Abstract
Developmental angiogenesis and the maintenance of the blood–brain barrier involve endothelial cell adhesion, which is linked to cytoskeletal dynamics. GPR124 (also known as TEM5/ADGRA2) is an adhesion G protein–coupled receptor family member that plays a pivotal role in brain angiogenesis and in ensuring a tight blood–brain barrier. However, the signaling properties of GPR124 remain poorly defined. Here, we show that ectopic expression of GPR124 promotes cell adhesion, additive to extracellular matrix–dependent effect, coupled with filopodia and lamellipodia formation and an enrichment of a pool of the G protein–coupled receptor at actin-rich cellular protrusions containing VASP, a filopodial marker. Accordingly, GPR124-expressing cells also displayed increased activation of both Rac and Cdc42 GTPases. Mechanistically, we uncover novel direct interactions between endogenous GPR124 and the Rho guanine nucleotide exchange factors Elmo/Dock and intersectin (ITSN). Small fragments of either Elmo or ITSN1 that bind GPR124 blocked GPR124-induced cell adhesion. In addition, Gβγ interacts with the C-terminal tail of GPR124 and promotes the formation of a GPR124–Elmo complex. Furthermore, GPR124 also promotes the activation of the Elmo–Dock complex, as measured by Elmo phosphorylation on a conserved C-terminal tyrosine residue. Interestingly, Elmo and ITSN1 also interact with each other independently of their GPR124-recognition regions. Moreover, endogenous phospho-Elmo and ITSN1 co-localize with GPR124 at lamellipodia of adhering endothelial cells, where GPR124 expression contributes to polarity acquisition during wound healing. Collectively, our results indicate that GPR124 promotes cell adhesion via Elmo–Dock and ITSN. This constitutes a previously unrecognized complex formed of atypical and conventional Rho guanine nucleotide exchange factors for Rac and Cdc42 that is putatively involved in GPR124-dependent angiogenic responses.
Keywords: adhesion, cell signaling, G protein-coupled receptor (GPCR), guanine nucleotide exchange factor (GEF), signal transduction
Introduction
Essential functions of blood vessels in the brain, including the formation of a tight blood–brain barrier, are established during developmental angiogenesis. They involve coordinated actions of multiple receptors and signaling systems (1). Among them, recent evidence supports a fundamental role of GPR124 (G-protein–coupled receptor 124) in brain angiogenesis and in proper establishment of the blood–brain barrier. Fetal GPR124-knock-out mice show limited remodeling of their brain vasculature and present leakiness of cerebral blood capillaries, leading to deadly intracranial hemorrhages and formation of glomeruloid vascular structures (2–4). In such developmental process, GPR124 functions as a Wnt7a/b coreceptor, providing specificity to Frizzled4-dependent gene expression signaling pathways controlled by β-catenin (5, 6), as well as a promoter of endothelial cell remodeling via yet to be identified signaling pathways.
GPR124 was first identified as tumor endothelial marker 5 (TEM5, recently renamed as ADGRA2) detected as an up-regulated transcript in tumor endothelial capillaries from human colorectal cancers (7). This adhesion G protein–coupled receptor (ADGR)5 family member has been implicated in VEGF-dependent (8) and -independent (2) cell migration events, indicating its stimulatory effect on actin cytoskeleton remodeling signaling pathways. In this regard, interfering with the activity of Cdc42 (cell division cycle 42; a member of the Rho family of small GTPases, particularly linked to filopodia formation) decreases GPR124-dependent directional sprouting (2). However, it has not been directly addressed whether GPR124 does stimulate Cdc42 and what are the molecular effectors linking to this and perhaps other RhoGTPases, putatively involved in the morphological adjustments occurring in endothelial cells during angiogenic events.
Two unrelated groups of guanine nucleotide exchange factors (RhoGEFs) regulate the activity of Rho proteins: the Dbl homology (DH)- and dedicator of cytokinesis (Dock)-RhoGEFs, constituted by 70 and 11 members per family, respectively (9–11). Members of these two classes of RhoGEFs are reported to act downstream of some ADGRs (12–15). In the case of Dock proteins, they are atypical GEFs that form functional complexes with Elmo (engulfment and cell motility), which interacts with ADGRs, such as BAI1 and BAI3, promoting Rac activation (13, 14). The Elmo–Dock system also participates in chemotactic responses elicited by conventional GPCRs, such as CXCR4 (C-X-C chemokine receptor type 4) (16, 17), which also activate G protein–regulated RhoGEFs (18, 19). Thus, the C-terminal regions of GPCRs as well as an intermediary role of heterotrimeric G protein subunits, such as Gα12/13, Gαq, or Gβγ, have been mechanistically linked to the regulation of multiple RhoGEFs (20–23).
Intersectin 1/2 (ITSN1/2), DH-domain RhoGEFs, stand out as a particularly multifaceted group of signaling scaffolds. Both ITSNs are expressed in endothelial cells (10), where they contribute to maintain intercellular adhesions (24). They are best known as participants in the endocytic machinery (25), which affects GPCR signaling and trafficking (26). Recently, ITSN1 and ITSN2 have been identified as components of breast cancer cell invadopodia, where actin polymerization is highly dynamic (27). Structurally, ITSN1/2 are highly complex Cdc42-specific RhoGEFs composed by multiple domains, including two EH (Eps15 homology) one CC (coiled-coil region), five SH3 (Src homology 3), one DH-PH module (composed of a DH domain followed by a pleckstrin homology (PH) domain, characteristic of the group of DH-domain RhoGEFs), and a C2 C-terminal domain (25). In the present study, we explored the signaling mechanisms by which GPR124 regulates cell adhesion and found that two distinct families of RhoGEFs, Elmo–Dock180 and ITSN1/2, form a complex together and also directly interact with GPR124, forming a complex involved in GPR124 signaling pathways during cell adhesion.
Results
GPR124 promotes cell adhesion, polarity, filopodia formation, and activation of Cdc42 and Rac1 (Ras-related C3 botulinum toxin substrate 1) GTPases
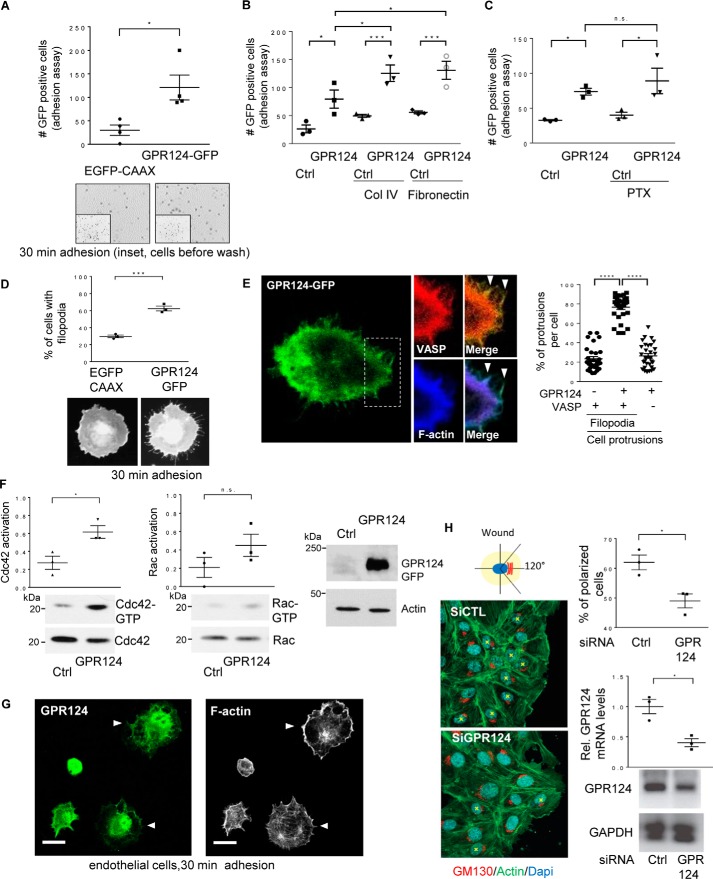
Accumulating evidence highlights the importance of GPR124 in developmental angiogenesis as well as tumor-induced angiogenesis, processes where this orphan adhesion GPCR modulates endothelial permeability and cell migration, indicating its contribution to adhesive properties and shape adjustments of endothelial cells (2–4, 7, 8, 28). To address the molecular mechanisms by which GPR124 controls cytoskeletal dynamics and cell adhesion, we hypothesized that this orphan receptor recruits RhoGEFs. First, we used a gain-of-function approach, in COS7 cells lacking GPR124 expression, to investigate whether ectopic expression of EGFP-tagged GPR124 modulates cell adhesion compared with EGFP–CAAX–expressing control cells. As shown in Fig. 1A, the number of EGFP-positive adherent COS7 cells was significantly higher in the group expressing GPR124. Additionally, GPR124 had an additive effect that increased the number of adherent cells plated on collagen IV and fibronectin substrates (Fig. 1B). Besides, it was insensitive to pertussis toxin (Fig. 1C, PTX), indicating that GPR124 promotes cell adhesion via a Gi-independent pathway. We then monitored the morphology of the adherent cells and found that GPR124 expression promoted filopodia formation (Fig. 1D). Moreover, by confocal microscopy, we detected a pool of GPR124 that localized at actin-rich protrusions also containing VASP, a filopodial marker (29) (Fig. 1E). Because filopodia are known to be formed as a consequence of the activation of Cdc42 (30, 31), we assayed whether this morphological effect of GPR124 expression correlated with the activation of the Rho GTPases Rac1 and Cdc42. Using a pulldown assay to capture GTP-bound Cdc42 and Rac, based on their affinity for the CRIB region of PAK fused to GST, we revealed that GPR124 expression led to a significant activation of Cdc42 in cells left to adhere for 30 min (Fig. 1F, left). In the case of Rac, we detected a consistent increase on its activation in GPR124-expressing cells; however, the difference was not statistically significant when compared with control cells expressing EGFP–CAAX (Fig. 1F, center). The expression of GPR124 in total cell lysates of these cells was confirmed by Western blotting (Fig. 1F, right). To investigate whether these effects of GPR124 were conserved in endothelial cells, we first investigated the localization of endogenous GPR124 in human umbilical vein endothelial cells (HUVECs) left to adhere for 30 min and its potential co-localization with filamentous actin. As shown in Fig. 1G (left), we detected a pool of endogenous GPR124 localized at extensions of adhering cells by confocal microscopy. Consistent with a role of GPR124 in cell adhesion and cytoskeletal dynamics, polymerized actin was also prominent in endothelial cell projections observed in adhering cells (Fig. 1G, right). Next, to determine the potential role of GPR124 in endothelial cell polarity acquired during migration, we performed wound-healing assays in control and GPR124-knockdown HUVECs. The perinuclear localization of the Golgi apparatus, oriented toward the wound within the area limited by a 120º angle, was considered as a positive indicator of polarized morphology (Fig. 1H, left; asterisk used to mark polarized cells). Interestingly, in GPR124 knockdown cells, we found a significant decrease in the number of polarized cells at the edge of the wound (Fig. 1H, right, top graph). Consequently, we next aimed to investigate the molecular mechanisms underlying GPR124-dependent cell adhesion.
