Abstract
Cellular heterogeneity represents a fundamental principle of cell biology for which a readily available single-cell research tool is urgently required. Here, we present a novel method combining cell-sized well arrays with sequential inkjet printing. Briefly, K562 cells with phosphate buffer saline buffer were captured at high efficiency (74.5%) in a cell-sized well as a “primary droplet” and sealed using fluorinated oil. Then, piezoelectric inkjet printing technology was adapted to precisely inject the cell lysis buffer and the fluorogenic substrate, fluorescein-di-β-D-galactopyranoside, as a “secondary droplet” to penetrate the sealing oil and fuse with the “primary droplet.” We thereby successfully measured the intracellular β-galactosidase activity of K562 cells at the single-cell level. Our method allows, for the first time, the ability to simultaneously accommodate the high occupancy rate of single cells and sequential addition of reagents while retaining an open structure. We believe that the feasibility and flexibility of our method will enhance its use as a universal single-cell research tool as well as accelerate the adoption of inkjet printing in the study of cellular heterogeneity.
I. INTRODUCTION
In traditional cell biology research, cells belonging to the same category are cultured, treated, and observed in groups. The experimental results thus stand for the average state of the community. However, over the past few decades, many studies1 have shown that cells differ with respect to size2 or gene expression,3 even within isogenic cultures. These differences between individual cells are termed cellular heterogeneity,4 which is regarded as a fundamental principle of cell biology. Cellular heterogeneity arises from the stochastic expression of genes, proteins, and metabolites5 and may hinder the design of accurate disease models and provide challenges in medical discovery.6 For example, intra-tumor heterogeneity,7 a consequence of imperfect DNA replication during tumor cell division, may induce a bottleneck effect upon medical treatment, such that resistant sub-clones survive and propagate to reform a heterogeneous tumor.8
In order to study cellular heterogeneity in further detail, there is an urgent need to develop a practical analysis tool at the single-cell level. Microfluidics has been considered as a potential tool for single cell analysis with its accurate control of fluid volume at the picoliter scale to realize low cost and high precision. At present, single-cell microfluidic devices can be divided into four categories: those that generate droplets-in-oil system,9 cell-sized well arrays,10 certain complex micro-fabricated devices,11,12 and even bio-printing technology,13 and have been successfully applied to some types of single-cell analysis. A detailed comparison of these is shown in Table I. However, various limitations still restrict their applications.
TABLE I.
Comparison of single cell analytic techniques.
| Droplet-in-oil system14–18,29 | Cell-sized well19,30 | Complex devices31–33 | Bio-printing24–26,34 | |
|---|---|---|---|---|
| Poisson distribution | Restriction | Overcome | Overcome | Restriction/overcome |
| Occupancy rate of single cells | Low | High | High-medium | High-low |
| Throughput | High | High | High-medium | Low |
| Addition of reagents | Hard | Hard | Medium | Medium |
| Manufacture | Easy | Easy | Hard | Easy |
| Equipment cost | Low | Low | Low | High |
| Reagent consumption | High | Low | Medium | High |
| Advantages | High-throughput, simple structure | High-throughput, high occupancy, simple structure | Superior performance | Define location |
| Disadvantages | Low occupancy, difficulty in adding reagents | Difficulty in adding reagents | Complex manufacturing process, narrow application | Expensive equipment and software, low throughput |
Microfluidic chips that generate water-in-oil micro-droplets such as T-junctions have been successfully applied in single-cell culture,14 genome sequencing,15 and enzyme analysis.16 However, the encapsulation of randomly distributed cells is in accordance with Poisson restriction,17 which cannot achieve uniform or even predominantly single-cell occupancy in droplets.18 Furthermore, cells in the closed micro-channels are also less available for re-treatment such as the addition of liquid.
In comparison, the manufacturing process of single-cell-sized arrays is simple and the occupancy of single cells may be as high as 92% (Ref. 19) for overcoming the Poisson restriction principle. However, the volume of a standard cell-sized well is too small to accommodate sufficient liquid for subsequent experiments such as single-cell reverse transcription polymerase chain reaction (PCR), which for reliable analysis requires ample dilution (>5 nl/cell) of the cell suspension.20 Furthermore, for preventing the pL reagent volume in a well from evaporating, an oil seal is added to the reagent droplet, which leads to difficulties in secondary liquid addition. To address these problems, Zhu et al.21,22 have invented a Microfluidic Droplet Robot to add reagents to oil-sealed droplets on two-dimensional droplet arrays. However, the liquid addition step includes contact, which may result in cross-contamination.
Alternatively, micro-fabricated devices that are manufactured with fine structures for capturing cells in isolated reaction chambers have been successfully applied to real time-PCR (RT-PCR).20,23 However, the complex and special design of the structure only meets particular applications and therefore cannot serve as an open tool for other single-cell experiments.
Finally, bio-printing technology adopts a noncontact nozzle to print living cells in the form of droplets as “bioink,” which is comprised of cells and medium. The principle of single-cell printing is similar to that of droplets generated in a micro-channel. Notably, the occupancy of a single-cell is limited by the Poisson restriction. Even if an optical particle detection mechanism and a sorting algorithm are used to select droplets that only include an individual cell,24 the blockage of the nozzle resulting from gravity and adhesion of cells in the precursor solution will reduce printing stability and throughput.25,26
Thus, developing a novel device that overcomes the aforementioned limitations remains an urgent priority. An ideal single-cell analysis tool should meet the following three conditions concurrently: (1) high single-cell occupancy rate that reduces the required reagent dosage; (2) flexibility that allows sequential addition of reagents; and (3) wide applicability with a simple and open device structure. In the current study, we present a novel type of open-pattern droplet-in-oil planar array system (ODOAS), which combines cell-sized well arrays with inkjet printing. Specifically, a cell-sized well array was used to capture single cells, overcoming the Poisson restriction, each of which, along with phosphate buffer saline (PBS) buffer in the micro-well as the “primary droplet,” was sealed with oil. Next, we adapted sequential inkjet printing27 to fabricate a “secondary droplet” including a surfactant for lysing the cell membrane and reagents for intracellular detection, which penetrated the oil layer and fused with the “primary droplet” in the cell-sized well. This reagent addition process is no-contact and the droplet exists at a pL reagent volume without evaporation.28 Moreover, the flexibility of inkjet printing for changing reagents allows ODOAS to function as an open tool for other single-cell experiments. Thus, this is the first presentation of an open single-cell analysis tool that satisfies the proposed three conditions of “ideal single-cell analysis tools.”
