Abstract
Individuals with Hajdu-Cheney syndrome (HCS) present with osteoporosis, and HCS is associated with NOTCH2 mutations causing deletions of the proline-, glutamic acid-, serine-, and threonine-rich (PEST) domain that are predicted to enhance NOTCH2 stability and cause gain-of-function. Previously, we demonstrated that mice harboring Notch2 mutations analogous to those in HCS (Notch2HCS) are severely osteopenic because of enhanced bone resorption. We attributed this phenotype to osteoclastic sensitization to the receptor activator of nuclear factor-κB ligand and increased osteoblastic tumor necrosis factor superfamily member 11 (Tnfsf11) expression. Here, to determine the individual contributions of osteoclasts and osteoblasts to HCS osteopenia, we created a conditional-by-inversion (Notch2COIN) model in which Cre recombination generates a Notch2ΔPEST allele expressing a Notch2 mutant lacking the PEST domain. Germ line Notch2COIN inversion phenocopied the Notch2HCS mutant, validating the model. To activate Notch2 in osteoclasts or osteoblasts, Notch2COIN mice were bred with mice expressing Cre from the Lyz2 or the BGLAP promoter, respectively. These crosses created experimental mice harboring a Notch2ΔPEST allele in Cre-expressing cells and control littermates expressing a wild-type Notch2 transcript. Notch2COIN inversion in Lyz2-expressing cells had no skeletal consequences and did not affect the capacity of bone marrow macrophages to form osteoclasts in vitro. In contrast, Notch2COIN inversion in osteoblasts led to generalized osteopenia associated with enhanced bone resorption in the cancellous bone compartment and with suppressed endocortical mineral apposition rate. Accordingly, Notch2 activation in osteoblast-enriched cultures from Notch2COIN mice induced Tnfsf11 expression. In conclusion, introduction of the HCS mutation in osteoblasts, but not in osteoclasts, causes osteopenia.
Keywords: genetic disease, mouse genetics, Notch pathway, osteoblast, osteoclast, Hajdu Cheney Syndrome, Notch2, bone remodeling, conditional by inversion
Introduction
Notch signaling plays a fundamental role in cell fate determination (1). Interactions of the four Notch receptors with cognate ligands of the Jagged and Delta-like families lead to the proteolytic cleavage of the receptor and the release of the Notch intracellular domain (NICD)2 from the cellular membrane (2). Subsequently, the NICD translocates to the nucleus and forms a complex with recombination signal-binding protein for the immunoglobulin κJ region (Rbpjκ), mastermind-like (Maml), and additional DNA-associated proteins to elicit a transcriptional response (3). These events result in the induction of Notch target genes, such as Hes1, Hey1, Hey2, and HeyL (4). Although this signaling mechanism is shared by the Notch paralogs, each receptor has distinct functions (5). The reason appears to be related to the differential cellular pattern of expression of the receptors, structural differences between the paralogs, and interactions of the individual NICDs with Rbpjκ (6–8).
Bone remodeling is the process whereby the coordinated activities of osteoclasts and osteoblasts preserve skeletal integrity (9). Osteoclasts are multinucleated bone-resorbing cells formed by the fusion of mononuclear myeloid precursors in the presence of receptor activator of nuclear factor κB ligand (Rankl), a protein encoded by Tnfsf11, and macrophage colony-stimulating factor (M-CSF) (10). Osteoblasts are bone-forming cells of mesenchymal origin that regulate bone resorption by secreting Rankl and its decoy receptor, osteoprotegerin (9, 11). Notch1 and Notch2 exhibit distinct functions in skeletal cells, and tight regulation of their activity is essential to maintain bone remodeling (12). Notch1 inhibits osteoclastogenesis and osteoblastogenesis, whereas Notch2 inhibits osteoblast differentiation/function but stimulates osteoclastogenesis (13–19).
Hajdu-Cheney syndrome (HCS) is a rare and devastating disease with multiple systemic manifestations, including osteoporosis, short stature, craniofacial deformities, and acroosteolysis (20, 21). The condition is associated with mutations in exon 34 of NOTCH2 that create a premature stop codon immediately upstream of the sequences coding for the proline- (P), glutamic acid- (E), serine- (S), and (T) threonine-rich (PEST) domain (22–26). The latter is required for the proteasomal degradation of NOTCH2, so that the mutations are predicted to lead to the translation of a stable NOTCH2 protein with sustained activity. Recently, we established a murine model of HCS by introducing the mutation found in a subject with severe osteoporosis into the mouse genome. The mutant, termed Notch2HCS, expresses a Notch2 protein of 2318 amino acids that lacks the PEST domain. Heterozygous Notch2HCS mice exhibit Notch2 gain-of-function and generalized osteopenia secondary to enhanced bone resorption, which was ascribed to the sensitization of osteoclast precursors to Rankl and increased Tnfsf11 expression in osteoblasts (27). However, the individual contribution of cells of the osteoclast and osteoblast lineages to the osteopenic phenotype of Notch2HCS mice remains to be determined.
In this study, a conditional by inversion (COIN) approach was utilized to create a conditional mouse model of HCS (Notch2COIN) (28, 29). This system was designed to introduce a premature STOP codon in exon 34 of Notch2 following Cre-mediated recombination leading to the translation of a truncated Notch2 protein, thus mimicking the genetic defect associated with HCS. To study the consequences of the Notch2 truncation in specific skeletal cell lineages, Notch2 conditional mice were crossed with appropriate Cre drivers to introduce the mutation in cells of the osteoclast (Lyz2Cre) or osteoblast (BGLAP-Cre) lineages. Mutant and control mice were examined for skeletal phenotypic changes by microcomputed tomography (μCT) and bone histomorphometry, and the potential mechanisms of Notch2 action were explored.
Results
Generation of a conditional HCS mouse model
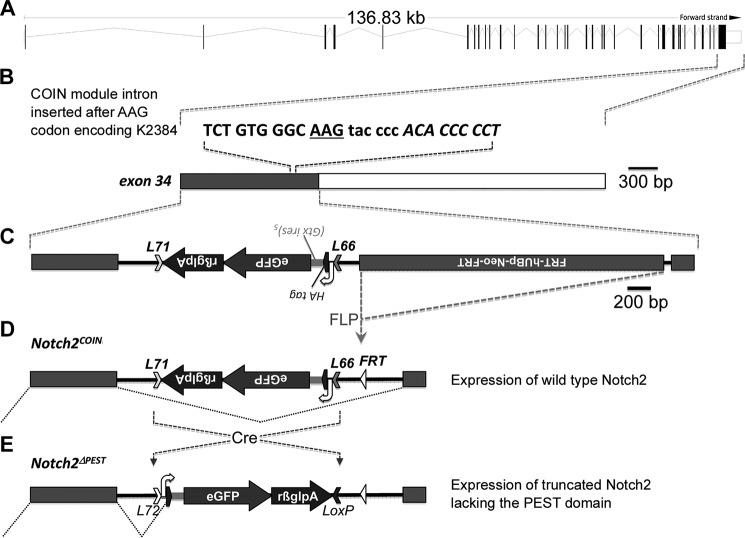
To induce the HCS mutation in selected cell populations, Notch2COIN mice were created by inserting an artificial COIN intron into exon 34 of the murine Notch2 locus (Fig. 1A). As a result, exon 34 was split into two exons at a position corresponding to lysine 2384, which is upstream of the PEST domain and downstream of the domains required for the transcriptional activation of Notch2 (NCBI protein database NP035058; Fig. 1B). The COIN module is composed of a gene trap-like lox66_HA-egfp-polyA_lox71 cassette encoding for a hemagglutinin (HA)-internal ribosome entry site and enhanced green fluorescent protein (eGFP) and placed in the antisense strand. The cassette is preceded by a 3′-splice region derived from the second intron of rabbit HBB2 and followed by the polyadenylation region of the same gene. The COIN element contains a neo selection cassette downstream of the UBp promoter and the EM7 prokaryotic promoter and upstream of the polyadenylation region of Pgk1 flanked by flippase (FLP) recognition target (FRT) sequences (Frt-neo-Pgk1polyA-Frt) (Fig. 1C) (30–32). Prior to Cre recombination, the COIN module is removed by splicing of the precursor mRNA to generate a Notch2COIN transcript that is indistinguishable from the Notch2WT mRNA (Fig. 1D). In the presence of Cre recombinase, which recognizes the lox71 and lox66 mutant sites in a mirror image configuration, the lox66_HA-egfp-polyA_lox71 cassette is brought into the sense strand, causing the irreversible conversion of the COIN allele. The resulting allele encodes for a bicistronic message that is translated into an HA-tagged Notch2 mutant truncated at lysine 2384 and thereby lacking the PEST domain and eGFP (Fig. 1E). This allele was termed Notch2ΔPEST.
Figure 1.
Engineering of the Notch2COIN allele. A, genomic structure and size of the Notch2 locus with the position of the 34 exons indicated by vertical black bars for coding sequences or white boxes for untranslated regions (UTR). B, position of the AAG codon (underlined) for lysine 2384 in exon 34. The sequence of the insertion site of the COIN module is in lowercase, and gray and white boxes indicate the coding sequence and the 3′-UTR (rβglpA), respectively. C, structure of exon 34 and of the targeting construct correctly integrated. From 5′ to 3′: lox71 (L71), rabbit β-globin polyadenylation signal, eGFP-coding sequence, internal ribosome entry site (Gtx ires)5, human influenza hemagglutinin (HA) tag coding sequence, 3′-splice region from the second intron of the rabbit β-globin gene (white curved arrow), and lox66 (L66) that constitute the COIN module and a flippase (FLP) recognition site (FRT)-flanked neo cassette downstream of the human UBp promoter (FRT-hUBp-neo-FRT). Removal of the neo cassette by FLP recombination is indicated (gray dotted lines). D, representation of the silent COIN module in the antisense orientation and of the splicing event (black dotted lines) that excises the COIN module from the nascent transcript, allowing expression of a wild-type Notch2 mRNA and protein. E, generation of the Notch2ΔPEST allele by Cre recombinase-mediated permanent inversion of the COIN module, and illustration of the splicing event (black dotted lines) that occurs during the maturation of the Notch2ΔPEST transcript. The latter is translated into a Notch2 mutant lacking the PEST domain. The position of the silent lox72 (L72) sequence and of the wild-type loxP site created by Cre recombination of L71 and L66 is indicated. Images are scaled either in kilobase (kb) or bp.
To ensure skeletal equivalency of the Notch2COIN and Notch2WT alleles, the microarchitecture of the distal femur in 1-month-old Notch2COIN/COIN male and female mice and wild-type C57BL/6J controls of the same sex and age was analyzed. Cancellous bone volume and cortical thickness were not different between Notch2COIN/COIN mice and controls, demonstrating that homozygosity for the Notch2COIN allele has no appreciable effect on femoral microarchitecture (data not shown).
