessary for control of this inflammasome. As the inflammasome is an early event in innate immune activation, it would be intriguing to explore whether this SIRT3-mediated control of the inflammasome plays a broader role in caloric restriction and or fasting mimetic diet–mediated blunting of inflammation (19, 20).
Experimental procedures
Animal studies
Wild-type or SIRT3 KO mice in the C57BL/6 background were used in this study. Mice were fasted for up to 48 h as described previously (29). Animal experiments were approved by the NHLBI, National Institutes of Health Animal Care and Use Committee.
Cell culture and transfection
THP-1 human monocyte cells obtained from the ATCC were cultured in RPMI 1640 plus 25 mm Hepes and 10% heat-inactivated FBS. They were differentiated into macrophages by incubation with 5 ng/ml phorbol 12-myristate 13-acetate for 48 h (51). THP-1 cells were transiently transfected with siRNA 18 h before differentiation as described previously (30). Lentiviral SOD2 vectors were generated in HEK293T cells using lentiviral packaging mix (Sigma). Viral particle infection to generate stable SOD2-overexpressing THP-1 cells occurred under puromycin (2 μg/ml) selection. J774A.1 mouse macrophages obtained from the ATCC were cultured in DMEM plus 10% heat-inactivated FBS. Stable control and SIRT3 KO J774A.1 cells were generated using puromycin-selective lentiviral shRNA constructs from Sigma as described previously (29). Transfection of poly(I:C) (Sigma) into the cytosol of J774A.1 cells was performed using Lipofectamine 3000 (Invitrogen).
Mouse peritoneal macrophages were collected by established methods from non-manipulated mice (52), allowed to adhere for 30 min in serum-free RPMI medium, washed, and incubated in RPMI 1640 plus 25 mm Hepes and 10% heat-inactivated FBS. Primary BMDMs were prepared using established methods. Briefly, bone marrow collected from mouse femora and tibiae was plated on sterile dishes and incubated for 7 days in DMEM plus 25 mm Hepes, 10% heat-inactivated FBS, and 20 ng/ml M-CSF.
Cell stimulation and cytokine assays
THP-1 macrophages were incubated at 1.5 × 106 cells/ml in 96-well plates in RPMI medium plus 25 mm Hepes and 10% heat-inactivated FBS with or without 10 ng/ml LPS (ultrapure Salmonella minnesota R595, Enzo Life Sciences) for 4 h. J774A.1 cells were incubated at 1.5 × 106 cells/ml in 96-well plates in DMEM plus 10% heat-inactivated FBS with or without 100 ng/ml LPS for 4 h. Peritoneal macrophages were incubated at 106 cells/ml in 96-well plates in RPMI medium plus 25 mm Hepes and 10% heat-inactivated FBS with or without 10 ng/ml LPS for 6 h. BMDMs were incubated at 1.5 × 106 cells/ml in 96-well plates in DMEM plus 25 mm Hepes and 10% heat-inactivated FBS with or without 100 ng/ml LPS for 6 h. To stimulate the release of IL-1β, 3–5 mm ATP (Sigma-Aldrich) or 10 μm nigericin was added for the last 30 min of incubation. Supernatants were collected, centrifuged to remove cells and debris, and stored at −80 °C for later analysis. IL-1β, IL-6, and TNFα cytokine analysis was performed by ELISA (R&D Systems). Results were normalized to cell number, as determined by the CyQuant cell proliferation assay (Invitrogen).
Immunoblot analysis
For Western blots, cell lysates were prepared using radioimmune precipitation assay buffer, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Images were captured using the Odyssey system (Li-Cor). The following antibodies were used: caspase-1 (2225, Cell Signaling Technology; sc-514, Santa Cruz Biotechnology; AG-20B-0042-C100, Adipogen), IL-1β (ab9722, Abcam), NLRP3 (AG-20B-0014-C100, Adipogen), ASC (sc-22514, Santa Cruz Biotechnology), SOD2 (sc-18504, Santa Cruz Biotechnology), SIRT3 (5490S, Cell Signaling Technology), β-actin (A1978, Sigma-Aldrich), β-tubulin (2146S, Cell Signaling Technology), and Tom20 (sc-11415, Santa Cruz Biotechnology).