II. MATERIALS AND METHODS
A. Cells and reagents
The K562 cells used in this study were purchased from China Infrastructure of Cell Line Resources. The cells were cultured in a humidified incubator (37 °C in an atmosphere of 5% CO2) and RPMI 1640 (Wisent, Canada) medium supplemented with fetal bovine serum (10%, Gibco, USA) and penicillin-streptomycin (1%, Gibco). We utilized two fluorescent probes. Hoechst 33342 Solution (Hoechst, Life Technologies, USA) is a cell-permeable DNA probe that is excited by ultraviolet light and emits blue fluorescence at 460–490 nm. Cells in the well stained by Hoechst dye can be identified under a fluorescence microscope. Propidium iodide (PI, Life Technologies, USA) is another nucleic acid probe with a fluorescence excitation maximum at 535 nm and an emission maximum at 617 nm that can identify cells with damaged membranes. An enzymatic assay for intracellular β-gal was carried out using a fluorogenic β-gal substrate, fluorescein-di-β-D-galactopyranoside (FDG, AAT Bioquest, USA).
B. Cell-size array design and fabrication
The cell-size arrays were fabricated with p-type (100) single silicon wafers using photolithography. First, positive photoresist AZ 6130 (thickness 5 μm) was spin-coated onto the silicon wafer for subsequent lithography using a photolithography mask. Round holes (1600) were designed over a 40 × 40 array on a 1.5 cm × 1.5 cm wafer spaced with a 150-μm distance between the centers of each hole (20-μm in diameter). Under the protection of the photoresist, a cell-sized well array (18-μm in deepness) was formed using reactive-ion etching. Finally, the remaining photoresist was removed with acetone. The manufactured cell-sized array was chemically cleaned by sequential immersion in acetone, ethanol, and deionized (DI) water for 15 min each and then used for experiments.
C. Single-cell capture process
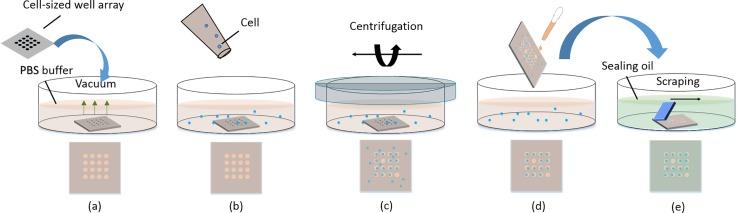
Prior to the capture experiment, cells were centrifuged at 800 rpm for 3 min. The culture medium was removed and phosphate buffer saline (PBS) was added to re-suspend cells at a concentration of 2500–4500 cells/ml. Then, the cells were pre-stained with Hoechst 33342 for 30 min to obtain a clear identification of the cells in the capture-well. Figure 1 shows the operating process of single-cell capture by gravity. A cleaned cell-sized array wafer was attached onto the bottom of a culture dish. PBS (2 ml) was added to the dish to flood the wafer. The dish was then placed into a vacuum chamber to evacuate the small volume of gas in the wells [Fig. 1(a)], after which 1 ml K562 cell suspension liquid was transferred to the dish [Fig. 1(b)]. After sealing with tape, the dish was centrifuged at 2500 rpm for 2 min to force the cells into the wells; owing to the size of the well, only a single cell could be inserted into each well [Fig. 1(c)]. Then, PBS was used to wash the residual cells out of the wells [Fig. 1(d)]) and the wafer was dipped into FC-40 oil (FC-40 Fluorinated (TM) Electronic Liquid, 3 M, USA). Using silica gel as a liquid scraping blade [Fig. 1(e)], the residual PBS reagent between the wells was scraped off. The FC-40 oil sealed the small reagent volume in each well.
FIG. 1.
Schematic representation of single-cell capture by the cell-sized well array wafer. (a) The cell-sized array wafer was attached on the bottom of a culture dish containing PBS buffer. The air in the well was removed with a vacuumizer. (b) Cell suspension solution (PBS as solvent) was added to the dish. (c) Cells were centrifuged into the wells. (d) Residual cells were washed out of the wafer. (e) The wafer was dipped into the oil. The residual PBS buffer between wells was then scraped off using silica gel; single cells in droplets in the wells were isolated by the oil.