Inversion of the Notch2COIN allele in the germ line causes osteopenia
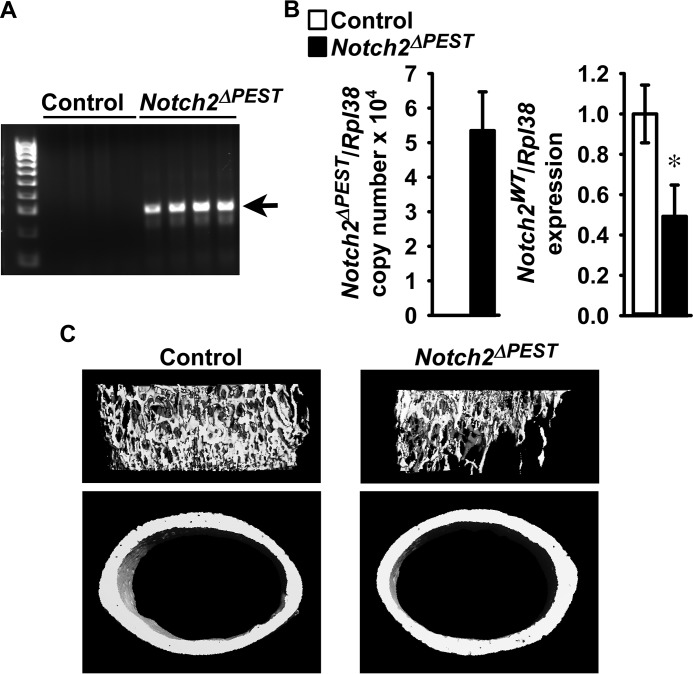
To validate the Notch2COIN mouse as a model of HCS, the skeletal phenotype of Notch2ΔPEST/WT mice created by inversion of the COIN module in the germ line was determined. To this end, Notch2COIN/WT male mice were crossed with Hprt-Cre female mice to create Notch2ΔPEST/WT mice; these were crossed with wild-type mice to create Notch2ΔPEST/WT heterozygous and control wild-type littermates. COIN inversion was documented by the presence of the Notch2ΔPEST allele in DNA from tails of Notch2ΔPEST/WT mice, and quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis of total RNA from tibiae confirmed the expression of the Notch2ΔPEST transcript in mutant mice but not in control littermates (Fig. 2, A and B). Notch2WT transcript levels were ∼50% lower in Notch2ΔPEST/WT mice than in wild-type littermates, and this is consistent with a systemic heterozygous Notch2ΔPEST inversion and comparable expression levels of the Notch2ΔPEST and Notch2WT alleles (Fig. 2B).
Figure 2.
Inversion of the Notch2COIN allele in the germ line causes osteopenia. One-month-old male Notch2ΔPEST/WT mutants (black bars; Notch2ΔPEST) were compared with wild-type littermate controls (white bars) of the same sex. A, DNA was extracted from tail, and Notch2COIN inversion was documented by gel electrophoresis of PCR products obtained with primers specific for the Notch2ΔPEST allele. Arrows indicate the position of the 250-bp amplicon. B, total RNA was extracted from tibiae, and expression of the Notch2ΔPEST and Notch2WT mRNA was measured by qRT-PCR. Transcript levels are reported as copy number corrected for Rpl38 mRNA levels; data for Notch2WT were normalized to corrected expression in control. Values are means ± S.D.; n = 4 for control, n = 5 for Notch2ΔPEST, all biological replicates. Two technical replicates were used for each qPCR. *, significantly different between control and Notch2ΔPEST, p < 0.05 by t test. C, representative μCT images of femoral proximal trabecular bone and midshaft cortical bones of control and Notch2ΔPEST mice; complete data set in Table 1.
One-month-old germ line Notch2ΔPEST/WT male mice appeared normal, albeit a small reduction (∼5%; p < 0.05) in femoral length was noted. Analysis of the distal femur by μCT revealed that, compared with sex-matched littermate controls, Notch2ΔPEST/WT male mice had a 50% decrease in trabecular bone volume secondary to a reduced number and thickness of trabeculae. Connectivity density was lower, and structure model index (SMI) was higher in Notch2ΔPEST/WT mice than in controls, indicating a prevalence of rod-like trabeculae (Table 1 and Fig. 2C). Notch2ΔPEST/WT mice had a thin and porous cortical bone, and their femurs were smaller than those from controls, because total area, bone area, and periosteal as well as endocortical perimeters were reduced (Table 1 and Fig. 2C). These results mirror the phenotype reported for global Notch2HCS mutants and validate the Notch2COIN mouse as a model to study the contribution of selected cell lineages to the phenotypic manifestations of Notch2HCS mutant mice (27).
Table 1.
Femoral microarchitecture assessed by μCT of 1-month-old Notch2ΔPEST/WT (Notch2ΔPEST) mice and sex-matched wild-type littermates (control)
μCT was performed at the femoral distal end for trabecular or midshaft for cortical bone. Values are means ± S.D.
| Control | Notch2ΔPEST | |
|---|---|---|
| Distal femur trabecular bone | n = 4 | n = 5 |
| Bone volume/total volume (%) | 18.9 ± 2.1 | 9.8 ± 1.5a |
| Trabecular separation (μm) | 134 ± 19 | 171 ± 13a |
| Trabecular no. (1/mm) | 7.7 ± 1.0 | 5.9 ± 0.4a |
| Trabecular thickness (μm) | 31 ± 1 | 25 ± 2a |
| Connectivity density (1/mm3) | 1360 ± 152 | 720 ± 94a |
| Structure model index | 1.6 ± 0.2 | 2.5 ± 0.1a |
| Density of material (mg HA/cm3) | 923 ± 12 | 920 ± 27 |
| Femoral midshaft cortical bone | n = 4 | n = 5 |
| Bone volume/total volume (%) | 87.6 ± 0.5 | 85.6 ± 1.6a |
| Porosity (%) | 12.4 ± 0.5 | 14.4 ± 1.6a |
| Cortical thickness (μm) | 110 ± 5 | 97 ± 6a |
| Total area (mm2) | 1.76 ± 0.10 | 1.50 ± 0.09a |
| Bone area (mm2) | 0.59 ± 0.03 | 0.51 ± 0.04a |
| Periosteal perimeter (μm) | 4.7 ± 0.1 | 4.3 ± 0.1a |
| Endocortical perimeter (mm) | 3.8 ± 0.1 | 3.5 ± 0.1a |
| Density of material (mg HA/cm3) | 1001 ± 11 | 999 ± 8 |
a Data are significantly different between control and Notch2ΔPEST, p < 0.05 by unpaired t test.
Inversion of the Notch2COIN allele in the osteoclast lineage does not cause a skeletal phenotype
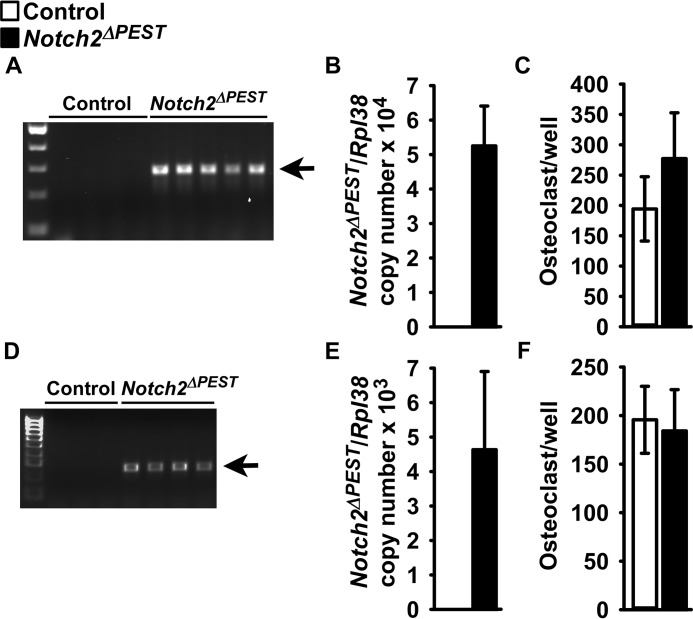
To establish whether the osteopenic phenotype of the Notch2HCS mutants is secondary to direct effects in cells of the osteoclast lineage, the Notch2COIN allele was introduced into Lyz2Cre/WT heterozygous mice. Subsequently, Lyz2Cre/WT;Notch2COIN/COIN mice were crossed with Notch2COIN/COIN mice for the creation of Lyz2Cre/WT;Notch2ΔPEST/ΔPEST experimental mice and Notch2COIN/COIN littermate controls. In an alternative mating scheme, the Notch2ΔPEST inversion was carried out in the context of Lyz2Cre homozygosity. To this end, Lyz2Cre/Cre;Notch2COIN/WT mice were crossed with Lyz2Cre/Cre;Notch2COIN/WT mice for the creation of Lyz2Cre/CreNotch2ΔPEST/ΔPEST experimental and Lyz2Cre/Cre;Notch2WT/WT control mice. In preliminary studies, we documented that 1- and 4-month-old Lyz2Cre and 1-month-old Lyz2Cre/Cre mice did not have a skeletal phenotype as determined by μCT of distal femurs, when compared with wild-type controls (data not shown). COIN inversion was demonstrated in cultures of bone marrow-derived macrophages (BMMs) from 1-month-old Lyz2Cre/WT;Notch2ΔPEST/ΔPEST and Lyz2Cre/Cre;Notch2ΔPEST/ΔPESTmice, and expression of the Notch2ΔPEST mRNA was detected in total RNA from their parietal bones (Fig. 3, A, B, D, and E). These results demonstrate that the Hajdu-Cheney mutation was introduced and transcribed in Lyz2-expressing cells. Femoral microarchitecture of male and female at 1- or 4-month-old Lyz2Cre/WT;Notch2ΔPEST/ΔPEST mice or 1-month-oldLyz2Cre/Cre;Notch2ΔPEST/ΔPEST mice was not different from that of wild-type sex-matched littermate controls (Tables 2 and 3). In addition, BMM cultures from either Lyz2Cre/WT;Notch2ΔPEST/ΔPEST or Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST mice and control littermates formed a similar number of osteoclasts in vitro (Fig. 3, C and F).
Figure 3.
Inversion of the Notch2COIN allele in Lyz2-expressing cells has no skeletal consequences. Documentation of Notch2COIN inversion, analysis of gene expression, and osteoclastogenesis in 1-month-old Lyz2Cre/WT;Notch2ΔPEST/ΔPEST or Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST (black bars; Notch2ΔPEST) and sex-matched Notch2COIN/COIN or Lyz2Cre/Cre;Notch2WT/WT (white bars) controls, respectively. A and D, BMMs from 1-month-old Lyz2Cre/WT;Notch2ΔPEST/ΔPEST (A) or Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST (D) mice and respective controls were cultured for 72 h in the presence of M-CSF at 30 ng/ml. DNA was extracted, and Notch2COIN inversion was demonstrated by gel electrophoresis of PCR products obtained with primers specific for the Notch2ΔPEST allele. The arrows indicate the position of the 250-bp amplicon. B and E, Notch2ΔPEST transcript levels were measured by qRT-PCR in total RNA from the parietal bones of Lyz2Cre/WT;Notch2ΔPEST/ΔPEST (B) or Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST (E) mice and respective controls. Transcript levels are reported as copy number corrected for Rpl38 mRNA levels. Values are means ± S.D.; n = 4–6 biological replicates. Values are means ± S.D.; n = 4 for both controls, n = 4 for Lyz2Cre/WT;Notch2ΔPEST/ΔPEST, n = 6 for Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST, all biological replicates. Two technical replicates were used for each qPCR. C and F, BMMs from 1-month-old Lyz2Cre/WT;Notch2ΔPEST/ΔPEST (C) or Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST (F) mice and respective controls were cultured for 72 h in the presence of M-CSF at 30 ng/ml and then with the addition of Rankl at 10 ng/ml until the formation of osteoclasts. Trap activity was assessed by enzyme histochemistry, and data are expressed as number of osteoclasts per well. Values are means ± S.D.; n = 4 for Notch2COIN/COIN, n = 3 for Lyz2Cre/Cre;Notch2WT/WT, n = 5 for Lyz2Cre/WT;Notch2ΔPEST/ΔPEST, and n = 4 for Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST, all biological replicates.
