Abstract
Increasing evidence indicates that alternative processing of mRNA, including alternative splicing, 3′ alternative polyadenylation, and regulation of mRNA stability/translation, represents a major mechanism contributing to protein diversification. For example, in alternative polyadenylation, the 3′ end of the immunoglobulin heavy chain mRNA is processed during B cell differentiation, and this processing involves RNA-binding proteins. hnRNPLL (heterogeneous nuclear ribonucleoprotein L-like protein) is an RNA-binding protein expressed in terminally differentiated lymphocytes, such as memory T cells and plasma cells. hnRNPLL regulates various processes of RNA metabolism, including alternative pre-mRNA splicing and RNA stability. In plasma cells, hnRNPLL also regulates the transition from the membrane isoform of the immunoglobulin heavy-chain (mIgH) to the secreted isoform (sIgH), but the precise mechanism remains to be identified. In this study, we report that hnRNPLL specifically associates with cytoplasmic PABPC1 (poly(A)-binding protein 1) in both T cells and plasma cells. We found that although PABPC1 is not required for the alternative splicing of CD45, a primary target of hnRNPLL in lymphocytes, PABPC1 does promote the binding of hnRNPLL to the immunoglobulin mRNA and regulates switching from mIgH to sIgH in plasma cells. Given the recently identified role of PABPC1 in mRNA alternative polyadenylation, our findings suggest that PABPC1 recruits hnRNPLL to the 3′-end of RNA and regulates the transition from membrane Ig to secreted Ig through mRNA alternative polyadenylation. In conclusion, our study has revealed a mechanism that regulates immunoglobulin secretion in B cells via cooperation between a plasma cell-specific RBP (hnRNPLL) and a universally expressed RBP (PABPC1).
Keywords: alternative splicing, immunoglobulin G (IgG), lymphocyte, RNA binding protein, RNA processing, plasma cells
Introduction
Increasing evidence has indicated that a major mechanism diversifying the proteasome during cellular differentiation is the alternative processing of mRNA, which includes alternative splicing, 3′ alternative polyadenylation, and mRNA stability/translation regulation (1–4). As the first discovered example of alternative polyadenylation, the 3′ end of immunoglobulin heavy chain (IgH) mRNA is processed in a cell-type-specific way during B cells differentiation (5, 6). Plasma cells, which are terminally differentiated B cells specialized in producing large amounts of secreted immunoglobulin, utilize an alternative poly(A) site (pAs)2 in the intron between the last exon of the constant region (e.g. Cμ4 of IgM) and the first exon of the transmembrane domain (the M1 exon), thus producing an mRNA coding for the secreted isoform (5, 6). In B cells, on the other hand, a distal pAs in the second coding exon of the transmembrane domain (the M2 exon) is used to generate the mRNA coding for the membrane-bound IgM. Thus, the ratios of membrane-associated to secreted IgH transcripts are modulated by the selective use of a splice site versus a cleavage/polyadenylation site at the 3′ end of the IgH pre-mRNA transcript (7).
Various RNA-binding proteins (RBPs) have been identified as regulating the cell-type-specific selection of secreted IgH versus membrane-bound IgH. These RBPs mainly include polyadenylation/cleavage factors (i.e. CstF64) (8), splicing factors (9, 10), and elongation factors (10, 11). The specific mechanisms of cooperation between these factors in regulating selection between mIg and sIg, however, remain largely unknown.
hnRNPLL (heterogeneous nuclear ribonucleoprotein L-like protein), encoded by HNRNPLL (12), is predominantly expressed in terminally differentiated lymphocytes, including memory T cells and plasma cells. Elevated hnRNPLL expression is usually accompanied by changes in CD45 splicing, switching from its higher molecular weight isoforms (e.g. CD45RABC or CD45RB) to its lower molecular weight isoforms (CD45RO) (10, 13–18). In plasma cells, hnRNPLL also regulates the ratio of membrane Igγ2b to secreted Igγ2b (10), although the molecular mechanism of this regulation remains to be fully understood.
In this study, we used mass spectrometry to identify PABPC1 as a binding partner of hnRNPLL in plasma cells and T cells. As a major component of cytoplasmic poly(A)-binding proteins, PABPC1 regulates mRNA translation (19, 20), hyperadenylation (21), non-sense-mediated decay (22) and alternative polyadenylation (23). We found that hnRNPLL bound to the RRM1 domain of PABPC1, which was distinct from the sites of PABPC1 that bound to the components of the translation initiation complex. Although PABPC1 was not necessary for CD45 splicing, it interacted with IgH mRNA and regulated the transition from mIg to sIg in plasma cells. We further demonstrated that PABPC1 could promote the binding of hnRNPLL to IgH mRNA. Thus, our study revealed a mechanism regulating immunoglobulin secretion in B cells through cooperation between a plasma cell-specific RBP (hnRNPLL) and a universally expressed RBP (PABPC1).
Results
hnRNPLL interacts with PABPC1 in plasma cells
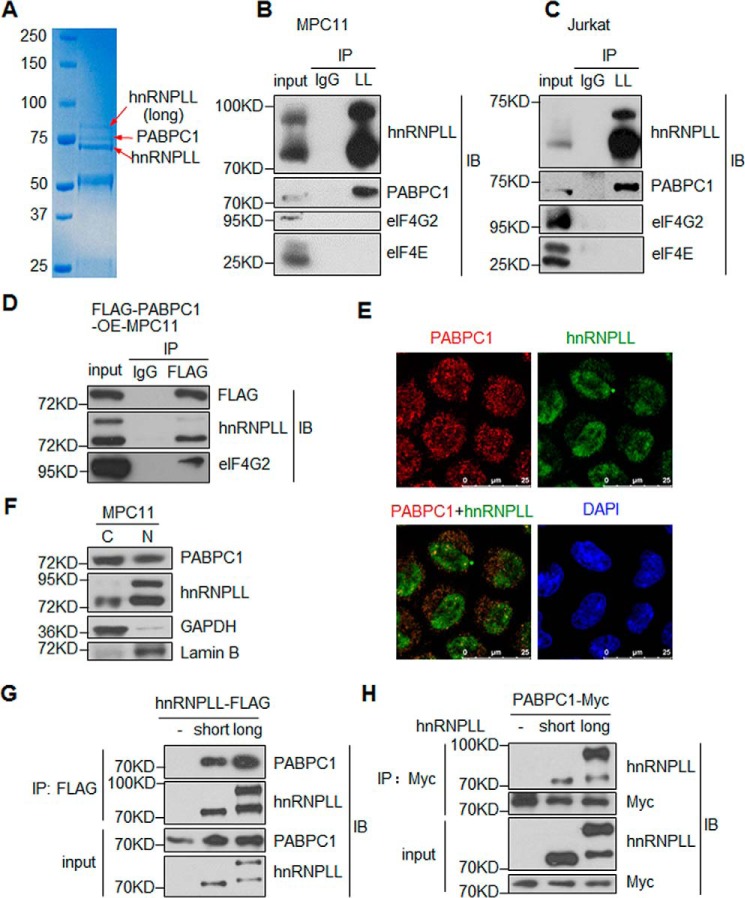
To explore the molecular mechanisms by which hnRNPLL regulates mRNA processing, we carried out an immunoprecipitation experiment in murine plasmacytoma cells MPC11 and then used mass spectrometry to identify its binding proteins. In addition to the two isoforms of the hnRNPLL protein, we found that a protein of ∼70 kDa was co-immunoprecipitated with hnRNPLL (Fig. 1A). Mass spectrometry analysis revealed that the protein band contained PABPC1, a major component of the cytoplasmic poly(A)-binding protein (PABP) in mammalian cells. It has been suggested that it promotes mRNA translation by binding to the 3′-poly(A) tail of the mRNA. Given the predominant 3′-UTR binding of hnRNPLL identified in previous PAR-CLIP experiments (17), we postulated that hnRNPLL and PABPC1 might cooperate to regulate certain aspects of the mRNA 3′-end processing.