D. Inkjet print technology
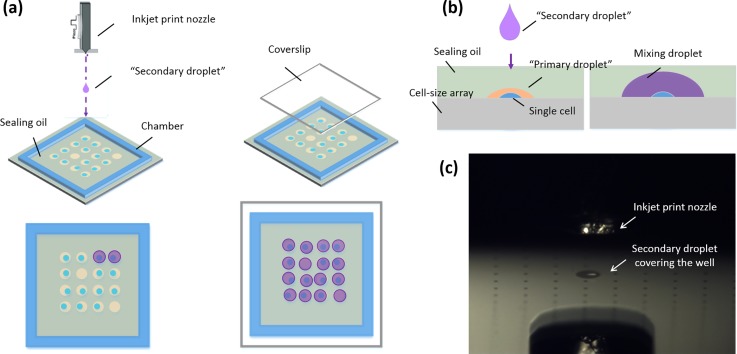
We built an inkjet-based printing system27 that is an improvement upon the basic Jetlab® inkjet platform (MicroFab). A single cell with PBS buffer, as a “primary droplet,” was isolated and sealed in a well using FC-40 fluorinated oil. Then the wafer was topped with a spacer made using double-sided adhesive tape (3 M) to provide a square-shaped wall and create a tablet-like 20 mm × 20 mm chamber for the enzymatic assay, and taken out of the dish. An FC-40 oil layer with 150-μm thickness, which was the same as that of the double-sided adhesive tape, was used to prevent the evaporation of the “primary droplet.” As the next step, inkjet print technology was applied to dispense the reagents for cell lysis and intracellular material detection to each well, as a “secondary droplet.” During this step, the “secondary droplet” penetrates the oil layer and fuses with the “primary droplet.” After the dispensing process, a glass coverslip was placed as a cover on the top of the wafer. The nozzle orifice diameter used for this process was 50 μm. Figures 2(a) and 2(b) show the working scheme and Fig. 2(c) is an image of the actual working state. Movie S1 (supplementary material) shows the intuitive printing process.
FIG. 2.
Schematic representation of nozzle printing of the “secondary droplet” for penetrating the oil layer. (a) Sealing oil is surrounded by the chamber. The inkjet nozzle prints the reagents as a secondary droplet, which penetrates the oil layer to precisely cover the micro-well and fuse with the “primary droplet.” The device then then packaged via a coverslip like a sandwich. Image (b) shows the side view of the printing process. (c) Bright-field image showing the actual working state.
E. Intracellular enzymatic assay
ODOAS platform functionality was demonstrated by measuring the intracellular β-gal activity of the trapped single K562 cells. The fluorogenic substrate FDG was stored at −18 °C as a 20 mM stock solution in Dimethyl sulfoxide (DMSO). Prior to the enzymatic assay, the solution was diluted with PBS reagent to a 200 μM working solution. The lysis reagent comprised PBS buffer containing 0.2% TritonX-100 as a surfactant. The reagent printed as a “secondary droplet” consisted of FDG, Triton X-100, and PBS, added as 1 volume working solution with 1 volume lysis reagent.
III. RESULTS AND DISCUSSION
A. Cell-sized well array for high single-cell capture efficiency
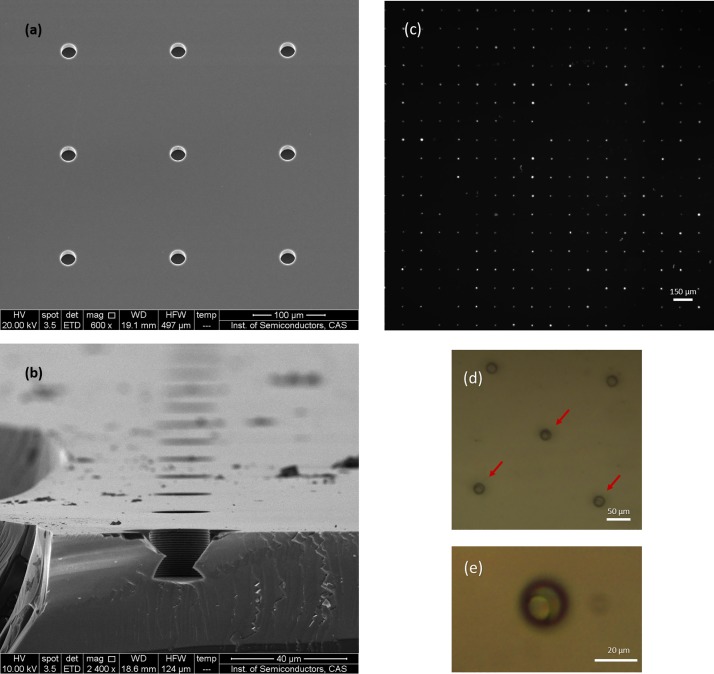
Following the sequential operations of cell capture as shown in Fig. 1, the K562 cells pre-stained with Hoechst dye were identified in wells. Based on the available, cell-sized space in each well (20-μm diameter; 18-μm deep) [Figs. 3(a) and 3(b)], only a single cell could have been trapped in each well. Therefore, each bright spot in the fluorescence images represented an individual cell [Fig. 3(c)]. We counted the number of bright spots using the ImageJ software (National Institutes of Health, USA). Dividing the total number of wells by the number of bright spots, we could obtain single-cell occupancy of approximately 74.5% (Fig. S1, supplementary material). The bright-field images in Figs. 3(d) and 3(e) show the cells in the individual wells.
FIG. 3.
Scanning electron microscope (SEM) image of the cell-sized well array (a) and cross-sectional view (b) of a single well. Fluorescence and bright-field images of cells captured in the micro-well array. (c) The bright spots represent cells in the wells. Image (d) and (e) show bright-field images of single cells in individual micro-wells. Owing to the space limitation of cell-sized well, only one cell can be trapped. Image (e) shows the cell with a well integrative morphology.
B. Inkjet printing technology
Using the cell-sized array, single cells were captured in wells and sealed by the oil. However, subsequent treatments for biological analysis are difficult for several reasons. For example, measurement of the intracellular β-gal activity of K562 cells, as performed in the current study, requires isolating the β-gal enzyme in one droplet for one cell. Yet, the space of each well is too small to contain sufficient reagents for subsequent experiments. The principle of the cell-sized array, to overcome the Poisson restriction, is that the space of the well is similar to the volume of a single cell. By expanding the dimensions of the well, more than one cell would likely be captured in each well and the single-cell occupancy rate would decrease.19 Moreover, a cell lysing process should be performed after isolation to prevent cross-contamination. However, no method yet exists for solving these issues.