Table 2.
Femoral microarchitecture assessed by μCT of 1- and 4-month-old Lyz2Cre/WT;Notch2ΔPEST/ΔPEST (NotchΔPEST) mice and sex-matched Notch2COIN/COIN littermates (control)
μCT was performed at the femoral distal end for trabecular or midshaft for cortical bone. Values are means ± S.D.
| 1 Month |
4 Months |
|||
|---|---|---|---|---|
| Control | Notch2ΔPEST | Control | Notch2ΔPEST | |
| Males | ||||
| Distal femur trabecular bone | n = 4 | n = 5 | n = 4 | n = 6 |
| Bone volume/total volume (%) | 5.7 ± 1.8 | 6.8 ± 2.5 | 16.6 ± 3.8 | 15.9 ± 8.1 |
| Trabecular separation (μm) | 224 ± 36 | 213 ± 71 | 201 ± 18 | 214 ± 55 |
| Trabecular no. (1/mm) | 4.6 ± 0.7 | 5.0 ± 1.3 | 4.9 ± 0.4 | 4.8 ± 1.1 |
| Trabecular thickness (μm) | 24 ± 2 | 25 ± 1 | 45 ± 5 | 43 ± 4 |
| Connectivity density (1/mm3) | 306 ± 130 | 360 ± 191 | 228 ± 31 | 234 ± 120 |
| Structure model index | 2.8 ± 0.2 | 2.7 ± 0.2 | 1.2 ± 0.3 | 1.4 ± 1.0 |
| Density of material (mg HA/cm3) | 787 ± 13 | 798 ± 8 | 968 ± 29 | 968 ± 18 |
| Femoral midshaft cortical bone | n = 4 | n = 5 | n = 5 | n = 5 |
| Bone volume/total volume (%) | 84.0 ± 2.5 | 85.1 ± 1.3 | 99.6 ± 0.0 | 99.6 ± 0.2 |
| Porosity (%) | 16.0 ± 2.5 | 14.9 ± 1.3 | 0.4 ± 0.0 | 0.4 ± 0.2 |
| Cortical thickness (μm) | 84 ± 12 | 89 ± 8 | 169 ± 14 | 175 ± 9 |
| Total area (mm2) | 1.43 ± 0.17 | 1.49 ± 0.22 | 2.87 ± 0.79 | 3.54 ± 1.99 |
| Bone area (mm2) | 0.41 ± 0.07 | 0.44 ± 0.06 | 1.55 ± 0.47 | 2.26 ± 1.84 |
| Periosteal perimeter (mm) | 4.2 ± 0.3 | 4.3 ± 0.3 | 6.0 ± 0.8 | 6.5 ± 1.7 |
| Endocortical perimeter (mm) | 3.6 ± 0.2 | 3.6 ± 0.3 | 4.0 ± 0.5 | 4.0 ± 0.4 |
| Density of material (mg HA/cm3) | 952 ± 25 | 968 ± 8 | 1187 ± 19 | 1218 ± 23 |
| Females | ||||
| Distal femur trabecular bone | n = 4 | n = 5 | n = 5 | n = 5 |
| Bone volume/total volume (%) | 6.2 ± 1.5 | 5.8 ± 1.6 | 6.7 ± 1.4 | 5.4 ± 1.8 |
| Trabecular separation (μm) | 220 ± 21 | 226 ± 27 | 290 ± 16 | 298 ± 23 |
| Trabecular no. (1/mm) | 4.6 ± 0.5 | 4.5 ± 0.5 | 3.5 ± 0.2 | 3.4 ± 0.2 |
| Trabecular thickness (μm) | 25 ± 1 | 25 ± 1 | 42 ± 3 | 38 ± 4 |
| Connectivity density (1/mm3) | 263 ± 101 | 257 ± 97 | 117 ± 27 | 96 ± 55 |
| Structure model index | 2.7 ± 0.2 | 2.8 ± 0.2 | 2.6 ± 0.3 | 2.7 ± 0.4 |
| Density of material (mg HA/cm3) | 783 ± 15 | 781 ± 15 | 973 ± 15 | 971 ± 22 |
| Femoral midshaft cortical bone | n = 4 | n = 4 | n = 6 | n = 4 |
| Bone volume/total volume (%) | 82.7 ± 3.3 | 83.3 ± 3.0 | 99.5 ± 0.2 | 99.4 ± 0.2 |
| Porosity (%) | 17.3 ± 3.3 | 16.7 ± 3.0 | 0.5 ± 0.2 | 0.6 ± 0.2 |
| Cortical thickness (μm) | 83 ± 15 | 87 ± 10 | 170 ± 7 | 170 ± 4 |
| Total area (mm2) | 1.45 ± 0.10 | 1.56 ± 0.09 | 1.97 ± 0.13 | 2.10 ± 0.10 |
| Bone area (mm2) | 0.42 ± 0.07 | 0.45 ± 0.05 | 1.05 ± 0.07 | 1.17 ± 0.09 |
| Periosteal perimeter (mm) | 4.3 ± 0.1 | 4.4 ± 0.1 | 5.0 ± 0.2 | 5.1 ± 0.1 |
| Endocortical perimeter (mm) | 3.6 ± 0.1 | 3.7 ± 0.1 | 3.4 ± 0.1 | 3.4 ± 0.1 |
| Density of material (mg HA/cm3) | 960 ± 27 | 958 ± 29 | 1217 ± 15 | 1226 ± 20 |
Table 3.
Femoral microarchitecture assessed by μCT of 1-month-old Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST (Notch2ΔPEST) and Notch2COIN/COIN mice (control) of the same sex and age
μCT was performed at the femoral distal end for trabecular or midshaft for cortical bone. Values are means ± S.D.
| 1 Month | Males |
Females |
||
|---|---|---|---|---|
| Control | Notch2ΔPEST | Control | Notch2ΔPEST | |
| Distal femur trabecular bone | n = 3 | n = 4 | n = 3 | n = 3 |
| Bone volume/total Volume (%) | 8.6 ± 3.2 | 7.6 ± 4.3 | 5.7 ± 1.3 | 9.6 ± 4.4 |
| Trabecular separation (μm) | 172 ± 31 | 194 ± 39 | 212 ± 18 | 175 ± 30 |
| Trabecular no. (1/mm) | 6.0 ± 1.1 | 5.3 ± 1.2 | 4.8 ± 0.4 | 5.9 ± 1.1 |
| Trabecular thickness (μm) | 27 ± 1 | 26 ± 3 | 26 ± 1 | 27 ± 4 |
| Connectivity density (1/mm3) | 376 ± 290 | 304 ± 328 | 176 ± 101 | 404 ± 337 |
| Structure model index | 2.8 ± 0.2 | 2.8 ± 0.1 | 2.7 ± 0.3 | 2.5 ± 0.1 |
| Density of material (mg HA/cm3) | 1011 ± 6 | 987 ± 15 | 979 ± 34 | 992 ± 16 |
| Femoral midshaft cortical bone | n = 3 | n = 4 | n = 3 | n = 3 |
| Bone volume/total volume (%) | 81.6 ± 3.9 | 84.1 ± 2.2 | 84.1 ± 0.6 | 85.6 ± 2.5 |
| Porosity (%) | 18.5 ± 3.9 | 15.9 ± 2.2 | 15.9 ± 0.6 | 14.4 ± 2.5 |
| Cortical thickness (μm) | 97 ± 13 | 105 ± 10 | 97 ± 6 | 111 ± 16 |
| Total area (mm2) | 1.55 ± 0.07 | 1. 51 ± 0.13 | 1.50 ± 0.17 | 1.55 ± 0.15 |
| Bone area (mm2) | 0.54 ± 0.03 | 0.53 ± 0.08 | 0.49 ± 0.01 | 0.57 ± 0.06 |
| Periosteal perimeter (mm) | 4.4 ± 0.1 | 4.3 ± 0.2 | 4.3 ± 0.2 | 4.4 ± 0.2 |
| Endocortical perimeter (mm) | 3.6 ± 0.1 | 3.5 ± 0.1 | 3.5 ± 0.3 | 3.5 ± 0.3 |
| Density of material (mg HA/cm3) | 1047 ± 3 | 1066 ± 7 | 1061 ± 14 | 1077 ± 49 |
These results demonstrate that the induction of a dual Notch2 mutant allele in cells of the osteoclastic lineage has no skeletal consequences and that the osteopenic phenotype of the global Notch2HCS mutant mice should be attributed to an effect in alternate cells (27).
Inversion of the Notch2COIN allele in osteoblasts causes osteopenia
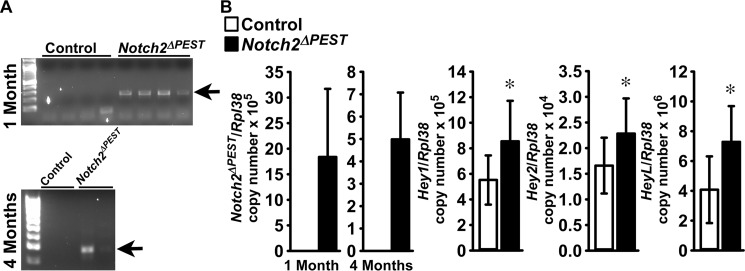
To determine whether the osteopenia observed in mice carrying the HCS mutation is driven by an effect in cells of the osteoblastic lineage, the Notch2ΔPEST mutation was created in Bglap-expressing cells. For this purpose, BGLAP-Cre+/−;Notch2COIN/COIN and Notch2COIN/COIN mice were crossed to create BGLAP-Cre;Notch2ΔPEST/ΔPEST mice and littermate Notch2COIN/COIN controls. As reported previously, BGLAP-Cre transgenics do not have a skeletal phenotype when compared with wild-type mice (15). Inversion of the COIN allele was detected in DNA from parietal bones of BGLAP-Cre;Notch2ΔPEST/ΔPEST mice at 1 and 4 months of age but not in littermate controls (Fig. 4A). Accordingly, the Notch2ΔPEST transcript was detected only in bones from BGLAP-Cre;Notch2ΔPEST/ΔPEST mice, documenting the induction of the HCS mutation in cells that express BGLAP. The presence of the Notch2ΔPEST mRNA was associated with increased transcript levels for Hey1, Hey2, and HeyL, demonstrating increased Notch2 signaling (Fig. 4B).
Figure 4.
Inversion of the Notch2COIN allele in osteoblasts leads to Notch2 activation in vivo. Documentation of Notch2COIN inversion and analysis of gene expression in BGLAP-Cre;Notch2ΔPEST/ΔPEST (black bars; Notch2ΔPEST) and Notch2COIN/COIN littermate controls (white bars). A, DNA was extracted from the parietal bones of 1- and 4-month-old male mice, and Notch2COIN inversion was demonstrated by gel electrophoresis of PCR products obtained with primers specific for the Notch2ΔPEST allele. The arrows indicate the position of the 250-bp amplicon. B, gene expression was measured by qRT-PCR in total RNA from tibiae of 4-month-old mice. Transcript levels are reported as Notch2ΔPEST, Hey1, Hey2, and HeyL mRNA copy number corrected for Rpl38 expression. Values are means ± S.D.; n = 11 biological replicates for both groups. Two technical replicates were used for each qPCR. *, significantly different between Notch2ΔPEST and control, p < 0.05 by t test.