Figure 1.
hnRNPLL interacts with PABPC1 A, mass spectrometric analysis identifies interaction between hnRNPLL and PABPC1. hnRNPLL was immunoprecipitated (IP) from MPC11 cells, and precipitated proteins were resolved on SDS-PAGE and stained with Coomassie Blue. The indicated protein bands were then exercised and analyzed by mass spectrometry. The identified proteins are as follows: top band, the hnRNPLL long isoform, which utilizes an alternative translational start site (17); middle band, PABPC1; lower band, the canonical hnRNPLL protein. B and C, PABPC1 interacts with hnRNPLL in lymphocytes. As in A, hnRNPLL (LL) was immunoprecipitated from MPC11 (B) or Jurkat cells (C), and the immunoprecipitate was analyzed for the presence of hnRNPLL, PABPC1, eIF4G2, and eIF4E by immunoblotting (IB) as indicated. Normal rabbit IgG (IgG) was used as a negative control for immunoprecipitation, and 5% of the total protein lysate was included as input control. D, hnRNPLL co-immunoprecipitates with PABPC1. FLAG-tagged PABPC1 was transduced into MPC11 cells, FLAG-PABPC1 was immunoprecipitated with an anti-FLAG antibody (M2), and the immunoprecipitate was analyzed for FLAG, hnRNPLL, and eIF4G2 by immunoblotting as indicated. Normal mouse IgG was used as negative control, and 3% of the total protein lysate was included as input control. E, hnRNPLL interacts with PABPC1 both in the nucleus and in the cytoplasm. MPC11 cells were co-stained with anti-PABPC1 (red) and anti-hnRNPLL (green) and analyzed by confocal immunofluorescence microscopy. Note that hnRNPLL was predominately observed in the nucleus with notable cytoplasmic localization; in contrast, PABPC1 was mainly in the cytoplasmic and was also presented in the nucleus. F, PABPC1 is located in both the nucleus and the cytoplasm of MPC11 cells. MPC11 cells were fractionated into cytoplasmic (C) and nuclear (N) fractions, and the expression of PABPC1 and hnRNPLL was analyzed by immunoblotting. GAPDH and lamin B were served as the controls for cytoplasmic and nucleus proteins, respectively. C, cytoplasmic fraction; N, nucleus fraction. G, both isoforms of hnRNPLL can interact with PABPC1. The FLAG-tagged hnRNPLL constructs were transfected into HEK 293 T cells, and hnRNPLL was immunoprecipitated from the cell lysate with the anti-FLAG (M2) antibody. The immunoprecipitate was analyzed for PABPC1 and hnRNPLL. 1% of the total protein lysate was included as input control. H, PABPC1 interacts with both isoforms of hnRNPLL. PABPC1 fused with a Myc tag was transfected into HEK 293 T cells alone or with either of the hnRNPLL isoforms. PABPC1 was immunoprecipitated from the cell lysate with an anti-Myc antibody, and the immunoprecipitate was analyzed for hnRNPLL and Myc. 1% of the total protein lysate was included as input control. B–H, data are representative of at least three independent experiments.
To test the interaction between hnRNPLL and PABPC1, we performed co-immunoprecipitation (co-IP) experiments. In both the MPC11 cells and the Jurkat human T cell line, PABPC1 was consistently co-immunoprecipitated with hnRNPLL (Fig. 1, B and C). Indeed, PABPC1 enhances protein translation by forming the translation initiation complex. However, in the same co-IP experiments, anti-hnRNPLL did not pull down other components of the translation initiation complex, including eIF4E and eIF4G2 (Fig. 1, B and C), suggesting that hnRNPLL and PABPC1 may form a novel protein complex specialized for other types of mRNA processing besides translation. The interaction between hnRNPLL and PABPC1 was supported by the immunoprecipitation of FLAG-tagged PABPC1 in MPC11 cells (Fig. 1D).
To further investigate the interaction between hnRNPLL and PABPC1 in situ, we performed immunofluorescence staining of these two proteins in MPC11 cells. Although PABPC1 was mainly localized in the cytoplasm as expected, a substantial amount was also present in the nucleus (Fig. 1E). The presence of PABPC1 in the nucleus was confirmed with cytoplasm/nucleus fractionation (Fig. 1F). In contrast, hnRNPLL, although also present in the cytoplasm, was primarily found in the nucleus and was highly enriched in nuclear bodies, such as Raji bodies (Fig. 1E). In both the nucleus and in the cytoplasm, we observed areas of overlapping fluorescences, indicating that hnRNPLL and PABPC1 partially co-localize (Fig. 1E). Our data thus strongly suggest that hnRNPLL and PAPBC1 interact with each other in plasma cells, either in the cytoplasm or in the nucleus.
An alternative translational start site gives rise to a longer isoform of hnRNPLL with an extra sequence at its N terminus (17). We found that both the longer and the canonical (short) isoforms of hnRNPLL were associated with PABPC1. When expressed in HEK 293 T cells, either isoform of hnRNPLL was able to pull down endogenous PABPC1 (Fig. 1G). In addition, Myc-tagged PABPC1 was able to pull down both the canonical and the longer isoform of hnRNPLL (Fig. 1H), with a stronger affinity for the longer isoform.
RRM1 domain of PABPC1 mediates interaction with hnRNPLL
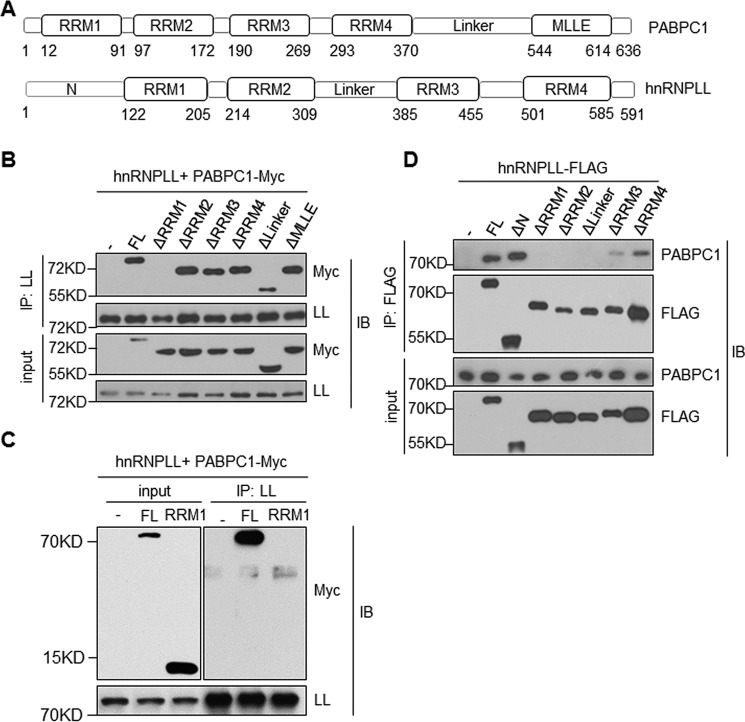
To understand how PABPC1 interacts with hnRNPLL, we deleted each individual functional domain of PABPC1 (Fig. 2A) and co-expressed the truncated proteins with hnRNPLL in HEK 293 T cells. Only one modified PABPC1, lacking the RRM1 domain, completely failed to interact with hnRNPLL (Fig. 2B). We observed a partial reduction in the interactions between hnRNPLL and a mutant PABPC1 missing the linker domain (Fig. 2B). Deletion of all other domains, including RRM2, RRM3, RRM4, and MLLE, did not disrupt the interaction of PABPC1 with hnRNPLL. However, the RRM1 domain alone was not sufficient for the binding of PABPC1 to hnRNPLL, indicating that cooperation between the RRM1 domain and other domains is necessary for association with hnRNPLL (Fig. 2C). The RRM2 domain of PABPC1 is known to interact with the eIF4G2, whereas the MLLE domain is required for the binding of PABPC1 to other PABP interaction motifs containing proteins (24, 25). The binding domains of PABPC1 that are important for the interactions with hnRNPLL suggest that their interaction may regulate other RNA processes besides protein translation.