In our previous work, we invented a double-inkjet printing technique that utilized piezoelectric inkjet printing equipment to first generate oil droplets of specified volumes on a planar substrate and then precisely deliver the water droplets into the oil droplets by jet action.28 Our group then proposed a sequential inkjet printing model that was successfully applied to enzyme inhibition assays.27 These developments solved two fundamental problems: reducing the evaporation of reagent droplets during array fabrication and multistep pL-scale droplet printing without cross-contamination. Therefore, in this study we combined the cell-sized array with sequential inkjet printing technology and ultimately solved the contradiction between the cell-sized well and reagent required. Briefly, the sequential inkjet printing model was developed to dispense a “secondary droplet” for cell lysing and intracellular material detection. The principle of the inkjet printing for droplet-in-oil formation is schematically illustrated in Fig. 2.
1. Droplet penetration into the oil layer
The key to successfully print the “secondary droplets” to penetrate the oil layer, like collisions between immiscible drops,35,36 is to match parameters on the Weber and Reynolds number of the water-oil system. To investigate this correlation, a test was performed based on a p-type (100) single silicon wafer without a well array as substrate with the DI water droplets printed using a 30–μm nozzle to observe droplet dynamics via our inkjet-printing visualizing platform.27 By tuning the parameters of pulse width and amplitude, different Weber numbers of droplets could be obtained.27,28 We utilized three kinds of oil, FC-40 fluorosilicon oil, mineral oil (M8410, Sigma-Aldrich, USA), and silicon oil (AR20, Sigma-Aldrich), which have a good biological compatibility, to explore the influence of the Reynolds numbers of oil to obtain a reliable printing process. Moreover, considering the energy conversion; i.e., the kinetic energy of the droplet that is transferred to the liquid flow, the thickness of the oil layer constitutes another important variable. For each test, the thickness of the oil layer was adjusted by adding different volumes of oil into a 2 cm × 2 cm chamber.
We could define a few classical parameters associated with this topic. The Weber and Reynolds numbers, We and Re, are, respectively, defined as the ratio of inertia forces to surface tension or viscous forces: We = ρUD2/σ and Re = ρUD/μ. In these equations, U is the impact velocity of the drop. μ, σ, and ρ are, respectively, the dynamic viscosity, the surface tension, and the density of the liquid. Generally, kinematic viscosity (ν0) is used to represent viscous force, ν0=μ/ρ. The dimensionless film thickness, H*, is defined as the ratio of the oil film thickness (h) to the water drop diameter (D). The subscripts “w” and “o” represent “water” and “oil.” Therefore, the physical property of the water droplet is defined as Wew = ρwUwDw2/σw and that of oil is Reo =ρoUwDw/μo = UwDw/ν0. The inherent and measuring parameters for this test are listed in Table II. Parameter characterization methods for droplets are shown in Fig. S2 (supplementary material).
TABLE II.
Inherent and measured parameters for analyzing the dynamics of droplet penetration. Water droplets: ρw = 0.998 g/cm3, σw = 0.072 N m−1.
| Experiment | A | B | C | D | E |
|---|---|---|---|---|---|
| Oil film | Fluorinated oil | Fluorinated oil | Fluorinated oil | Silicon oil | Mineral oil |
| ρ0 (g/cm3) | 1.86 | 1.86 | 1.86 | 1.01 | 0.85 |
| μ0 (cps) | 4.1 | 4.1 | 4.1 | 20 | 26 |
| ν0 (cst) | 2.2 | 2.2 | 2.2 | 19.8 | 30.6 |
| Dw (μm) | 40.28 | 43.41 | 44.58 | 40.28 | 40.28 |
| Uw (m/s) | 3.29 | 3.83 | 4.96 | 3.29 | 3.29 |
| Wew | 6.06 | 8.84 | 15.23 | 6.06 | 6.06 |
| Reo | 60.2 | 75.6 | 100.50 | 6.69 | 4.33 |
| h (μm) | 275 | 325 | 375 | 175 | 150 |
| H* | 6.82 | 7.49 | 8.41 | 4.34 | 3.72 |
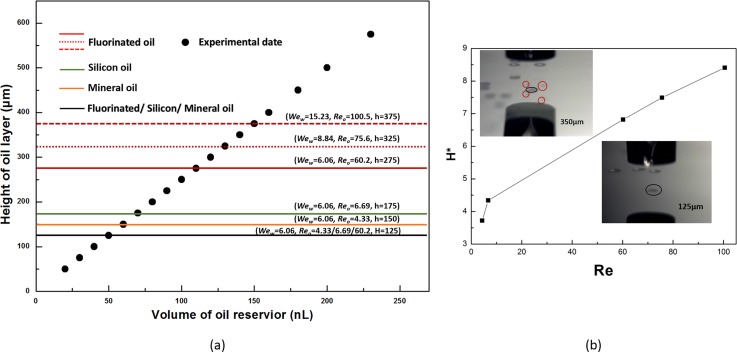
We observe three droplet dynamics for the water-oil system with a specific Weber and Reynolds number, non-coverage, complete-coverage, and partial-coverage [Movie S2 (supplementary material)], whose boundaries are expressed by the oil film thickness (h) shown in Fig. 4(a). For the condition under Wew = 6.06 and Reo = 60.2, the non-coverage region is h < H = 125 μm, where the water droplet penetrates the oil layer and then adheres onto the wafer, whereas the oil just slowly packages the droplet resulting in evaporation. As the height of the FC-40 oil layer increases, 125 μm <h < 275 μm, representing complete-coverage; in this range, the oil layer can rapidly seal the droplet upon penetration. However, h > 275 μm represents a partial-coverage region where numerous small drops float on the oil layer surface that are separated from the host droplet. The appearance of these three different regions can be explained in terms of energy conversion. When the droplet impacts the oil film, the kinetic energy of the droplet is transferred to that of the oil to cause flux at the radial direction and different features of the collision plane will be formed.37–39 At the non-coverage area, dramatic fluctuation of the oil level leads to difficult oscillation back to completely coat the water droplet. For a thicker oil layer, in a complete-coverage region, a thicker oil layer would consume more kinetic energy of the droplet and the velocity of the oil stream in the radial direction would be smaller than those of a thin oil layer. Therefore, the oil can quickly cover the penetrated droplets without evaporation. However, for partial-coverage, the oil surface may oscillate back too quickly to shear the water droplet as there is a deformation of drops.36,40
FIG. 4.