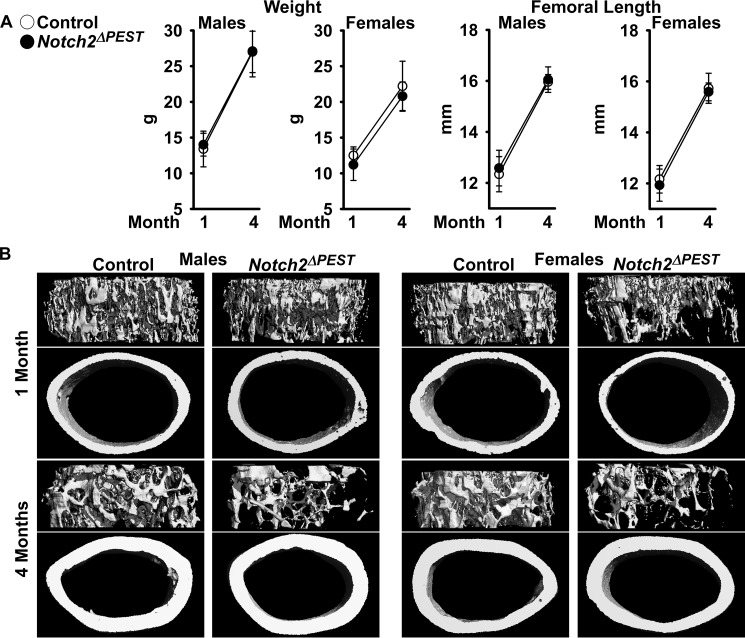
The general appearance, weight, and femoral length of 1- and 4-month-old BGLAP-Cre;Notch2ΔPEST/ΔPEST mice were not different from those of control sex-matched littermates (Fig. 5A). At 1 month of age, μCT revealed cancellous and cortical bone osteopenia in BGLAP-Cre;Notch2ΔPEST/ΔPEST female but not male mice. BGLAP-Cre/Rpl38 copy number was (mean ± S.D.; n = 5–6) 1.6 ± 0.7 in male and 3.5 ± 1.2 (p < 0.05) in female littermates, possibly explaining the absence of a phenotype in BGLAP-Cre;Notch2ΔPEST/ΔPEST male mice. One month old BGLAP-Cre;Notch2ΔPEST/ΔPEST female mice had an ∼50% reduction in cancellous bone volume secondary to a reduced number of trabeculae and connectivity density, associated with increased SMI, indicating a prevalence of rod-like over plate-like trabeculae. Cortical bone thickness and bone area were decreased in female mutant mice, and cortical bone was porous (Fig. 5B and Table 4). At 4 months of age, the skeletal phenotype of BGLAP-Cre;Notch2ΔPEST/ΔPEST female mice was less pronounced, and cancellous bone volume/total volume was 30% lower than in control littermates (p < 0.071). A modest cortical osteopenia with cortical thinning and increased porosity was observed in BGLAP-Cre;Notch2ΔPEST/ΔPEST 4-month-old mice of both sexes (Fig. 6B and Table 4).
Figure 5.
Notch2 activation in osteoblasts causes osteopenia. One- and 4-month-old male and female BGLAP-Cre;Notch2ΔPEST/ΔPEST (black dots; Notch2ΔPEST) were compared with sex-matched littermate Notch2COIN/COIN controls (open circles). A, weight and femoral length. Values are means ± S.D.; in males at 1 month of age n = 7 for control, n = 12 for Notch2ΔPEST, and at 4 months of age n = 6 for control, n = 6 for Notch2ΔPEST; in females at 1 month of age n = 7 for control, n = 7 for Notch2ΔPEST, and at 4 months of age n = 5 for control, n = 6 for Notch2ΔPEST, all biological replicates. B, representative μCT images of femoral proximal trabecular bone and midshaft. Complete data set in Table 4.
Table 4.
Femoral microarchitecture assessed by μCT of 1- and 4-month-old BGLAP-Cre;Notch2ΔPEST/ΔPEST (Notch2ΔPEST) mice and sex-matched Notch2COIN/COIN littermates (control)
μCT was performed at the femoral distal end for trabecular or midshaft for cortical bone. Values are means ± S.D.
| 1 Month |
4 Months |
|||
|---|---|---|---|---|
| Control | Notch2ΔPEST | Control | Notch2ΔPEST | |
| Males | ||||
| Distal femur trabecular bone | n = 7 | n = 12 | n = 6 | n = 6 |
| Bone volume/total volume (%) | 10.0 ± 4.4 | 10.4 ± 5.5 | 13.3 ± 2.5 | 11.7 ± 2.8 |
| Trabecular separation (μm) | 176 ± 32 | 186 ± 29 | 207 ± 30 | 226 ± 48 |
| Trabecular no. (1/mm) | 5.9 ± 1.0 | 5.6 ± 0.9 | 4.9 ± 0.7 | 4.6 ± 0.9 |
| Trabecular thickness (μm) | 28 ± 4 | 29 ± 6 | 44 ± 5 | 40 ± 3 |
| Connectivity density (1/mm3) | 500 ± 262 | 532 ± 224 | 341 ± 124 | 361 ± 159 |
| Structure model index | 2.6 ± 0.5 | 2.4 ± 0.5 | 2.0 ± 0.2 | 2.1 ± 0.3 |
| Density of material (mg HA/cm3) | 799 ± 15 | 791 ± 11 | 941 ± 10 | 927 ± 11a |
| Femoral midshaft cortical bone | n = 7 | n = 12 | n = 6 | n = 6 |
| Bone volume/total volume (%) | 83.7 ± 2.9 | 83.6 ± 1.5 | 88.5 ± 1.0 | 87.0 ± 0.9a |
| Porosity (%) | 16.3 ± 2.9 | 16.5 ± 1.5 | 11.5 ± 1.0 | 13.0 ± 0.9a |
| Cortical thickness (μm) | 95 ± 13 | 93 ± 14 | 179 ± 7 | 158 ± 9a |
| Total area (mm2) | 1.52 ± 0.14 | 1.65 ± 0.16 | 2.2 ± 0.2 | 2.2 ± 0.3 |
| Bone area (mm2) | 0.48 ± 0.07 | 0.51 ± 0.10 | 1.00 ± 0.08 | 1.03 ± 0.33 |
| Periosteal perimeter (mm) | 4.4 ± 0.2 | 4.5 ± 0.2 | 5.2 ± 0.2 | 5.3 ± 0.4 |
| Endocortical perimeter (mm) | 3.6 ± 0.1 | 3.8 ± 0.1a | 3.8 ± 0.2 | 3.9 ± 0.4 |
| Density of material (mg HA/cm3) | 967 ± 27 | 961 ± 29 | 1198 ± 12 | 1182 ± 11a |
| Females | ||||
| Distal femur trabecular bone | n = 7 | n = 7 | n = 5 | n = 6 |
| Bone volume/total volume (%) | 9.3 ± 3.0 | 4.3 ± 2.3a | 5.7 ± 1.0 | 4.0 ± 1.7b |
| Trabecular separation (μm) | 175 ± 32 | 293 ± 56a | 309 ± 37 | 377 ± 64b |
| Trabecular no. (1/mm) | 5.9 ± 1.0 | 3.6 ± 0.8a | 3.3 ± 0.4 | 2.8 ± 0.5b |
| Trabecular thickness (μm) | 27 ± 1 | 25 ± 4 | 43 ± 3 | 39 ± 4b |
| Connectivity density (1/mm3) | 517 ± 244 | 187 ± 178a | 103 ± 32 | 79 ± 45 |
| Structure model index | 2.6 ± 0.2 | 3.0 ± 0.3a | 2.7 ± 0.1 | 2.7 ± 0.3 |
| Density of material (mg HA/cm3) | 791 ± 11 | 782 ± 11a | 947 ± 16 | 921 ± 13a |
| Femoral midshaft cortical bone | n = 7 | n = 7 | n = 5 | n = 6 |
| Bone volume/total volume (%) | 83.3 ± 1.3 | 79.6 ± 3.2a | 87.6 ± 1.1 | 86.3 ± 0.7a |
| Porosity (%) | 16.7 ± 1.3 | 20.4 ± 3.2a | 12.4 ± 1.1 | 13.7 ± 0.7a |
| Cortical thickness (μm) | 93 ± 7 | 78 ± 8a | 171 ± 16 | 152 ± 7a |
| Total area (mm2) | 1.54 ± 0.15 | 1.44 ± 0.18 | 1.73 ± 0.14 | 1.60 ± 0.07 |
| Bone area (mm2) | 0.48 ± 0.06 | 0.40 ± 0.05a | 0.85 ± 0.12 | 0.74 ± 0.05b |
| Periosteal perimeter (mm) | 4.4 ± 0.2 | 4.2 ± 0.3 | 4.7 ± 0.2 | 4.5 ± 0.1 |
| Endocortical perimeter (mm) | 3.7 ± 0.2 | 3.6 ± 0.2 | 3.3 ± 0.1 | 3.3 ± 0.1 |
| Density of material (mg HA/cm3) | 965 ± 29 | 941 ± 34 | 1235 ± 7 | 1211 ± 15a |
a Data are significantly different between control and Notch2ΔPEST, p < 0.05 by unpaired t test.
b p < 0.071 done by unpaired t test.
Figure 6.
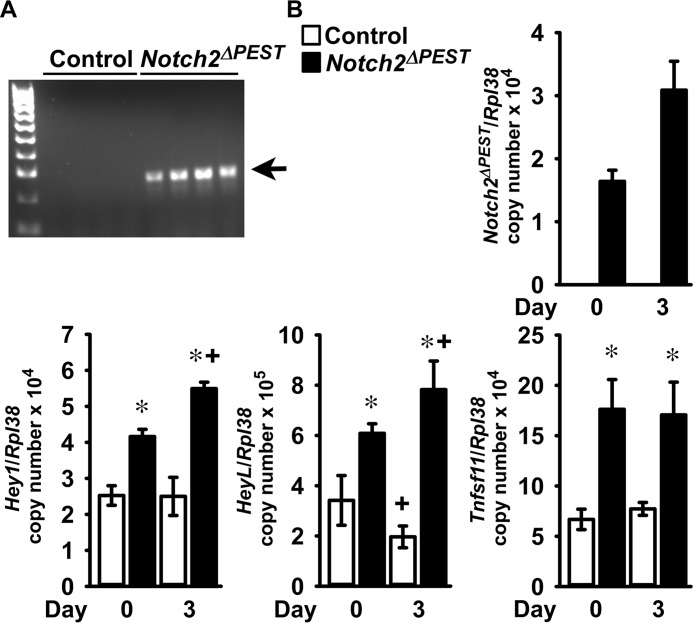
Notch2 activation in osteoblasts induces Tnfsf11 expression. Calvarial osteoblast-enriched cells from 3- to 5-day-old Notch2COIN/COIN mice of both sexes were infected with Ad-CMV-Cre (Notch2ΔPEST; black bars) or Ad-CMV-GFP (control, white bars). A, DNA was extracted, and Notch2COIN inversion was documented by gel electrophoresis of PCR products obtained with primers specific for the Notch2ΔPEST allele. The arrow indicates the position of the 250-bp amplicon. B, total RNA was extracted, and gene expression was measured by qRT-PCR in the presence of specific primers. Transcript levels are reported as Notch2ΔPEST, Hey1, HeyL, and Tnfsf11, corrected for Rpl38 expression. Values are means ± S.D.; n = 4 for all groups, all technical replicates from the same cell preparation. Two technical replicates were used for each qPCR. *, significantly different between Notch2ΔPEST and control, p < 0.05; +, significantly different from day 0, p < 0.05; two-way analysis of variance with Holm-Šídák post-hoc analysis.