Figure 2.
Domain requirement for the hnRNPLL and PABPC1 interaction. A, a diagram shows domain structures of PABPC1 (upper panel) and hnRNPLL (lower panel). The location of each domain was indicated at the bottom. B, RRM1 domain of PABPC1 is required for its interaction with hnRNPLL (LL). HEK 293 T cells were transfected with indicated mutants of PABPC1 together with hnRNPLL. hnRNPLL was immunoprecipitated (IP), and its interaction with PABPC1 was analyzed by immunoblotting (IB). 1% of total protein lysates was used as an input control and analyzed for PABPC1 and hnRNPLL. C, the RRM1 domain of PABPC1 alone is not sufficient for interacting with hnRNPLL. HEK 293 T cells were transfected with Myc-tagged full-length or the RRM1 domain of PABPC1 together with hnRNPLL, respectively. hnRNPLL was immunoprecipitated with anti-mouse hnRNPLL antibody, and the immunoprecipitate was analyzed by immunoblotting. 1% of total protein lysates were used as input control. D, RRM1, RRM2, Linker, and RRM3 domains of hnRNPLL are required for its interaction with PABPC1. As in B, HEK 293 T cells were transfected with plasmids expressing FLAG-tagged hnRNPLL proteins lacking the indicated domains. The FLAG-tagged hnRNPLL proteins were immunoprecipitated with anti-FLAG, and their interactions with PABPC1 were determined by immunoblotting analysis. The data are representative of at least three independent experiments.
In parallel, we created a series of domain-depleted mutants of hnRNPLL (Fig. 2A) and expressed the truncated proteins in HEK 293 T cells. Modified hnRNPLL lacking RRM1, RRM2, or the linker domains failed to interact with PABPC1 entirely, whereas RRM3 and RRM4 domains were partially required for the binding of hnRNPLL to PABPC1 (Fig. 2D). The N-terminal region of hnRNPLL was not required for interactions with PABPC1. Interestingly, through a tethering assay, it was shown that the N-terminal region and the linker region of hnRNPLL were together sufficient to induce exon splicing in CD45 (26). Dependence on different hnRNPLL protein domain for CD45 splicing and for interaction with PABPC1 suggests that a distinct functional module of hnRNPLL mediates its interaction with PABPC1.
PABPC1 is not required for hnRNPLL-mediated splicing of CD45
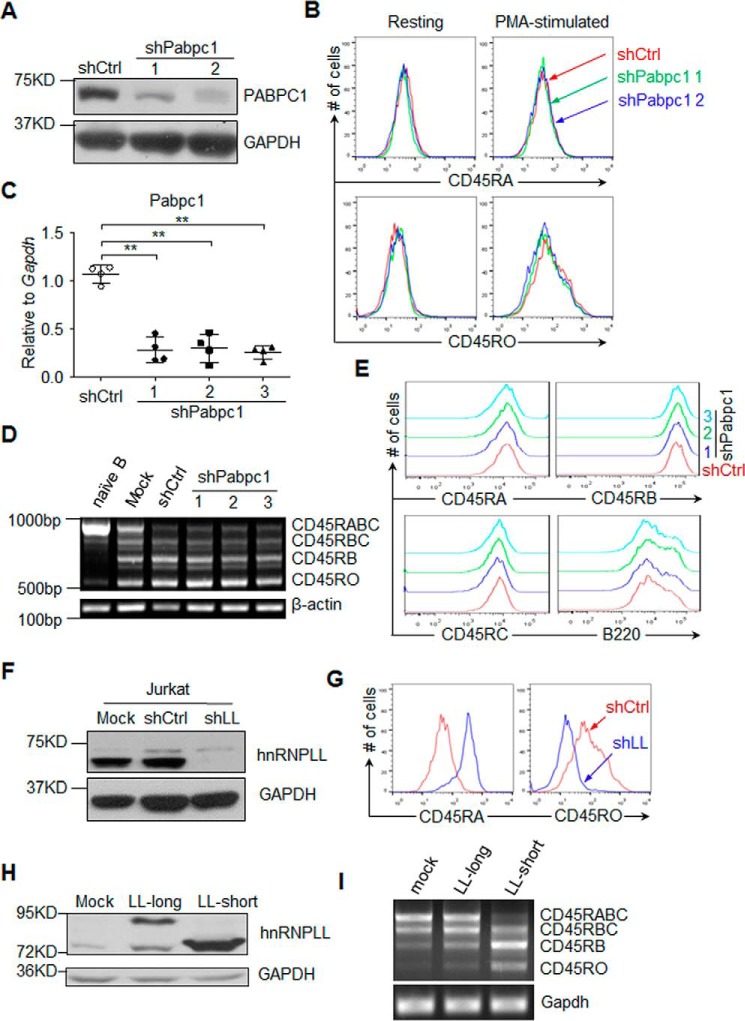
hnRNPLL is highly expressed in terminally differentiated lymphocytes, including memory B cells and plasma cells. One major function of hnRNPLL is to induce switching from the CD45RA to the CD45RO isoform.
To determine whether PABPC1 is involved in the alternative splicing of CD45, we knocked down PABPC1 in Jurkat T cells with two independent shRNAs (Fig. 3A). As expected, PMA stimulation of the Jurkat T cells decreased the expression of the CD45RA isoform and increased the expression of the CD45RO isoform (Fig. 3B). Knockdown of PABPC1, however, did not significantly alter the expression of either CD45RA or CD45RO expression after PMA stimulation (Fig. 3B). In contrast, when we knocked down hnRNPLL in Jurkat cells, we observed significantly increased CD45RA expression and decreased CD45RO after the PMA stimulation (Fig. 3, F and G).
Figure 3.