(a) Effect of the Weber and Reynolds number on the result of droplet penetration. The black spots represent the experimental date points at a definite thickness of oil layer. The lines in the map represent a change of droplet penetrating dynamic with special Wew and Reo. The black solid line is a public split line for FC-40, silicon oil, and mineral oil to distinguish the un-coverage and the complete-coverage region at the condition of Wew = 6.06. The red, green, and orange color solid lines are the split line complete-coverage and the partial coverage region separately contains FC-40, silicon oil, and mineral oil at Wew = 6.06. The red broken line stand of split line complete-coverage and partial-coverage region just for FC-40 oil with different Wew. (b) The variations of the dimensionless film thickness H* to the Reynolds number Reo. The inset images are bright-field images show the actual working states at Wew = 6.06 and Reo = 60.2 separately for partial-coverage, whose height of oil is 350-μm, up the curve, and complete- coverage, 125-μm down the curve. The red circle points out the small drops floating on the oil surface.
We then changed the Wew and Reo to explore the variation trend of oil film thickness (h) in greater depth. At first, with the same Wew = 6.06, oil with different dynamic viscosity, Reo = 4.33 and 6.69, the thickness that distinguished non-coverage and complete-coverage was unchanged [Fig. 4(a), black line]. Because the intensely impact cause the thin oil film dramatic fluctuation as usual even with bigger dynamic viscosity. In comparison, the height for distinguishing the complete-coverage and partial-coverage decreases as the Reo is reduced [Fig. 4(a), red, green, and orange color solid lines]. As in the previous analysis, the case of partial-coverage results from oil surface droplet shearing. The smaller Reynolds number means that viscous force has a more important impact on droplet penetration of the oil layer. A larger viscous force leads to more difficulty in causing an oil stream and finally a smaller oscillation of the oil film, which results in a lower height of the boundary that distinguishes the complete-coverage from partial-coverage. Consequently, FC-40 oil with the smallest kinematic viscosity possesses the largest range for complete-coverage, which is beneficial for practical application. Second, FC-40 oil is used independently, changing the Weber number of the droplet. The height for distinguishing the complete-coverage from partial-coverage increases as the Wew and Reo increases [Fig. 4(a), red solid and broken lines]. In this test, increasing the Wew indicates that the inertia force is also increasing, such that the droplet has more kinetic energy to penetrate the oil layer. With respect to Reo, it is easier to understand this physical process. For the same oil, the viscosity remaining unchanged while the Reo increasing indicates that the inertia force has a greater impact on this system, which in turn leads to a bigger impact on the oscillation to increase the height of the oil film that distinguishes complete-coverage from partial-coverage.
According to our experimental results, the Reynolds numbers play a key role for successful droplet penetration. We then defined dimensionless film thickness, H*, as the ratio of the film thickness (h), which distinguishes the complete-coverage from partial-coverage, to the water drop diameter (D). Figure 4(b) is a plot of H* vs Reo and it is linear. Higher up the curve represents the partial-coverage region whereas lower down the curve represents the region of complete-coverage. As the Reynolds numbers get larger, the complete-coverage range increases as well. Although limited to the accuracy of our measurement, to address the difficulty of accurately controlling the height of the oil layer, the correlation between H* and Reo clearly provides a guide to improve the printing success rate when the operator needs to adopt different specimens. For example, we use the 50–μm nozzle to print the bio-medium of intracellular enzymatic assay for obtaining bigger volume of one droplet which can increase the Weber and Reynolds numbers.