Figure 1.
GPR124 promotes cell adhesion, polarity, filopodia formation, and activation of Cdc42 and Rac1 GTPases. A, GPR124-dependent cell adhesion. Adhesion assays were performed using serum-starved COS7 cells expressing GFP-tagged GPR124 or EGFP–CAAX, used as control. Cells were fixed with 4% PFA, and images were taken with a Nikon Eclipse Ti inverted microscope and analyzed with NIS-Elements software. The graph shows the mean ± S.E. (error bars) of adherent GFP-positive cells for each condition (Student's t test; *, p < 0.05; n = 4). Representative fields below the graph show adherent cells at 30 min, and the inset shows all EGFP-positive cells before non-adherent cells were washed away. B, GPR124 shows an additive effect on cell adhesion to collagen IV and fibronectin. COS7 cells transfected with EGFP–CAAX (control plasmid (Ctrl)) or GPR124–GFP were plated on plastic, collagen IV, or fibronectin for 30 min. Adherent cells were analyzed using ImageJ software. The graph shows the mean ± S.E. of adherent GFP-positive cells (*, p < 0.05; ***, p < 0.001). Statistics were performed using one-way ANOVA followed by Tukey's multiple-comparison post hoc test (n = 3). C, inhibition of Gi heterotrimeric proteins does not alter GPR124-dependent cell adhesion. Control (Ctrl; EGFP–CAAX) or GPR124–GFP–transfected COS7 cells were treated overnight and during 30 min adhesion assays with vehicle or pertussis toxin (PTX; 200 ng/μl). Fixed adherent cells were analyzed using ImageJ software. The graph shows the mean ± S.E. of GFP-positive cells and was analyzed by one-way ANOVA followed by Tukey's multiple-comparison post hoc test (*, p < 0.05; n = 3). D, GPR124 induces filopodia formation. The morphology of COS7 cells expressing GPR124–GFP was monitored in cells left adhering for 30 min and compared with control COS7 cells expressing EGFP–CAAX. Statistics were performed by Student's t test (***, p < 0.0005; n = 3). Representative cells are shown at the bottom of the graph. E, GPR124 localizes at VASP-positive filopodia in adhering cells. COS7 cells transfected with GPR124–GFP and mCherry–VASP, a filopodial marker, were subjected to adhere for 30 min, fixed, stained for F-actin, and analyzed by confocal microscopy. About 75% of VASP-positive filopodia protrusions per cell were also positive for GPR124–GFP (graph in the right panel). A representative cell is shown at the left. Cellular protrusions positive for either GPR124 and VASP or GPR124 and F-actin are indicated by arrowheads in the merged images. At least 30 cells were analyzed by confocal Leica TCS SP8 using a ×100 objective. The graph shows the mean ± S.E. (n = 3). One-way ANOVA followed by Tukey's multiple-comparison post hoc test was performed for statistics (****, p < 0.0001). F, GPR124 promotes activation of Cdc42 and Rac GTPases. Serum-starved cells expressing either GPR124–GFP or control EGFP–CAAX cells were lysed and incubated with PAK-N beads to capture active Cdc42 and Rac1. Cdc42-GTP and Rac1-GTP were identified by Western blotting. Cdc42-GTP was increased in COS7 cells expressing GPR124. The graph shows the mean ± S.E. of normalized Cdc42 and Rac activation (Student's t test; *, p < 0.05; n = 3). G, GPR124 is localized at cell projections of adhering endothelial cells. Endogenous GPR124 (top) was localized with filamentous actin (bottom) at cell projections of HUVECs (white arrowheads) left to adhere for 30 min (similar results were observed in 36 of 60 cells). Bar, 20 μm. H, GPR124 is required to polarize HUVECs. HUVECs with siRNA knockdown of GPR124 (siGPR124) were less polarized at the edge of the wound than siRNA control (siCTL). Nuclei were stained with DAPI (blue), and filamentous actin was visualized with green fluorescent phalloidin. Cells in which GM130 (a Golgi marker; red) did not localize within a 120° angle facing the wound were considered as non-polarized cells. At least 100 cells near the wound edge were randomly examined. The percentage of polarized cells is shown in the top graph (right). Data are represented as mean ± S.E. (Student's t test; *, p < 0.05; n = 3). GPR124 knockdown was confirmed by quantitative RT-PCR (right, bottom graph).
GPR124 interacts with Gβγ and Elmo–Dock, promoting the activation of this atypical RhoGEF
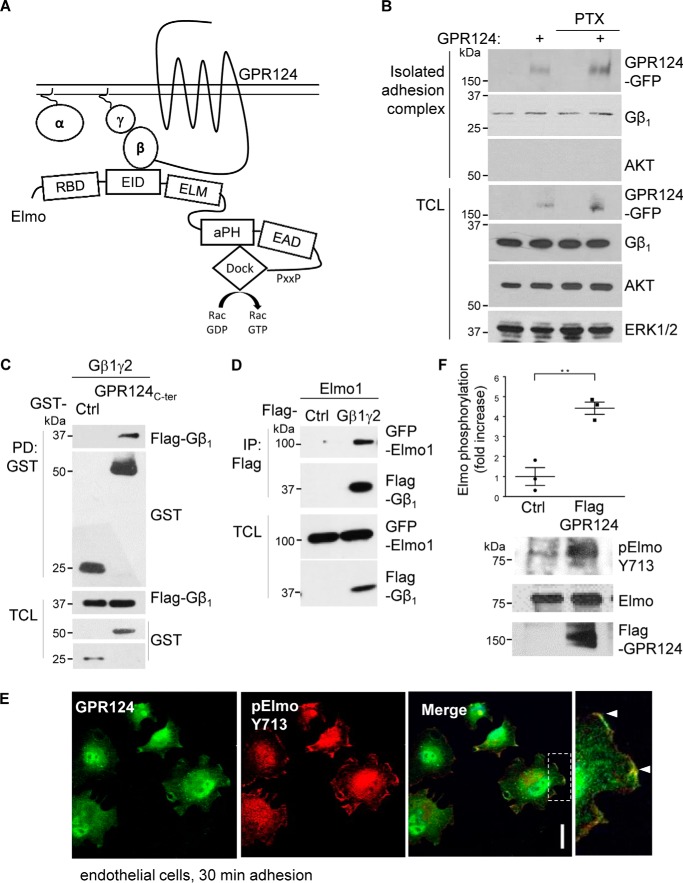
The signaling mechanisms by which most ADGRs promote cell adhesion and migration are poorly understood. Emerging evidence points to a paramount role of their C-terminal tail connecting to molecular scaffolds and guanine nucleotide exchange factors of the Elmo–Dock family, as well as heterotrimeric G protein–dependent pathways linking to the activation of Rho GTPases (32, 33). Considering the fundamental roles played by Gβγ as a GPCR transducer directly activating guanine nucleotide factors for Rho GTPases, we hypothesized that GPR124 recruits this heterodimeric transducer and the Elmo–Dock complex to activate it (Fig. 2A).
Figure 2.
GPR124 interacts with Gβγ and Elmo promoting its activation. A, schematic model showing that GPR124 recruits Elmo via Gβγ and direct interactions. B, detection of GPR124 and Gβ1γ2 in isolated cell adhesion complexes. Control and GPR124-expressing COS7 cells left to adhere for 30 min were lysed, and proteins at adhesion complexes that remained attached to the plates were washed and recovered with Laemmli sample buffer. FLAG–GPR124–GFP and Gβγ were detected by Western blotting in the isolated adhesion complexes as well as in total cell lysates (TCL). AKT was used as a cytosolic marker (absent in the isolated adhesion complex) and, together with ERK, served as loading controls in total cell lysates. C, GPR124 C-terminal tail interacts with Gβ1γ2 subunits. HEK293T cells were co-transfected with GST–GPR124 C-terminal tail (G124Ct) and FLAG–Gβ1γ2, and the interaction between GPR124 C terminus and Gβ1γ2 was detected by GST pulldown assays. D, Elmo interacts with Gβ1γ2. HEK293T cells were co-transfected with GFP–Elmo and FLAG–Gβγ or pCEFL-vector as control. The interaction between Elmo and Gβ1γ2 was confirmed by coimmunoprecipitation. E, phospho-Elmo is localized with GPR124 at cell projections during endothelial cell adhesion. Localization of phospho-Elmo and GPR124 was visualized by confocal microscopy in HUVECs left to adhere for 30 min, using specific antibodies. Phospho-Elmo (Tyr-713) and endogenous GPR124 were enriched at cell protrusions (white arrowheads) of adhering cells. Similar results were observed in 95 of 142 cells. Co-localization was confirmed by assessing three different points in cell protrusions of three independent experiments. Scale bar, 20 μm. F, GPR124 promotes phosphorylation of Elmo at Tyr-713. Phospho-active Elmo (ElmoY713) was found in GPR124-expressing HEK293T cells compared with control HEK293T cells transfected with pCEFL vector (Ctl). The graph shows the mean ± S.E. (error bars) of Elmo phosphorylation (Student's t test; **, p < 0.01; n = 3).
