Abstract
Most cancer patients die from metastasis. Recent studies have shown that gold nanoparticles (AuNPs) can slow down the migration/invasion speed of cancer cells and suppress metastasis. Since nuclear stiffness of the cell largely decreases cell migration, our hypothesis is that targeting AuNPs to the cell nucleus region could enhance nuclear stiffness, and therefore inhibit cell migration and invasion. Our results showed that upon nuclear targeting of AuNPs, the ovarian cancer cell motilities decrease significantly, compared with nontargeted AuNPs. Furthermore, using atomic force microscopy, we observed an enhanced cell nuclear stiffness. In order to understand the mechanism of cancer cell migration/invasion inhibition, the exact locations of the targeted AuNPs were clearly imaged using a high-resolution three-dimensional imaging microscope, which showed that the AuNPs were trapped at the nuclear membrane. In addition, we observed a greatly increased expression level of lamin A/C protein, which is located in the inner nuclear membrane and functions as a structural component of the nuclear lamina to enhance nuclear stiffness. We propose that the AuNPs that are trapped at the nuclear membrane both (1) add to the mechanical stiffness of the nucleus and (2) stimulate the overexpression of lamin A/C located around the nuclear membrane, thus increasing nuclear stiffness and slowing cancer cell migration and invasion.
Keywords: gold nanoparticles, cancer cell migration and invasion, metastasis, nuclear stiffness, AFM, lamin A/C, three-dimensional DIC microscopy
Graphical Abstract
Metastasis, a process in which cancer cells migrate to other locations of the human body, is responsible for most cancer-related mortality. It usually begins with local invasion to the surrounding tissues, followed by intravasation into the lymph and blood microvasculature before the cancer cells finally colonize within the microenvironment of other locations in the patient’s body.1,2 Many treatments for inhibiting metastasis are based on drugs that target specific proteins that promote the cell migration or invasion process; however, past attempts to develop antimetastasis drugs have not been efficacious in clinical trials.3 In many cases, the anticancer drugs that target specific proteins on the cancer cells might lose their efficacy after several months of treatment due to protein mutations thus conferring drug resistance to cancer cells.4 Moreover, the anticancer drugs could cause side effects to appear in healthy tissues.5
Recent advances in nanomedicine provide us with a great opportunity to avoid the drawbacks of current drugs.4,6–10 Nanoparticles have been widely used in cancer diagnosis and cancer therapy thanks to their intrinsic chemical, physical and optical properties.9,11–13 Nanoparticles with proper surface modifications can target tumors selectively,7,14–17 and their effects on cancer cell migration or metastasis have drawn attention from many researchers.18–21 In 2013, Murphy et al. reported that gold nanoparticles (AuNPs) with different surface charges and sizes can affect cancer cell migration.18 In 2014, Chor Yong Tay et al.20 found that after incubation with nanoceramics, such as titania, silica, and hydroxyapatite, cells showed significantly impaired wound healing capability because of the disruption of the intracellular microtubule assembly. In the same year, Zhou et al.21 showed that gold nanorods (AuNRs) coated with bovine serum albumin (BSA) exhibited reduced cell migration and invasion by impairing ATP synthesis, which subsequently inhibits the F-actin cytoskeletal assembly and decreases metastatic ability of tumor.21 Arvizo et al. used nonspecific targeted gold nanospheres (AuNSs) to inhibit tumor growth and metastasis by abrogating MAPK signaling and reversing the epithelial-mesenchymal transition.16 For most of the related works, nonspecific targeted nanoparticles have been used. For instance, Zhou et al.21 used BSA coated AuNRs that showed inhibitory effects on cancer cell migration, but the high concentration of AuNRs (50–200 μM) used might be an obstacle for clinical usage. To maintain the nanoparticle effect on slowing the cancer cell migration and invasion with a minimized amount of nanoparticles, the intracellular locations of nanoparticles could be an important factor to consider. It is thus promising to design nanoparticles that can target specific intracellular regions to enhance the inhibition of cancer cell migration and invasion.
Mechanical stiffness of cancer cells has been shown to grade metastatic potential in patient tumor cells,22 as well as in cultured cancer cell lines.23,24 Lower stiffness is related to more invasive cells.25 In eukaryotic cells, the nucleus contains most of the cell’s genetic material and controls cell activities by transcriptional regulation. It is the largest and stiffest organelle in most cells and largely determines the cell migration ability.26–28 Lamin A/C (LMNA) proteins are an important factor in nuclear stiffness. They form a dense protein network that connects the nuclear membrane and chromatin structures on the interior of the nuclear membrane. Recent studies have shown that nuclear lamin A protein scales with tissue stiffness29 and generates a barrier to cells migrating through three-dimensional (3D) environments.30 It is shown that lamin A/C deficiency hampers cell mechanics, polarization, migration and invasion.31–33
Here, targeted AuNPs were used to locate the AuNPs to the cells, because of their specific physical and chemical properties and better biocompatibility than other nanomaterials such as nanoceramics or silver nanoparticles.17,34 By targeting and locating the AuNPs to the cell in a manner to modulate the stiffness of its nucleus, we could improve the inhibition effect on cell migration and invasion. In our experiment, we used three ligands, methoxy-polyethylene glycol thiol (PEG) for increasing the biocompatibility of AuNPs, RGD (RGDRGDRGDRGDPGC) peptides for binding to the surface integrin of cancer cells and enhancing endocytosis, and nuclear localization signal (NLS, CGGGPKKKRKVGG) peptides for targeting the AuNPs to the nucleus. Cell migration or invasion abilities have been measured, and the results show a clear decrease in these functions after the nuclear targeting of the nanoparticles. For studying the cell mechanical response, atomic force microscope (AFM) showed that the two types of AuNPs (i.e., AuNRs and AuNSs) both significantly enhanced the nuclear stiffness. A high-resolution 3D optical imaging system showed the exact location of the nanoparticles, which were trapped at the nuclear membrane. The levels of lamin A/C were found to be elevated upon nanoparticle incubation, which could be an explanation for the observed enhanced nuclear stiffness causing inhibition of cell motility upon gold nanoparticle treatment.
RESULTS AND DISCUSSION
Au Nanoparticles Synthesis, Conjugation, Cellular Uptake and Cytotoxicity Measurement
The AuNRs with an average size of 25 (±2) × 5 (±0.5) nm were synthesized using a seedless method,35 as shown in Figure 1a (TEM figure) and a surface plasmon resonance (SPR) peak centered ~800 nm (UV–vis spectrum in Figure 1d). The AuNSs with an average size of 35 ± 2 nm that absorb at 535 nm wavelength of light were synthesized using citrate reduction method and were shown in Figure 1b and 1e (TEM and UV–vis spectrum). Both of the two types of the nanoparticles are widely used in the biological studies.34,36,37
Figure 1.
Characterization of Au nanoparticles. (a) Transmission electron microscopic (TEM) image of gold nanorods (AuNRs) and (b) TEM image of gold nanospheres (AuNSs), Scale bar = 100 nm. (c) Schematic figure of Au nanoparticle conjugation with PEG, RGD and NLS peptides. (d) UV–vis extinction spectra of the unconjugated AuNRs (black spectrum), AuNRs@PEG (red spectrum), and AuNRs@PEG@RGD/NLS (green spectrum), (e) UV–vis extinction spectra of the unconjugated AuNSs (black spectrum), AuNSs@PEG (red spectrum), and AuNSs@PEG@ RGD/NLS (green spectrum).