PABPC1 is not required for CD45 alternative splicing. A, Jurkat cells were transduced with PLKO.1 lentivirus containing either shPabpc1 or a Scrambled sequence, and expression of PABPC1 was determined by immunoblotting analysis. GAPDH was served as a loading control. B, PABPC1 was not required for activation-induced CD45 isoforms switching in Jurkat cells. As in A, Jurkat cells stably expressing the shRNAs against PABPC1 or Scramble were stimulated with PMA, and expression of CD45RA and CD45RO was determined by flow cytometry. C, naïve B cells were isolated from C57BL/6 mice and stimulated with 10 μg/ml LPS in the presence of IL-4 (10 ng/ml) and IL-5 (10 ng/ml). The activated B cells were then transduced with retrovirus containing three independent shRNAs against Pabpc1 (shPabpc1) or a scrambled sequence (shCtrl) at 16 and 40 h after stimulation, and 1 μg/ml puromycin was added into cultured cells at 48 h after stimulation. At 96 h after stimulation, the expression of PABPC1 was analyzed by qRT-PCR. D and E, PABPC1 was not required for CD45 splicing in activated B cells. As in C, RNA was extracted from unstimulated B cells, LPS-stimulated B cells (mock), LPS-stimulated B cells transduced with retrovirus containing a scrambled sequence (shCtrl), or three independent shRNAs against Pabpc1 (shPabpc1). CD45 splicing was analyzed by RT-PCR; PCR products corresponding to each splicing isoform of CD45 are indicated (D); and cell surface expression of CD45RA, CD45RB, CD45RC, and B220 (CD45RABC) was determined by flow cytometry (E). E, red indicates the cells transduced with shCtrl, and green, blue, and cyan indicate the cells transduced with one of the shPabpc1 shRNAs. F and G, hnRNPLL was required for exclusion of CD45RA exon. Jurkat cells were stably transduced with shRNA against either hnRNPLL (shLL) or a Scrambled sequence (shCtrl). Expression of hnRNPLL was determined by immunoblotting (F), and expression of CD45RA and CD45RO was determined by flow cytometry (G). H and I, the short but not the long isoform of hnRNPLL regulates CD45 splicing. A20 cells were transduced with control construct (mock) or long or short isoform of hnRNPLL (LL-long or LL-short), respectively, and expression of the hnRNPLL was analyzed by immunoblotting (H). CD45 splicing was analyzed by RT-PCR, and the PCR products corresponding to each splicing isoform of CD45 are indicated (I). The data are representative of at least three independent experiments.
Similar to the results that we observed in T cells, PABPC1 was not required for the splicing of B220 (CD45RABC) after LPS stimulation in B cells. To induce plasma cell differentiation, we purified total naïve B cells and stimulated them with LPS along with IL-4 and IL-5. The cells were then transduced with either shPabpc1 or control shRNAs (Fig. 3C). The expression of CD45 isoforms was similar between PABPC1-deficient cells and control cells as determined by either RT-PCR or flow cytometry (Fig. 3, D and E). These results suggest that PABPC1 is dispensable for CD45 alternative splicing in both T and B cells.
We also compared the activity of the two hnRNPLL isoforms in inducing alternative splicing of CD45. We found that after being overexpressed in A20 cells, only the canonical (short) isoform promoted the exclusion of the CD45RA exon (Fig. 3, H and I, and supplemental Fig. S1). These data indicate that only the canonical isoform of hnRNPLL is able to induce CD45 splicing.
PABPC1 regulates switching between membrane Ig and secreted Ig in plasma cells
In addition to regulating mRNA alternative splicing, we showed in previous work that hnRNPLL regulates the switching of mIg to sIg in plasma cells (10). Switching between mIg and sIg is a tightly controlled process regulated by both mRNA alternative splicing and alternative polyadenylation (8, 27, 28). PABP proteins, including PABPC1 and PABPN1, have recently been identified as critical regulators of alternative polyadenylation (23, 29).
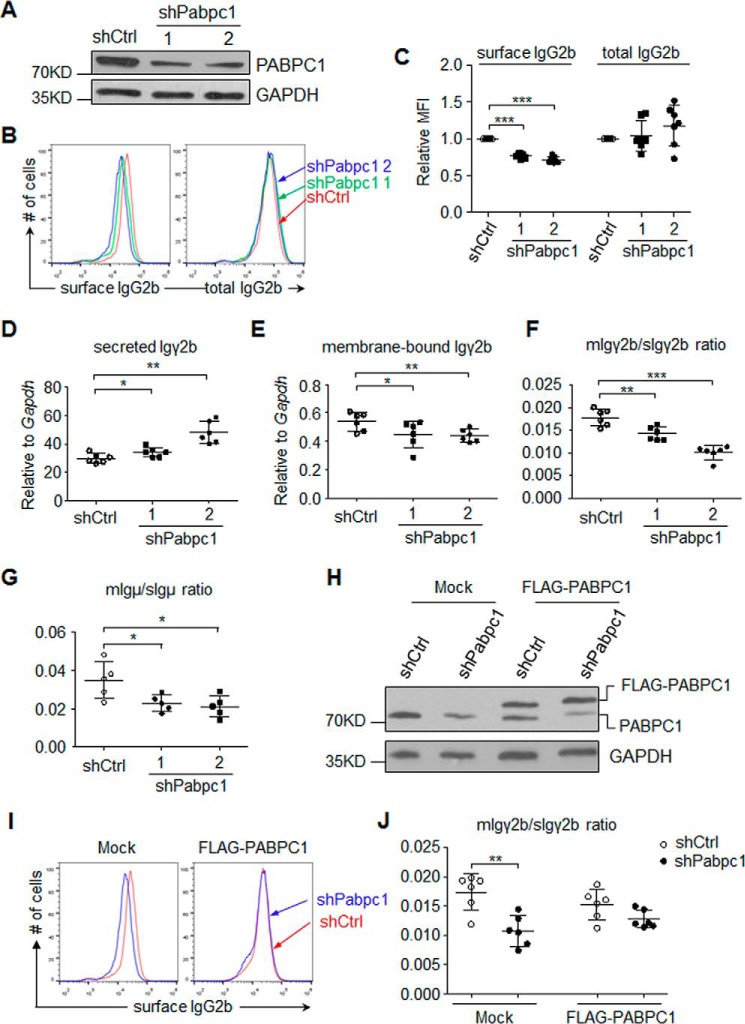
We thus postulated that hnRNPLL and PABPC1 might cooperate to regulate the alternative processing of IgH mRNA in B cells. To test this hypothesis, we first knocked down PABPC1 in MPC11 cells and then examined both the surface expression of IgG2b and the total IgG2b (Fig. 4A). We used flow cytometry to show that surface IgG2b expression decreased significantly in the PABPC1-deficient cells compared with the control cells, whereas total IgG2b level (as determined by intracellular staining) was not significantly altered (Fig. 4, B and C). This result indicates that PABPC1 is critical for switching between mIg and sIg but is not required for translation of immunoglobulin in MPC11 cells.
Figure 4.
PABPC1 regulates the production of secreted Ig in plasma cells. A, MPC11 cells were transduced with control shRNA (shCtrl) or two different shRNAs against Pabpc1 (shPabpc1.1 and shPabpc1.2, respectively), and expression of PABPC1 was analyzed by immunoblotting. B and C, knockdown PABPC1 decreased expression of the surface IgG2b in MPC11 cells. Expression of IgG2b on the cell surface or total IgG2b was analyzed through cell surface staining or intracellular staining in MPC11 cells, respectively. B, representative flow cytometry plots; C, summary of MFI of IgG2b staining from seven experiments. D–F, PABPC1 enhanced membrane Igγ2b while suppressing secreted Igγ2b expression in MPC11 cells. As in A, expression of secreted Igγ2b (D) or membrane Igγ2b (E) was analyzed by qRT-PCR, and ratios of the membrane Igγ2b to the secreted Igγ2b were calculated (F). G, PABPC1 promoted the membrane IgM expression in LPS-activated primary B cells. As in Fig. 3A, RNA was extracted from LPS-stimulated B cells transduced with retrovirus containing a scrambled sequence (shCtrl) or two independent shRNAs against Pabpc1 (shPabpc1) at 96 h after LPS stimulation. The expression of membrane Igμ was analyzed by qRT-PCR, and the ratios of membrane Igμ to secreted Igμ were calculated as indicated. H–J, shRNA-resistant PABPC1 reversed the defects of mIg/sIg ratio in the PABPC1-deficient MPC11 cells. H, Flag-tagged shRNA-resistant PABPC1 was transduced into MPC11 cells with Pabpc1 knockdown, and expression of exogenous (3xFLAG-PABPC1) and endogenous PABPC1 was determined by immunoblotting. I, expression of surface IgG2b was determined by flow cytometry analysis. J, the ratio of the membrane Igγ2b to the secreted Igγ2b mRNA was determined by qRT-PCR. The data are representative (A, B, H, and I) or summary (C–G and J) of at least three independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05 in Student's t test.