Based on the demonstrated effect of GPR124 promoting cell adhesion, we predicted that this receptor might remain as a component of an isolated adhesion complex in which its signaling effectors might also be detected. To start addressing this possibility, COS7 cells adhering for 30 min were lysed, and then adhesion complexes were washed, and proteins that remained bound to the plate were recovered with Laemmli sample buffer. As predicted, FLAG–GPR124–GFP was detected in the isolated adhesion complex that also contained Gβγ (Fig. 2B, first and second blots). The absence of AKT, used as a cytosolic marker, indicated the lack of cytosolic contaminants in the isolated adhesion complex (Fig. 2B, third blot). The presence of Gβ1, AKT, and ERK in total cell lysates served as loading controls, whereas the presence of transfected GPR124 was confirmed with antibodies against GFP (Fig. 2B, bottom, TCL). Consistent with the lack of effect of pertussis toxin on GPR124-dependent cell adhesion, GPR124 was detected in the isolated adhesion complex irrespective of the treatment with this toxin (Fig. 2B, top). Interestingly, we found a novel interaction between the GPR124 C-terminal tail and Gβ1γ2 (Fig. 2C). Then we explored whether the Gβγ heterodimer, identified as a putative transducer of GPR124, could directly interact with Elmo. A recent report described that stromal cell–derived factor 1 (SDF-1) induces the interaction between Gβγ and Elmo by the chemotactic CXCR4 GPCR in HeLa cells (16). We confirmed that Gβ1γ2 interacts with Elmo1 in serum-starved HEK293T cells (Fig. 2D). This interaction was reduced when Gαi subunit was co-expressed (data not shown), raising the possibility that Gβ1γ2 recognizes Elmo1 as an effector. Then we examined the cellular localization of endogenously activated Elmo–Dock180 in endothelial cells and whether it co-localized with endogenous GPR124 during this process. Using confocal microscopy to address this, we found that Elmo phosphorylated at Tyr-713, which is indicative of its active state (34), co-localized with GPR124 at cell projections (Fig. 2E). We next confirmed these cellular data by demonstrating that GPR124 overexpression is sufficient to promote Elmo phosphorylation at Tyr-713 by Western blotting (Fig. 2F). These data suggest that GPR124 interacts with Elmo–Dock180 to promote its activity.
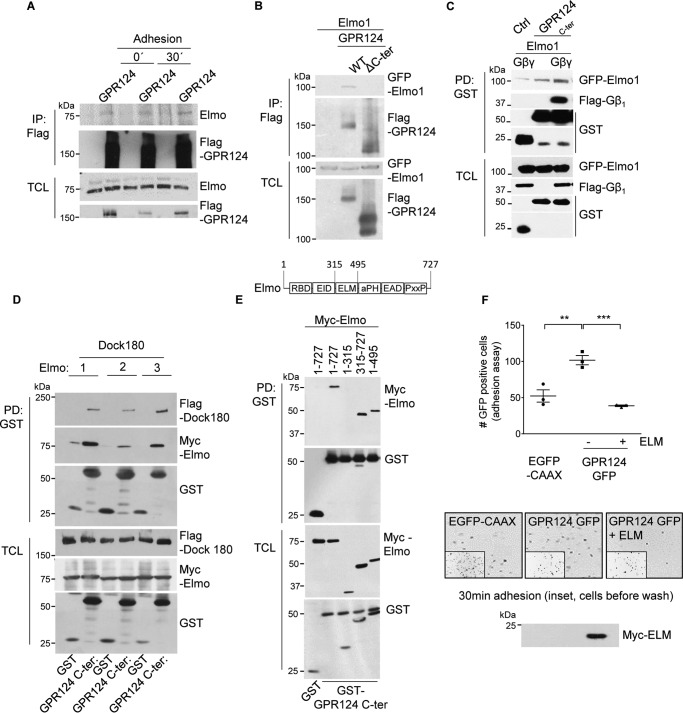
GPR124 C-terminal tail interacts with Elmo–Dock
Considering that BAI1 and BAI3, ADGRs homologous to GPR124, function in part via their interaction with Elmo–Dock180 (13, 14), we explored whether GPR124 interacts directly with Elmo. First, full-length FLAG–GPR124 expressed in HEK293T cells was immunoprecipitated, and endogenous Elmo was revealed as an interactor in basal conditions, as well as during cell adhesion (Fig. 3A). In addition, GPR124 lacking its C-terminal tail lost its ability to interact with Elmo (Fig. 3B). Because Gβ1γ2 heterodimer interacted with both GPR124 and Elmo, we assessed its effect on the interaction between GPR124 C-terminal tail and Elmo and found that this association was positively modulated by Gβ1γ2 (Fig. 3C). Additionally, the GPR124 C-terminal tail was able to associate with the three variants of Elmo in complex with Dock180 (Fig. 3D). We characterized this interaction using truncated forms of Elmo to map the region recognized by the GPR124 C-terminal tail. Pulldown assays revealed that the central region of Elmo contains the minimal binding site required to associate with the C-terminal tail of GPR124, which is different from the N-terminal region of Elmo binding to Gβγ (16) (Fig. 3E). This minimal region, corresponding to the ELM domain, was tagged with Myc and used to test its effect on GPR124-dependent adhesion of transfected COS7 cells. Consistent with a role of Elmo in GPR124-dependent cell adhesion, this module blocked the effect of this receptor on cell adhesion, suggesting a competition with endogenous Elmo (Fig. 3F). Collectively, these results demonstrate a direct interaction between Elmo, via its ELM domain, and GPR124 that is essential for cell adhesion.
Figure 3.
GPR124 C-terminal tail interacts with Elmo–Dock. A, interaction between GPR124 and endogenous Elmo in basal conditions and during cell attachment. FLAG-tagged GPR124 was immunoprecipitated from total cell lysates of HEK293T cells in basal and cell adhesion conditions, and endogenous Elmo associated with GPR124 was detected by Western blotting. B, the GPR124 C terminus is required to bind Elmo. Cell lysates from HEK293T cells expressing GFP–Elmo1 and either FLAG–GPR124 (WT) or FLAG–GPR124 lacking its C terminus (ΔC-ter) were subjected to immunoprecipitation with FLAG antibody. GFP–Elmo1 was able to interact with FLAG–GPR124 WT but not with the FLAG–GPR124 truncated version (ΔC-ter). C, association between Elmo and the GPR124 C-terminal region is increased in the presence of Gβ1γ2. HEK293T cells were co-transfected with the GST–GPR124 C-terminal construct (GPR124C-ter) and GFP–Elmo1 in the presence or absence of FLAG–Gβ1γ2. GST–vector was used as control (Ctrl). GST pulldown assays were performed, and the effect of Gβγ on the interaction between GFP–Elmo and GST–GPR124 C terminus was analyzed by Western blotting. D, GPR124 C terminus interacts with three variants of Elmo. GST pulldown assays were performed with cell lysates from HEK293T cells expressing FLAG–Dock180 and GST–C terminus of GPR124 (GPR124C-ter) together with each variant of Elmo tagged with the Myc epitope. All three variants of Elmo, in complex with Dock180, were detected in association with GST–GPR124 C terminus by Western blotting. E, GPR124 interacts with the central region of Elmo. Different Myc-tagged fragments of Elmo1 and GST–GPR124 C terminus (GST–GPR124C-ter) were co-transfected in HEK293T cells. GST pulldown experiments were performed, and full-length Elmo1 and the Myc-Elmo1 fragments interacting with GST–GPR124C-ter were detected by Western blotting (top). The fragments of Elmo1 containing the ELM domain interacted with the GPR124 C terminus, whereas Elmo1(1–315) did not. F, Elmo1-ELM domain negatively regulates cell adhesion promoted by GPR124. GPR124-dependent cell adhesion was reduced in COS7 cells expressing the central region of Elmo1 (ELM domain). Data are represented as mean ± S.E. (error bars) Statistics were performed by one-way ANOVA followed by Tukey's multiple-comparison post hoc test (**, p < 0.01; ***, p < 0.001; n = 3). Representative pictures showing adhering cells are shown at the bottom (the inset shows a field of fluorescent cells before washing out non-adherent cells).
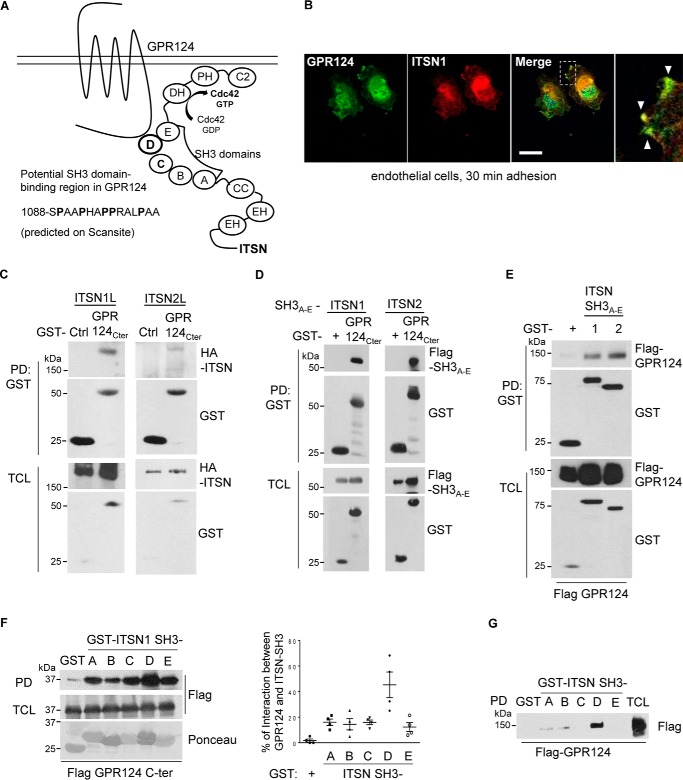
GPR124 interacts with intersectins via its C-terminal tail, which exhibits affinity for ITSN SH3 modules
Exploiting the Scansite 2.0 bioinformatic platform, we found that the GPR124 C-terminal tail contains a predicted ITSN1 interaction site with putative affinity for one of the SH3 domains of this Cdc42-specific RhoGEF (schematized in Fig. 4A). We therefore investigated by confocal immunofluorescence the localization of endogenous ITSN and GPR124 in HUVEC endothelial cells left to adhere for 30 min. Interestingly, we found that ITSN localizes with GPR124 at the extensions of adhering cells (Fig. 4B). To directly test the predicted interaction between GPR124 C-terminal tail and full-length ITSN, we transfected HEK293T cells and performed GST–GPR124 C-terminal tail pulldown assays. We found that the GPR124 C-terminal region interacted with both ITSN1 and ITSN2 (Fig. 4C). To confirm that the predicted SH3A–E module of ITSNs was involved in the interaction with GPR124, we generated constructs containing in tandem the five SH3 domains of ITSN1 and ITSN2. As predicted by in silico analysis, full-length GPR124 as well as the fragment corresponding to its C-terminal tail interacted with both ITSN1/2 SH3A–E modules (Fig. 4, D and E). Next, using individual recombinant SH3 domains of ITSN1, we explored the potential selectivity of the GPR124 C-terminal tail for any of them, finding that it associates with all of them (Fig. 4F). However, full-length GPR124 preferentially interacted with the ITSN1-SH3D domain (Fig. 4G). These results demonstrate a novel interaction between GPR124 and the Cdc42-specific RhoGEFs ITSN1/2.