2. Droplet fusion
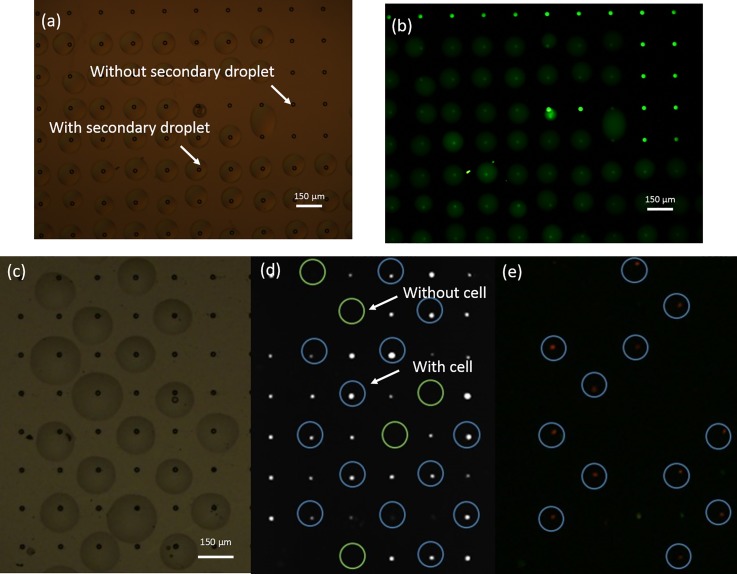
A droplet fusion experiment was carried out to investigate the fusion between the “primary droplet” in the well and the printed “secondary droplet.” An Alexa Fluor 488 solution was first isolated in the well and sealed using the FC-40 oil layer as the “primary droplet.” Next, the nozzle printed the water droplet. Figure 5 shows the experimental result. From the bright-field images [Fig. 5(a)], the water droplet was able to cover the well. Some wells without secondary droplets were used as the references. From the corresponding fluorescence images [Fig. 5(b)], the large droplet emitted green fluorescence with an intensity weaker than that of the references. This indicates that the large water droplet fused with the Alexa Fluor 488 solution in the well and that the dye solution diffused into the large water droplet. From this droplet fusion experiment, we considered that the “secondary droplet” could fuse with the “primary droplet” in the cell-sized well, demonstrating the successful addition of reagents to micro-droplets in the oil.
FIG. 5.
Fusion between a “primary droplet” and a “secondary droplet.” Image (a) clearly shows the wells with or without a “secondary droplet.” In the fluorescence (b) image, the green fluorescence signal emitted from the Alexa Fluor 488 dye is smaller for the well with a secondary droplet compared to that without the large droplet. The reason is that the secondary droplet, DI water, coalesces with the small volume of solution containing the Alexa Fluor 488 dye in the well and then dilutes the dye to decrease the fluorescence intensity. Bright-field (c) and fluorescence (d) and (e) images show the experimental phenomenon of a single-cell lysed in a micro-well. Image (c) shows the wells with or without a “secondary droplet” containing lysis reagent. Bright spots in the fluorescence image (d) represent K562 cells that were pre-stained by Hoechst dye. The wells both with a cell and covering lysis droplet are circled in blue. In comparison, wells without cells are circled by green. Following excitation, the cells with cytomembranes disrupted by the surfactant emitted red light, circled by blue in (e).
3. Single cell lysis in an individual droplet
K562 cells with PBS were sealed by the FC-40 oil in the wells as the “primary droplet.” For lysing the cell in the well, we printed the lysis reagent (PBS containing 0.2% TritonX-100 surfactant and PI dye) as the “secondary droplet.” For clearly displaying the experimental phenomenon, we printed the reagent droplets at intervals. Figure 5(c) shows the bright-field image. In the fluorescence image [Fig. 5(d)], the bright spots represent single cells in the well, which are circled by blue rings whereas the empty wells are circled by green rings. After a 10-min incubation at room temperature (approximately 20 °C), the cell membrane was destroyed by the surfactant and the nucleic acids were stained by PI as shown Fig. 5(e). Fluorescent microscopy showed that all but one of the cells treated by the lysis reagent emitted red fluorescence, with the latter potentially not having finished the lysing process. From these results, we concluded that our printing method could efficiently lyse a single cell in a cell-sized well.
C. Intracellular enzymatic assay at the single-cell level
We next applied ODOAS to measure the intracellular β-gal activity at a single-cell level. Diluted K562 cells were stained with Hoechst dye and introduced into the single-cell capture system. Then, the nozzle printed the reaction droplet (approximately 200 pL) as a “secondary droplet” that contained FDG (100 μM) and TritonX-100 (0.1%), with PBS as a solvent. During the droplet-array generation process, the microchip was maintained at a relatively low temperature (4 °C) to minimize the enzyme reaction prior to incubation. We then transferred the enzymatic assay to the room temperature environment and located the cell. We obtained a final array of single-cell packed droplets in oil; the bright-field and fluorescence images are shown in Fig. 6(a). The time of duration between the moment of transfer and completion of cell location was <10 min. During this time, the surfactant also disrupted the cell membrane to release the intracellular β-gal to catalyze the FDG substrate.
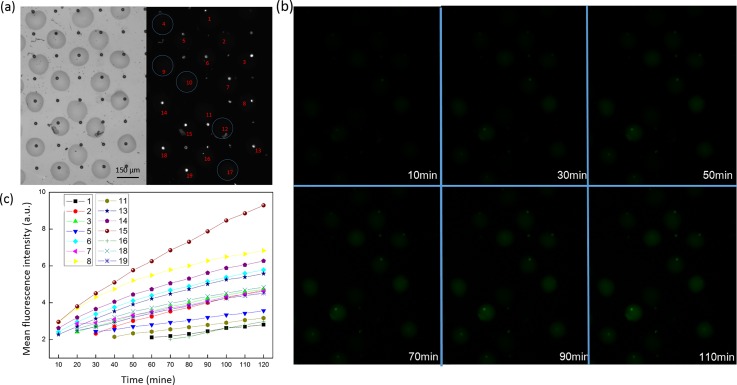
FIG. 6.
Single cell enzymatic assay of K562 cells using ODOAS. Bright-field and fluorescence images (a) determined the presence of a cell in an enzymatic detection solution. We generated 19 droplets using our sequential-inkjet printing technology. Of these, five wells (4, 9, 10, 12, and 17) without cells are circled in blue. The trapped cells were lysed inside the wells in the presence of FDG. (b) Post-lysis fluorescence images of the wells were acquired using a green emission filter (515 nm) to monitor the presence of the fluorescent product (fluorescein) from FDG hydrolysis by intracellular β-gal. (c) Kinetics of the average fluorescence intensity from the wells containing a cell.