Cancellous bone histomorphometry of the distal femur of 1-month-old female BGLAP-Cre;Notch2ΔPEST/ΔPEST mice confirmed the decreased bone volume/tissue volume secondary to a reduced number of trabeculae. Eroded surface and osteoclast numbers were increased, whereas the numbers of osteoblasts and bone formation rates were not different from control littermates (Table 5). Cortical bone histomorphometry revealed a suppressed endocortical mineral apposition rate in BGLAP-Cre;Notch2ΔPEST/ΔPEST mice (Table 6).
Table 5.
Cancellous bone histomorphometry of 1-month-old BGLAP-Cre;Notch2ΔPEST/ΔPEST (Notch2ΔPEST) female mice and sex-matched Notch2COIN/COIN littermates (control)
Histomorphometry was carried out on sagittal sections of the distal femur. Values are means ± S.D.
| Distal femur trabecular bone | Control | Notch2ΔPEST |
|---|---|---|
| Static histomorphometry | n = 6 | n = 5 |
| Bone volume/tissue volume (%) | 11.1 ± 1.8 | 6.2 ± 1.3a |
| Trabecular separation (μm) | 274 ± 57 | 542 ± 348 |
| Trabecular no. (1/mm) | 3.4 ± 0.7 | 2.2 ± 0.9a |
| Trabecular thickness (μm) | 34 ± 7 | 32 ± 11 |
| Osteoblast surface/bone surface (%) | 13.0 ± 6.0 | 15.5 ± 10.3 |
| Osteoblasts/bone perimeter (1/mm) | 12.0 ± 5.1 | 13.5 ± 8.2 |
| Osteoid surface/bone surface (%) | 1.4 ± 1.4 | 1.4 ± 1.9 |
| Osteoclast surface/bone surface (%) | 17.2 ± 3.8 | 27.0 ± 9.3a |
| Osteoclasts/bone perimeter (1/mm) | 5.8 ± 1.0 | 9.8 ± 3.8a |
| Eroded surface/bone surface (%) | 8.2 ± 1.4 | 14.2 ± 4.2a |
| Dynamic histomorphometry | n = 3 | n = 3 |
| Mineral apposition rate (μm/day) | 2.8 ± 1.1 | 2.5 ± 0.5 |
| Mineralizing surface/bone surface (%) | 2.5 ± 1.0 | 4.0 ± 2.2 |
| Bone formation rate (μm3/μm2/day) | 0.08 ± 0.05 | 0.11 ± 0.07 |
a Data are significantly different between control and Notch2ΔPEST, p < 0.05 by unpaired t test.
Table 6.
Cortical bone histomorphometry of 1-month-old BGLAP-Cre;Notch2ΔPEST/ΔPEST (Notch2ΔPEST) female mice and sex-matched Notch2COIN/COIN littermates (control)
Cortical bone histomorphometry was performed at the femoral mid-diaphysis. Values are means ± S.D.
| Control | Notch2ΔPEST | |
|---|---|---|
| Cortical bone | n = 6 | n = 6 |
| Cortical thickness (μm) | 199 ± 19 | 190 ± 23 |
| Bone area (mm2) | 0.48 ± 0.04 | 0.45 ± 0.08 |
| Endocortical surface | ||
| Static histomorphometry | n = 6 | n = 6 |
| Osteoblasts/bone perimeter (1/mm) | 10.3 ± 5.0 | 6.0 ± 2.3 |
| Osteoclasts/bone perimeter (1/mm) | 2.4 ± 0.6 | 2.7 ± 0.7 |
| Eroded surface/bone surface (%) | 3.9 ± 0.9 | 4.1 ± 0.9 |
| Dynamic histomorphometry | n = 4 | n = 5 |
| Mineral apposition rate (μm/day) | 2.5 ± 0.3 | 1.7 ± 0.2a |
a Data are significantly different between control and Notch2ΔPEST, p < 0.05 by unpaired t test.
Inversion of the Notch2COIN allele in osteoblasts induces Tnfsf11
To determine the mechanisms responsible for the skeletal phenotype of the BGLAP-Cre;Notch2ΔPEST/ΔPEST mice, osteoblast-enriched cells were obtained from the parietal bones of Notch2COIN/COIN newborns. Cultures were infected with an adenoviral vector expressing Cre recombinase under the control of the cytomegalovirus (CMV) promoter, and parallel cultures infected with an adenoviral vector where the CMV promoter governs GFP expression (Ad-CMV-GFP) served as controls. Ad-CMV-Cre, but not Ad-CMV-GFP, infection led to the inversion of the COIN module and expression of the Notch2ΔPEST mRNA associated with induction of Hey1 and HeyL, demonstrating activation of Notch signaling (Fig. 6, A and B). In accordance with the enhanced bone resorption observed in BGLAP-Cre;Notch2ΔPEST/ΔPEST mice, expression of Tnfsf11 was induced in Notch2ΔPEST cells (27).
Discussion
In this study, the individual contributions of the osteoclast and osteoblast lineages to the bone loss observed in Notch2HCS mutant mice were explored by the conditional introduction of the HCS genetic defect in selected cell lineages. The mutations associated with the disease occur within exon 34 of NOTCH2, and conditional insertion of a premature STOP codon in the homologous region of the murine Notch2 locus was achieved by the creation of a COIN allele. The COIN module can be introduced directly into coding exons without disrupting the expression or function of the targeted allele, a goal that cannot be accomplished with traditional Cre-loxP approaches (28). Absence of an appreciable phenotype in Notch2COIN/COIN mice documented the skeletal equivalency of the wild-type and engineered Notch2 alleles prior to Cre-mediated inversion. The Notch2ΔPEST mutants generated by germ line inversion of the COIN module expressed the Notch2ΔPEST transcript and exhibited a 50% reduction in wild-type Notch2 mRNA, indicating comparable expression levels of maternal and paternal Notch2. Notch2ΔPEST germ line mice exhibited generalized osteopenia and reduced bone size and length, phenocopying global Notch2HCS mutants. These results validated the COIN strategy and confirmed that generalized expression of a Notch2 mutant lacking the PEST domain causes bone loss (27). Although these findings should be extrapolated with caution to the human condition, they support the concept that de novo or inherited dominant NOTCH2 gain-of-function mutations are responsible for the bone loss in subjects with HCS (33).
Selective introduction of the HCS mutation in osteoblasts, but not in cells of the myeloid lineage, led to generalized bone loss. The reduction in cancellous bone volume was observed only in female mice and was more pronounced in younger BGLAP-Cre;Notch2ΔPEST/ΔPEST mice. The bone loss was attributed to enhanced bone resorption uncoupled from a bone-forming response and suppressed endocortical bone formation. These features are consistent with the skeletal phenotype of global Notch2HCS mutants and demonstrate that a direct effect in osteoblasts is largely responsible for the osteopenia associated with the HCS mutation in mice (27). Absence of a phenotype in Lyz2Cre/WT;Notch2ΔPEST/ΔPEST and Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST mice is congruent with the observation that the Notch2 deletion in Lyz2-expressing cells has no consequences on skeletal homeostasis (18). These results indicate that either the activation or inactivation of Notch2 in myeloid cells in vivo has no skeletal consequences and that the effect of Notch2 on bone resorption is secondary to its actions on alternate cells (17, 27, 34). However, the in vivo observations are in contrast with in vitro studies demonstrating that Notch2 enhances osteoclastogenesis directly and as a result bone resorption (17). This would suggest that the overall effect of Notch2 in osteoclastogenesis is complex and derived from its actions in various cellular lineages.
In agreement with previous work demonstrating increased expression of Tnfs11 in bone extracts from Notch2HCS mutant mice, Notch2ΔPEST/ΔPEST osteoblasts expressed increased levels of Tnfs11 mRNA suggesting that osteoblast-derived Rankl is responsible for the enhanced bone resorption in vivo in HCS mutant mice. These findings are in agreement with those in a subject with HCS and severe osteoporosis who was reported to present with elevated levels of circulating RANKL (27, 35). However, a limitation of this work was the inability to detect Rankl protein by Western blot analysis in either control or Notch2ΔPEST/ΔPEST osteoblasts. This is possibly related to low levels of Rankl expression and the lack of available antibodies with sufficient sensitivity to detect murine Rankl in osteoblasts. There was an absence of a bone-forming response to the increased bone resorption implying that Notch2 inhibits bone formation. Moreover, Notch2 gain-of-function suppresses endocortical mineral apposition rate, an effect that possibly contributes to the cortical osteopenic phenotype. The role of Notch2 as an inhibitor of bone formation is supported by previous studies demonstrating that deletion of Notch2 in Runx2-expressing cells increases trabecular bone volume due to enhanced osteoblast differentiation and activity (18). Further support for an inhibitory role of Notch2 on bone formation is derived from studies showing that the dual inactivation of Notch1 and Notch2 in cells of the osteoblastic lineage increases bone mass (36, 37).
It is important to mention that some discrepancies exist between the phenotypes of the BGLAP-Cre;Notch2ΔPEST/ΔPEST mice and of the global Notch2HCS mutants (27). The osteoblast-selective mutation did not affect femoral length, and this was expected because the BGLAP-Cre transgene is not expressed in chondrocytes, cells that govern longitudinal bone growth. Direct inhibitory effects of Notch2 on endochondral bone formation are accountable for the reduced femoral length of the Notch2HCS mutants (38, 39). Cancellous bone osteopenia was detected only in female BGLAP-Cre;Notch2ΔPEST/ΔPEST mice, although both sexes were affected by the global Notch2HCS mutation (27). These sex-related differences may be secondary to the more pronounced expression of the BGLAP-Cre transgene in female than in male mice. Alternatively, a higher rate of bone remodeling in young female than in male mice, a known attribute of the C57BL/6 genetic background, might have sensitized female mice to a greater activation of Notch2 in osteoblasts (40, 41). The cortical bone osteopenia was milder in BGLAP-Cre;Notch2ΔPEST/ΔPEST than in the Notch2HCS mice, and low expression of the BGLAP-Cre transgene during embryonic skeletal development might account for the less pronounced phenotype of the conditional mice (42). It is of interest that the BGLAP-Cre;Notch2ΔPEST/ΔPEST mice did not display the increase in endocortical bone resorption observed in the global Notch2HCS mutants. This difference may also account for the modest cortical bone phenotype of the conditional mice and suggests that the presence of the HCS mutation in both osteoclasts and osteoblasts might be necessary to recapitulate the cortical bone-resorptive phenotype and osteopenia of the Notch2HCS mouse (27).
The conditional HCS model described in this study reaffirmed that Notch2, like Notch1, increases the transcript levels of Hey1, Hey2, and HeyL, thereby confirming that both paralogs are able to activate Rbpjκ-mediated Notch signaling in skeletal cells. The increase in mRNA levels for the Notch target genes reflects activation of the Notch canonical pathway but does not imply that Hey proteins mediate the effects of Notch2 in bone. In fact, either generalized or skeletal misexpression of Heys has a small impact on skeletal microarchitecture (43–46). The current observations also indicate that Notch1 and Notch2 have distinct skeletal functions because Notch1 induces osteoprotegerin and inhibits bone resorption, whereas Notch2 induces Rankl and stimulates the resorptive event.