AuNPs were functioned with three ligands, methoxypolyethylene glycol thiol (PEG), RGD (RGDRGDRGDRGDPGC) peptides and nuclear localization signal (NLS, CGGGPKKKRKVGG) peptides, as shown in schematic Figure 1c. First, we conjugated the PEG to the surface of the AuNPs surface in order to enhance the biocompatibility.38 The second ligand, RGD bound to surface integrins, which are highly expressed on the surface of the cancer cells when compared to healthy cells to enhance the receptor-mediated endocytosis of the nanoparticles selectively to the cancer cells.39 The third ligand, NLS, was recognized by importin and translocate into the nucleus.40 Successful surface modification of AuNRs@ PEG@RGD/NLS (AuNRs@NLS) is evident in the red-shift of the plasmon peak of AuNRs to longer wavelengths, from 800 nm for the as-synthesized cetyl-trimethy ammonium bromide (CTAB) coated AuNRs to 805 nm for PEGylated AuNRs, and finally to 825 nm for the RGD and NLS modified AuNRs (Figure 1d). Similarly, the surface plasma peak of AuNSs@ PEG@RGD/NLS (AuNSs@NLS) was also red-shifted (Figure 1e), which is in agreement with our former publication.41
The zeta potential of the AuNRs at the different conjugating stages of the three ligands were measured (Table S1) to confirm the surface modifications. The as-synthesized CTAB coated AuNRs has a highly positive surface charge (50.9 ± 7.97 mV); this makes sense as CTAB is a highly cationic surfactant. After PEG modification, the AuNRs become negatively charged (−13.6 ± 11.8 mV). The zeta potential of the AuNRs becomes positive again (14.9 ± 3.13 mV) after further modification with RGD and NLS peptides (Table S1). Also, the zeta potential of the AuNSs@NLS proved their successful surface modification (Table S1), similar to previous studies.41
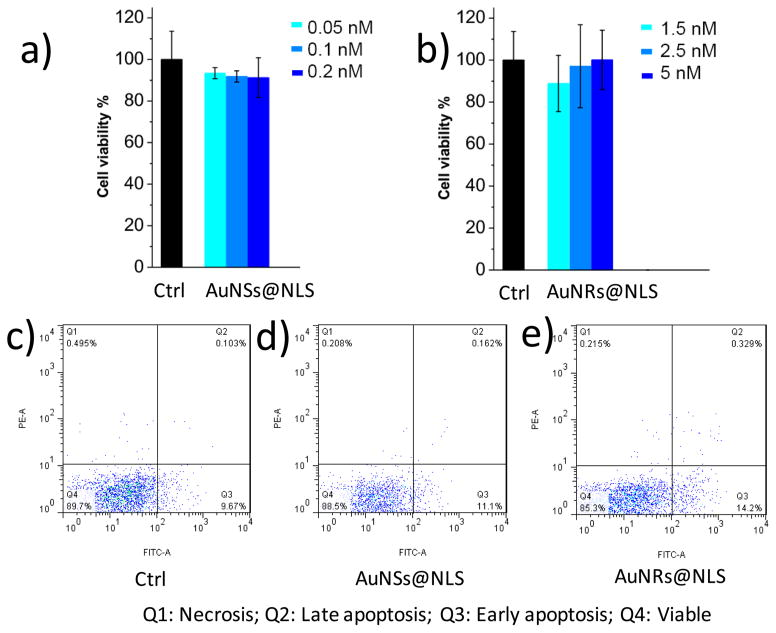
To examine the cytotoxicity of the AuNPs, the XTT cell proliferation assay was conducted, and no significant change of the cell viability was observed for nanoparticles at frequently used concentrations 0.5, 2.5, and 5 nM (for AuNRs)17,42 (Figure 2a and 2b), and 0.05, 0.1, and 0.2 nM concentrations (for AuNSs).41,43 Apoptosis/necrosis assay was also conducted for 5 nM of the AuNRs or 0.2 nM of the AuNSs (Figure 2c, 2d and 2e) using flow cytometry. The results indicate that the concentrations of the AuNRs used in this study are lower than those affecting cell viability or inducing apoptosis.
Figure 2.
Au nanoparticles cytotoxicity measurements and cellular uptake. (a) Cell viability measurement (XTT assay, n = 3) of HEY A8 cells after 24 h incubation with AuNSs@NLS at concentrations 0.05 nM (light blue), 0.1 nM (medium blue) and 0.2 nM (dark blue). (b) Cell viability (XTT, n = 3) assay for cells after 1.5 nM (light blue), 2.5 nM (medium blue) and 5 nM (dark blue) of AuNRs@NLS incubation with HEY-A8 cells for 24h. (c, d, and e) Flow cytometry experiment for apoptosis/necrosis assay (c, Ctrl; d, cells incubated with 0.2 nM of AuNSs@NLS; e, cells incubated with 5 nM of AuNRs@NLS).
The mass concentration (gram/L) of the two types of particles are very similar (SI, eq 1). The uptake of AuNPs@ NLS was monitored using dark-field (DF) microscopy and UV–vis absorption. The HEY A8 cells, which were previously identified to be highly invasive cell line,44 were incubated with 2.5 nM of AuNRs@NLS or 0.05 nM of AuNSs@NLS for 24 h. As shown in the DF image (Figure S1a and S1b), clear internalization of both AuNPs (AuNRs@NLS and AuNSs@ NLS) was observed. To evaluate the AuNPs uptake to the HEY A8 cells, UV–vis spectra were collected for the AuNPs in culture media before incubation with cells and compared with the ones after 24 h cell incubation (Figure S1c and S1d). According to the Beer’s law, the concentration of gold nanoparticles is linearly correlated with the absorbance at their localized surface plasmon resonance (LSPR) wavelength. Therefore, the decrease of the absorbance indicates the portion of AuNRs internalized in cells.45,46
Nontargeted AuNPs with bovine serum albumin (BSA) coating were also fabricated. Successful surface modification of AuNPs@BSA (both AuNRs@BSA and AuNSs@BSA) was evident in the red-shift of the surface plasmon peak of AuNPs to longer wavelengths (Figure S2a and S2b). Zeta potential of AuNRs after BSA modification became negatively charged (−19.6 ± 9.89 mV, Table S1) due to the negative charge of BSA, while the as-synthesized CTAB coated AuNRs has highly positive surface charge (50.9 ± 7.97 mV, Table S1). The AuNSs@BSA also has a negative zeta potential of −15.2 ± 12.5 mV (Table S1). No toxicity effect of AuNPs@BSA was observed, as shown in Figure S2c and S2d.
Nuclear Targeting Gold Nanoparticles Inhibit Cancer Cell Migration and Invasion
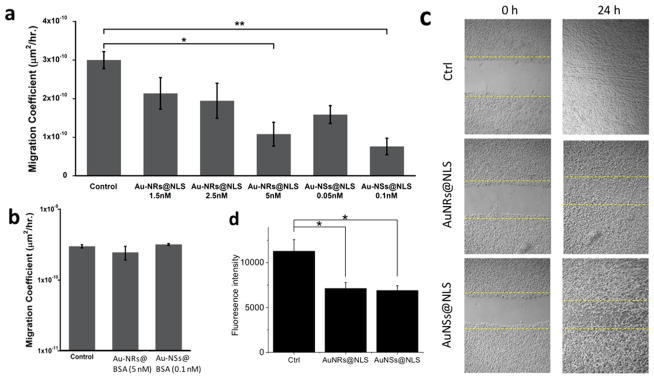
To test the cell motility, HEYA8 cells were incubated with AuNPs for 12 h before staining with fluorescent nuclear dye. Cells were then placed on an inverted epi-fluorescent microscope equipped with a cell culture chamber for continuous bright field and fluorescence imaging. The cell migration coefficients were then determined from the images. As shown in Figure 3a, both nuclear-targeted AuNRs and AuNSs inhibit the motility of HEY A8 cells. The average migration coefficient of the cells decreases from 3 × 10−10 by a factor of 3–10. (Figure 3a and Supporting Information Videos for motility). We conducted a control experiment of nontargeted AuNPs coated with BSA, (AuNPs@BSA, characterization information in the Supporting Information and Figure S2). The motility assay shows that there is no apparent inhibition of AuNRs@BSA or AuNSs@BSA on cell migration (Figure 3b).
Figure 3.