To measure caspase-1 and IL-1β release into the supernatant, THP-1 macrophages or peritoneal cells were seeded in 24-well plates, and BMDMs or J774A.1 cells were seeded in 6-well plates to a cell density of 1 × 106 cells/well. Cells were primed with LPS for 4–6 h at 37 °C, followed by washing once with PBS and transfer to serum-free medium with or without nigericin or ATP for 30 min. Supernatants were collected and centrifuged at 10,000 × g for 10 s to remove any detached cells, followed by transfer to a fresh tube. The supernatant was concentrated by TCA precipitation. Then the precipitated pellets were washed with acetone, dissolved in 10 μl of 0.2 m NaOH, diluted with 40 μl of H2O, supplemented with 10 μl of 6× SDS-PAGE sample buffer, boiled for 5 min, and analyzed by Western blotting.
Determination of ASC oligomerization
THP-1 macrophages were seeded in 24-well plates to a cell density of 1 × 106 cells/well. Cells were primed with LPS and then incubated with or without nigericin for 30 min. They were then rinsed in PBS and treated with 50 mm ice-cold Tris-HCl containing 0.5% Triton X-100, followed by passage through a 21-gauge needle 10 times. Lysates were centrifuged at 330 × g for 10 min at 4 °C. The pellets were washed twice in 1 ml of ice-cold PBS, resuspended in 500 μl of PBS with 2 mm disuccinimidyl suberate (DSS), and incubated at room temperature for 30 min. Samples were then centrifuged at 330 × g for 10 min at 4 °C, and the cross-linked pellets were resuspended in 1× SDS-PAGE sample buffer, boiled, and analyzed by Western blotting.
Coimmunoprecipitation
THP-1 macrophages were seeded in 15-cm dishes. Cells were primed with LPS and then incubated with or without ATP or nigericin for 30 min. Then they were washed with PBS and lysed with lysis buffer (20 mm Tris-HCl (pH 7.5), 137 mm NaCl, 5 mm EDTA, and 0.5% Nonidet P-40, and protease inhibitor mixture) for 30 min at 4 °C. The homogenates were centrifuged for 15 min at 14,000 × g. at 4 °C, and equal amounts of cell supernatants were then incubated overnight at 4 °C with no antibody or with anti-ASC or irrelevant rabbit IgG. Samples were then incubated for 4 h at 4 °C with protein A/G Plus-agarose beads (Santa Cruz Biotechnology), washed three times, and finally boiled in 1× SDS-PAGE sample buffer for immunoblotting.
Quantitative PCR analysis
mRNA was isolated using Tripure (Roche), and cDNA was produced using a first-strand synthesis kit (Invitrogen). Transcript levels were measured using validated gene-specific primers (Qiagen).
To measure mtDNA in the cytosol, 8 × 106 cells were homogenized with a Dounce homogenizer in 10 mm Tris solution (pH 7.4) containing 0.25 m sucrose, 25 mm KCl, 5 mm MgCl2, and protease inhibitor and then centrifuged at 700 × g for 10 min at 4 °C. Cytosolic fractions were prepared by centrifugation at 10,000 × g for 30 min at 4 °C, and DNA was isolated from them using the DNeasy blood and tissue kit (Qiagen). The copy number of the DNA encoding cytochrome c oxidase I (in human cells) or cytochrome b (in mouse cells) was measured by quantitative real-time PCR using specific primers (Qiagen).
ROS detection
BMDM or J774A.1 LPS-primed cells were incubated with 5 μm MitoSOX Red (to measure mitochondrial ROS production) or 2.5 μm CM-H2DCFDA (chloromethyl H2-DCFDA) (to measure total cellular ROS production) (Life Technologies) for 30 min in Hanks' balanced salt solution (Life Technologies), washed, and analyzed by flow cytometry on a BD FACSCanto (BD Biosciences).