In conclusion, osteoblast expression of a Notch2 mutant lacking the PEST domain causes osteopenia in mice.
Experimental procedures
Creation of the Notch2COIN mouse
The targeting vector containing the COIN element was electroporated into embryonic stem (ES) cells, and the cassette was used for the selection of G418-resistant cells from 129SvJ/C57BL/6J embryos at the Gene Targeting and Transgenic Facility of UConn Health. Targeted clones were verified by long-range PCR of genomic DNA. Correct integration of the 5′-homology arm was tested with forward F1 5′-GGGAGGTGCTTACCGACCTCTC-3′ and reverse R1 5′-CACCCTGAAAACTTTGCCCCCTCC-3′ primers followed by nested forward F2 5′-CTGTTCTTGGATACCGAGGTACAC-3′ and reverse R2 5′-CAATCAAGGGTCCCCAAACTCAC-3′ primers. Proper integration of the 3′-homology arm was ensured with forward F3 5′-CCAAAACCCGGCGCGGAGGCCATGC-3′ and reverse R3 5′-CACTTGAGAGCAAGGCTGCAAGGC-3′ primers followed by nested forward F4 5′-CCTTCTTCTCTTTCCTACAGTACCCC-3′ and reverse R4 5′-GGTGCAAGGGCAGGAGATCAACAG-3′ primers (all primers from Integrated DNA Technologies, IDT, Coralville, IA). Positive ES clones were used for morula aggregations and the creation of chimeras, and the Frt-neo-Pgk1polyA-Frt cassette was removed by FLP recombination following crosses of male chimeras with mice expressing FLP under the control of the Rosa26 promoter (Rosa26FLP; The Jackson Laboratory, Bar Harbor, ME) (47, 48). Excision of the cassette was verified by PCR in ear punches of F1 pups, and the Rosa26FLP allele segregated by breeding with C57BL/6J wild-type mice. Correct integration of the COIN module into the Notch2 locus was confirmed in the progeny by loss of wild-type allele assay.
Induction of the HCS mutation in the germ line, osteoclasts, or osteoblasts
To test whether the Notch2COIN and Notch2WT alleles are functionally equivalent, the skeletal phenotype of Notch2COIN/COIN mice was compared with the phenotype of wild-type C57BL/6J controls of the same age and sex. To achieve systemic inversion of the Notch2COIN allele, F1 heterozygous Notch2COIN/WT male mice were bred with female mice expressing Cre under the control of the Hprt promoter (HprtCre) (49). This resulted in the germ line inversion of the COIN module and consequent creation of mice heterozygous for the Notch2ΔPEST allele (Notch2ΔPEST/WT). The latter were crossed with wild-type C57BL/6J mice to generate Notch2ΔPEST/WT experimental and wild-type control cohorts.
C57BL/6J mice where the Cre coding sequence was inserted into the endogenous Lyz2 locus (Lyz2Cre; The Jackson Laboratory) were used to express Cre recombinase in cells of the myeloid lineage (50, 51). To induce inversion of the COIN module in osteoclast precursor, homozygous Notch2COIN mice heterozygous for the Lyz2Cre allele (Lyz2Cre/WT; Notch2COIN/COIN) were bred with Notch2COIN/COIN mice to create Lyz2Cre/WT;Notch2ΔPEST/ΔPEST mice. In an alternate mating scheme, heterozygous Notch2COIN mice homozygous for the Lyz2Cre allele (Lyz2Cre/Cre;Notch2COIN/WT) were inter-mated to create Lyz2Cre/Cre;Notch2ΔPEST/ΔPEST experimental and Lyz2Cre/Cre;Notch2WT/WT control mice.
C57BL/6J mice harboring a transgene where the Cre recombinase coding sequence was cloned downstream a 3.9-kb human BGLAP promoter fragment (BGLAP-Cre; The Jackson Laboratory) were used to induce inversion of the COIN module in osteoblasts (42). Hemizygous BGLAP-Cre transgenics homozygous for the Notch2COIN allele (BGLAP-Cre;Notch2COIN/COIN) were bred with Notch2COIN/COIN mice to generate BGLAP-Cre;Notch2ΔPEST/ΔPEST experimental and Notch2COIN/COIN littermate control cohorts.
Allelic composition was determined by PCR analysis in tail DNA with primers specific for the HprtWT, HprtCre, Notch2WT, Notch2COIN, Notch2ΔPEST, Lyz2Cre, and Lyz2WT alleles and for the BGLAP-Cre transgene. Inversion of the COIN module was documented by PCR analysis in DNA from BMMs or parietal bones (all primers were from IDT; Table 7). The generation and establishment of the Notch2COIN mouse line were approved by the Institutional Animal Care and Use Committees of UConn Health and of Saint Francis Hospital Medical Center. All other studies were approved by the Institutional Animal Care and Use Committee of UConn Health.
Table 7.
Primers used for genotyping and determination of the COIN module inversion by PCR
| Allele | Strand | Sequence 5′–3′ | Amplicon size (bp) |
|---|---|---|---|
| Notch2COIN | Forward | CCGGGCCGCGACTGAAACCCTAG | 330 |
| Reverse | CCACCACCTCCAGGAGTTGGGC | ||
| Notch2WT | Forward | GCTCAGACCATTGTGCCAACCTAT | 100 |
| Reverse | CAGCAGCATTTGAGGAGGCGTAA | ||
| HprtWT | Forward | TTTCTATAGGACTGAAAGACTTGCTC | 200 |
| Reverse | CACAGTAGCTCTTCAGTCTGATAAAA | ||
| HprtCre | Forward | GCGGTCTGGCAGTAAAAACTATC | 100 |
| Reverse | GTGAAACAGCATTGCTGTCACTT | ||
| Lyz2Cre | Forward1 | TTACAGTCGGCCAGGCTGAC | Lyz2WT = 350 |
| Forward2 | CCCAGAAATGCCAGATTACG | Lyz2Cre = 700 | |
| Reverse | CTTGGGCTGCCAGAATTTCTC | ||
| BGLAP-Cre | Forward | CAAATAGCCCTGGCAGAT | 300 |
| Reverse | TGATACAAGGGACATCTTCC | ||
| Fabp1 | Forward | TGGACAGGACTGGACCTCTGCTTTCC | 200 |
| Reverse | TAGAGCTTTGCCACATCACAGGTCAT | ||
| Notch2ΔPEST | Forward | GTACTTCAGCACAGTTTTAGAGAAC | 250 |
| Reverse | GTGAGTCACCCGCCGGATGTC |
Microcomputed tomography
Femoral microarchitecture was determined using a microcomputed tomography instrument (Scanco μCT 40; Scanco Medical AG, Bassersdorf, Switzerland), which was calibrated periodically using a phantom provided by the manufacturer (41, 52). Femurs were scanned in 70% ethanol at high resolution, energy level of 55 peak kV, intensity of 145 μA, and integration time of 200 ms. A total of 100 slices at midshaft and 160 slices at the distal metaphysis was acquired at an isotropic voxel size of 216 μm3 and a slice thickness of 6 μm and chosen for analysis. Trabecular bone volume fraction (bone volume/total volume) and microarchitecture were evaluated starting ∼1.0-mm proximal from the femoral condyles. Contours were manually drawn every 10 slices, a few voxels away from the endocortical boundary, to define the region of interest for analysis, whereas the remaining slice contours were iterated automatically. Total volume, bone volume, bone volume fraction, trabecular thickness, trabecular number, connectivity density, SMI, and material density were measured in trabecular regions using a Gaussian filter (σ = 0.8) and user-defined thresholds (41, 52). For analysis of cortical bone, contours were iterated across 100 slices along the cortical shell of the femoral midshaft, excluding the marrow cavity. Analysis of bone volume/total volume, porosity, cortical thickness, total cross-sectional and cortical bone area, periosteal and endosteal perimeter, and material density were conducted using a Gaussian filter (σ = 0.8, support = 1) with operator-defined thresholds.
Bone histomorphometric analysis
Bone histomorphometry was carried out in 1-month-old mice injected with 20 mg/kg calcein and 50 mg/kg demeclocycline at a 2-day interval and sacrificed 2 days after demeclocycline administration. Femurs were dissected, fixed in 70% ethanol, and embedded in methyl methacrylate. For cancellous bone analysis, bones were sectioned at a thickness of 5 μm along the sagittal plane on a Microm microtome (Richards-Allan Scientific, Kalamazoo, MI) and stained with 0.1% toluidine blue. Static and dynamic parameters of bone morphometry were measured in a defined area between 0.35 and 2.16 mm from the growth plate at a magnification of ×100 using an OsteoMeasure morphometry system (Osteometrics, Atlanta, GA). Stained sections were used to draw the bone and to measure trabecular separation, number, and thickness, osteoid and eroded surface, as well as to count osteoblast and osteoclast surface and number. Mineralizing surface per bone surface and mineral apposition rate were measured on unstained sections visualized under UV light and a triple diamidino-2-phenylindole/fluorescein/Texas Red set long pass filter, and bone formation rate was calculated.
For cortical histomorphometry, femurs were embedded in methyl methacrylate and cut through the mid-diaphysis along the transverse plane with an EXAKT Precision Saw, ground using an EXAKT 400 CS Micro Grinding System (Exakt Technologies, Oklahoma City, OK), and surface-polished to a thickness of ∼15 μm (Alizee Pathology, Baltimore, MD). Parameters of cortical bone morphometry were measured at a magnification of ×400 using OsteoMeasureXP software (Osteometrix). Stained sections were used to draw the cortical bone, marrow space, and cell surfaces, as well as to measure osteoblasts and osteoclasts along the endocortical surface. Mineral apposition rate was measured in unstained sections under UV light, using a triple diamidino-2-phenylindole/fluorescein/Texas Red set long pass filter. Terminology and units used for cancellous and cortical bone histomorphometry are those recommended by the Histomorphometry Nomenclature Committee of the American Society for Bone and Mineral Research (53, 54).
Culture of BMMs and osteoclast formation
To obtain BMMs, the marrow was removed by flushing with a 26-gauge needle, and erythrocytes were lysed in 150 mm NH4Cl, 10 mm KHCO3, and 0.1 mm EDTA (pH 7.4). Cells were centrifuged, and the sediment was suspended in α-minimum essential medium (α-MEM) in the presence of 10% fetal bovine serum (FBS; both from Thermo Fisher Scientific, Waltham, MA) and recombinant human M-CSF at 30 ng/ml. M-CSF cDNA and expression vector were obtained from D. Fremont (St. Louis, MO), and M-CSF was purified as reported previously (55). Cells were seeded at a density of 300,000 cells/cm2 and cultured for 3–4 days. Inversion of the COIN module was documented by PCR of genomic DNA using primers specific for the Notch2ΔPEST allele (Table 7). For osteoclast formation, cells were collected following treatment with 0.05% trypsin/EDTA for 5 min and seeded at a density of 47,000 cells/cm2 in α-MEM with 10% FBS, M-CSF at 30 ng/ml, and recombinant murine Rankl at 10 ng/ml. Rankl cDNA and expression vector were obtained from M. Glogauer (Toronto, Canada), and GST-tagged Rankl was expressed and purified as described (56). Cultures were carried out until formation of multinucleated tartrate-resistant acid phosphatase (Trap)-positive cells. Trap enzyme histochemistry was conducted using a commercial kit (Sigma), in accordance with manufacturer's instructions. Trap-positive cells containing more than three nuclei were considered osteoclasts.