Effect of AuNPs (2.5 nM AuNRs@NLS and 0.1 nM AuNSs@NLS if not mentioned) on motility and invasion of HEY A8 cells. Cell migration study was performed to determine the effects of both AuNRs@NLS and AuNSs@NLS (a), and AuNRs@BSA (5 nM) and AuNSs@ BSA (0.1 nM) (b) on the HEY A8 cells motility (error bar ± SEM, n = 2). (c) Scratch assay of cells incubated with AuNRs@NLS and AuNSs@ NLS displayed arrested healing/closing of the scratch (representative pictures from 3 repeated experiments). (d) Invasion assay of cells without AuNPs or with AuNRs@NLS and AuNSs@NLS treatment (error bar ± SD, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
The scratch assay was conducted to evaluate the migration ability. Results (Figure 3c) indicate that the control cells had a completely healed “wound” after 24 h following AuNPs incubation, while the ones treated with AuNRs@NLS and AuNSs@NLS were not completely healed after 24 h. No obvious change in cell proliferation rate were observed after 24 h (Figure 2a and 2b), thus the scratch assay result merely reflects the migration ability of the cells.
In order to examine the invasion ability of HEY A8 cells after their treatment with the nuclear membrane-targeted AuNPs, the transwell invasion assay was performed. The cells that invaded the basement membrane extract (BME) after 32 h were dissociated and stained with Calcein AM, a fluorescent dye that labels living cells. A control experiment has been performed to eliminate the possibility that gold nanoparticles could quench the fluorescence from Calcein AM (Figure S3). A significant decrease in fluorescence intensity was observed in the AuNPs treated groups, indicating the inhibition of the invasion ability of HEY A8 cells particles (Figure 3d).
In general, the cell migration and invasion abilities of HEY A8 cells were inhibited effectively by both AuNRs@NLS and AuNSs@NLS.
Nuclear Targeting Gold Nanoparticles Enhance Nuclear Stiffness
Next, we tested our hypothesis that the nuclear-targeted AuNPs can enhance nuclear stiffness. Cell stiffness as quantified by the Young’s modulus has been used as a biomarker of the metastatic potential of cancer cells.19 For AFM measurements (Figure 4a), a beaded cantilever was lowered on top of the individual cells, producing an indentation in those cells and corresponding deflection of the AFM cantilever, which allowed for the measurement of cell stiffness. An overhead image of AFM cantilever tip next to HEY A8 cells with nanoparticles was shown in Figure 4b. The distribution of Young’s moduli of individual cells, as well as that of the cell nucleus, from different nanoparticle treatments and the control is depicted in Figure 4c and 4d. In our study, both AuNSs@ NLS and AuNRs@NLS exhibit significant increase in the cell stiffness (Figure 4c), which is similar to previous observations that nanoparticles could increase cell stiffness.47,48 For the nuclear stiffness, as shown in Figure 4d, the mean nuclear Young’s modulus of the cells treated with AuNPs (AuNRs and AuNSs) were also significantly higher than the mean nuclear Young’s modulus of the untreated cells, in agreement with the results of the overall cell stiffness. In addition, we observed the increase of gold nanoparticle amount could increase the nuclear stiffness, as shown in Figure S4.
Figure 4.
Stiffness distribution of cells. (a) Schematic of measurements on cells with AFM; δ is indentation, Δx is cantilever deflection. To measure bulk cellular stiffness, a beaded cantilever was used to increase cell-probe surface area. (b) Overhead image of AFM cantilever tip next to HEY A8 cells with nanoparticles. (c) Box-and-whisker plots of stiffness of single cells for different nanoparticles treatment, the percentiles are 10%, 25%, 50%, 75%, and 90%. Overall difference between means is significant (p-value calculated from ANOVA). (d) Box-and-whisker plots of nuclear stiffness. *P < 0.05, **P < 0.01, ***P < 0.001, n = 3, cell counts >20 for each sample.
AuNPs Accumulate at Nuclear Membrane Resolved by Three-Dimensional Microscopy
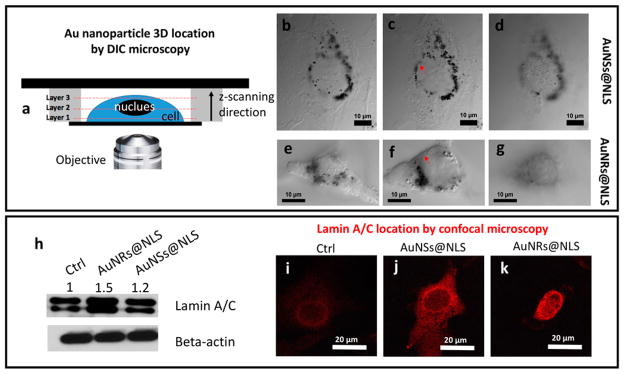
Resolving the exact localizations of AuNPs with regard to the nuclear membranes49,50 is a crucial, yet highly challenging, step in our attempt to understand the effects of AuNPs on the inhibition of cell migration/invasion. Most commonly used optical microscopy methods, such as confocal fluorescence microscopy and dark field (DF) microscopy, do not offer the accurate locations of the nuclear membranes and AuNPs simultaneously, and they usually suffer from high background. On the other hand, TEM, despite its high resolving power, is limited by the high costs and tedious sample preparation to gain the full 3D distribution of AuNPs inside the cells. To circumvent these challenges, we employed a recently developed differential interference contrast (DIC) microscopy-based 3D imaging method to visualize and locate plasmonic AuNPs inside the cells.51–53 As shown in Figure 5a, the cells were placed in a sandwiched chamber. DIC optical sectioning was performed on the whole cell thickness. The arrow indicates the scanning optical sectioning of the cell, directed from layer 1 (close to the surface of the cover glass) to layer 2 (middle of the cell) and to layer 3 (top of the cell) (Figure 5 and S5, and Videos S1 and S2 in the Supporting Information). The DIC microscope, which was equipped with a set of high numerical aperture (NA = 1.4), oil-immersion condenser and objective, features a shallow depth of field in optical sectioning of a 3D specimen to generate sharply focused images. More importantly, the nuclear membranes are clearly visible under the DIC microscope to allow the determination of the relative positions of the nuclear membranes and AuNPs.
Figure 5.
Locations of AuNPs inside the HEYA8 cell (up) and lamin A/C protein location/expression (down) inside the HEY A8 cell. (a) Scheme of the cell sample in sandwiched chamber for 3-dimensional DIC microscope imaging. Z-axis scanning step is 65 nm from the bottom (close to the attached glass surface) to the top of the cell. Three layers from the bottom, middle, and the top of the cell, for cells incubated with 0.1 nM of AuNSs@NLS (b–d) and 2.5 nM of AuNRs@NLS (e–g) were imaged, corresponding to frame 75, 235, and 395 (AuNSs@ NLS) and frame 49, 179, and 285 (AuNRs@NLS) of Videos S1 and S2 in the Supporting Information. (h) Western-blot results of lamin A/C, with beta-actin as reference protein. (i, j and k) Lamin A/C localization by confocal microscope of (i) cells without or (j) with AuNSs@NLS or (k) AuNSs@NLS incubation. The red arrows in (c) and (f) indicate the nuclear membrane of the cells.
Figure 5b–d shows the AuNSs@NLS locations inside the HEY A8 cells. The black spots in the figures are the nanoparticle aggregates, which are shown clearly surrounding the nucleus from difference optical sections. Similarly, Figure 5e–g shows a similar distribution of AuNRs@NLS inside HEY A8 cells. Both AuNRs@NLS and AuNSs@NLS aggregates were located predominantly on the nuclear membranes (indicated by the red arrows in Figure 5c and 5f, more evidence in Figure S6), while the internalization of nanoparticles inside the nucleus was rarely found, which was likely due to the large sizes of the nanoparticles and their aggregates compared to the nuclear pores (around 9–12 nm54).
Nuclear Targeting Gold Nanoparticles Cause Lamin A/C Protein Increase
Lamins, especially lamin A/C, are intermediate filament proteins found at nearly all cell nuclei and contribute to nuclear stiffness and stability.55,56 Nuclear lamins interact with the membrane-associated proteins to form the nuclear lamina (30–100 nm thick), which is located in the interior of the nuclear membrane. It has been reported that lamin A/C-deficient cells exhibit severely reduced nuclear stiffness.29,56 To further understand the biological mechanism for why nuclear membrane-targeting AuNPs increase nuclear stiffness and inhibit cancer cell migration/invasion, we measured the expression level of lamin A/C in Western-blots (Figure 5h) and confocal microscopy imaging after immunostaining (Figure 5). The results indicate a clear overexpression of lamin A/C after incubation with AuNSs@NLS or AuNRs@ NLS. As shown in Figure 5i, 5j and 5k, the fluorescence signal from lamin A/C was increased as a circle-surrounding the nucleus, which is in agreement with the location of nuclear lamina.