We collected fluorescence images over the course of a 10-min period using a color CCD camera and extracted the mean fluorescence intensity of each well-point using the ImageJ software. The values are plotted in Fig. 6(c). Figure 6(b) shows the fluorescence images only over the course of a 20 min period. This demonstrated that (1) no increase in fluorescence intensity was observed in empty wells (denoted as 4, 9, 10, 12, and 17). Leakage of fluorescein to the outside of the droplet was not observed. This result indicated that the intracellular materials from single cells were efficiently isolated. No cross contamination was detected between two wells. Thus, the droplet-in-oil system generated by our inkjet technology is stable. (2) As shown in Figs. 6(b) and 6(c), the fluorescence intensity increased as the time for catalytic reaction was extended. At the same time points, signal values arising from different wells also differ. The slope of the plot for each individual cell was also distinct, which may be attributed to the cellular heterogeneity. A similar phenomenon has also been reported in previous studies.41 Therefore, our method allowed highly sensitive measurement of intracellular β-gal activity in the lysates of single cells.
IV. CONCLUSIONS
We have developed an open-pattern droplet-in-oil planar array system based on inkjet printing technology for single cell research. Using this system, we successfully measured the intracellular β-gal activity in single K562 cells and revealed the inter-cellular heterogeneity. Compared to other single cell analysis tools, ODOAS concurrently satisfies the requirements of high occupancy rate of single cells, sequential addition of reagents, and open structure. Specifically, (1) using cell-sized wells to capture individual cells could achieve high occupancy rates of single cells for overcoming the Poisson restriction principle. (2) Inkjet printing was used to sequentially add liquid to the wells. An individual cell with PBS buffer was isolated in a single well and sealed with oil. Lysing reagent or intracellular material detection reagent was then printed to penetrate the oil layer and fuse with the liquid in the well. (3) As an ODOAS providing reliable multistep droplet printing of different reagents, our method exhibits an expanded range of applications without changes to the device structure. In our previous studies, we had successfully applied inkjet printing technology to real-time PCR and enzyme inhibition assays.27,28 In the current study, we expand the application range to single-cell intracellular material measurement. In future studies, we will apply ODOAS to RT-PCR at the single-cell level for circulating tumor cell screening. Furthermore, we illustrate the correlation between stable printing arrangement and the Reynolds number of an oil layer, which can guide other operators when adopting this method. We believe that our ODOAS will likely constitute new and effective biotechnology for single cell research.
V. SUPPLEMENTARY MATERIAL
See supplementary material for the statistical data of single-cell occupancy rate and the videos for the dynamic process of sequential inkjet printing.
ACKNOWLEDGMENTS
This work was supported by grants from the Instrument Development Project of the Chinese Academy of Sciences (Grant No. 61334008), the National Natural Science Foundation of China (Grant No. 31200643), and the Instrument Developing Project of the Chinese Academy of Sciences (Grant No. YZ201301).
References
- 1. Carlo D. D. and Lee L. P., Anal. Chem. 78(23), 7918–7925 (2006). 10.1021/ac069490p [DOI] [PubMed] [Google Scholar]
- 2. Newman J. R. S., Ghaemmaghami S., Ihmels J., Breslow D. K., Noble M., Derisi J. L., and Weissman J. S., Nature 441(7095), 840 (2006). 10.1038/nature04785 [DOI] [PubMed] [Google Scholar]
- 3. Toriello N. M., Douglas E. S., Thaitrong N., Hsiao S. C., Francis M. B., Bertozzi C. R., and Mathies R. A., Proc. Nat. Acad. Sci. U.S.A. 105(51), 20173–20178 (2008). 10.1073/pnas.0806355106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Altschuler S. J. and Wu L. F., Cell 141(4), 559 (2010). 10.1016/j.cell.2010.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang D. and Bodovitz S., Trends Biotechnol. 28(6), 281 (2010). 10.1016/j.tibtech.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Heath J. R., Ribas A., and Mischel P. S., Nat. Rev. Drug Discovery 15(3), 204 (2016). 10.1038/nrd.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marusyk A., Almendro V., and Polyak K., Nat. Rev. Cancer 12(5), 323 (2012). 10.1038/nrc3261 [DOI] [PubMed] [Google Scholar]
- 8. Marusyk A. and Polyak K., Biochim. Biophys. Acta 1805(1), 105–117 (2009). 10.1016/j.bbcan.2009.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumaresan P., Yang C. J., Cronier S. A., Blazej R. G., and Mathies R. A., Anal. Chem. 80(10), 3522–3529 (2008). 10.1021/ac800327d [DOI] [PubMed] [Google Scholar]
- 10. Lindström S. and Anderssonsvahn H., Biochim. Biophys. Acta 1810(3), 308–316 (2011). 10.1016/j.bbagen.2010.04.009 [DOI] [PubMed] [Google Scholar]