We next tested whether reduced surface IgG2b expression in the PABPC1-deficient cells was caused by the alteration of Ig heavy chain processing. Using qRT-PCR assays to specifically detect either membrane or secreted IgG2b, we found that in PABPC1-deficient cells, the expression of the secreted Igγ2b isoform increased, whereas the expression of the membrane isoform decreased (Fig. 4, D and E). As a result, the ratio of membrane Ig to secreted Ig was significantly lower in the PABPC1-deficient cells than in control cells (Fig. 4F). Similar to the results we observed in the MPC11 cells, the ratio of membrane Igμ to secreted Igμ in the PABPC1-deficient primary B cells decreased significantly after LPS stimulation in vitro (Fig. 4G).
To exclude any off-target effects of the shRNAs on PABPC1, we expressed a shRNA-resistant PABPC1 in the PABPC1-deficient MPC11 cells (Fig. 4H). This overexpression of PABPC1 increased the expression of mIgG2b and completely reversed the effects of PABPC1 deficiency (decreased expression of surface IgG2b and lowered the mIg/sIg ratio) in the PABPC1-deficient MPC11 cells, as determined by flow cytometry (Fig. 4I) and qRT-PCR (Fig. 4J). These results suggest that PABPC1 promotes membrane Ig expression while suppressing secreted Ig expression in plasma cells, a role similar to hnRNPLL. Cooperation between hnRNPLL and PABPC1 may thus promote the selection of the distal polyadenylation site over the proximal polyadenylation site.
PABPC1 binds to IgH mRNA and promotes interaction between hnRNPLL and IgH mRNA
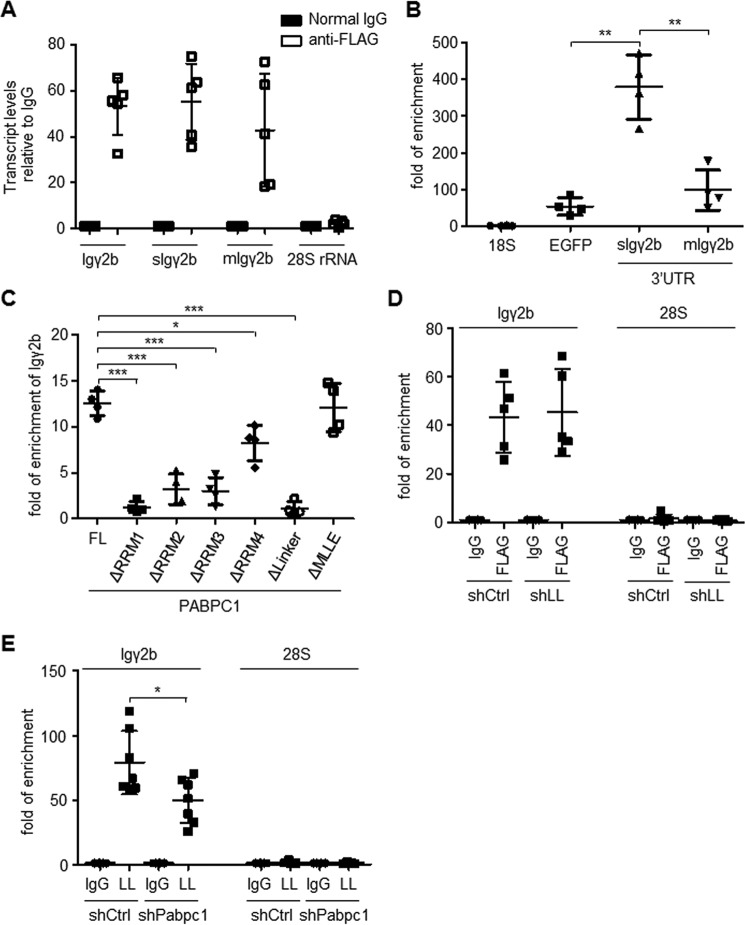
Our results strongly suggest that PABPC1 directly regulates 3′ alternative processing of immunoglobulin mRNA. Further supporting this notion, we found that Igγ2b transcripts bound strongly to PABPC1, as determined by RNA immunoprecipitation (Fig. 5A). Using a construct that fused EGFP to the 3′-UTR of Igγ2b, we found that PABPC1 was associated with the 3′-UTR of the Igγ2b transcript and bound preferentially to the 3′-UTR of secreted Igγ2b (Fig. 5B).
Figure 5.
PABPC1 promotes hnRNPLL binding to the Ig mRNA. A, PABPC1 bound to the immunoglobulin mRNA. FLAG-tagged PABPC1 was stably expressed in MPC11 cells, and PABPC1 was immunoprecipitated with an anti-FLAG antibody. RNA was extracted with TRIzol reagent. The abundance of Igγ2b mRNA in the immunoprecipitates was determined by qRT-PCR after normalized to mouse IgG control. B, PABPC1 was associated with the 3′-UTR of Igγ2b mRNA. 3′-UTR sequences of the secIgγ2b or the memIgγ2b mRNA was cloned downstream of EGFP coding sequence in pEGFP-N1 vector. The plasmids were co-transfected into HEK 293 T cells with the FLAG-tagged PABPC1. The FLAG-tagged PABPC1 protein was immunoprecipitated with an anti-FLAG antibody, and the enrichment of indicated 3′-UTR sequences in the immunoprecipitates was determined by qRT-PCR after normalized to mouse IgG control. Enrichment of EGFP and 18S rRNA in the immunoprecipitates from the cells transfected with empty pEGFP-N1 vector was included as negative controls. Note, despite some enrichment of EGFP mRNA, secIgγ2b mRNA was significantly more abundant in the PABPC1 immunoprecipitates. C, FLAG-tagged PABPC1 proteins lacking the indicated domains (as in Fig. 2B) were stably expressed in MPC11 cells and were immunoprecipitated with the anti-FLAG antibody. The abundance of the Igγ2b mRNA in the immunoprecipitates was determined by qRT-PCR after RNA extraction with TRIzol reagent. D, hnRNPLL deficiency did not affect PABPC1 binding to the Igγ2b mRNA. MPC11 cells with FLAG-PABPC1 were transduced with shRNAs against hnRNPLL (shLL) or a Scrambled sequence, PABPC1 was immunoprecipitated with an anti-FLAG antibody, and the PABPC1-associated RNA was extracted with TRIzol reagent. The abundance of Igγ2b in the immunoprecipitates was determined by qRT-PCR after normalized to mouse IgG control. E, PABPC1 promotes hnRNPLL (LL) binding to the Igγ2b mRNA. As in D, MPC11 cells stably expressing shRNAs against Pabpc1 or Scramble were lysed with Nonidet P-40 lysis buffer, and hnRNPLL was immunoprecipitated with the anti-mouse hnRNPLL antibody. The abundance of Igγ2b in the immunoprecipitates was determined by qRT-PCR after RNA extraction. The data are summary of at least three independent experiments. Error bars stand for standard deviation of the mean. ***, p < 0.001; **, p < 0.01; *, p < 0.05 in Student's t test.