AuNPs conjugated with a nuclear localization signal were thought to be able to internalize into the nucleus.49,57 In our study we clearly observed most of the NLS conjugated gold nanoparticles aggregated around the nuclear membrane. Without NLS, the nanoparticles (AuNPs@RGD) spread in the cytoplasm, instead of accumulating around the cell nuclear region, which has been discussed in our previous reports).49 Western blot experiment showed that the endocytosis and nuclear transportation has been activated upon nanoparticle incubation (Figure S7), due to the increased expression level of dynamin protein (a GTPase responsible for endocytosis in the eukaryotic cell) and GTP-binding nuclear protein Ran (involved in the transport into and out of the cell nucleus). Since the cellular and nuclear transportation are all activated, the trapping of AuNPs at the nuclear membrane was most likely due to the large size of the gold nanoparticles aggregates compared to the nuclear pores.
It has been widely reported that following the entry of nanoparticles, they traffic through early endosomes to late endosomes and lysosomes (endolysosomal trafficking).58,59 To achieve nuclear membrane targeting, nanoparticles need to escape from the endosome and/or lysosomes. There are several well-established mechanisms explaining the cytosolic release of the NPs from endosomes or lysosomes. One of the most popular mechanism is through the charge interactions. The cationic nanoparticles could interact with the negatively charged phosphor lipid membrane, followed by “proton sponge” effect, causing endosomal membrane rupturing60,61 and nanoparticle escaping. In our study, the positively charged AuNRs@NLS62 could have the similar mechanism to escape from the endosome. In our results, most of the nanoparticles finally locates surrounding the cell nucleolus after incubation with cells overnight (Figure 5b–g and Supporting Videos S1 and S2), indicating a good efficacy that gold nanoparticles escape from the endosome/lysosomes and target the nuclear membrane. Meanwhile, the above results show that the effect of endosome degradation63 of the surface conjugated peptides might be very minor.
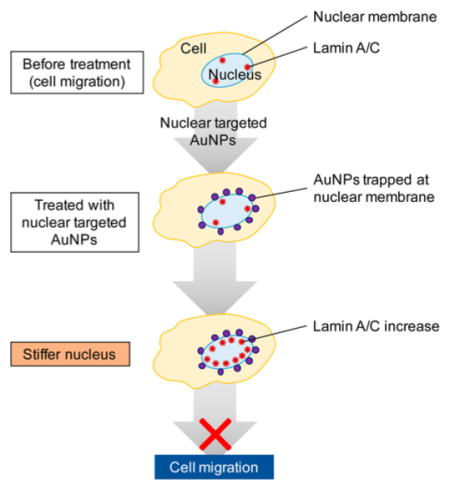
Coincident with the increased nuclear stiffness by the AuNPs is the aggregation of the AuNPs at the nuclear membrane and the increase of the Lamin A/C expression, which is located at the inner side of nuclear membrane. Lamin A/C is known to maintain the mechanical strength of the nucleus,64 and is thus consistent with an inhibited cell migration or invasion. In our results, a clearly increased expression level of Lamin A/C was observed. Thus, we propose that the increase of nuclear stiffness not only due to the mechanical contribution of the presence of gold nanoparticles, but could also due to the increase of Lamin A/C. How AuNPs increase Lamin A/C is not yet well explored in literature. Figure 5C shows AuNPs closely contact with the nuclear membrane, which could potentially disturb the membrane integrity. Therefore, we propose it could be a cellular defense mechanism as lamin is known to remain the mechanical strength of nucleus. Interestingly, nuclear lamin-associated proteins, such as emerin, which stabilizes nuclear architecture for maintaining the structural integrity,63,65,66 are also increased as the AuNPs are added at the nuclear membrane as shown in Figure S7. To investigate this mechanism, further studies such as proteomics and high-resolution imaging could be fruitful to elucidate the exact role of the AuNPs in inhibiting cell migration and invasion.
The clearance of nanoparticles from body after treatment has great importance to the evaluation of long-term effect of nanoparticles. While small nanoparticles (hydrodynamic diameter less than 5.5 nm) can be discard rapidly and efficiently through renal/urinary excretion.67 big nanoparticles (over 18 nm) tend to accumulate in liver and spleen.68 Such body deposition of metallic NPs over a long time period raises significant concerns regarding their long-term safety. A decrease of the liver content of gold has been reported after 1 month from 0.54% to the 0.07%.69 The ultimate fate and the body elimination pattern of gold nanoparticles are not well studied. Future work will be focused on studying the effect of gold nanoparticles for preventing and treating the metastasis in animals.
CONCLUSIONS
This study shows that nuclear membrane-targeting AuNPs can increase nuclear stiffness and thereby inhibit cell migration and invasion. Compared with the previous studies with nontargeted AuNPs at relatively high amount (50–200 μM of AuNRs@ BSA21 and 5–20 μg AuNSs@Citrate19), the nuclear membrane-targeted AuNPs showed higher inhibition effects at significantly lower concentrations (0.1 nM for 35 nm AuNSs and 2.5 nM for 25 × 5 nm AuNRs). The AuNPs were found to be trapped on the nuclear membranes from mapping the 3D distributions of the AuNPs under a DIC microscope. The trapping of AuNPs at the nuclear membranes could possibly (1) add to the mechanical stiffness of the nucleus, and (2) stimulate the overexpression of lamin A/C, which is known to lead to nuclear stiffness and thus slows down cancer cell migration and invasion. This insight takes us one step closer to fully understanding the effects on AuNPs on the inhibition of metastasis.
METHODS
Materials
Tetrachloroauric acid trihydrate (HAuCl4.3H2O), trisodium citrate, NaBH4, ascorbic acid, cetyltrimethylammonium bromide (CTAB), AgNO3, and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (USA). Methoxypoly(ethylene glycol)-thiol (mPEG-SH, MW 5000) was purchased from Laysan Bio, Inc. Cell penetrating peptide RGD (RGDRGDRGDRGDPGC) and nuclear localization signal (NLS, CGGGPKKKRKVGG) peptides were obtained from GenScript,Inc. Dulbecco’s phosphate buffered saline (PBS), RPMI-1640 cell culture media, fetal bovine serum (FBS), antibiotic solution, and 0.25% trypsin/2.2 mM EDTA solution were purchased from VWR. 8.0 μm polycarbonate membrane inserts were bought from Costar. Hoechst 33342 solution was purchased from Thermo Fisher Scientific (20 mM solution)
Instrumentation
Gold nanoparticles were imaged using a JEOL 100CX-2 transmission electron microscope (TEM) microscope and their average size was then measured by ImageJ software. UV–vis spectra were obtained using an Ocean Optics HR4000CG UV-NIR spectrometer. Cell stiffness was obtained using a MFP-3D AFM (Asylum Research, Santa Barbara, CA) with a combined Nikon Ti inverted optical microscope (Nikon, Melville, NY) for optically aligning the probe (MCST-AUHW, Bruker, Camarillo, CA) with a nominal spring constant of 0.03 N/m) to the cells. Confocal images were taken with a Zeiss LSM 700–405 confocal microscope.
Gold Nanoparticle Synthesis, Conjugation and Characterization
Gold nanospheres (AuNSs) with an average diameter of 35 nm were synthesized using the citrate reduction method.70 Briefly, 100 mL of 0.254 mM HAuCl4.3H2O solution was heated until boiling, and then reduced by adding 2.5 mL of 0.35% of trisodium citrate. The solution was then left heating until it turned wine red, followed by cooling under water flow. The citrate stabilized AuNSs were centrifuged under 5000g for 10 min and redispersed in deionized (DI) water to remove extra citrate and be ready for conjugation.