Figure 4.
GPR124 interacts with intersectins via its C-terminal tail, which exhibits affinity for ITSN SH3 modules. A, schematic representation GPR124 in complex with ITSN. B, GPR124 localizes with ITSN at cell projections of adherent cells. Endogenous GPR124 and ITSN were visualized by immunostaining in HUVECs left to adhere for 30 min. Analysis by confocal microscopy showed that GPR124 localized with ITSN at the cell membrane during cell attachment (white arrowheads). Co-localization was measured, selecting three different areas at cell protrusions from three independent experiments. Similar results were observed in 65 of 124 cells. Scale bar, 20 μm. C, GPR124 interacts with ITSN1 and -2. HEK293T cells were co-transfected with GST–GPR124 C terminus (GPR124C-ter) and full-length HA–intersectin 1 (HA–ITSN1L) or 2 (HA–ITSN2L). GST pulldown assays were performed, and ITSN1 and ITSN2 associated with GPR124 were revealed by Western blotting. GST was used as a control (Ctrl). D, the GPR124 C terminus interacts with SH3 domains of ITSNs. HEK293T cells were co-transfected with SH3A–E domains of ITSN1 or ITSN2 and GST–GPR124 C terminus. GST pulldown assays were performed, and SH3A–E domains of both ITSNs were able to specifically associate with the GPR124 C terminus. GST–vector was used as a control (GST +). E, full-length GPR124 interacts with ITSN1/2-SH3A–E domains in HEK293T cells. Cells were transfected with FLAG-tagged full-length GPR124 and GST–ITSN1–SH3A–E or GST–ITSN2–SH3A–E. GST pulldown experiments were performed, and full-length GPR124 was detected associated with SH3A–E domains of both ITSNs. GST was used as a control. Expression of transfected proteins was confirmed in total cell lysates (TCL). F, the GPR124 C terminus directly interacts with all five SH3 domains of ITSN1. HEK293T cells were transfected with FLAG–GPR124 C terminus. GST pull-down assays were performed using individual GST–recombinant SH3 domains (A–E; shown at the bottom, stained with Ponceau) of ITSN1 or GST–vector, used as control. All ITSN1-SH3 domains interacted with the GPR124 C terminus, the ITSN1-SH3D domain being a more effective interactor. G, full-length GPR124 preferentially interacts with ITSN1-SH3D domain. Cells were transfected with full-length FLAG–GPR124. GST pull down assays using the individual recombinant GST–ITSN1-SH3 domains (A–E) or GST–vector, used as control, showed that GPR124 preferentially interacts with ITSN1-SH3D. Error bars, S.E.
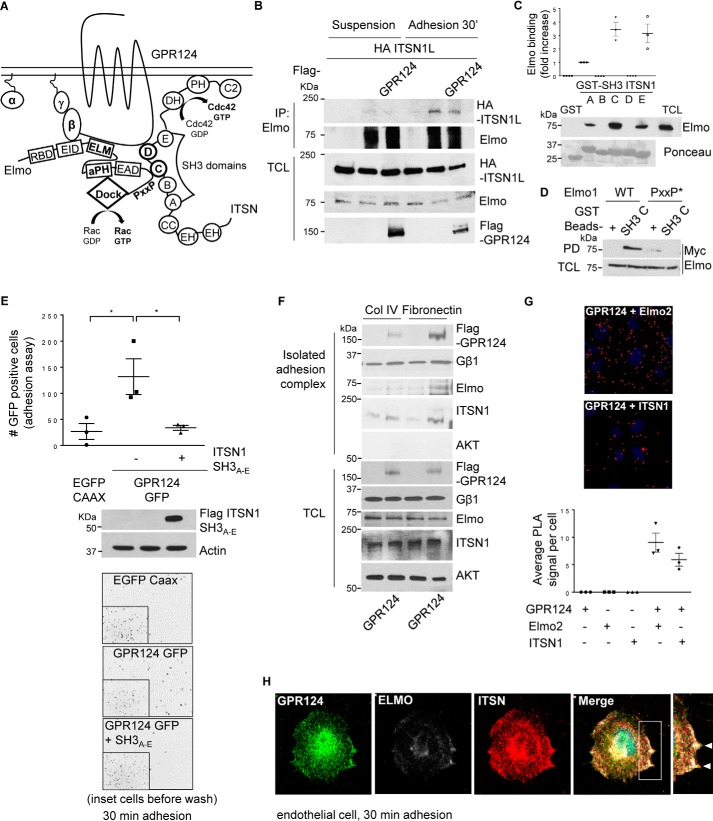
GPR124-dependent cell adhesion is mediated by its interaction with ITSN, which directly interacts with Elmo–Dock180, forming a signaling complex that co-localizes with GPR124
We next hypothesized that ITSN directly interacts with Elmo to integrate GPR124 signaling (Fig. 5A). By immunoprecipitating Elmo followed by immunoblotting against ITSN, we revealed the existence of a complex between these signaling proteins in adhering COS7 cells (Fig. 5B). To further characterize the interaction between Elmo and ITSN, we did pulldown assays with the individual recombinant SH3 domains of ITSN1 and found that ITSN1-SH3C and ITSN1-SH3E were the best Elmo interactors (Fig. 5C). In contrast, a mutant Elmo with a modified PXXP motif at its C terminus did not interact with ITSN1-SH3C (Fig. 5D). Because the ITSN–SH3A–E region plays a central role in the interaction with Elmo as well as GPR124, we investigated the effect of this ITSN1-SH3 module as a potential inhibitor of GPR124-dependent cell adhesion in COS7 cells. As predicted, the ITSN1-SH3A–E module inhibited the pro-adhesion effect of ectopically expressed GPR124 (Fig. 5E). Next, we investigated the presence of Elmo and ITSN in the adhesion complex isolated from adhering COS7 cells previously shown to contain GPR124 and Gβγ. Similar to the results shown in Fig. 2B, FLAG–GPR124 and endogenous Gβγ were detected in the isolated adhesion complex, which also contained endogenous Elmo and ITSN (Fig. 5F). Furthermore, using the proximity ligation assay (PLA), we demonstrated that endogenous GPR124 interacts with endogenous Elmo2 as well as with endogenous ITSN1 in endothelial cells (HUVECs). The PLA signals were revealed as red dots per field, which were next normalized to the number of cells in the field (DAPI signal) to obtain the average PLA counts per cell (Fig. 5G). No background signal was detected when only GPR124, Elmo2, or ITSN1 antibodies were used (Fig. 5G, graph at the bottom). To visualize the localization of Elmo, ITSN, and GPR124 in endothelial cells left to adhere for 30 min, we used specific antibodies to detect these endogenous proteins. By immunostaining, we found that GPR124 localizes with ITSN and Elmo at the extensions of adhering cells (Fig. 5H). Altogether, these results are consistent with the idea that GPR124 promotes cell adhesion via direct interactions with a signaling complex composed of Elmo–Dock and ITSN.
Figure 5.