Moreover, similar to the results we have shown regarding the interaction of modified forms of hnRNPLL and PABPC1, PABPC1 missing either the RRM1 domain or the linker domain was completely unable to bind to Igγ2b mRNA (Fig. 5C). RRM2- and RRM3-deficient PABPC1 also displayed reduced binding to Igγ2b mRNA, and interactions between RRM4-deficient PABPC1 and Igγ2b mRNA also decreased somewhat. In contrast, the binding of MLLE-deficient PABPC1 to Igγ2b mRNA was unaffected.
We further investigated whether the ability of PABPC1 to bind to IgH mRNA was dependent on its interaction with hnRNPLL. In hnRNPLL-deficient MPC11 cells, the ability of PABPC1 to bind to Igγ2b was not significantly affected as compared with the control cells (Fig. 5D). In contrast, in PABPC1-deficient cells, the ability of hnRNPLL to bind to the Igγ2b transcript decreased significantly as compared with the control cells (Fig. 5E). Our results thus suggest that hnRNPLL is recruited to Igγ2b mRNA partially via its interaction with PABPC1.
Discussion
In this study, we identified PABPC1 as a protein that interacts with hnRNPLL to regulating mIg to sIg switching in plasma cells. Although PABPC1 is not required for CD45 splicing in either T cells or plasma cells, it enhances binding of hnRNPLL to IgH mRNA and regulates switching from mIg to sIg. Given the recently identified role of PABPC1 in mRNA alternative polyadenylation (23), our results suggest that the interaction between hnRNPLL and PABPC1 regulates alternative polyadenylation of immunoglobulin mRNA, a process critical for switching between mIg and sIg in plasma cells. Our study thus provides an example of lineage-specific mRNA processing that is achieved through cooperation between a universal RBP (e.g. PABPC1) and a lineage-specific RBP (e.g. hnRNPLL).
Alternative cleavage and polyadenylation (APA) in eukaryotic cells generates multiple mRNA isoforms of various 3′-UTR lengths from the same gene (30, 31). PABPN1 has been recognized as a critical RBP regulating APA: recent work using an siRNA knockdown with deep sequencing indicated that PABPC1 enhances the usage of pre-mRNA distal pAs during APA (23). Our data suggest that PABPC1 facilitates the expression of membrane isoforms of IgH mRNA similarly to other APA regulators (e.g. CstF-64).
Notably, deletion of either hnRNPLL or PABPC1 resulted in only a moderate reduction in the expression of the membrane Ig isoform, indicating that other proteins may regulate the mIg to sIg switching independently. One of such proteins may be ELL2 (11), a transcription elongation factor that is up-regulated in plasma cells and enhances the recognition of the secretory IgH polyadenylation site. In co-immunoprecipitation experiments, we found that ELL2 did not interact with hnRNPLL in MPC11 cells (supplemental Fig. S2), suggesting that hnRNPLL functions independent of ELL2. Thus, the poly(A) site selection of IgH mRNA is under control of multiple protein complexes.
Results from our previous study indicate that translation of the hnRNPLL protein can start from either the canonical start codon (AUG) or an alternative translational start codon (CUG) at its 5′-UTR (17). Interestingly, only the canonical (short) isoform of hnRNPLL can induce splicing of CD45 when expressed in A20 cells, suggesting that the long isoform of hnRNPLL lacks the ability to mediating splicing. In contrast, the long isoform of hnRNPLL binds to PABPC1 with higher affinity than does the canonical isoform. These results raise the possibility that hnRNPLL activities can be modulated through the selection of alternative translational start sites.
RNA-binding proteins often have overlapping functions in various aspects of RNA processing. Some splicing factors also regulate RNA stability (32) and RBPs initially believed to be primarily involved in RNA-stability regulation are now also known to regulate alternative splicing (33, 34). To mediate various steps of RNA processing, many RBPs show nucleus/cytoplasm shuttling, thus mediating RNA splicing and polyadenylation in the nucleus and regulating stability and translation in the cytoplasm (35–37). Although predominantly found in the nucleus, hnRNPLL is also present in the cytoplasm; conversely, PABPC1 is primarily distributed in the cytoplasm but is also present in the nucleus. Interactions between these two RBPs are observed in both the nucleus and the cytoplasm, suggesting that they form a complex and thus co-regulate their shuttling. Finally, the tissue-specific alternative processing of mRNA is believed to mediate lineage differentiation. As we have shown, cooperation between lineage-specific RBPs and universal RBPs may account for the diverse functions of RBPs in cellular differentiation. Future studies will concentrate on the interactomes of RNA-binding proteins, which will help to clarify the regulatory mechanisms of post-transcriptional regulation in cellular differentiation and lineage commitment.
Experimental procedures
Plasmids and cloning
The long and short isoform (canonical) of mouse hnRNPLL cDNA with the FLAG tag at the C terminus were cloned into an MSCV retrovirus vector, and human PABPC1 cDNA with FLAG tag at the N terminus was cloned into a MSCV-IRES-Thy1.1 retrovirus vector. These constructs together with pCL-10A1 were transfected into HEK 293 T cells to produce retrovirus. The domain deletion mutants of mouse hnRNPLL and human PABPC1 were cloned into pCMV6-Entry vector or MSCV-IRES-Thy1.1 vector. The breakpoints of the domains used in these fusion proteins are listed in supplemental Table S1, and the RRM1 cDNA sequence of human PABPC1 was also cloned into pCMV6-Entry vector. A shRNA-resistant PABPC1 was created by inducing the following synonymous mutations: 1275 T → C, 1276 A → T, 1277 G → C, 1278 C → T, 1281 A → G, 1284 T → C, 1287 T → C, 1290 A → G, and 1293 A → G.
Generation of stable cell lines
MPC11 cells (3 × 105) were transduced with pLKO.1 shScramble, pLKO.1 shPabpc1-1, or pLKO.1 shPabpc1-2 lentivirus with 8 μg/ml Polybrene (H9268; Sigma), and transduced cells were selected with 5 μg/ml puromycin (Merck/Millipore). Selected cells were maintained in RPMI 1640 containing 1 μg/ml puromycin. pLKO.1 shPabpc1-1 and shPabpc1-2 correspond to TRCN0000054948 and TRCN0000054952, respectively. Jurkat cells were transduced with pLKO.1 lentivirus against human PABPC1 with two shRNAs corresponding to TRCN0000286207 and TRCN0000293649, respectively, and against human HNRNPLL with shRNA corresponding to TRCN0000075101.