Gold nanorods (AuNRs) with an average size of 25 × 6 nm (length × width) were synthesized using a seedless growth method.35 Briefly, 5 mL of 1.0 mM HAuCl4 was added to a mixture of 5 mL of 0.20 M CTAB, 250 ul of 4.0 mM AgNO3 and 8 μL of 37% HCl. For reduction, 70 μL of 78.8 mM ascorbic acid was added, followed by immediate injection of 15 μL of 0.01 M of ice-cold NaBH4. The solution was left undisturbed for 12 h, then centrifuged at 21 000 rpm for 50 min and redispersed in DI water followed by a second centrifugation at 19 000 rpm for 40 min to remove the extra CTAB. TEM was used to measure the sizes and homogeneity of the nanoparticles.
AuNSs and AuNRs were then conjugated according to previous work,41,15 to achieve nuclear and cytoplasmic targeting. For nuclear targeting, first, mPEG-SH (1 mM) was added to the nanoparticles for overnight to achieve about 1000 ligands on each particle. Then, the PEGylated nanoparticles (1 nM) were treated with RGD (1 mM) and NLS (1 mM) to achieve 104 and 105 molar excess, respectively. The solution was then allowed to shake overnight at room temperature. Excess ligands were removed by centrifugation. For preparing BSA conjugated nanoparticles, BSA (4.5 mM) was added to the nanoparticles and left for 3 h to incubate. UV–vis spectrometer and zetasizer was used to test the conjugation. Surface modification causes red shift of UV–vis spectra due to the change in the dielectric constant of the surrounding environment of Au nanoparticles.
Cell Culture and AuNPs Incubation
The ovarian cancer HEY A8 cell lines were provided by Dr. G. Mills (MD Anderson Cancer Center, Houston, TX) and were grown in RPMI-1640 media supplemented with 10% FBS and 1% antibiotic-antimycotic solution. Cells were kept in a humidified incubator at 37 °C and under 5% CO2. After achieving 50% confluence, the cells were incubated with functionalized AuNSs or AuNRs in supplemented DMEM cell culture medium for 24 h. The concentration of nanoparticles was carefully chosen to avoid cytotoxicity or perturbation to the cell cycle.71
Apoptosis/Necrosis Assay
The HEY A8 cells were collected by trypsinization and washed with cold PBS twice. Then, cells were the dispersed in 493 μL of Annexin V binding buffer before labeling by 5 μL of Annexin V FITC (BioLegend) and 2 μL of PI (BioLegend, 100 μg/mL) according to previous reports.9 The mixture was then incubated for 15 min at room temperature. The cells were subjected to flow cytometry analysis using a BSR LSR II flow cytometer (BD Biosciences). For excitation, a 488 nm laser was applied. FITC and PI were detected in FL-1 and FL-2 using 525/30 and 575/30 BP filters, separately. Standard compensation using unstained and single-stained cells was conducted before performing actual experiments. FlowJo software (Tree Star Inc.) was used for data analysis. At least 10 000 events were collected for each experiment.57
Cell Motility Assay
HEY A8 cells were seeded on uncoated 24-well plate at a subconfluent density for 24 h. Then the cells were treated with nanoparticles of varying shapes and conjugated motifs before returning them to incubator for 12 h to facilitate particle uptake. After the incubation period, cells were stained with nuclear dye Hoechst 33342 (dilution 1:10 000) for 30–60 min. Cells were maintained at 37 °C and 5% carbon dioxide throughout the experiment using an environmental cell chamber (InVivo Scientific). For observation, a Nikon Eclipse Ti inverted epifluorescent microscope was used and both bright field (BF) and DAPI images were taken at multiple xy positions at 12 min time interval for 6–8 h at 10× magnification. The locations of cell nuclei, segmented from fluorescent images, were tracked in MATLAB to define cell traces. The cell migration coefficients and directional velocities were determined by fitting the traces to the persistent random walk model. Briefly, mean square displacements were calculated from the two-dimensional tracking data and was used for fitting the following equation
| (1) |
where P = persistence time and μ = migration coefficient.
Transwell Invasion Assay
The Cultured 24 Well BME Cell Invasion Assay kit (Trevigen) was used according to the manufacturer’s instructions. For these studies, HEY A8 cells were seeded and grown in a 6 well plate to 60–80% confluency before treating with nanoparticles in serum free media for 24 h. Cells were then detached, spun down and resuspended in serum free media. We also counted the cell number at this time to adjust the density to 500 000 cells/mL. Then 50 000 cells (100 μL) were added for each condition to the top surface of transwell inserts with 8 μm membrane pores coated with basement membrane matrix (BME). Cells were allowed to migrate toward the 10% FBS containing media in bottom chamber acting as the chemo attractant for a period of 32 h. After the desired incubation time, nonmigratory cells were gently removed from the top of each transwell using q-tips and the migrated cells at the bottom surface were detached using detaching buffer and incubated with Calcein AM. A plate reader was used to measure the fluorescence intensity, which is positively related to the number of transwell cells.
Atomic Force Microscopy
AFM mechanical measurements72,73 of HEY A8 cells were obtained using an MFP 3-D AFM (Asylum Research, Santa Barbara, CA) on a vibration isolation table (Herzan, Laguna Hills, CA). A silicon nitride cantilever (Bruker, Camarillo, CA) was used for the experiments. The pyramidal tip had a half angle of 35° and the radius of curvature of the point of the tip was 20 nm. Measurements were performed on cells plated to the glass bottom of the Fluorodish and in culture media at room temperature. For eliminating the effect of the overlapping neighboring cells on the stiffness, single cells were measured. Thermal calibration74 yielded the cantilever spring constant, k = 28.01pN/nm. A measurement rate of 0.39 Hz was used. The 5 nN force trigger resulted in indentations of approximately 4 μm for typical cells. Cells were optically located using a Nikon Eclipse Ti microscope (Nikon, Melville, NY). Force–displacement curves were recorded to obtain the Young’s modulus of each cell. Two distinct sets of measurements were performed with the AFM. The first investigated changes in mean cell stiffness between populations treated with AuNPs@NLS and an untreated control population. The second set of measurements investigated subcellular elasticity of nucleus. For the first set of measurements, the cantilever probe was positioned over the individual cells for indentation and measurement. For the second set, the probe was positioned over the perinuclear region.
Cell Imaging Using DIC Microscopy
An inverted Nikon Eclipse Ti-E microscope equipped with Perfect Focus System (PFS, 25 nm z-axial resolution) was used for imaging and z-stacks acquisitions under differential interference contrast (DIC) microscopy. The DIC mode utilized a pair of DIC polarizer and analyzer, a high resolution 100 × IR DIC slider, a high numerical aperture (N.A., 1.40) oil immersion condenser lens, a Nikon CFI Apo TIRF 100× (N.A., 1.49) oil immersion objective, and a 12 V/100 W halogen lamp as light source. Appropriate bandpass filters were placed in the light path. The z-stack movies were taken by a Hamamatsu ORCA-Flash 4.0 V2 CMOS camera (C11440–22CU, pixel size: 6.5 μm × 6.5 μm) with Camera Link interface using Micro-Manager and analyzed using NIH ImageJ and reconstructed in Amira. Fixed HEYA8 cells on 22 mm × 22 mm glass coverslips were rinsed with DPBS at pH 7.4 and fabricated into a sandwiched chamber with two pieces of double-sided tape and a cleaned glass slide. PBS solution was added into the chamber to fill the space and the chamber was then sealed by clear nail polish. The so-formed sample slide was then place under the microscope for observation. Z-stacks were acquired using the Multi-Dimensional Acquisition function in Micro-Manger. More specifically, the DIC optical sectioning through the whole cell thickness was achieved by moving the objective on the motorized nosepiece using PFS at 65 nm/step at 33 ms (30 fps) exposure time.
Scratch Assay
The scratch assay has been performed according to former report.75 Cells were cultured in a 6 well plate to a confluent monolayer. A p200 pipet tip was used to scrape the cell monolayer in a straight line to create an empty gap. The debris was then remove by washing the cells once with culture medium and then replace with 2 mL of fresh medium. Then the cells were imaged shortly after and 12 h after scratch.