GPR124-dependent cell adhesion is mediated by its interaction with ITSN, which directly interacts with Elmo–Dock180, forming a signaling complex that co-localizes with GPR124. A, working model. GPR124 interacts with the Gβγ–Elmo–Dock complex as well as with ITSN1/2. These atypical and conventional guanine nucleotide exchange factors also directly interact with each other, constituting a novel signaling complex that mediates GPR124-dependent cell adhesion. B, Elmo interacts with ITSN during cell adhesion. COS7 cells expressing full-length HA–ITSN1 and FLAG–GPR124 (as indicated) were left in suspension or adhering for 30 min. Then endogenous Elmo was immunoprecipitated, and interacting HA-ITSN1 was revealed by Western blotting. Preimmune rabbit IgG was used as a negative control for immunoprecipitation (first lane for suspension and adhesion conditions). C, endogenous Elmo directly interacts with ITSN1-SH3A, ITSN1-SH3C, and ITSN1-SH3E domains. HEK293T cell lysates were incubated for 3 h at 4 °C with individual recombinant GST–ITSN1-SH3 domains (A–E). GST pulldown assays were performed as described under “Experimental procedures.” Endogenous Elmo specifically binds to SH3A, SH3C, and SH3E domains of ITSN1 (PD, Western blotting, anti-Elmo). Recombinant GST–ITSN-SH3 domains are shown stained with Ponceau. D, the proline-rich (PXXP) motif in Elmo is required to bind the ITSN1-SH3C domain. Cell lysates from HEK293T cells expressing either wild-type Myc–Elmo or PXXP* mutant Myc–Elmo were incubated for 3 h at 4 °C with the recombinant GST–ITSN1-SH3C domain, and the interacting Elmo was revealed by Western blotting with anti-Myc (top). The expression of Elmo in total cell lysates (TCL) is shown at the bottom. GST was used as a negative control in the pulldown assays. The interaction between Elmo and ITSN1-SH3C domain was lost when the proline-rich region of Elmo was mutated. E, GPR124-dependent cell adhesion is inhibited by the ITSN1-SH3A–E module. COS7 cells were transfected with GPR124–GFP either with or without the FLAG–ITSN1-SH3A–E module. Cell adhesion assays were performed for 30 min. GPR124-dependent cell adhesion was decreased in FLAG–ITSN1-SH3A–E module-expressing cells. Basal adhesion of EGFP–CAAX–expressing cells was used as a reference. Bars, mean ± S.E. (error bars). Statistics were performed by one-way ANOVA followed by Tukey's multiple-comparison post hoc test (*, p < 0.05; n = 3). The middle panel shows the expression of FLAG–ITSN1-SH3A–E module in total cell lysates, and actin was used as a loading control. Representative images showing adherent cells are shown at the bottom; insets show all fluorescent cells in the field before washing out non-adherent cells. F, GPR124, Gβγ, Elmo, and ITSN are detected in the isolated adhesion complex. Control and GPR124-transfected COS7 cells were left to adhere for 30 min on collagen IV or fibronectin-coated plates. Adherent cells were lysed, and proteins that remained attached to the plates were washed and recovered with Laemmli sample buffer. GPR124, Gβγ, Elmo, and ITSN were detected by Western blotting in the isolated adhesion complex as well as in total cell lysates. AKT was used to confirm that isolated adhesion complexes were not contaminated by nonspecifically bound cytosolic proteins and as a loading control in TCL. G, endogenous interaction between GPR124 and Elmo2 as well as between GPR124 and intersectin 1 in endothelial cells. Proximity ligation assays were performed in endothelial cells (HUVECs) using the indicated pairs of antibodies to detect endogenous GPR124 interacting with endogenous Elmo or with endogenous ITSN1. Individual antibodies were used alone as a control. The PLA signal per cell was quantified by ImageJ software. At least 30 cells were analyzed. The graph represents three independent experiments (mean ± S.E., n = 3). Representative pictures of PLA signals, depicted as red dots, are shown in the top and middle panels. Cell nuclei were stained with DAPI. H, endogenous GPR124 co-localizes with Elmo and ITSN at cell protrusions of adhering endothelial cells. Cell adhesion assays were performed on gelatin-coated glass coverslips for 30 min, followed by immunostaining to determine the localization of endogenous GPR124, Elmo, and ITSN proteins in HUVECs (white arrows). A representative cell observed by confocal microscopy is shown. Similar results were observed in 29 of 61 cells.
Discussion
GPR124 plays a critical role during developmental brain angiogenesis and the establishment of the blood–brain barrier (2–4). These processes are probably mediated by the role of this orphan ADGR as a Frizzled4 co-receptor specifying Wnt7a/b signaling to promote β-catenin–regulated gene expression (5, 6) and its ability to dynamically control cell morphology via undefined mechanisms. In vitro, GPR124 has been linked to cell sprouting, therefore suggesting that this ADGR engages RhoGEFs to activate Rac and Cdc42 (2, 35–38). Here, we tested the hypothesis, schematically represented in Fig. 5A, that GPR124 promotes cell adhesion via direct interactions with Elmo–Dock180 and ITSNs, RhoGEFs for Rac and Cdc42 GTPases, respectively (9, 25).
Because the identity of GPR124 agonist is unidentified, we used a gain-of-function approach to investigate whether its expression in cells that normally lack this ADGR stimulates Cdc42 and Rac members of the family of Rho GTPases and promotes cell adhesion via interactions with Elmo–Dock and ITSNs. Furthermore, we investigated whether these molecular events correlated with the localization of endogenously expressed GPR124, Elmo, and ITSN in endothelial cells during adhesion. We found that ectopically expressed GPR124 promotes cell adhesion with the concurrent activation of Cdc42 and Rac small GTPases. Furthermore, consistent with a previous report in Myc–GPR124–expressing HEK293T cells (4), this receptor increases filopodia formation. Compatible with its potential role regulating the actin cytoskeleton, GPR124 localizes to these cellular protrusions, known as prominent cellular readouts of Cdc42 activity (30, 31), identified as filopodia by the presence of VASP (29).
According to our results, cell adhesion promoted by GPR124 is additive to the effect of extracellular matrix proteins, such as collagen IV and fibronectin. Interestingly, a soluble N-terminal construct of GPR124, including an RGD motif, serves as substrate for endothelial cell adhesion putatively being recognized by integrins (39). These findings support an extended role of GPR124 signaling from its importance at cell–cell adhesions, relevant to maintain the blood–brain barrier, to a dynamic role during adhesion, critical for capillary remodeling, which is one of the processes with observed defects in GPR124-knock-out mice (4). Consistently, fibronectin and collagen IV are known as efficient adhesion substrates for the rat brain endothelial cell line (GPNT) as well as for primary rat brain endothelial cells. These adhesive glycoproteins are highly expressed after cerebral ischemia (40). In this pathological condition, GPR124 positively regulates the expression of laminin and collagen IV as well as Wnt signaling (41).
In adhering endothelial cells, endogenously expressed GPR124 co-localizes with polymerized actin at lamellipodia extending at the cell edges, indicative of Rac activity (42). Furthermore, knockdown of GPR124 decreased endothelial cell polarization induced by scratching the cell monolayer. These findings, and the reported requirement of Cdc42 in the process of orientated migration of GPR124-expressing bEND3 endothelial cells following a gradient of forebrain conditioned medium (2), prompted us to directly address the interaction of GPR124 with Elmo–Dock, known to be critical transducers of the BAI group of ADGR receptors (13, 14); the potential role of Gβγ, recently described as an Elmo interactor (16), and a significant transducer of GPCR signaling to Rac (23, 43); and the potential interaction with ITSNs, which constitute a family of Cdc42-specific RhoGEFs of the Dbl domain group linked to vesicle trafficking and cell polarity (44, 45). ITSNs were postulated as GPR124 interactors because the ITSN1-SH3 module, in particular, was identified by bioinformatics as a potential interactor of the GPR124 C-terminal tail. Consistent with these possibilities, we found the existence of a signaling complex between Elmo–Dock180 and ITSNs, representing, to the best of our knowledge, a completely new concept in terms of signaling integration in which two distinct classes of atypical and conventional guanine nucleotide exchange factors specific for Rho GTPases, members of the Dock and Dbl families, respectively, are integrated into a functional complex.
Most GPCRs activate heterotrimeric G proteins as transducers of adhesion-related and chemotactic signaling pathways. It is known that Gβγ subunits activate the Rac GTPase through its interaction with GEFs of the Dbl family, contributing to recruit them to the cell membrane (23, 43, 46). In addition, recent evidence indicates that Elmo is a direct effector of Gβγ stimulated by CXCR4 chemotactic receptors (16). In the current work, we confirmed that Gβγ associates with Elmo (16) and also demonstrated that Gβγ directly interacts with the GPR124 C-terminal tail, increasing the association of this region with Elmo. The GPR124 C-terminal tail interacts with Elmo1, Elmo2, and Elmo3, in all cases in complex with Dock180, indicating a possible redundancy of these complexes in GPR124-dependent cell adhesion. Thus, we used the central region of Elmo, mapped as the minimal interacting domain, to assess its effect on cell adhesion. Consistent with a role of the Elmo–Dock system in GPR124-stimulated cell adhesion, the ELM domain of Elmo inhibited this effect, indicating that GPR124 uses the Gβγ–Elmo–Dock system to promote cell adhesion. Furthermore, GPR124 co-localizes with phospho-Elmo during endothelial cell adhesion, and, as revealed by a gain-of-function approach in HEK293T, GPR124 expression promotes Elmo activation, as evidenced by its phosphorylation at Tyr-713/720. The identity of the kinase involved in GPR124-dependent Elmo phosphorylation remains to be elucidated. Putatively, receptor tyrosine kinases may be involved. Emerging evidence indicating a role of GPR124 in VEGF-dependent cell migration (8) raises the possibility that this receptor tyrosine kinase might be involved in Elmo regulation downstream of GPR124 in endothelial cells. In Drosophila border cells, the Elmo–Dock system is an essential player downstream of PDGF- and VEGF-related receptors during the initial phase of collective migration (47). In addition, previous work demonstrated that Axl, a receptor tyrosine kinase, leads to the phosphorylation of Elmo, essential for Dock180-mediated Rac activation, in breast cancer cells (34).
Independently of its interaction with Elmo, GPR124 also directly interacts with ITSNs, which constitute a particularly complex subgroup of DH-domain RhoGEFs specific for Cdc42. The GPR124 C-terminal tail interacts with the five individual SH3 domains of ITSN1. Nevertheless, full-length GPR124 preferentially interacts with the ITSN1-SH3D domain. Because it is known that the ITSN1-SH3 region interacts with multiple signaling proteins (48–51), our results point to the interesting possibility that GPR124 brings about a macromolecular signaling complex including the Gβγ–Elmo–Dock and ITSN systems, each of them exhibiting specific GPR124-interacting domains and also showing direct interactions between them. Specifically, the Elmo C-terminal proline-rich region interacts with ITSN-SH3A, -SH3C, and -SH3E but not ITSN-SH3D, which is the preferred GPR124 binding domain. These signaling complexes mediate GPR124-dependent cell adhesion, as indicated by the inhibitory effect of the GPR124-interacting domains derived from both Elmo and ITSN. In addition, the Elmo–ITSN interaction is established during cell adhesion, even in the absence of GPR124, indicating that this signaling complex might be available for other signaling pathways in addition to the one elicited by GPR124. Altogether, our studies sustain an important role for GPR124 in cell adhesion via its interaction with Elmo–Dock and ITSNs. Further studies are warranted to assess these findings in different systems under diverse pathophysiological conditions, including brain microvascular endothelial cells known to show phenotypic peculiarities.