Co-immunoprecipitation
The cells were lysed in Nonidet P-40 lysis buffer (50 mm HEPES-KOH, pH 7.5, 250 mm KCl, 2 mm EDTA, 1 mm NaF, 0.5% Nonidet P-40, supplemented with protease inhibitors (Roche) and 1 mm DTT), and target protein was immunoprecipitated with corresponding antibody and protein A (for rabbit antibody) or protein G (for mouse antibody) Dynabeads (Invitrogen) by incubating at 4 °C for 4 h. After being washed once with high-salt IP wash buffer (50 mm HEPES-KOH, pH 7.5, 500 mm KCl, 0.05% Nonidet P-40) and three times with low-salt IP wash buffer (50 mm HEPES-KOH, pH 7.5, 300 mm KCl, 0.05% Nonidet P-40), immunoprecipitate was eluted by boiling at 95 °C for 10 min in 1× SDS loading buffer (50 mm Tris, pH 6.8, 2% SDS, 6% glycerol, 10 mm DTT, 0.02% bromphenol blue).
hnRNPLL was immunoprecipitated with an anti-mouse hnRNPLL antibody in MPC11 cells or with an anti-human hnRNPLL antibody (ab74063; Abcam) in Jurkat cells. PABPC1 was immunoprecipitated with anti-FLAG M2 antibody (F1804; Sigma) in MPC11 cells that were stably transduced with a FLAG-tagged PABPC1, and normal IgG was used as immunoprecipitate control.
Western blot
The cells were lysed in RIPA buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 2 mm EDTA, 1% Nonidet P-40, 0.1% SDS, supplemented with protease inhibitors and 1 mm DTT). Proteins were separated on 8 or 10% SDS-PAGE gel, and immunoblotting was performed with anti-mouse hnRNPLL, anti-human hnRNPLL (ab74063; Abcam), anti-PABPC1 (orb34329; biorbyt), anti-eIF4G2 (AB60254a; BBI), anti-ELL2 (12727-1-AP; Proteintech), anti-eIF4E (AB55221; BBI), anti-lamin B (sc-365962; Santa Cruz), anti-FLAG (F7425; Sigma), anti-Myc (AT0023; CMCTAG), and anti-GAPDH (sc-48166; Santa Cruz).
RNA immunoprecipitation
MPC11 cells stably transduced with a FLAG-tagged PABPC1 or domain-deletion mutations of PABPC1 were lysed as in the co-immunoprecipitation experiment and 200 units/ml Recombinant RNase Inhibitor (2313A; TaKaRa) was added into the lysis buffer. The PABPC1–RNA complex was immunoprecipitated with anti-FLAG M2 antibody and protein G Dynabeads (Invitrogen), and RNA was extracted using TRIzol reagent. Purified RNA was reverse-transcribed using PrimeScriptTM RT reagent kit after depleting DNA with gDNA Eraser (RR047A; TaKaRa). The enrichment of target mRNA was analyzed by qRT-PCR and normalized to the Normal mouse IgG control.
Mouse 3′-UTR sequences of secIgγ2b and memIgγ2b mRNA were cloned at the downstream of EGFP coding sequence in pEGFP-N1 vector, and they were co-expressed with FLAG-tagged PABPC1 in HEK 293 T cells separately, and then the 3′-UTR RNA-immunoprecipitation was performed with anti-FLAG M2 antibody and qRT-PCR as described above. The 3′-UTR sequence corresponds to mouse Dec.2011 (GRCm38/mm10) of UCSC Genome Browser, the secIgγ2b 3′-UTR sequence corresponds to chr12:113,306,289–113,306,388, and the memIgγ2b 3′-UTR sequence corresponds to chr12:113,302,967–113,304,397.
Primary B cells isolation and activation
Total B cells were isolated from the spleen of C57BL/6 mice. Briefly, splenocytes (108 cells/ml) were incubated with Biotin anti-mouse CD19 (Biolegend) for 15 min and then incubated with Biotin selection mixture for 15 min according to the manufacturer's instructions (18556; StemCell). Isolated B cells were cultured in RPMI 1640 medium (10% FBS, 100 units/ml penicillin, 100 units/ml streptomycin, 50 μm β-mercaptoethanol, and 1% l-glutamine) and stimulated with 10 μg/ml LPS (ALX-581-008-L002, ENZO), 10 ng/ml IL-4, and 10 ng/ml IL-5 (215-15-25; Peprotech). Activated B cells were infected twice with retrovirus containing indicated shRNAs at 16 and 40 h after activation and were selected with 1 μg/ml puromycin at 48 h. LMP shPabpc1-1 corresponds to TRCN0000054948, shPabpc1-2 corresponds to TRCN0000054951, and shPabpc1-3 corresponds to TRCN0000054952.
Flow cytometry
Primary B cells were activated and transduced shRNA retrovirus as described above and were stained with PE anti-mouse CD45RA (BD Pharmingen), PE anti-mouse CD45RC (BD Pharmingen), Alexa Fluro 647 anti-mouse CD45RC (Biolegend), and APC anti-mouse CD45R/B220 (Biolegend) at 96 h after activation. A20 cells stably expressing the hnRNPLL long or short isoform were stained as primary B cells for CD45 splicing analysis. Jurkat cells were stained with FITC anti-human CD45RA (Biolegend) and APC anti-human CD45RO (Biolegend). To analyze immunoglobulin expression, MPC11 cells were stained with FITC anti-mouse IgG2b (Biolegend) for surface IgG2b expression or were first permeabilized with the fixation/permeabilization solution (554722; BD Pharmingen) for total IgG2b staining.
RNA extraction, cDNA synthesis, and (q)RT-PCR analysis
RNA was purified with TRIzol reagent or Qiagen RNeasy mini kit, and cDNA was synthesized using PrimeScriptTM RT reagent kit with gDNA Eraser according to the manufacturer's instructions. qRT-PCR was performed by using SYBR green reagent (RR420A; TaKaRa) in 10-μl reactions on the Fast Real-Time PCR System (ABI 7900HT, ViiATM 7; Applied Biosystems). The primers are listed in supplemental Table S2. CD45 splicing in primary B cells and A20 cells was analyzed by RT-PCR using rTag polymerase (R001B; TaKaRa), β-actin as loading control, and primers used for amplify CD45 are CD45-F, 5′-CCCTATTTCTTAGGGGCACA; CD45-R, 5′-CCTTTTCTTTTGGTGTGCAG; β-actin-F, 5′-AGAGGGAAATCGTGCGTGAC; and β-actin-R, 5′-CAATAGTGATGACCTGGCCGT. Products of RT-PCR were separated on 2% agarose gel.
Immunofluorescence and confocal microscopy
MPC11 cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 1% Triton X-100 in PBS for 30 min at room temperature, and blocked with 1% BSA + 1% goat serum for 1 h at room temperature. The fixed cells were then incubated with anti-PABPC1 at 4 °C overnight, followed by incubation with Alexa Fluor 488-conjugated anti-rabbit IgG for 2 h at room temperature together with Alexa Fluor 647-conjugated anti-mouse hnRNPLL antibody (which was prepared using the APEXTM antibody labeling kit (A 10475; Invitrogen)) at 4 °C overnight. After being washed with PBS, the nuclei of cells were stained with DAPI. The images were acquired with a Leica TCS SP5 confocal microscope.
Statistical analysis
The data were analyzed using Student's t test and plotted as the means ± standard deviation. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author contributions
Y. P., J. Y., and Z. Z. conducted experiments and analyzed the data. X. C. supervised the study and wrote the paper.
Supplementary Material
This work was supported by Grants 31370858 and 81671552 from the National Science Foundation of China, 2014CB943600 from Ministry of Science and Technology, China, 13PJ1409300 and 17ZR449700 from Science and Technology Commission of Shanghai Municipality, and National Thousand Talents Program for Distinguished Young Scholars (China). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
- pAs
- poly(A) site
- RBP
- RNA-binding protein
- PABP
- poly(A)-binding protein
- IP
- immunoprecipitation
- qRT-PCR
- quantitative RT-PCR
- APA
- Alternative cleavage and polyadenylation.