Western Blot
Briefly, cells were lysed in RIPA buffer (20 mM Tris pH 7.4, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 0.1% Sodium Deoxycholate, 1% Triton X-100, 0.1% SDS) containing protease inhibitors (Sigma-Aldrich). BCA assay (Pierce) was performed to measure the protein concentration and equal amounts of protein were loaded on a SDS-PAGE gel. After SDS-PAGE, the resulting gels were transferred to PVDF membranes (Millipore) overnight. Afterward, the gel was blocked with 5% milk in TBS (20 mM Tris, 150 mM NaCl). A rabbit polyclonal antibody to Lamin A/C was used as the primary antibody (Bethyl Laboratories, Inc.) overnight in 4 °C with shaking. A goat anti rabbit HRP labeled antibody was used as the secondary antibody (Jackson Immuno Research Laboratories). Blots were washed 3 times for 20 m in TBS after primary and secondary antibodies. Konica Minolta developer and Hyglo enhanced chemiluminescence (Denville) were used to develop the immunoblots.
Immunofluorescence Labeling and Confocal Microscopy
Cells were cultured on confocal chamber slides (MATECH Co. USA). After gold nanoparticle treatment,17 cells were fixed in 4% Paraformaldehyde/0.1% Glutaraldehyde for 10 min at room temperature the wash with PBS. Cells were then permeabilized with 0.1% Triton X-100 for 5 min at room temperature. Cells were then blocked with 5% BSA and incubated with the primary antibody as stated in the Western-blot method for overnight. Cells were then incubated for 1 h with an Alexa Fluor 488 secondary antibody (Invitrogen) for 1 h before mounting with Prolong Gold (Invitrogen). Lastly images were taken with a Zeiss LSM 700–405 confocal microscope and the fluorescence intensity was quantified in ImageJ.
Data Analysis
To determine the Young’s modulus, IGOR Pro software (Wavemetrics, Portland, OR) was used to apply the Hertzian contact model76–78 from 10 to 90% of the maximum indentation of the extension force–displacement curve. Due to the unequal sample size and heteroscedasticity of the AFM data, overall statistical significance of differences in mean cell stiffness and nuclear stiffness between cells treated with AuNPs@NLS was tested using Welch’s analysis of variance (ANOVA). Posthoc analysis was performed using the Games–Howell test. For the rest of the studies, we used t test. The analyses were performed with the alpha type error set at 0.05.
Supplementary Material
Acknowledgments
We acknowledge the NSF Division of Chemistry (CHE) grant 1608801 for its support of this work. We also acknowledge NSF CMMI 153816; Georgia Tech’s Presidential Undergraduate Research Award. KC and NF acknowledge the funding support from NIH (–1R01GM115763). We acknowledge Tiegang Han and Yan Tang for their advice, Vy H.T. Nguyen for her helping with the DIC microscopic measurement, and Savita C. Chapman and Kamillah J. Kassam for helping in AuNPs fabrication and in culturing the cells, Sarah Ghalayini and Cassidy Tobin for their proofreading, and Jean-Guillaume D.S. Durand for his helping in designing the figure abstract.
ABBREVIATIONS
- AuNPs
gold nanoparticles
- AuNSs
gold nanospheres
- AuNRs
gold nanorods
- AFM
atomic force microscopey
- DIC
differential interference contrast microscopy
- PEG
methoxypolyethylene glycol thiol
- NLS
nuclear localization signal
- DF
dark-field microscopy
- TEM
transmission electron microscopy
- BSA
bovine serum albumin
Footnotes
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.6b08345.
Experimental methods, results and figures/tables of AuNPs fabrication and characterization, cellular uptake and cytotoxicity examination, videos for cell motility assay and 3D imaging scanning, etc. (PDF)
The authors declare no competing financial interest.
References
- 1.Fidler IJ. The Pathogenesis of Cancer Metastasis: the ’Seed and Soil’ Hypothesis Revisited. Nat Rev Cancer. 2003;3:453–8. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of Cancer: the Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Weber GF. Why Does Cancer Therapy Lack Effective Anti-Metastasis Drugs? Cancer Lett. 2013;328:207–11. doi: 10.1016/j.canlet.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 4.Morgillo F, Lee HY. Resistance to Epidermal Growth Factor Receptor-Targeted Therapy. Drug Resist Updates. 2005;8:298–310. doi: 10.1016/j.drup.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 5.Early Breast Cancer Trialists’ Collaborative Group (EBCTCG) Effects of Chemotherapy and Hormonal Therapy for Early Breast Cancer on Recurrence and 15-Year Survival: an Overview of the Randomised Trials. Lancet. 2005;365:1687–717. doi: 10.1016/S0140-6736(05)66544-0. [DOI] [PubMed] [Google Scholar]
- 6.Murphy CJ, Gole AM, Stone JW, Sisco PN, Alkilany AM, Goldsmith EC, Baxter SC. Gold Nanoparticles in Biology: Beyond Toxicity to Cellular Imaging. Acc Chem Res. 2008;41:1721–1730. doi: 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch LR, Stafford RJ, Bankson JA, Sershen SR, Rivera B, Price RE, Hazle JD, Halas NJ, West JL. Nanoshell-Mediated Near-Infrared Thermal Therapy of Tumors Under Magnetic Resonance Guidance. Proc Natl Acad Sci U S A. 2003;100:13549–13554. doi: 10.1073/pnas.2232479100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nanocarriers As an Emerging Platform for Cancer Therapy. Nat Nanotechnol. 2007;2:751–60. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 9.Ali MR, Ibrahim IM, Ali HR, Selim SA, El-Sayed MA. Treatment of Natural Mammary Gland Tumors in Canines and Felines Using Gold Nanorods-Assisted Plasmonic Photothermal Therapy to Induce Tumor Apoptosis. Int J Nanomed. 2016;11:4849. doi: 10.2147/IJN.S109470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi J, Kantoff PW, Wooster R, Farokhzad OC. Cancer Nanomedicine: Progress, Challenges and Opportunities. Nat Rev Cancer. 2017;17:20–37. doi: 10.1038/nrc.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun YW, Huh YM, Choi JS, Lee JH, Song HT, Kimkim, Yoon S, Kim KS, Shin JS, Suh JS, Cheon J. Nanoscale Size Effect of Magnetic Nanocrystals and Their Utilization for Cancer Diagnosis via Magnetic Resonance Imaging. J Am Chem Soc. 2005;127:5732–5733. doi: 10.1021/ja0422155. [DOI] [PubMed] [Google Scholar]
- 12.Jain PK, Lee KS, El-Sayed IH, El-Sayed MA. Calculated Absorption and Scattering Properties of Gold Nanoparticles of Different Size, Shape, and Composition: Applications in Biological Imaging and Biomedicine. J Phys Chem B. 2006;110:7238–7248. doi: 10.1021/jp057170o. [DOI] [PubMed] [Google Scholar]
- 13.Ali MR, Panikkanvalappil SR, El-Sayed MA. Enhancing the Efficiency of Gold Nanoparticles Treatment of Cancer by Increasing Their Rate of Endocytosis and Cell Accumulation Using Rifampicin. J Am Chem Soc. 2014;136:4464–4467. doi: 10.1021/ja4124412. [DOI] [PubMed] [Google Scholar]
- 14.Danhier F, Feron O, Preat V. To Exploit the Tumor Microenvironment: Passive and Active Tumor Targeting of Nanocarriers for Anti-Cancer Drug Delivery. J Controlled Release. 2010;148:135–46. doi: 10.1016/j.jconrel.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 15.Ali MRK, Panikkanvalappil SR, El-Sayed MA. Enhancing the Efficiency of Gold Nanoparticles Treatment of Cancer by Increasing Their Rate of Endocytosis and Cell Accumulation Using Rifampicin. J Am Chem Soc. 2014;136:4464–4467. doi: 10.1021/ja4124412. [DOI] [PubMed] [Google Scholar]