The signaling properties of GPR124 and most other adhesion GPCRs are much less characterized than those of other GPCR families (52–54). Here, we show that GPR124 promotes cell adhesion via direct interactions with the Gβγ–Elmo–Dock and ITSN systems. These RhoGEFs form a previously uncharacterized signaling complex, which, based on our current findings, might have a potentially broad role in the signaling of ADGRs.
Experimental procedures
Antibodies
Anti-HA (H-3663), FLAG M2 (F-3165), AKT (P-2482), and c-Myc (clone 9E10, M-5546) were purchased from Sigma; GST (B-14, sc-138), GFP (B-2, sc-9996), Cdc42 (B-8, sc-8401), Gβ (M-14, sc-261), and ERK2 (C-14, sc-154) from Santa Cruz Biotechnology, Inc.; and Elmo2 from Novus Biologicals (catalog no. 100-879). Elmo2 and Tyr(P)-713 Elmo2 were described previously (34); GPR124 was kindly provided by Dr. Brad St. Croix (NCI-Frederick) and described previously (4); Rac1 (610650) was from BD Transduction Laboratories, and ITSN antibodies were from Abcam (ab-118262) and Millipore (ABN-1378). Secondary antibodies used for Western blotting were monoclonal goat anti-mouse (KPL 074-1802) and goat anti-rabbit (Rockland Immunochemicals (catalog no. 611-1323) or KPL (catalog no. 074-1802)). Immobilon membranes were purchased from Millipore.
Plasmid constructs
The pcDNA3.1 plasmids with the Elmo constructs GFP–Elmo1, Myc–Elmo1, Myc–Elmo2, Myc–Elmo3, Myc–Elmo1 (1–315), Myc–Elmo1(1–495), Myc–Elmo1(315–727), and Myc–Elm1 (ELM domain) were described previously (55). pCXN2-FLAG Dock180 was kindly provided by Dr. Michiyuki Matsuda (Kyoto University, Japan). 3×FLAG GPR124 GFP and 3×FLAG GPR124 were kindly donated by Dr. Brad St. Croix (4) (NCI-Frederick). pCDNA3-HA-Intersectin 1 and 2 were kindly donated by Dr. Susana de la Luna (56) (Medical and Molecular Genetics Center, IRO, Barcelona, Spain). cDNA encoding individual SH3 domains of intersectin 1 into pGEX-4T1 were kind gifts from Dr. Peter McPherson (48) (McGill University, Montreal, Canada) (Addgene, catalog nos. 47413, 47414, 47415, 47416, and 47417). mCherry VASP was kindly provided by Dr. Melanie Barzik (29). SH3 domains of ITSN1/2 in tandem (from Val-743 to Thr-1213 for ITSN1 and from Val-760 to Thr-1185 for ITSN2) were cloned as 5′-BamHI/3′-EcoRI into pCEFL-GST and pCEFL-3×FLAG mammalian expression vectors. Intersectin 1/2 SH3 primers were 5′-ataGGATCCGTGTATTACCGGGCACTGTACC and 3′-ataGAATTCTGTGGTCAGCTTCACATAATTGG (intersectin 1) and 5′-ataGGATCCGTGAATTATAGAGCATTATACC and 3′-ataGAATTCTGTCGTCATCTTAACGTAGTTTGAAGG (intersectin 2). The GPR124 C terminus was generated by cloning GPR124-Cter (from Arg-1077 to Val-1338) as 5′-BamHI/3′-EcoRI enzyme restriction sites into pCEFL-GST vector and GPR124-ΔC-terminal (from Met-1 to Cys-1083) cloning as 5′-HindIII/3′-EcoRI into pCEFL-3×FLAG vector. The set of primers to obtain these constructs was as follows: 5′-ataGGATCCAGAGCCTCGTGGCGCGCCTGC and 3′-ataGAATTCTTAGACGGTAGTTTCGCTCTTCC; 5′-ataAAGCTTATGGGCGCCGGGGGACGCAGG and 3′-ataGAATTCTTAGCAGGCGCGCCACGAGGCTCTCAC, respectively.
Cell culture and transfections
HEK293T, COS7, and HUVEC-CS cell lines were routinely grown in DMEM (Sigma) supplemented with 10 or 20% FBS (Gibco). HUVECs were cultured in M199 medium containing 20% FBS, endothelial cell growth supplement (0.05 mg/ml), and heparin (0.1 mg/ml). HUVECs were used at passages <5. HEK293T and COS7 cells were transfected using Lipofectamine and Plus reagents (Invitrogen), according to the manufacturer's instructions. The assays were performed 48 h after transfection, and cells were serum-starved for 16 h before each experiment. Transfection efficiency was between 20 and 30%, and only EGFP-positive cells were counted in the adhesion assays. In the case of endothelial cells, they were starved for just 3 h before the experiments. Knockdown experiments were done by transfecting cells with 50 nm GPR124 siRNA, containing a pool of four siRNA sequences, siGenome human (M-005540-00-0005 siGENOME Human ADGRA2 25960) GPR124 or siRNA CTL SMARTpool (Dharmacon, Lafayette, CO), using Lipofectamine 2000 (Invitrogen). Transfected cells were used 48 h post-transfection. Knockdown efficiency was validated by quantitative PCR.
Immunoprecipitation, pulldowns, and immunoblotting
Cells grown in 10-cm dishes were washed with PBS and incubated on ice for 10 min with 1 ml of ice-cold lysis buffer (50 mm Tris, 150 mm NaCl, pH 7.5, 1% Triton X-100, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 1 mm NaF, 1 mm sodium orthovanadate, and 1 mm β-glycerol phosphate). Cell lysates were centrifuged at 13,000 rpm for 10 min at 4 °C. The lysates were precleared with protein A/G-agarose beads before the addition of specific IgG antibodies or a nonspecific antibody (Jackson ImmunoResearch, 011-000-003), used as a control. Supernatants were incubated with 5 μl of the indicated antibodies and incubated overnight at 4 °C. The immune complexes were recovered by incubation for 3 h at 4 °C with Protein G-agarose beads (30 μl; Millipore 16-266). Beads were washed three times with ice-cold lysis buffer and boiled for 5 min in 1× Laemmli sample buffer containing 2-mercaptoethanol. Immunoprecipitated proteins were resolved by SDS-PAGE and detected by Western blotting. Pulldown assays were performed with lysates from cells expressing GST-tagged proteins, which were incubated for 30 min with 25 μl of glutathione–Sepharose beads (catalog no. 17-5279-01, GE Healthcare). The beads were collected by centrifugation and processed as described for immunoprecipitations. In the case of individual recombinant ITSN1-SH3 domains fused to GST, they were expressed in DH5α Escherichia coli and purified using glutathione–Sepharose beads (20 μl), incubated for 3 h at 4 °C, and washed three times with lysis buffer. Recombinant GST was used as a control. Proteins were quantified, resolved on SDS-PAGE, and stained with Coomassie Blue.
Rac1 and Cdc42 activation assays
Activation of Cdc42 and Rac1 was analyzed by using an affinity pulldown assay with the CRIB domain of PAK as a GST fusion protein (57). Briefly, cells were washed with PBS containing 10 mm MgCl2 and lysed with 1× Triton X-100 buffer containing 10 mm MgCl2, phosphatases, and protease inhibitors. Cell lysates were incubated for 45 min with 30 μl of GST–PAK CRIB beads at 4 °C. Beads were centrifuged at 5,000 rpm for 1 min and washed three times with lysis buffer. Beads were then boiled for 5 min in 30 μl of 1× Laemmli buffer. Rac1 and Cdc42 proteins were detected by Western blotting.
Adhesion assay
COS7 cells expressing GPR124–GFP were serum-starved overnight, and HUVECs were starved for 3 h with 2% FBS. Then cells were trypsinized for 5 min, collected with serum-free medium, and centrifuged at 1,000 rpm for 5 min. The pellet was suspended with serum-free medium plus 15 mm HEPES, and cells were incubated (2 h for COS7 cells and 20 min for HUVECs) in a rocking platform at 50–75 rpm at 37 °C. Then HUVECs at a density of 7 × 105 cells were plated on Petri dishes precoated with 0.1% gelatin, and COS7 cells at a density of 3 × 105 cells were plated on plastic Petri dishes. After 30 min, cells were washed three times with PBS and fixed with 4% paraformaldehyde (PFA). Images were taken to assess confluence and transfection efficiency before starting each experiment. COS7 cells transfected with GPR124–GFP or EGFP–CAAX were quantified from five images using a ×10 objective with a Nikon Eclipse Ti inverted microscope and captured with a Digital Sight DS-Qi1Mc Nikon camera. HUVECs were subjected to GPR124 immunostaining, and images were taken with a LSM 710 confocal laser-scanning microscope (Carl Zeiss). To measure fluorescence intensity (pixels) at the cell periphery, three areas of adherent cells were selected. At least 45 areas were analyzed (n = 3) by using Fiji ImageJ software, NIS-Elements software, and Zen software.
Polarity assay
Confluent HUVECs grown on coverslips were transfected with 50 mm siRNA against endogenous GPR124. 48 h post-transfection, cells were washed and starved for 5 h in M199 medium containing 1% FBS. Then a wound was made by using a 100-μl pipette tip, and complete M199 medium containing 20% FBS was added for 30 min. Cells were fixed with 4% PFA and permeabilized in 0.1% Triton X-100. Nonspecific staining was blocked by incubation for 1 h with 1% BSA. Then cells were incubated with anti-GM130 antibody for 2 h at room temperature, followed by incubation for 1 h with Alexa Fluor 568–conjugated goat anti-mouse antibody (Invitrogen) and Alexa Fluor 488 phalloidin. The samples were then counterstained with DAPI to stain the nuclei and analyzed with a Carl Zeiss confocal microscope. The orientation of the Golgi was assessed as described previously (58, 59). Cells in which GM130 staining did not localize within a 120° angle facing the wound were considered as non-polarized cells. At least 100 cells near the wound edge were randomly examined, and the percentage of polarized cells was determined (n = 3). Data are represented as mean ± S.E. *, p < 0.05.