SOD activity
Mitochondrial fractions were obtained from 8 × 106 LPS-treated BMDMs using the Q-proteome mitochondrion isolation kit (Qiagen). SOD activity was tested in 0.5 μg of BMDM mitochondria using the SOD assay kit (Cayman Chemical), and to ensure the detection of only Mn-SOD (SOD2) activity, 5 mm potassium cyanide was added to the assay to inhibit cytosolic Cu/Zn-SOD (SOD1) and extracellular Cu/Zn-SOD (SOD3) activities.
In vivo experiments
Fed or 48-h–fasted mice were challenged with a sublethal intraperitoneal injection of 1 mg/kg LPS (Escherichia coli 055:B5, Sigma-Aldrich) or PBS. After 2 h, the mice were euthanized, serum was collected for ALT/aspartate aminotransferase (AST) enzymatic activity determination, and the cells of the peritoneal cavity were collected for quantitative PCR analysis of cytokines.
Statistical analysis
Data are expressed as the mean ± S.E. Two-tailed Student's t tests were performed between groups, and multiple comparisons analysis was performed by analysis of variance. p < 0.05 was considered statistically significant.
Author contributions
J. T. and M. N. S. conceived the project. J. T., S. S. G., M. K. S., K. H., and O. H. R. performed the experiments, analyzed the data, and interpreted the results. J. T., S. S. G., D. G., R. M. S., and M. N. S. designed the research and interpreted the results. J. T. and M. N. S. wrote the paper, which was reviewed by all authors
Supplementary Material
This research was supported by Grants HL006047-06 and HL005102-11 from the NHLBI, National Institutes of Health Division of Intramural Research (to M. N. S.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains supplemental Figs. S1–S4.
- NLPR3
- Nod-like receptor pyrin domain–containing 3
- ROS
- reactive oxygen species
- DAMP
- damage-associated molecular pattern
- SOD
- superoxide dismutase
- BMDM
- bone marrow–derived macrophage
- KD
- knockdown
- TFAM
- transcription factor A of mitochondria
- cGAS
- cyclic GAMP synthetase
- ASC
- apoptosis-associated speck-like protein containing a CARD
- ALT
- alanine aminotransferase
- BHB
- β-hydroxybutyrate
- DSS
- disuccinimidyl suberate
- DCFDA
- 2′,7′-dichlorodihydrofluorescein diacetate
- GAMP
- guanosine monophosphate–adenosine monophosphate.
References
- 1. Vandanmagsar B., Youm Y. H., Ravussin A., Galgani J. E., Stadler K., Mynatt R. L., Ravussin E., Stephens J. M., and Dixit V. D. (2011) The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 17, 179–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee H. M., Kim J. J., Kim H. J., Shong M., Ku B. J., and Jo E. K. (2013) Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes 62, 194–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim H. Y., Lee H. J., Chang Y. J., Pichavant M., Shore S. A., Fitzgerald K. A., Iwakura Y., Israel E., Bolger K., Faul J., DeKruyff R. H., and Umetsu D. T. (2014) Interleukin-17-producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat. Med. 20, 54–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Osborn O., and Olefsky J. M. (2012) The cellular and signaling networks linking the immune system and metabolism in disease. Nat. Med. 18, 363–374 [DOI] [PubMed] [Google Scholar]
- 5. Kheirandish-Gozal L., Peris E., Wang Y., Tamae Kakazu M., Khalyfa A., Carreras A., and Gozal D. (2014) Lipopolysaccharide-binding protein plasma levels in children: effects of obstructive sleep apnea and obesity. J. Clin. Endocrinol. Metab. 99, 656–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Goossens G. H., Blaak E. E., Theunissen R., Duijvestijn A. M., Clément K., Tervaert J. W., and Thewissen M. M. (2012) Expression of NLRP3 inflammasome and T cell population markers in adipose tissue are associated with insulin resistance and impaired glucose metabolism in humans. Mol. Immunol. 50, 142–149 [DOI] [PubMed] [Google Scholar]