- 16.Dickerson EB, Blackburn WH, Smith MH, Kapa LB, Lyon LA, Mcdonald JF. Chemosensitization of Cancer Cells by Sirna Using Targeted Nanogel Delivery. BMC Cancer. 2010;10:10. doi: 10.1186/1471-2407-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali MRK, Ali HR, Rankin CR, El-Sayed MA. Targeting Heat Shock Protein 70 Using Gold Nanorods Enhances Cancer Cell Apoptosis in Low Dose Plasmonic Photothermal Therapy. Biomaterials. 2016;102:1–8. doi: 10.1016/j.biomaterials.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Yang JA, Phan HT, Vaidya S, Murphy CJ. Nanovacuums: Nanoparticle Uptake and Differential Cellular Migration on A Carpet of Nanoparticles. Nano Lett. 2013;13:2295–2302. doi: 10.1021/nl400972r. [DOI] [PubMed] [Google Scholar]
- 19.Arvizo RR, Saha S, Wang E, Robertson JD, Bhattacharya R, Mukherjee P. Inhibition of Tumor Growth and Metastasis by a Self-Therapeutic Nanoparticle. Proc Natl Acad Sci U S A. 2013;110:6700–6705. doi: 10.1073/pnas.1214547110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tay CY, Cai P, Setyawati MI, Fang W, Tan LP, Hong CH, Chen X, Leong DT. Nanoparticles Strengthen Intracellular Tension and Retard Cellular Migration. Nano Lett. 2014;14:83–8. doi: 10.1021/nl4032549. [DOI] [PubMed] [Google Scholar]
- 21.Zhou T, Yu MF, Zhang B, Wang LM, Wu XC, Zhou HJ, Du YP, Hao JF, Tu YP, Chen CY, et al. Inhibition of Cancer Cell Migration by Gold Nanorods: Molecular Mechanisms and Implications for Cancer Therapy. Adv Funct Mater. 2014;24:6922–6932. [Google Scholar]
- 22.Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R. Mechanical Stiffness Grades Metastatic Potential in Patient Tumor Cells and in Cancer Cell Lines. Cancer Res. 2011;71:5075–5080. doi: 10.1158/0008-5472.CAN-11-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cross SE, Jin YS, Tondre J, Wong R, Rao J, Gimzewski JK. AFM-Based Analysis of Human Metastatic Cancer Cells. Nanotechnology. 2008;19:384003. doi: 10.1088/0957-4484/19/38/384003. [DOI] [PubMed] [Google Scholar]
- 24.Wirtz D, Konstantopoulos K, Searson PC. The Physics of Cancer: the Role of Physical Interactions and Mechanical Forces in Metastasis. Nat Rev Cancer. 2011;11:512–522. doi: 10.1038/nrc3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lautscham LA, Kammerer C, Lange JR, Kolb T, Mark C, Schilling A, Strissel PL, Strick R, Gluth C, Rowat AC, et al. Migration in Confined 3D Environments Is Determined by a Combination of Adhesiveness, Nuclear Volume, Contractility, and Cell Stiffness. Biophys J. 2015;109:900–13. doi: 10.1016/j.bpj.2015.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahl KN, Ribeiro AJS, Lammerding J. Nuclear Shape, Mechanics, and Mechanotransduction. Circ Res. 2008;102:1307. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Isermann P, Lammerding J. Nuclear Mechanics and Mechanotransduction in Health and Disease. Curr Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mcgregor AL, Hsia CR, Lammerding J. Squish and Squeeze-the Nucleus As a Physical Barrier During Migration in Confined Environments. Curr Opin Cell Biol. 2016;40:32–40. doi: 10.1016/j.ceb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al. Nuclear Lamin-A Scales With Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science. 2013;341:1240104. doi: 10.1126/science.1240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada T, Swift J, Irianto J, Shin JW, Spinler KR, Athirasala A, Diegmiller R, Dingal PC, Ivanovska IL, Discher DE. Nuclear Lamin Stiffness Is a Barrier to 3D Migration, but Softness Can Limit Survival. J Cell Biol. 2014;204:669–82. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JS, Hale CM, Panorchan P, Khatau SB, George JP, Tseng Y, Stewart CL, Hodzic D, Wirtz D. Nuclear Lamin A/C Deficiency Induces Defects in Cell Mechanics, Polarization, and Migration. Biophys J. 2007;93:2542–52. doi: 10.1529/biophysj.106.102426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davidson PM, Denais C, Bakshi MC, Lammerding J. Nuclear Deformability Constitutes a Rate-Limiting Step During Cell Migration in 3-D Environments. Cell Mol Bioeng. 2014;7:293–306. doi: 10.1007/s12195-014-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong L, Schafer G, Bu HJ, Zhang Y, Zhang YX, Klocker H. Lamin A/C Protein Is Overexpressed in Tissue-Invading Prostate Cancer and Promotes Prostate Cancer Cell Growth, Migration and Invasion Through the PI3K/AKT/PTEN Pathway. Carcinogenesis. 2012;33:751–759. doi: 10.1093/carcin/bgs022. [DOI] [PubMed] [Google Scholar]
- 34.Mackey MA, Ali MRK, Austin LA, Near RD, El-Sayed MA. The Most Effective Gold Nanorod Size for Plasmonic Photothermal Therapy: Theory and in vitro Experiments. J Phys Chem B. 2014;118:1319–1326. doi: 10.1021/jp409298f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ali MRK, Snyder B, El-Sayed MA. Synthesis and Optical Properties of Small Au Nanorods Using A Seedless Growth Technique. Langmuir. 2012;28:9807–9815. doi: 10.1021/la301387p. [DOI] [PubMed] [Google Scholar]
- 36.Chithrani BD, Ghazani AA, Chan WCW. Determining the Size and Shape Dependence of Gold Nanoparticle Uptake into Mammalian Cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 37.El-Sayed IH, Huang XH, El-Sayed MA. Surface Plasmon Resonance Scattering and Absorption of Anti-EGFR Antibody Conjugated Gold Nanoparticles in Cancer Diagnostics: Applications in Oral Cancer. Nano Lett. 2005;5:829–834. doi: 10.1021/nl050074e. [DOI] [PubMed] [Google Scholar]
- 38.Prencipe G, Tabakman SM, Welsher K, Liu Z, Goodwin AP, Zhang L, Henry J, Dai HJ. PEG Branched Polymer for Functionalization of Nanomaterials With Ultralong Blood Circulation. J Am Chem Soc. 2009;131:4783–4787. doi: 10.1021/ja809086q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruoslahti E, Pierschbacher MD. Arg-Gly-Asp - A Versatile Cell Recognition Signal. Cell. 1986;44:517–518. doi: 10.1016/0092-8674(86)90259-x. [DOI] [PubMed] [Google Scholar]
- 40.Kalderon D, Roberts BL, Richardson WD, Smith AE. A Short Amino-Acid Sequence Able to Specify Nuclear Location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 41.Kang B, Austin LA, El-Sayed MA. Observing Real-Time Molecular Event Dynamics of Apoptosis In Living Cancer Cells Using Nuclear-Targeted Plasmonically Enhanced Raman Nanoprobes. ACS Nano. 2014;8:4883–92. doi: 10.1021/nn500840x. [DOI] [PubMed] [Google Scholar]
- 42.Ali HR, Ali MRK, Wu Y, Selim SA, Abdelaal HFM, Nasr EA, El-Sayed MA. Gold Nanorods As Drug Delivery Vehicles for Rifampicin Greatly Improve the Efficacy of Combating Mycobacterium Tuberculosis With Good Biocompatibility With the Host Cells. Bioconjugate Chem. 2016;27:2486–2492. doi: 10.1021/acs.bioconjchem.6b00430. [DOI] [PubMed] [Google Scholar]