Immunofluorescence confocal microscopy
Glass coverslips were coated with 20 μg/ml fibronectin or 0.1% gelatin for 30 min at room temperature. HUVECs were prepared for the adhesion assay as described above. Cells were fixed, permeabilized, and stained with primary antibodies at room temperature for 1 h at a 1:100 dilution. Cells were then incubated with species-specific appropriate secondary antibodies at a 1:100 dilution (anti-mouse 488, anti-rabbit 568, and anti-goat 688; Molecular Probes) and then stained with Alex Fluor 633 phalloidin 1:200 (Molecular Probes) for 20 min. Cells were then mounted using Fluoromont (Sigma-Aldrich) with DAPI and observed in an LSM 710 confocal laser-scanning microscope (Carl Zeiss). Images were acquired by using Zen software with a ×63/1.4 numeric aperture oil objective.
PLA
HUVECs were plated on a glass coverslip and used to perform PLAs, following the manufacturer's protocol (catalog no. DUO92004-30RXN Duolink In Situ, Sigma-Aldrich). The dilutions of the antibodies were as follows: GPR124 (1:100), ITSN1 (1:100), and Elmo2 (1:50). For quantification of the PLA signal, the numbers of red dots per field were divided by the number of cells (DAPI) to give an average of dots per cell. At least 30 cells/condition were analyzed in three independent experiments, and this was achieved using Fiji ImageJ software.
mRNA isolation and quantitative RT-PCR
Total RNA was extracted with an RNeasy minikit (Qiagen). 1 μg of RNA was reverse-transcribed into cDNA by using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. RT-PCR was done using Taq DNA polymerase. GAPDH was used as a reference gene. The sequences of the primers are as follows: human GPR124, 5′-GGCACTGAGGTGAAGGGATA-3′ (forward primer) and 5′-AGAAGGTGGAGATCGTGGTG-3′ (reverse primer); human GAPDH, 5′-CCACTCCTCCACCTTTGAC-3′ (forward primer) and 5′-ACCCTGTTGCTGTAGCCA-3′ (reverse primer).
Statistical analysis
Polarity assays were analyzed using Student's t test. All other results were subjected to analysis by Kruskal–Wallis one-way ANOVA or Tukey test using GraphPad Prism software version 6.0. A value of p < 0.05 was considered significant.
Author contributions
M. N. H.-V. designed, performed, and analyzed most of the experiments in Figs. 1 (A, E, and G), 2, 3, 4, and 5 (A–F and H). S. R. A.-G. designed, performed, and analyzed the experiment shown in Fig. 1F. N. H. designed, performed, and analyzed the experiment shown in Fig. 5G. R. C. designed, performed, and analyzed the experiment shown in Fig. 1H. G. R.-C., J. P. G., and J.-F. C. provided technical assistance, analyzed data, and reviewed the manuscript. J. V.-P. designed and coordinated the study and wrote the manuscript with M. N. H.-V.
Acknowledgments
Technical assistance provided by Estanislao Escobar Islas, Margarita Valadez, David Pérez, and Jaime Estrada Trejo is acknowledged. We especially thank the CINVESTAV Confocal Unit (LaNSE) and Iván J. Galván MSc for support of confocal microscopy image acquisition.
This work was supported by CONACyT (Consejo Nacional de Ciencia y Tecnología, Mexico) Grants 152434 (to J. V.-P.) and 240119 (to G. R.-C.), a Canadian Institutes of Health Research grant (to J. F. C.), and PNPC (en el marco del “Programa de Fortalecimiento Académico del Posgrado de Alta Calidad”). The authors declare that they have no conflicts of interest with the contents of this article.
- ADGR
- adhesion G-protein–coupled receptor
- COS7
- CV-1 (simian) in origin and carrying the SV40 genetic material
- HUVEC
- human umbilical vein endothelial cell
- ITSN
- intersectin
- GEF
- guanine nucleotide exchange factor
- SH3
- Src homology 3
- GPCR
- G protein-coupled receptor
- PLA
- proximity ligation assay
- PH
- pleckstrin homology
- DH
- Dbl homology
- PFA
- paraformaldehyde
- ANOVA
- analysis of variance.
References
- 1. Zhao Z., Nelson A. R., Betsholtz C., and Zlokovic B. V. (2015) Establishment and dysfunction of the blood-brain barrier. Cell 163, 1064–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuhnert F., Mancuso M. R., Shamloo A., Wang H. T., Choksi V., Florek M., Su H., Fruttiger M., Young W. L., Heilshorn S. C., and Kuo C. J. (2010) Essential regulation of CNS angiogenesis by the orphan G protein-coupled receptor GPR124. Science 330, 985–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson K. D., Pan L., Yang X. M., Hughes V. C., Walls J. R., Dominguez M. G., Simmons M. V., Burfeind P., Xue Y., Wei Y., Macdonald L. E., Thurston G., Daly C., Lin H. C., Economides A. N., et al. (2011) Angiogenic sprouting into neural tissue requires Gpr124, an orphan G protein-coupled receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 2807–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cullen M., Elzarrad M. K., Seaman S., Zudaire E., Stevens J., Yang M. Y., Li X., Chaudhary A., Xu L., Hilton M. B., Logsdon D., Hsiao E., Stein E. V., Cuttitta F., Haines D. C., et al. (2011) GPR124, an orphan G protein-coupled receptor, is required for CNS-specific vascularization and establishment of the blood-brain barrier. Proc. Natl. Acad. Sci. U.S.A. 108, 5759–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhou Y., and Nathans J. (2014) Gpr124 controls CNS angiogenesis and blood-brain barrier integrity by promoting ligand-specific canonical wnt signaling. Dev. Cell 31, 248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Posokhova E., Shukla A., Seaman S., Volate S., Hilton M. B., Wu B., Morris H., Swing D. A., Zhou M., Zudaire E., Rubin J. S., and St Croix B. (2015) GPR124 functions as a WNT7-specific coactivator of canonical β-catenin signaling. Cell Rep. 10, 123–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. St Croix B., Rago C., Velculescu V., Traverso G., Romans K. E., Montgomery E., Lal A., Riggins G. J., Lengauer C., Vogelstein B., and Kinzler K. W. (2000) Genes expressed in human tumor endothelium. Science 289, 1197–1202 [DOI] [PubMed] [Google Scholar]
- 8. Wang Y., Cho S. G., Wu X., Siwko S., and Liu M. (2014) G-protein coupled receptor 124 (GPR124) in endothelial cells regulates vascular endothelial growth factor (VEGF)-induced tumor angiogenesis. Curr. Mol. Med. 14, 543–554 [DOI] [PubMed] [Google Scholar]
- 9. Laurin M., and Côté J. F. (2014) Insights into the biological functions of Dock family guanine nucleotide exchange factors. Genes Dev. 28, 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hernández-García R., Iruela-Arispe M. L., Reyes-Cruz G., and Vázquez-Prado J. (2015) Endothelial RhoGEFs: a systematic analysis of their expression profiles in VEGF-stimulated and tumor endothelial cells. Vascul. Pharmacol. 74, 60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cook D. R., Rossman K. L., and Der C. J. (2014) Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene 33, 4021–4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tang X., Jin R., Qu G., Wang X., Li Z., Yuan Z., Zhao C., Siwko S., Shi T., Wang P., Xiao J., Liu M., and Luo J. (2013) GPR116, an adhesion G-protein-coupled receptor, promotes breast cancer metastasis via the Gαq-p63RhoGEF-Rho GTPase pathway. Cancer Res. 73, 6206–6218 [DOI] [PubMed] [Google Scholar]
- 13. Hamoud N., Tran V., Croteau L. P., Kania A., and Côté J. F. (2014) G-protein coupled receptor BAI3 promotes myoblast fusion in vertebrates. Proc. Natl. Acad. Sci. U.S.A. 111, 3745–3750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park D., Tosello-Trampont A. C., Elliott M. R., Lu M., Haney L. B., Ma Z., Klibanov A. L., Mandell J. W., and Ravichandran K. S. (2007) BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature 450, 430–434 [DOI] [PubMed] [Google Scholar]
- 15. Peeters M. C., Fokkelman M., Boogaard B., Egerod K. L., van de Water B., IJzerman A. P., and Schwartz T. W. (2015) The adhesion G protein-coupled receptor G2 (ADGRG2/GPR64) constitutively activates SRE and NFκB and is involved in cell adhesion and migration. Cell. Signal. 27, 2579–2588 [DOI] [PubMed] [Google Scholar]
- 16. Wang Y., Xu X., Pan M., and Jin T. (2016) ELMO1 directly interacts with Gβγ subunit to transduce GPCR signaling to Rac1 activation in chemotaxis. J. Cancer 7, 973–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanematsu F., Hirashima M., Laurin M., Takii R., Nishikimi A., Kitajima K., Ding G., Noda M., Murata Y., Tanaka Y., Masuko S., Suda T., Meno C., Côté J. F., Nagasawa T., and Fukui Y. (2010) DOCK180 is a Rac activator that regulates cardiovascular development by acting downstream of CXCR4. Circ. Res. 107, 1102–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carretero-Ortega J., Walsh C. T., Hernández-García R., Reyes-Cruz G., Brown J. H., and Vázquez-Prado J. (2010) Phosphatidylinositol 3,4,5-triphosphate-dependent Rac exchanger 1 (P-Rex-1), a guanine nucleotide exchange factor for Rac, mediates angiogenic responses to stromal cell-derived factor-1/chemokine stromal cell-derived factor-1 (SDF-1/CXCL-12) linked to Rac activation, endothelial cell migration, and in vitro angiogenesis. Mol. Pharmacol. 77, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yagi H., Tan W., Dillenburg-Pilla P., Armando S., Amornphimoltham P., Simaan M., Weigert R., Molinolo A. A., Bouvier M., and Gutkind J. S. (2011) A synthetic biology approach reveals a CXCR4-G13-Rho signaling axis driving transendothelial migration of metastatic breast cancer cells. Sci. Signal. 4, ra60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aittaleb M., Boguth C. A., and Tesmer J. J. (2010) Structure and function of heterotrimeric G protein-regulated Rho guanine nucleotide exchange factors. Mol. Pharmacol. 77, 111–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ledezma-Sánchez B. A., García-Regalado A., Guzmán-Hernández M. L., and Vázquez-Prado J. (2010) Sphingosine-1-phosphate receptor S1P1 is regulated by direct interactions with P-Rex1, a Rac guanine nucleotide exchange factor. Biochem. Biophys. Res. Commun. 391, 1647–1652 [DOI] [PubMed] [Google Scholar]