- 7. Stienstra R., van Diepen J. A., Tack C. J., Zaki M. H., van de Veerdonk F. L., Perera D., Neale G. A., Hooiveld G. J., Hijmans A., Vroegrijk I., van den Berg S., Romijn J., Rensen P. C., Joosten L. A., Netea M. G., and Kanneganti T. D. (2011) Inflammasome is a central player in the induction of obesity and insulin resistance. Proc. Natl. Acad. Sci. U.S.A. 108, 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bauernfeind F., Bartok E., Rieger A., Franchi L., Núñez G., and Hornung V. (2011) Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol. 187, 613–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Won J. H., Park S., Hong S., Son S., and Yu J. W. (2015) Rotenone-induced impairment of mitochondrial electron transport chain confers a selective priming signal for NLRP3 inflammasome activation. J. Biol. Chem. 290, 27425–27437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Iyer S. S., He Q., Janczy J. R., Elliott E. I., Zhong Z., Olivier A. K., Sadler J. J., Knepper-Adrian V., Han R., Qiao L., Eisenbarth S. C., Nauseef W. M., Cassel S. L., and Sutterwala F. S. (2013) Mitochondrial cardiolipin is required for Nlrp3 inflammasome activation. Immunity 39, 311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., and Choi A. M. (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. West A. P., Koblansky A. A., and Ghosh S. (2006) Recognition and signaling by Toll-like receptors. Annu. Rev. Cell Dev. Biol. 22, 409–437 [DOI] [PubMed] [Google Scholar]
- 13. Subramanian N., Natarajan K., Clatworthy M. R., Wang Z., and Germain R. N. (2013) The adaptor MAVS promotes NLRP3 mitochondrial localization and inflammasome activation. Cell 153, 348–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Misawa T., Takahama M., Kozaki T., Lee H., Zou J., Saitoh T., and Akira S. (2013) Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14, 454–460 [DOI] [PubMed] [Google Scholar]
- 15. Zhou R., Yazdi A. S., Menu P., and Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 16. Shimada K., Crother T. R., Karlin J., Dagvadorj J., Chiba N., Chen S., Ramanujan V. K., Wolf A. J., Vergnes L., Ojcius D. M., Rentsendorj A., Vargas M., Guerrero C., Wang Y., Fitzgerald K. A., et al. (2012) Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity 36, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henao-Mejia J., Elinav E., Strowig T., and Flavell R. A. (2012) Inflammasomes: far beyond inflammation. Nat. Immunol. 13, 321–324 [DOI] [PubMed] [Google Scholar]
- 18. Strowig T., Henao-Mejia J., Elinav E., and Flavell R. (2012) Inflammasomes in health and disease. Nature 481, 278–286 [DOI] [PubMed] [Google Scholar]
- 19. Fontana L., Meyer T. E., Klein S., and Holloszy J. O. (2004) Long-term calorie restriction is highly effective in reducing the risk for atherosclerosis in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 6659–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brandhorst S., Choi I. Y., Wei M., Cheng C. W., Sedrakyan S., Navarrete G., Dubeau L., Yap L. P., Park R., Vinciguerra M., Di Biase S., Mirzaei H., Mirisola M. G., Childress P., Ji L., et al. (2015) A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 22, 86–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nisoli E., Tonello C., Cardile A., Cozzi V., Bracale R., Tedesco L., Falcone S., Valerio A., Cantoni O., Clementi E., Moncada S., and Carruba M. O. (2005) Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 310, 314–317 [DOI] [PubMed] [Google Scholar]
- 22. Civitarese A. E., Carling S., Heilbronn L. K., Hulver M. H., Ukropcova B., Deutsch W. A., Smith S. R., Ravussin E., and CALERIE Pennington Team (2007) Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 4, e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bevilacqua L., Ramsey J. J., Hagopian K., Weindruch R., and Harper M. E. (2004) Effects of short- and medium-term calorie restriction on muscle mitochondrial proton leak and reactive oxygen species production. Am. J. Physiol. Endocrinol. Metab. 286, E852–E861 [DOI] [PubMed] [Google Scholar]