Osteoblast-enriched cell cultures
The parietal bones of 3–5-day-old Notch2COIN/COIN mice were exposed to 1.2 units/ml LiberaseTM TL (Sigma) for 20 min at 37 °C, and cells were extracted in five consecutive reactions (57). Cells from the last three digestions were pooled and seeded at a density of 10,000 cells/cm2, as described (40). Osteoblast-enriched cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with non-essential amino acids (both from Thermo Fisher Scientific), 20 mm HEPES, 100 μg/ml ascorbic acid (both from Sigma), and 10% heat-inactivated FBS (Atlanta Biologicals, Norcross, GA) in a humidified 5% CO2 incubator at 37 °C. To induce inversion of the COIN allele, cells were infected with Ad-CMV-Cre, and parallel cultures infected with Ad-CMV-GFP (both from Vector Biolabs, Philadelphia, PA) served as controls (58). To this end, sub-confluent osteoblast-enriched cells were transferred to culture medium containing 2% heat-inactivated FBS for 1 h and exposed overnight to 100 multiplicity of infection of replication-defective recombinant adenoviruses. Cells were allowed to recover for 24 h in DMEM containing 10% heat-inactivated FBS and then seeded at a density of 22,000 cells/cm2. Confluent cultures were exposed to medium supplemented with 5 mm β-glycerophosphate (Sigma) to induce osteoblast maturation. To document inversion of the COIN module, the presence of the Notch2ΔPEST allele was determined by PCR in genomic DNA using specific primers (Table 7).
RNA integrity and qRT-PCR
Total RNA was extracted from osteoblast-enriched cells with the RNeasy kit (Qiagen, Valencia, CA) and from homogenized bones with the micro RNeasy kit (Qiagen), in accordance with manufacturer's instructions. The integrity of the RNA was assessed by microfluidic electrophoresis on an Experion system (Bio-Rad), and only RNA with a quality indicator number equal to or higher than 7.0 was used for subsequent analysis (59, 60). Equal amounts of RNA were reverse-transcribed using the iScript RT-PCR kit (Bio-Rad) and amplified in the presence of specific primers (all primers from IDT; Table 8) with the iQ SYBR Green Supermix (Bio-Rad) at 60 °C for 35 cycles. Transcript copy number was estimated by comparison with a serial dilution of cDNA for Hes1 (from American Type Culture Collection, ATCC; Manassas, VA), Hey1 or Hey2 (T. Iso, Los Angeles, CA), HeyL (D. Srivastava, Dallas, TX), or Tnfsf11 (Source BioScience, Nottingham, UK) (61–64).
Table 8.
Primers used for qRT-PCR determinations
GenBankTM accession numbers identify the transcripts recognized by primer pairs.
| Gene | Strand | Sequence 5′–3′ | GenBankTM accession no. |
|---|---|---|---|
| Hes1 | Forward | ACCAAAGACGGCCTCTGAGCACAGAAAGT | NM_008235 |
| Reverse | ATTCTTGCCCTTCGCCTCTT | ||
| Hey1 | Forward | ATCTCAACAACTACGCATCCCAGC | NM_010423 |
| Reverse | GTGTGGGTGATGTCCGAAGG | ||
| Hey2 | Forward | AGCGAGAACAATTACCCTGGGCAC | NM_013904 |
| Reverse | GGTAGTTGTCGGTGAATTGGACCT | ||
| HeyL | Forward | CAGTAGCCTTTCTGAATTGCGAC | NM_013905 |
| Reverse | AGCTTGGAGGAGCCCTGTTTC | ||
| Notch2WT | Forward | CCATTGTGCCAACCTATCAT | NM_010928a |
| Reverse | TTGAGGAGGCGTAACTGT | ||
| Notch2ΔPEST | Forward | GGCTTTCCCACCTACCAT | Not applicable |
| Reverse | TAGTCGGGCACGTCGTAG | ||
| Rpl38 | Forward | AGAACAAGGATAATGTGAAGTTCAAGGTTC | NM_001048057; NM_001048058; NM_023372 |
| Reverse | CTGCTTCAGCTTCTCTGCCTTT | ||
| Tnfsf11 | Forward | TATAGAATCCTGAGACTCCATGAAAAC | NM_011613 |
| Reverse | CCCTGAAAGGCTTGTTTCATCC |
a This recognizes a fragment coding for the PEST domain of Notch2.
To monitor for the efficiency of the COIN inversion, primers designed to amplify a sequence of the Notch2 transcript coding for the PEST domain were used (Table 8). These primers allow detection by qRT-PCR of the transcripts for Notch2WT and Notch2COIN but not for Notch2ΔPEST, because the latter lacks the sequences coding for the PEST domain. Notch2WT and Notch2COIN copy numbers were measured by comparing with a serial dilution of Notch2 cDNA (Thermo Fisher Scientific). Notch2ΔPEST transcripts were detected with primers that generate an amplicon straddling the artificial splice junction generated within exon 34 of the targeted Notch2 locus upon inversion of the COIN module (Table 8). Primers are specific for the Notch2ΔPEST mRNA and do not recognize the wild-type Notch2 transcript or the Notch2COIN mRNA prior to the COIN inversion. Notch2ΔPEST copy number was estimated by comparison with a serial dilution of an ∼200 bp synthetic DNA template (IDT) cloned into pcDNA3.1(−) (Thermo Fisher Scientific) by isothermal single reaction assembly using commercially available reagents (New England Biolabs, Ipswich, MA) (65).
Amplification reactions were conducted in CFX96 qRT-PCR detection systems (Bio-Rad), and fluorescence was monitored during every PCR cycle at the annealing step. Data are expressed as copy number corrected for Rpl38 expression estimated by comparison with a serial dilution of Rpl38 (ATCC) (66).
Statistics
Data are expressed as means ± S.D. Statistical differences were determined by Student's t test or two-way analysis of variance with Holm-Šídák post hoc analysis for pairwise or multiple comparisons, respectively.
Author contributions
S. Z. designed research studies, conducted experiments, analyzed data, and wrote the manuscript. J. Y. conducted experiments and analyzed data. A. S. conducted experiments and analyzed data. L. S. conducted the analysis of skeletal phenotypes. C. S. and A. N. E. designed and created the Notch2COIN targeting construct. E. C. designed research studies, analyzed data, and wrote the manuscript.
Acknowledgments
We thank D. Fremont for M-CSF cDNA, M. Glogauer for Rankl cDNA, T. Iso for Hey1 and Hey2 cDNAs, D. Srivastava for HeyL cDNA, David Bridgewater and Tabitha Eller for technical assistance, and Mary Yurczak for secretarial support.
This work was supported by National Institutes of Health Grant DK045227 from the NIDDK. C. S. and A. E. receive stock options from Regeneron Pharmaceuticals. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- NICD
- Notch intracellular domain
- Ad
- adenovirus
- α-MEM
- α-minimum essential medium
- ATCC
- American Type Culture Collection
- BMM
- bone marrow-derived macrophage
- CMV
- cytomegalovirus
- eGFP
- enhanced green fluorescent protein
- FLP
- flippase
- FRT
- FLP recognition target
- HCS
- Hajdu-Cheney syndrome
- kb
- kilobase
- L66
- lox66
- L71
- lox71
- L72
- lox72
- IDT
- Integrated DNA Technologies
- M-CSF
- macrophage-colony-stimulating factor
- μCT
- microcomputed tomography
- PEST
- proline- (P), glutamic acid- (E), serine- (S), and (T) threonine-rich
- qRT-PCR
- quantitative reverse transcription-PCR
- rβglpA
- β-globin polyadenylation signal
- Rankl
- receptor activator of nuclear factor κB ligand
- Rbpjκ
- recombination signal binding protein for immunoglobulin κJ region
- Trap
- tartrate-resistant acid phosphatase
- SMI
- structure model index.
References
- 1. Fortini M. E. (2009) Notch signaling: the core pathway and its posttranslational regulation. Dev. Cell 16, 633–647 [DOI] [PubMed] [Google Scholar]
- 2. Kopan R., and Ilagan M. X. (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kovall R. A. (2007) Structures of CSL, notch and mastermind proteins: piecing together an active transcription complex. Curr. Opin. Struct. Biol. 17, 117–127 [DOI] [PubMed] [Google Scholar]
- 4. Iso T., Kedes L., and Hamamori Y. (2003) HES and HERP families: multiple effectors of the Notch signaling pathway. J. Cell. Physiol. 194, 237–255 [DOI] [PubMed] [Google Scholar]
- 5. Groot A. J., Habets R., Yahyanejad S., Hodin C. M., Reiss K., Saftig P., Theys J., and Vooijs M. (2014) Regulated proteolysis of NOTCH2 and NOTCH3 receptors by ADAM10 and presenilins. Mol. Cell. Biol. 34, 2822–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baeten J. T., and Lilly B. (2015) Differential regulation of NOTCH2 and NOTCH3 contribute to their unique functions in vascular smooth muscle cells. J. Biol. Chem. 290, 16226–16237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Z., Brunskill E., Varnum-Finney B., Zhang C., Zhang A., Jay P. Y., Bernstein I., Morimoto M., and Kopan R. (2015) The intracellular domains of Notch1 and Notch2 are functionally equivalent during development and carcinogenesis. Development 142, 2452–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yuan Z., Friedmann D. R., VanderWielen B. D., Collins K. J., and Kovall R. A. (2012) Characterization of CSL (CBF-1, Su(H), Lag-1) mutants reveals differences in signaling mediated by Notch1 and Notch2. J. Biol. Chem. 287, 34904–34916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Canalis E., Giustina A., and Bilezikian J. P. (2007) Mechanisms of anabolic therapies for osteoporosis. N. Engl. J. Med. 357, 905–916 [DOI] [PubMed] [Google Scholar]
- 10. Teitelbaum S. L. (2007) Osteoclasts: what do they do and how do they do it? Am. J. Pathol. 170, 427–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bianco P., and Gehron Robey P. (2000) Marrow stromal stem cells. J. Clin. Invest. 105, 1663–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zanotti S., and Canalis E. (2016) Notch signaling and the skeleton. Endocr. Rev. 37, 223–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai S., Kopan R., Zou W., Hilton M. J., Ong C. T., Long F., Ross F. P., and Teitelbaum S. L. (2008) NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J. Biol. Chem. 283, 6509–6518 [DOI] [PubMed] [Google Scholar]
- 14. Canalis E., Adams D. J., Boskey A., Parker K., Kranz L., and Zanotti S. (2013) Notch signaling in osteocytes differentially regulates cancellous and cortical bone remodeling. J. Biol. Chem. 288, 25614–25625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canalis E., Parker K., Feng J. Q., and Zanotti S. (2013) Osteoblast lineage-specific effects of Notch activation in the skeleton. Endocrinology 154, 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Engin F., Yao Z., Yang T., Zhou G., Bertin T., Jiang M. M., Chen Y., Wang L., Zheng H., Sutton R. E., Boyce B. F., and Lee B. (2008) Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 14, 299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fukushima H., Nakao A., Okamoto F., Shin M., Kajiya H., Sakano S., Bigas A., Jimi E., and Okabe K. (2008) The association of Notch2 and NF-κB accelerates RANKL-induced osteoclastogenesis. Mol. Cell. Biol. 28, 6402–6412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yorgan T., Vollersen N., Riedel C., Jeschke A., Peters S., Busse B., Amling M., and Schinke T. (2016) Osteoblast-specific Notch2 inactivation causes increased trabecular bone mass at specific sites of the appendicular skeleton. Bone 87, 136–146 [DOI] [PubMed] [Google Scholar]
- 19. Zanotti S., Smerdel-Ramoya A., Stadmeyer L., Durant D., Radtke F., and Canalis E. (2008) Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology 149, 3890–3899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheney W. D. (1965) Acro-osteolysis. Am. J. Roentgenol. Radium. Ther. Nucl. Med. 94, 595–607 [PubMed] [Google Scholar]