- 22. Stephenson J. R., Paavola K. J., Schaefer S. A., Kaur B., Van Meir E. G., and Hall R. A. (2013) Brain-specific angiogenesis inhibitor-1 signaling, regulation, and enrichment in the postsynaptic density. J. Biol. Chem. 288, 22248–22256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vázquez-Prado J., Bracho-Valdés I., Cervantes-Villagrana R. D., and Reyes-Cruz G. (2016) Gβγ pathways in cell polarity and migration linked to oncogenic GPCR signaling: potential relevance in tumor microenvironment. Mol. Pharmacol. 90, 573–586 [DOI] [PubMed] [Google Scholar]
- 24. Bardita C., Predescu D., and Predescu S. (2013) Long-term silencing of intersectin-1s in mouse lungs by repeated delivery of a specific siRNA via cationic liposomes. Evaluation of knockdown effects by electron microscopy. J. Vis. Exp. 10.3791/50316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrero-Garcia E., and O'Bryan J. P. (2017) Intersectin scaffold proteins and their role in cell signaling and endocytosis. Biochim. Biophys. Acta 1864, 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Savdie C., Ferguson S. S., Vincent J., Beaudet A., and Stroh T. (2006) Cell-type-specific pathways of neurotensin endocytosis. Cell Tissue Res. 324, 69–85 [DOI] [PubMed] [Google Scholar]
- 27. Gryaznova T., Kropyvko S., Burdyniuk M., Gubar O., Kryklyva V., Tsyba L., and Rynditch A. (2015) Intersectin adaptor proteins are associated with actin-regulating protein WIP in invadopodia. Cell. Signal. 27, 1499–1508 [DOI] [PubMed] [Google Scholar]
- 28. Carson-Walter E. B., Watkins D. N., Nanda A., Vogelstein B., Kinzler K. W., and St Croix B. (2001) Cell surface tumor endothelial markers are conserved in mice and humans. Cancer Res. 61, 6649–6655 [PubMed] [Google Scholar]
- 29. Barzik M., McClain L. M., Gupton S. L., and Gertler F. B. (2014) Ena/VASP regulates mDia2-initiated filopodial length, dynamics, and function. Mol. Biol. Cell 25, 2604–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nobes C. D., and Hall A. (1995) Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62 [DOI] [PubMed] [Google Scholar]
- 31. Kozma R., Ahmed S., Best A., and Lim L. (1995) The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol. Cell. Biol. 15, 1942–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Müller A., Winkler J., Fiedler F., Sastradihardja T., Binder C., Schnabel R., Kungel J., Rothemund S., Hennig C., Schöneberg T., and Prömel S. (2015) Oriented cell division in the C. elegans embryo is coordinated by G-protein signaling dependent on the adhesion GPCR LAT-1. PLoS Genet. 11, e1005624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Iguchi T., Sakata K., Yoshizaki K., Tago K., Mizuno N., and Itoh H. (2008) Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a Gα12/13 and Rho pathway. J. Biol. Chem. 283, 14469–14478 [DOI] [PubMed] [Google Scholar]
- 34. Abu-Thuraia A., Gauthier R., Chidiac R., Fukui Y., Screaton R. A., Gratton J. P., and Côté J. F. (2015) Axl phosphorylates Elmo scaffold proteins to promote Rac activation and cell invasion. Mol. Cell. Biol. 35, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hall A. (2012) Rho family GTPases. Biochem. Soc. Trans. 40, 1378–1382 [DOI] [PubMed] [Google Scholar]
- 36. Sadok A., and Marshall C. J. (2014) Rho GTPases: masters of cell migration. Small GTPases 5, e29710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wojciak-Stothard B., and Ridley A. J. (2002) Rho GTPases and the regulation of endothelial permeability. Vascul. Pharmacol. 39, 187–199 [DOI] [PubMed] [Google Scholar]
- 38. Beckers C. M., van Hinsbergh V. W., and van Nieuw Amerongen G. P. (2010) Driving Rho GTPase activity in endothelial cells regulates barrier integrity. Thromb. Haemost. 103, 40–55 [DOI] [PubMed] [Google Scholar]
- 39. Vallon M., and Essler M. (2006) Proteolytically processed soluble tumor endothelial marker (TEM) 5 mediates endothelial cell survival during angiogenesis by linking integrin α(v)β3 to glycosaminoglycans. J. Biol. Chem. 281, 34179–34188 [DOI] [PubMed] [Google Scholar]
- 40. Summers L., Kangwantas K., Rodriguez-Grande B., Denes A., Penny J., Kielty C., and Pinteaux E. (2013) Activation of brain endothelial cells by interleukin-1 is regulated by the extracellular matrix after acute brain injury. Mol. Cell Neurosci. 57, 93–103 [DOI] [PubMed] [Google Scholar]
- 41. Chang J., Mancuso M. R., Maier C., Liang X., Yuki K., Yang L., Kwong J. W., Wang J., Rao V., Vallon M., Kosinski C., Zhang J. J., Mah A. T., Xu L., Li L., et al. (2017) Gpr124 is essential for blood-brain barrier integrity in central nervous system disease. Nat. Med. 23, 450–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ridley A. J., Paterson H. F., Johnston C. L., Diekmann D., and Hall A. (1992) The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410 [DOI] [PubMed] [Google Scholar]
- 43. Welch H. C., Coadwell W. J., Ellson C. D., Ferguson G. J., Andrews S. R., Erdjument-Bromage H., Tempst P., Hawkins P. T., and Stephens L. R. (2002) P-Rex1, a PtdIns(3,4,5)P3- and Gβγ-regulated guanine-nucleotide exchange factor for Rac. Cell 108, 809–821 [DOI] [PubMed] [Google Scholar]
- 44. Friesland A., Zhao Y., Chen Y. H., Wang L., Zhou H., and Lu Q. (2013) Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc. Natl. Acad. Sci. U.S.A. 110, 1261–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rodriguez-Fraticelli A. E., Vergarajauregui S., Eastburn D. J., Datta A., Alonso M. A., Mostov K., and Martín-Belmonte F. (2010) The Cdc42 GEF Intersectin 2 controls mitotic spindle orientation to form the lumen during epithelial morphogenesis. J. Cell Biol. 189, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Barber M. A., Donald S., Thelen S., Anderson K. E., Thelen M., and Welch H. C. (2007) Membrane translocation of P-Rex1 is mediated by G protein βγ subunits and phosphoinositide 3-kinase. J. Biol. Chem. 282, 29967–29976 [DOI] [PubMed] [Google Scholar]
- 47. Bianco A., Poukkula M., Cliffe A., Mathieu J., Luque C. M., Fulga T. A., and Rørth P. (2007) Two distinct modes of guidance signalling during collective migration of border cells. Nature 448, 362–365 [DOI] [PubMed] [Google Scholar]
- 48. Yamabhai M., Hoffman N. G., Hardison N. L., McPherson P. S., Castagnoli L., Cesareni G., and Kay B. K. (1998) Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J. Biol. Chem. 273, 31401–31407 [DOI] [PubMed] [Google Scholar]
- 49. Hussain N. K., Jenna S., Glogauer M., Quinn C. C., Wasiak S., Guipponi M., Antonarakis S. E., Kay B. K., Stossel T. P., Lamarche-Vane N., and McPherson P. S. (2001) Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol. 3, 927–932 [DOI] [PubMed] [Google Scholar]
- 50. Tong X. K., Hussain N. K., de Heuvel E., Kurakin A., Abi-Jaoude E., Quinn C. C., Olson M. F., Marais R., Baranes D., Kay B. K., and McPherson P. S. (2000) The endocytic protein intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. EMBO J. 19, 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Roos J., and Kelly R. B. (1998) Dap160, a neural-specific Eps15 homology and multiple SH3 domain-containing protein that interacts with Drosophila dynamin. J. Biol. Chem. 273, 19108–19119 [DOI] [PubMed] [Google Scholar]
- 52. Monk K. R., Hamann J., Langenhan T., Nijmeijer S., Schöneberg T., and Liebscher I. (2015) Adhesion G protein-coupled receptors: from in vitro pharmacology to in vivo mechanisms. Mol. Pharmacol. 88, 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Langenhan T., Aust G., and Hamann J. (2013) Sticky signaling–adhesion class G protein-coupled receptors take the stage. Sci. Signal. 6, re3. [DOI] [PubMed] [Google Scholar]
- 54. Paavola K. J., and Hall R. A. (2012) Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol. Pharmacol. 82, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Patel M., Margaron Y., Fradet N., Yang Q., Wilkes B., Bouvier M., Hofmann K., and Côté J. F. (2010) An evolutionarily conserved autoinhibitory molecular switch in ELMO proteins regulates Rac signaling. Curr. Biol. 20, 2021–2027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pucharcos C., Estivill X., and de la Luna S. (2000) Intersectin 2, a new multimodular protein involved in clathrin-mediated endocytosis. FEBS Lett. 478, 43–51 [DOI] [PubMed] [Google Scholar]
- 57. Côté J. F., and Vuori K. (2002) Identification of an evolutionarily conserved superfamily of DOCK180-related proteins with guanine nucleotide exchange activity. J. Cell Sci. 115, 4901–4913 [DOI] [PubMed] [Google Scholar]
- 58. Nobes C. D., and Hall A. (1999) Rho GTPases control polarity, protrusion, and adhesion during cell movement. J. Cell Biol. 144, 1235–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Etienne-Manneville S., and Hall A. (2001) Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCζ. Cell 106, 489–498 [DOI] [PubMed] [Google Scholar]