- 24. Kume S., Uzu T., Horiike K., Chin-Kanasaki M., Isshiki K., Araki S., Sugimoto T., Haneda M., Kashiwagi A., and Koya D. (2010) Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J. Clin. Invest. 120, 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sack M. N., and Finkel T. (2012) Mitochondrial metabolism, sirtuins, and aging. Cold Spring Harb. Perspect. Biol. 4, a013102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wood J. G., Rogina B., Lavu S., Howitz K., Helfand S. L., Tatar M., and Sinclair D. (2004) Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430, 686–689 [DOI] [PubMed] [Google Scholar]
- 27. Guarente L. (2008) Mitochondria: a nexus for aging, calorie restriction, and sirtuins? Cell 132, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., Stevens R. D., Li Y., Saha A. K., Ruderman N. B., Bain J. R., Newgard C. B., et al. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464, 121–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lu Z., Chen Y., Aponte A. M., Battaglia V., Gucek M., and Sack M. N. (2015) Prolonged fasting identifies heat shock protein 10 as a Sirtuin 3 substrate: elucidating a new mechanism linking mitochondrial protein acetylation to fatty acid oxidation enzyme folding and function. J. Biol. Chem. 290, 2466–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Traba J., Kwarteng-Siaw M., Okoli T. C., Li J., Huffstutler R. D., Bray A., Waclawiw M. A., Han K., Pelletier M., Sauve A. A., Siegel R. M., and Sack M. N. (2015) Fasting and refeeding differentially regulate NLRP3 inflammasome activation in human subjects. J. Clin. Invest. 125, 4592–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tschopp J., and Schroder K. (2010) NLRP3 inflammasome activation: the convergence of multiple signalling pathways on ROS production? Nat. Rev. Immunol. 10, 210–215 [DOI] [PubMed] [Google Scholar]
- 32. Bronner D. N., Abuaita B. H., Chen X., Fitzgerald K. A., Nuñez G., He Y., Yin X. M., and O'Riordan M. X. (2015) Endoplasmic reticulum stress activates the inflammasome via NLRP3- and caspase-2-driven mitochondrial damage. Immunity 43, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. West A. P., Khoury-Hanold W., Staron M., Tal M. C., Pineda C. M., Lang S. M., Bestwick M., Duguay B. A., Raimundo N., MacDuff D. A., Kaech S. M., Smiley J. R., Means R. E., Iwasaki A., and Shadel G. S. (2015) Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Harvie M. N., Pegington M., Mattson M. P., Frystyk J., Dillon B., Evans G., Cuzick J., Jebb S. A., Martin B., Cutler R. G., Son T. G., Maudsley S., Carlson O. D., Egan J. M., Flyvbjerg A., and Howell A. (2011) The effects of intermittent or continuous energy restriction on weight loss and metabolic disease risk markers: a randomized trial in young overweight women. Int. J. Obes. (Lond.) 35, 714–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson J. B., Summer W., Cutler R. G., Martin B., Hyun D. H., Dixit V. D., Pearson M., Nassar M., Telljohann R., Tellejohan R., Maudsley S., Carlson O., John S., Laub D. R., and Mattson M. P. (2007) Alternate day calorie restriction improves clinical findings and reduces markers of oxidative stress and inflammation in overweight adults with moderate asthma. Free Radic. Biol. Med. 42, 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lu Z., Bourdi M., Li J. H., Aponte A. M., Chen Y., Lombard D. B., Gucek M., Pohl L. R., and Sack M. N. (2011) SIRT3-dependent deacetylation exacerbates acetaminophen hepatotoxicity. EMBO Rep. 12, 840–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Webster B. R., Lu Z., Sack M. N., and Scott I. (2012) The role of sirtuins in modulating redox stressors. Free Radic. Biol. Med. 52, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang J. Z., Liu Z., Liu J., Ren J. X., and Sun T. S. (2014) Mitochondrial DNA induces inflammation and increases TLR9/NF-κB expression in lung tissue. Int. J. Mol. Med. 