- 21. Hajdu N., and Kauntze R. (1948) Cranio-skeletal dysplasia. Br. J. Radiol. 21, 42–48 [DOI] [PubMed] [Google Scholar]
- 22. Gray M. J., Kim C. A., Bertola D. R., Arantes P. R., Stewart H., Simpson M. A., Irving M. D., and Robertson S. P. (2012) Serpentine fibula polycystic kidney syndrome is part of the phenotypic spectrum of Hajdu-Cheney syndrome. Eur. J. Hum. Genet. 20, 122–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Isidor B., Lindenbaum P., Pichon O., Bézieau S., Dina C., Jacquemont S., Martin-Coignard D., Thauvin-Robinet C., Le Merrer M., Mandel J. L., David A., Faivre L., Cormier-Daire V., Redon R., and Le Caignec C. (2011) Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 43, 306–308 [DOI] [PubMed] [Google Scholar]
- 24. Majewski J., Schwartzentruber J. A., Caqueret A., Patry L., Marcadier J., Fryns J. P., Boycott K. M., Ste-Marie L. G., McKiernan F. E., Marik I., Van Esch H., FORGE Canada Consortium, Michaud J. L., and Samuels M. E. (2011) Mutations in NOTCH2 in families with Hajdu-Cheney syndrome. Hum. Mutat. 32, 1114–1117 [DOI] [PubMed] [Google Scholar]
- 25. Simpson M. A., Irving M. D., Asilmaz E., Gray M. J., Dafou D., Elmslie F. V., Mansour S., Holder S. E., Brain C. E., Burton B. K., Kim K. H., Pauli R. M., Aftimos S., Stewart H., Kim C. A., et al. (2011) Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 43, 303–305 [DOI] [PubMed] [Google Scholar]
- 26. Zhao W., Petit E., Gafni R. I., Collins M. T., Robey P. G., Seton M., Miller K. K., and Mannstadt M. (2013) Mutations in NOTCH2 in patients with Hajdu-Cheney syndrome. Osteoporos. Int. 24, 2275–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Canalis E., Schilling L., Yee S. P., Lee S. K., and Zanotti S. (2016) Hajdu Cheney mouse mutants exhibit osteopenia, increased osteoclastogenesis and bone resorption. J. Biol. Chem. 291, 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Economides A. N., Frendewey D., Yang P., Dominguez M. G., Dore A. T., Lobov I. B., Persaud T., Rojas J., McClain J., Lengyel P., Droguett G., Chernomorsky R., Stevens S., Auerbach W., Dechiara T. M., et al. (2013) Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc. Natl. Acad. Sci. U.S.A. 110, E3179–E3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yang M., Trettel L. B., Adams D. J., Harrison J. R., Canalis E., and Kream B. E. (2010) Col3.6-HSD2 transgenic mice: A glucocorticoid loss-of-function model spanning early and late osteoblast differentiation. Bone 47, 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Adra C. N., Boer P. H., and McBurney M. W. (1987) Cloning and expression of the mouse pgk-1 gene and the nucleotide sequence of its promoter. Gene 60, 65–74 [DOI] [PubMed] [Google Scholar]
- 31. Beck E., Ludwig G., Auerswald E. A., Reiss B., and Schaller H. (1982) Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene 19, 327–336 [DOI] [PubMed] [Google Scholar]
- 32. Yoshikawa Y., Kode A., Xu L., Mosialou I., Silva B. C., Ferron M., Clemens T. L., Economides A. N., and Kousteni S. (2011) Genetic evidence points to an osteocalcin-independent influence of osteoblasts on energy metabolism. J. Bone Miner. Res. 26, 2012–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Canalis E., and Zanotti S. (2016) Hajdu-Cheney syndrome, a disease associated with NOTCH2 mutations. Curr. Osteoporos. Rep. 14, 126–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Canalis E., Sanjay A., Yu J., and Zanotti S. (2017) An antibody of Notch2 reverses the osteopenic phenotype of Hajdu Cheney mutant male mice. Endocrinology 158, 730–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Adami G., Rossini M., Gatti D., Orsolini G., Idolazzi L., Viapiana O., Scarpa A., and Canalis E. (2016) Hajdu Cheney syndrome; report of a novel NOTCH2 mutation and treatment with denosumab. Bone 92, 150–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hilton M. J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H. M., Teitelbaum S. L., Ross F. P., Kopan R., and Long F. (2008) Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 14, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zanotti S., and Canalis E. (2014) Notch1 and Notch2 expression in osteoblast precursors regulates femoral microarchitecture. Bone 62, 22–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dong Y., Jesse A. M., Kohn A., Gunnell L. M., Honjo T., Zuscik M. J., O'Keefe R. J., and Hilton M. J. (2010) RBPjκ-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development 137, 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mead T. J., and Yutzey K. E. (2009) Notch pathway regulation of chondrocyte differentiation and proliferation during appendicular and axial skeleton development. Proc. Natl. Acad. Sci. U.S.A. 106, 14420–14425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Canalis E., Zanotti S., and Smerdel-Ramoya A. (2014) Connective tissue growth factor is a target of Notch signaling in cells of the osteoblastic lineage. Bone 64, 273–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Glatt V., Canalis E., Stadmeyer L., and Bouxsein M. L. (2007) Age-related changes in trabecular architecture differ in female and male C57BL/6J mice. J. Bone Miner. Res. 22, 1197–1207 [DOI] [PubMed] [Google Scholar]
- 42. Zhang M., Xuan S., Bouxsein M. L., von Stechow D., Akeno N., Faugere M. C., Malluche H., Zhao G., Rosen C. J., Efstratiadis A., and Clemens T. L. (2002) Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J. Biol. Chem. 277, 44005–44012 [DOI] [PubMed] [Google Scholar]
- 43. Salie R., Kneissel M., Vukevic M., Zamurovic N., Kramer I., Evans G., Gerwin N., Mueller M., Kinzel B., and Susa M. (2010) HEY1 regulates bone mass and cartilage hypertrophy by linking BMP signaling with the PTH receptor. Bone 46, 680–694 [DOI] [PubMed] [Google Scholar]
- 44. Tu X., Chen J., Lim J., Karner C. M., Lee S. Y., Heisig J., Wiese C., Surendran K., Kopan R., Gessler M., and Long F. (2012) Physiological notch signaling maintains bone homeostasis via RBPjk and Hey upstream of NFATc1. PLoS Genet. 8, e1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zanotti S., and Canalis E. (2013) Hairy and enhancer of split-related with YRPW Motif (HEY)2 regulates bone remodeling in mice. J. Biol. Chem. 288, 21547–21557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Canalis E., and Zanotti S. (2017) Hairy and enhancer of split-related with YRPW Motif-like (HeyL) is dispensable for bone remodeling in mice. J. Cell. Biochem. 118, 1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Buchholz F., Angrand P. O., and Stewart A. F. (1996) A simple assay to determine the functionality of Cre or FLP recombination targets in genomic manipulation constructs. Nucleic Acids Res. 24, 3118–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Buchholz F., Angrand P. O., and Stewart A. F. (1998) Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16, 657–662 [DOI] [PubMed] [Google Scholar]
- 49. Tang S. H., Silva F. J., Tsark W. M., and Mann J. R. (2002) A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis 32, 199–202 [DOI] [PubMed] [Google Scholar]
- 50. Clausen B. E., Burkhardt C., Reith W., Renkawitz R., and Förster I. (1999) Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 [DOI] [PubMed] [Google Scholar]
- 51. Takeda K., Clausen B. E., Kaisho T., Tsujimura T., Terada N., Förster I., and Akira S. (1999) Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity 10, 39–49 [DOI] [PubMed] [Google Scholar]
- 52. Bouxsein M. L., Boyd S. K., Christiansen B. A., Guldberg R. E., Jepsen K. J., and Müller R. (2010) Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J. Bone Miner. Res. 25, 1468–1486 [DOI] [PubMed] [Google Scholar]
- 53. Dempster D. W., Compston J. E., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R., and Parfitt A. M. (2013) Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 28, 2–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., and Recker R. R. (1987) Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 55. Lee S. H., Rho J., Jeong D., Sul J. Y., Kim T., Kim N., Kang J. S., Miyamoto T., Suda T., Lee S. K., Pignolo R. J., Koczon-Jaremko B., Lorenzo J., and Choi Y. (2006) v-ATPase V0 subunit d2-deficient mice exhibit impaired osteoclast fusion and increased bone formation. Nat. Med. 12, 1403–1409 [DOI] [PubMed] [Google Scholar]
- 56. Wang Y., Lebowitz D., Sun C., Thang H., Grynpas M. D., and Glogauer M. (2008) Identifying the relative contributions of Rac1 and Rac2 to osteoclastogenesis. J. Bone Miner. Res. 23, 260–270 [DOI] [PubMed] [Google Scholar]
- 57. Yesil P., Michel M., Chwalek K., Pedack S., Jany C., Ludwig B., Bornstein S. R., and Lammert E. (2009) A new collagenase blend increases the number of islets isolated from mouse pancreas. Islets 1, 185–190 [DOI] [PubMed] [Google Scholar]
- 58. Zanotti S., Smerdel-Ramoya A., and Canalis E. (2013) Nuclear factor of activated T-cells (Nfat)c2 inhibits Notch signaling in osteoblasts. J. Biol. Chem. 288, 624–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nazarenko I., Lowe B., Darfler M., Ikonomi P., Schuster D., and Rashtchian A. (2002) Multiplex quantitative PCR using self-quenched primers labeled with a single fluorophore. Nucleic Acids Res. 30, e37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Nazarenko I., Pires R., Lowe B., Obaidy M., and Rashtchian A. (2002) Effect of primary and secondary structure of oligodeoxyribonucleotides on the fluorescent properties of conjugated dyes. Nucleic Acids Res. 30, 2089–2195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Glinka A., Wu W., Delius H., Monaghan A. P., Blumenstock C., and Niehrs C. (1998) Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature 391, 357–362 [DOI] [PubMed] [Google Scholar]
- 62. Iso T., Sartorelli V., Chung G., Shichinohe T., Kedes L., and Hamamori Y. (2001) HERP, a new primary target of Notch regulated by ligand binding. Mol. Cell. Biol. 21, 6071–6079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lian J., Stewart C., Puchacz E., Mackowiak S., Shalhoub V., Collart D., Zambetti G., and Stein G. (1989) Structure of the rat osteocalcin gene and regulation of vitamin D-dependent expression. Proc. Natl. Acad. Sci. U.S.A. 86, 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nakagawa O., Nakagawa M., Richardson J. A., Olson E. N., and Srivastava D. (1999) HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev. Biol. 216, 72–84 [DOI] [PubMed] [Google Scholar]
- 65. Gibson D. G., Young L., Chuang R. Y., Venter J. C., Hutchison C. A. 3rd., and Smith H. O. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 [DOI] [PubMed] [Google Scholar]
- 66. Kouadjo K. E., Nishida Y., Cadrin-Girard J. F., Yoshioka M., and St-Amand J. (2007) Housekeeping and tissue-specific genes in mouse tissues. BMC. Genomics 8, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]