References
- 1. Nilsen T. W., and Graveley B. R. (2010) Expansion of the eukaryotic proteome by alternative splicing. Nature 463, 457–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tian B., and Manley J. L. (2013) Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 38, 312–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Moore M. J., and Proudfoot N. J. (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136, 688–700 [DOI] [PubMed] [Google Scholar]
- 4. Kini H. K., Silverman I. M., Ji X., Gregory B. D., and Liebhaber S. A. (2016) Cytoplasmic poly(A) binding protein-1 binds to genomically encoded sequences within mammalian mRNAs. RNA 22, 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alt F. W., Bothwell A. L., Knapp M., Siden E., Mather E., Koshland M., and Baltimore D. (1980) Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3′ ends. Cell 20, 293–301 [DOI] [PubMed] [Google Scholar]
- 6. Early P., Rogers J., Davis M., Calame K., Bond M., Wall R., and Hood L. (1980) Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell 20, 313–319 [DOI] [PubMed] [Google Scholar]
- 7. Peterson M. L. (2007) Mechanisms controlling production of membrane and secreted immunoglobulin during B cell development. Immunol. Res. 37, 33–46 [DOI] [PubMed] [Google Scholar]
- 8. Takagaki Y., Seipelt R. L., Peterson M. L., and Manley J. L. (1996) The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87, 941–952 [DOI] [PubMed] [Google Scholar]
- 9. Ma J., Gunderson S. I., and Phillips C. (2006) Non-snRNP U1A levels decrease during mammalian B-cell differentiation and release the IgM secretory poly(A) site from repression. RNA 12, 122–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Benson M. J., Aijö T., Chang X., Gagnon J., Pape U. J., Anantharaman V., Aravind L., Pursiheimo J. P., Oberdoerffer S., Liu X. S., Lahesmaa R., Lähdesmäki H., and Rao A. (2012) Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc. Natl. Acad. Sci. U.S.A. 109, 16252–16257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martincic K., Alkan S. A., Cheatle A., Borghesi L., and Milcarek C. (2009) Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat. Immunol. 10, 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Topp J. D., Jackson J., Melton A. A., and Lynch K. W. (2008) A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14, 2038–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oberdoerffer S., Moita L. F., Neems D., Freitas R. P., Hacohen N., and Rao A. (2008) Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu Z., Jia X., de la Cruz L., Su X. C., Marzolf B., Troisch P., Zak D., Hamilton A., Whittle B., Yu D., Sheahan D., Bertram E., Aderem A., Otting G., Goodnow C. C., and Hoyne G. F. (2008) Memory T cell RNA rearrangement programmed by heterogeneous nuclear ribonucleoprotein hnRNPLL. Immunity 29, 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yabas M., Godfrey D. I., Goodnow C. C., and Hoyne G. F. (2011) Differential requirement for the CD45 splicing regulator hnRNPLL for accumulation of NKT and conventional T cells. PLoS One 6, e26440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cho V., Mei Y., Sanny A., Chan S., Enders A., Bertram E. M., Tan A., Goodnow C. C., and Andrews T. D. (2014) The RNA-binding protein hnRNPLL induces a T cell alternative splicing program delineated by differential intron retention in polyadenylated RNA. Genome Biol. 15, R26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chang X., Li B., and Rao A. (2015) RNA-binding protein hnRNPLL regulates mRNA splicing and stability during B-cell to plasma-cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 112, E1888–E1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chang X. (2016) RNA-binding protein hnRNPLL as a critical regulator of lymphocyte homeostasis and differentiation. Wiley Interdiscip. Rev. RNA 7, 295–302 [DOI] [PubMed] [Google Scholar]
- 19. Imataka H., Gradi A., and Sonenberg N. (1998) A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17, 7480–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wells S. E., Hillner P. E., Vale R. D., and Sachs A. B. (1998) Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2, 135–140 [DOI] [PubMed] [Google Scholar]
- 21. Kumar G. R., and Glaunsinger B. A. (2010) Nuclear import of cytoplasmic poly(A) binding protein restricts gene expression via hyperadenylation and nuclear retention of mRNA. Mol. Cell Biol. 30, 4996–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Behm-Ansmant I., Gatfield D., Rehwinkel J., Hilgers V., and Izaurralde E. (2007) A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 26, 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li W., You B., Hoque M., Zheng D., Luo W., Ji Z., Park J. Y., Gunderson S. I., Kalsotra A., Manley J. L., and Tian B. (2015) Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet. 11, e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Safaee N., Kozlov G., Noronha A. M., Xie J., Wilds C. J., and Gehring K. (2012) Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol. Cell 48, 375–386 [DOI] [PubMed] [Google Scholar]
- 25. Khaleghpour K., Kahvejian A., De Crescenzo G., Roy G., Svitkin Y. V., Imataka H., O'Connor-McCourt M., and Sonenberg N. (2001) Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 21, 5200–5213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shankarling G., and Lynch K. W. (2013) Minimal functional domains of paralogues hnRNP L and hnRNP LL exhibit mechanistic differences in exonic splicing repression. Biochem. J., 453, 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Peterson M. L. (1992) Balanced efficiencies of splicing and cleavage-polyadenylation are required for mu-s and mu-m mRNA regulation. Gene Expr. 2, 319–327 [PMC free article] [PubMed] [Google Scholar]
- 28. Bruce S. R., Dingle R. W., and Peterson M. L. (2003) B-cell and plasma-cell splicing differences: a potential role in regulated immunoglobulin RNA processing. RNA 9, 1264–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jenal M., Elkon R., Loayza-Puch F., van Haaften G., Kühn U., Menzies F. M., Oude Vrielink J. A., Bos A. J., Drost J., Rooijers K., Rubinsztein D. C., and Agami R. (2012) The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell 149, 538–553 [DOI] [PubMed] [Google Scholar]
- 30. Fu Y., Sun Y., Li Y., Li J., Rao X., Chen C., and Xu A. (2011) Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res. 21, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shepard P. J., Choi E. A., Lu J., Flanagan L. A., Hertel K. J., and Shi Y. (2011) Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17, 761–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang H., Wang H., Han L., Zhao G., Shen H., Wang P., Sun Z., Xu C., Su Y., Li G., Tong T., and Chen J. (2016) hnRNP A1 antagonizes cellular senescence and senescence-associated secretory phenotype via regulation of SIRT1 mRNA stability. Aging Cell, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pascale A., Amadio M., Scapagnini G., Lanni C., Racchi M., Provenzani A., Govoni S., Alkon D. L., and Quattrone A. (2005) Neuronal ELAV proteins enhance mRNA stability by a PKCα-dependent pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 12065–12070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bellavia D., Mecarozzi M., Campese A. F., Grazioli P., Talora C., Frati L., Gulino A., and Screpanti I. (2007) Notch3 and the Notch3-upregulated RNA-binding protein HuD regulate Ikaros alternative splicing. EMBO J. 26, 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cassola A., and Frasch A. C. (2009) An RNA recognition motif mediates the nucleocytoplasmic transport of a trypanosome RNA-binding protein. J. Biol. Chem. 284, 35015–35028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ye J., and Blelloch R. (2014) Regulation of pluripotency by RNA binding proteins. Cell Stem Cell 15, 271–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cho Y. S., Chennathukuzhi V. M., Handel M. A., Eppig J., and Hecht N. B. (2004) The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J. Biol. Chem. 279, 31514–31523 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.