- 11. Streets A. M., Zhang X., Cao C., Pang Y., Wu X., Xiong L., Yang L., Fu Y., Zhao L., and Tang F., Proc. Nat. Acad. Sci. 111(19), 7048 (2014). 10.1073/pnas.1402030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tehranirokh M., Kouzani A. Z., Francis P. S., and Kanwar J. R., Biomicrofluidics 7(5), 51502 (2013). 10.1063/1.4826935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dababneh A. B. and Ozbolat I. T., J. Manuf. Sci. Eng. 136(6), 061016 (2014). 10.1115/1.4028512 [DOI] [Google Scholar]
- 14. Holtze C., Rowat A. C., Agresti J. J., Hutchison J. B., Angilè F. E., Schmitz C. H., Köster S., Duan H., Humphry K. J., and Scanga R. A., Lab Chip 8(10), 1632 (2008). 10.1039/b806706f [DOI] [PubMed] [Google Scholar]
- 15. Gawad C., Koh W., and Quake S. R., Nat. Rev. Genet. 17(3), 175 (2016). 10.1038/nrg.2015.16 [DOI] [PubMed] [Google Scholar]
- 16. Baret J. C., Miller O. J., Taly V., Ryckelynck M., El-Harrak A., Frenz L., Rick C., Samuels M. L., Hutchison J. B., and Agresti J. J., Lab Chip 9(13), 1850–1858 (2009). 10.1039/b902504a [DOI] [PubMed] [Google Scholar]
- 17. Mary P., Dauphinot L., Bois N., Potier M. C., Studer V., and Tabeling P., Biomicrofluidics 5(2), 9 (2011). 10.1063/1.3596394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joensson H. N. and Andersson S. H., Angew. Chem. Int. Ed. 51(49), 12176 (2012). 10.1002/anie.201200460 [DOI] [PubMed] [Google Scholar]
- 19. Rettig J. R. and Folch A., Anal. Chem. 77(17), 5628–5634 (2005). 10.1021/ac0505977 [DOI] [PubMed] [Google Scholar]
- 20. White A. K., Vaninsberghe M., Petriv O. I., Hamidi M., Sikorski D., Marra M. A., Piret J., Aparicio S., and Hansen C. L., Proc. Nat. Acad. Sci. 108(34), 13999 (2011). 10.1073/pnas.1019446108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhu Y., Zhang Y. X., Liu W. W., Ma Y., Fang Q., and Yao B., Sci. Rep. 5, 9551 (2015). 10.1038/srep09551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu Y., Zhu L. N., Guo R., Cui H. J., Ye S., and Fang Q., Sci. Rep. 4(6186), 5046 (2014). 10.1038/srep05046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. White A. K., Heyries K. A., Doolin C., Vaninsberghe M., and Hansen C. L., Anal. Chem. 85(15), 7182 (2013). 10.1021/ac400896j [DOI] [PubMed] [Google Scholar]
- 24. Yusof A., Keegan H., Spillane C. D., Sheils O. M., Martin C. M., O'Leary J. J., Zengerle R., and Koltay P., Lab Chip 11(14), 2447 (2011). 10.1039/c1lc20176j [DOI] [PubMed] [Google Scholar]
- 25. Nakamura M., Kobayashi A., Takagi F., Watanabe A., Hiruma Y., Ohuchi K., Iwasaki Y., Horie M., Morita I., and Takatani S., Tissue Eng. 11(12), 1658–1666 (2005). 10.1089/ten.2005.11.1658 [DOI] [PubMed] [Google Scholar]
- 26. Leibacher I., Schoendube J., Dual J., Zengerle R., and Koltay P., Biomicrofluidics 9(2), 024109 (2015). 10.1063/1.4916780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sun Y., Chen X., Zhou X., Zhu J., and Yu Y., Lab Chip 15(11), 2429 (2015). 10.1039/C5LC00356C [DOI] [PubMed] [Google Scholar]
- 28. Sun Y., Zhou X., and Yu Y., Lab Chip 14(18), 3603–3610 (2014). 10.1039/C4LC00598H [DOI] [PubMed] [Google Scholar]
- 29. Huang L., Bian S., Cheng Y., Shi G., Liu P., Ye X., and Wang W., Biomicrofluidics 11(1), 011501 (2017). 10.1063/1.4975666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park M. C., Hur J. Y., Cho H. S., Park S. H., and Suh K. Y., Lab Chip 11(1), 79 (2011). 10.1039/C0LC00114G [DOI] [PubMed] [Google Scholar]
- 31. Declan R., Kangning R., and Hongkai W., Biomicrofluidics 5(2), 021501 (2011). 10.1063/1.3574448 [DOI] [Google Scholar]
- 32. Arakawa T., Noguchi M., Sumitomo K., Yamaguchi Y., and Shoji S., Biomicrofluidics 5(1), 14114 (2011). 10.1063/1.3567101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin D., Deng B., Li J. X., Cai W., Tu L., Chen J., Wu Q., and Wang W. H., Biomicrofluidics 9(1), 014101 (2015). 10.1063/1.4905428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schoendube J., Wright D., Zengerle R., and Koltay P., Biomicrofluidics 9(1), 014117 (2015). 10.1063/1.4907896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anilkumar A. V., Lee C. P., and Wang T. G., Phys. Fluids A 3(11), 2587–2591 (1991). 10.1063/1.858199 [DOI] [Google Scholar]
- 36. Chen R. H. and Chen C. T., Exp. Fluids 41(3), 453–461 (2006). 10.1007/s00348-006-0173-2 [DOI] [Google Scholar]
- 37. Xie H., Koshizuka S., and Oka Y., Int. J. Numer. Methods Fluids 45(9), 1009–1023 (2004). 10.1002/fld.736 [DOI] [Google Scholar]
- 38. Pan K. L., Cheng K. R., Chou P. C., and Wang C. H., Exp. Fluids 45(3), 435–446 (2008). 10.1007/s00348-008-0486-4 [DOI] [Google Scholar]
- 39. Rein M., Fluid Dyn. Res. 12(2), 61–93 (1993). 10.1016/0169-5983(93)90106-K [DOI] [Google Scholar]
- 40. Chen X., Sun Y., Xue C., Yu Y., and Hu G., Microfluid. Nanofluid. 21(6), 109 (2017). 10.1007/s10404-017-1944-1 [DOI] [Google Scholar]
- 41. Kim S. H. and Fujii T., Lab Chip 16(13), 2440 (2016). 10.1039/C6LC00241B [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See supplementary material for the statistical data of single-cell occupancy rate and the videos for the dynamic process of sequential inkjet printing.