- 43.Panikkanvalappil SR, Hira SM, Mahmoud MA, El-Sayed MA. Unraveling the Biomolecular Snapshots of Mitosis in Healthy and Cancer Cells Using Plasmonically-Enhanced Raman Spectroscopy. J Am Chem Soc. 2014;136:15961–15968. doi: 10.1021/ja506289u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu WW, Mezencev R, Kim B, Wang LJ, Mcdonald J, Sulchek T. Cell Stiffness Is A Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS One. 2012;7:7. doi: 10.1371/journal.pone.0046609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cho EC, Liu Y, Xia YN. A Simple Spectroscopic Method for Differentiating Cellular Uptakes of Gold Nanospheres and Nanorods From Their Mixtures. Angew Chem, Int Ed. 2010;49:1976–1980. doi: 10.1002/anie.200906584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho EC, Zhang Q, Xia YN. The Effect of Sedimentation and Diffusion on Cellular Uptake of Gold Nanoparticles. Nat Nanotechnol. 2011;6:385–391. doi: 10.1038/nnano.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pietuch A, Bruckner BR, Schneider D, Tarantola M, Rosman C, Sonnichsen C, Janshoff A. Mechanical Properties of MDCK II Cells Exposed to Gold Nanorods. Beilstein J Nanotechnol. 2015;6:223–231. doi: 10.3762/bjnano.6.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee CW, Jang LL, Pan HJ, Chen YR, Chen CC, Lee CH. Membrane Roughness As A Sensitive Parameter Reflecting the Status of Neuronal Cells in Response to Chemical and Nanoparticle Treatments. J Nanobiotechnol. 2016 doi: 10.1186/s12951-016-0161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang B, Mackey MA, El-Sayed MA. Nuclear Targeting of Gold Nanoparticles in Cancer Cells Induces DNA Damage, Causing Cytokinesis Arrest and Apoptosis. J Am Chem Soc. 2010;132:1517. doi: 10.1021/ja9102698. [DOI] [PubMed] [Google Scholar]
- 50.Patel S, Jung D, Yin PT, Carlton P, Yamamoto M, Bando T, Sugiyama H, Lee KB. Nanoscript: A Nanoparticle-Based Artificial Transcription Factor for Effective Gene Regulation. ACS Nano. 2014;8:8959–8967. doi: 10.1021/nn501589f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang GF, Stender AS, Sun W, Fang N. Optical Imaging of Non-Fluorescent Nanoparticle Probes in Live Cells. Analyst. 2010;135:215–221. doi: 10.1039/b916395f. [DOI] [PubMed] [Google Scholar]
- 52.Stender AS, Marchuk K, Liu C, Sander S, Meyer MW, Smith EA, Neupane B, Wang G, Li J, Cheng JX, et al. Single Cell Optical Imaging and Spectroscopy. Chem Rev. 2013;113:2469–2527. doi: 10.1021/cr300336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun W, Wang G, Fang N, Yeung ES. Wavelength-Dependent Differential Interference Contrast Microscopy: Selectively Imaging Nanoparticle Probes in Live Cells. Anal Chem. 2009;81:9203–9208. doi: 10.1021/ac901623b. [DOI] [PubMed] [Google Scholar]
- 54.Paine PL. Nucleocytoplasmic Movement of Fluorescent Tracers Microinjected into Living Salivary-Gland Cells. J Cell Biol. 1975;66:652–657. doi: 10.1083/jcb.66.3.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical Plasticity of the Nucleus in Stem Cell Differentiation. Proc Natl Acad Sci U S A. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C But Not Lamin B1 Regulate Nuclear Mechanics. J Biol Chem. 2006;281:25768–80. doi: 10.1074/jbc.M513511200. [DOI] [PubMed] [Google Scholar]
- 57.Dreaden EC, Mackey MA, Huang XH, Kang B, El-Sayed MA. Beating Cancer in Multiple Ways Using Nanogold. Chem Soc Rev. 2011;40:3391–3404. doi: 10.1039/c0cs00180e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandin P, Fitzpatrick LW, Simpson JC, Dawson KA. High-Speed Imaging of Rab Family Small Gtpases Reveals Rare Events in Nanoparticle Trafficking in Living Cells. ACS Nano. 2012;6:1513–1521. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
- 59.Kim AJ, Boylan NJ, Suk JS, Lai SK, Hanes J. Non-Degradative Intracellular Trafficking of Highly Compacted Polymeric DNA Nanoparticles. J Controlled Release. 2012;158:102–107. doi: 10.1016/j.jconrel.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chu ZQ, Miu KK, Lung PS, Zhang SL, Zhao SS, Chang HC, Lin G, Li Q. Rapid Endosomal Escape of Prickly Nanodiamonds: Implications for Gene Delivery. Sci Rep. 2015;5:11661. doi: 10.1038/srep11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit JP. Progress in Developing Cationic Vectors for Non-Viral Systemic Gene Therapy Against Cancer. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]
- 62.Ali MRK, Wu Y, Hang TG, Zang XL, Xiao HP, Tang Y, Wu RH, Fernandez FM, El-Sayed MA. Simultaneous Time-Dependent Surface-Enhanced Raman Spectroscopy, Metabolomics, and Proteomics Reveal Cancer Cell Death Mechanisms Associated With Gold Nanorod Photothermal Therapy. J Am Chem Soc. 2016;138:15434–15442. doi: 10.1021/jacs.6b08787. [DOI] [PubMed] [Google Scholar]
- 63.Rowat AC, Lammerding J, Ipsen JH. Mechanical Properties of the Cell Nucleus and the Effect of Emerin Deficiency. Biophys J. 2006;91:4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ge Y, Bruno M, Wallace K, Winnik W, Prasad RY. Proteome Profiling Reveals Potential Toxicity and Detoxification Pathways Following Exposure of BEAS-2B Cells to Engineered Nanoparticle Titanium Dioxide. Proteomics. 2011;11:2406–22. doi: 10.1002/pmic.201000741. [DOI] [PubMed] [Google Scholar]
- 65.Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT. Abnormal Nuclear Shape and Impaired Mechano-transduction in Emerin-Deficient Cells. J Cell Biol. 2005;170:781–791. doi: 10.1083/jcb.200502148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berk JM, Tifft KE, Wilson KL. The Nuclear Envelope LEM-Domain Protein Emerin. Nucleus-Austin. 2013;4:298–314. doi: 10.4161/nucl.25751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi HS, Liu W, Misra P, Tanaka E, Zimmer JP, Ipe BI, Bawendi MG, Frangioni JV. Renal Clearance of Quantum Dots. Nat Biotechnol. 2007;25:1165–1170. doi: 10.1038/nbt1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Semmler-Behnke M, Kreyling WG, Lipka J, Fertsch S, Wenk A, Takenaka S, Schmid G, Brandau W. Biodistribution of 1.4-and 18-Nm Gold Particles in Rats. Small. 2008;4:2108–2111. doi: 10.1002/smll.200800922. [DOI] [PubMed] [Google Scholar]
- 69.Sadauskas E, Danscher G, Stoltenberg M, Vogel U, Larsen A, Wallin H. Protracted Elimination of Gold Nanoparticles From Mouse Liver. Nanomedicine. 2009;5:162–169. doi: 10.1016/j.nano.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Turkevich J, Stevenson PC, Hillier J. A Study of the Nucleation and Growth Processes in the Synthesis of Colloidal Gold. Discuss Faraday Soc. 1951;11:55. [Google Scholar]
- 71.Austin LA, Ahmad S, Kang B, Rommel KR, Mahmoud M, Peek ME, El-Sayed MA. Cytotoxic Effects of Cytoplasmic-Targeted and Nuclear-Targeted Gold and Silver Nanoparticles in HSC-3 Cells - a Mechanistic Study. Toxicol In Vitro. 2015;29:694–705. doi: 10.1016/j.tiv.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Bao G, Suresh S. Cell and Molecular Mechanics of Biological Materials. Nat Mater. 2003;2:715–725. doi: 10.1038/nmat1001. [DOI] [PubMed] [Google Scholar]
- 73.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical Analysis of Cells From Cancer Patients. Nat Nanotechnol. 2007;2:780–783. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 74.Cross SE, Jin YS, Rao J, Gimzewski JK. Nanomechanical Analysis of Cells From Cancer Patients. Nat Nanotechnol. 2007;2:780–3. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 75.Liang CC, Park AY, Guan JL. In vitro Scratch Assay: A Convenient and Inexpensive Method for Analysis of Cell Migration in vitro. Nat Protoc. 2007;2:329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 76.Hutter JL, Bechhoefer J. Calibration of Atomic-Force Microscope Tips. Rev Sci Instrum. 1993;64:3342–3342. [Google Scholar]
- 77.Renger A, Johnson KL. Contact Mechanics. XII. Cambridge University Press; 1985. p. 452. [Google Scholar]; Renger A, Johnson KL. Z Angew Math Mech. 1989;69:214–214. [Google Scholar]
- 78.Xu WW, Chahine N, Suchek T. Extreme Hardening of PDMS Thin Films Due to High Compressive Strain and Confined Thickness. Langmuir. 2011;27:8470–8477. doi: 10.1021/la201122e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.