33, 817–824 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. van der Burgh R., Nijhuis L., Pervolaraki K., Compeer E. B., Jongeneel L. H., van Gijn M., Coffer P. J., Murphy M. P., Mastroberardino P. G., Frenkel J., and Boes M. (2014) Defects in mitochondrial clearance predispose human monocytes to interleukin-1β hypersecretion. J. Biol. Chem. 289, 5000–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Webster B. R., Scott I., Han K., Li J. H., Lu Z., Stevens M. V., Malide D., Chen Y., Samsel L., Connelly P. S., Daniels M. P., McCoy J. P. Jr., Combs C. A., Gucek M., and Sack M. N. (2013) Restricted mitochondrial protein acetylation initiates mitochondrial autophagy. J. Cell Sci. 126, 4843–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tseng A. H., Shieh S. S., and Wang D. L. (2013) SIRT3 deacetylates FOXO3 to protect mitochondria against oxidative damage. Free Radic. Biol. Med. 63, 222–234 [DOI] [PubMed] [Google Scholar]
- 42. Qiao A., Wang K., Yuan Y., Guan Y., Ren X., Li L., Chen X., Li F., Chen A. F., Zhou J., Yang J. M., and Cheng Y. (2016) Sirt3-mediated mitophagy protects tumor cells against apoptosis under hypoxia. Oncotarget 7, 43390–43400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rongvaux A., Jackson R., Harman C. C., Li T., West A. P., de Zoete M. R., Wu Y., Yordy B., Lakhani S. A., Kuan C. Y., Taniguchi T., Shadel G. S., Chen Z. J., Iwasaki A., and Flavell R. A. (2014) Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159, 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y., Ning X., Gao P., Wu S., Sha M., Lv M., Zhou X., Gao J., Fang R., Meng G., Su X., and Jiang Z. (2017) Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46, 393–404 [DOI] [PubMed] [Google Scholar]
- 45. Tao R., Coleman M. C., Pennington J. D., Ozden O., Park S. H., Jiang H., Kim H. S., Flynn C. R., Hill S., Hayes McDonald W., Olivier A. K., Spitz D. R., and Gius D. (2010) Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Mol. Cell 40, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tao R., Vassilopoulos A., Parisiadou L., Yan Y., and Gius D. (2014) Regulation of MnSOD enzymatic activity by Sirt3 connects the mitochondrial acetylome signaling networks to aging and carcinogenesis. Antioxid. Redox Signal. 20, 1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lu J., Cheng K., Zhang B., Xu H., Cao Y., Guo F., Feng X., and Xia Q. (2015) Novel mechanisms for superoxide-scavenging activity of human manganese superoxide dismutase determined by the K68 key acetylation site. Free Radic. Biol. Med. 85, 114–126 [DOI] [PubMed] [Google Scholar]
- 48. Youm Y. H., Nguyen K. Y., Grant R. W., Goldberg E. L., Bodogai M., Kim D., D'Agostino D., Planavsky N., Lupfer C., Kanneganti T. D., Kang S., Horvath T. L., Fahmy T. M., Crawford P. A., Biragyn A., et al. (2015) The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 21, 263–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schupp M., Chen F., Briggs E. R., Rao S., Pelzmann H. J., Pessentheiner A. R., Bogner-Strauss J. G., Lazar M. A., Baldwin D., and Prokesch A. (2013) Metabolite and transcriptome analysis during fasting suggest a role for the p53-Ddit4 axis in major metabolic tissues. BMC Genomics 14, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhao W. Y., Zhang L., Sui M. X., Zhu Y. H., and Zeng L. (2016) Protective effects of sirtuin 3 in a murine model of sepsis-induced acute kidney injury. Sci. Rep. 6, 33201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Park E. K., Jung H. S., Yang H. I., Yoo M. C., Kim C., and Kim K. S. (2007) Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 56, 45–50 [DOI] [PubMed] [Google Scholar]
- 52. Bulua A. C., Simon A., Maddipati R., Pelletier M., Park H., Kim K. Y., Sack M. N., Kastner D. L., and Siegel R. M. (2011) Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J. Exp. Med. 208, 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.