Abstract
Chronic lung allograft dysfunction (CLAD) is linked to rejection and limits survival following lung transplantation. HLA-Bw4 recipients of HLA-Bw6 grafts have enhanced host-versus-graft (HVG) NK cell activity mediated by KIR3DL1 ligand. Because natural killer (NK) cells may promote tolerance by depleting antigen-presenting cells, we hypothesized improved outcomes for HLA-Bw4 recipients of HLA-Bw6 grafts.
We evaluated differences in acute cellular rejection (ACR) and CLAD-free survival across 252 KIR3DL1+ recipients from UCSF. For validation, we assessed survival and freedom from BOS, retransplantation, or death in 12,845 non-KIR typed recipients from the UNOS registry. Cox proportional hazards models were adjusted for age, gender, ethnicity, transplant type, and HLA mismatching.
HVG-capable subjects in the UCSF cohort had a decreased risk of CLAD or death (HR 0.57, 95% CI 0.36–0.88) and decreased early lymphocytic bronchitis. The HVG effect was not significant in subjects with genotypes predicting low KIR3DL1 expression. In the UNOS cohort, HVG-capable subjects had a decreased risk of BOS, retransplant, or death (HR 0.95, 95% CI 0.91–0.99). Survival improved with the higher affinity Bw4-80I ligand and in Bw4 homozygotes.
Improved outcomes in HVG-capable recipients are consistent with a protective NK cell role. Augmentation of NK activity could supplement current immunosuppression techniques.
Introduction
Lung allograft recipients have poor survival outcomes relative to other solid organ transplants (1, 2). The primary cause of death after the first post-operative year is chronic lung allograft dysfunction (CLAD). CLAD can manifest as airway obstruction from constrictive bronchiolitis, defined spirometrically as bronchiolitis obliterans syndrome (BOS), or as pleuroparenchymal fibrosis causing restrictive allograft syndrome (RAS) (3). Multiple types of immune and non-immune responses are postulated to contribute to these pathologies (4).
NK cells are cytotoxic lymphocytes of the innate immune system with a variety of roles, including defense against virus-infected or malignant cells that lack surface expression of Human Leukocyte Antigen (HLA) class I antigen presenting molecules (5). In some contexts, the ability of an NK cell to recognize cells missing self-HLA molecules requires “licensing.” Self-HLA molecules license NK cells by stimulating inhibitory killer-cell immunoglobulin-like receptors (KIRs) during NK cell maturation. Licensing “educates” NK cells to self-antigens and arms them to activate upon encountering a cell missing those ligands (6, 7). This set of self-HLA molecules will then inhibit NK cell activation against self-cells. One important HLA-KIR interaction occurs between the KIR3DL1 inhibitory ligand, which is expressed in 93 to 97% of the general population (8), and HLA allotypes containing the Bw4 epitope. The Bw4 and Bw6 epitopes are present on most HLA-B and some HLA-A molecules and are binding sites for commonly occurring alloreactive antibodies. By contrast, Bw6 HLA alleles do not stimulate KIR3DL1. The strength of the KIR3DL1-Bw4 interaction is increased when residue 80 of the Bw4 epitope is isoleucine (Bw4-80I) rather than threonine (Bw4-80T) (9) and when both HLA-B alleles are Bw4 types rather than Bw4/Bw6 heterozygous (10).
Host-versus-graft (HVG) NK cell responses may be protective in lung allograft recipients (11). The KIR group A haplotype, in which the distribution of KIR receptors favors inhibition rather than activation, is a risk factor for CLAD (12), and percentages of NK cells in bronchoalveolar lavage fluid decrease during acute cellular rejection (13). In a mouse model of orthotopic lung transplantation, NK cell deficiency increased allograft rejection, while autologous NK cell expansion decreased alloreactive T cell activation and graft failure (14). Improved graft survival was associated with perforin-mediated NK cell depletion of host antigen presenting cells (APC), while potential anti-graft effect of the alloreactive NK cells appeared to be outweighed by the decrease in alloreactive T cells (14). An analogous process is observed in human stem cell transplantation, where KIR ligand incompatibility resulting in depletion of host APC protects against graft-versus host disease, at least in the context of certain conditioning regimens (15).
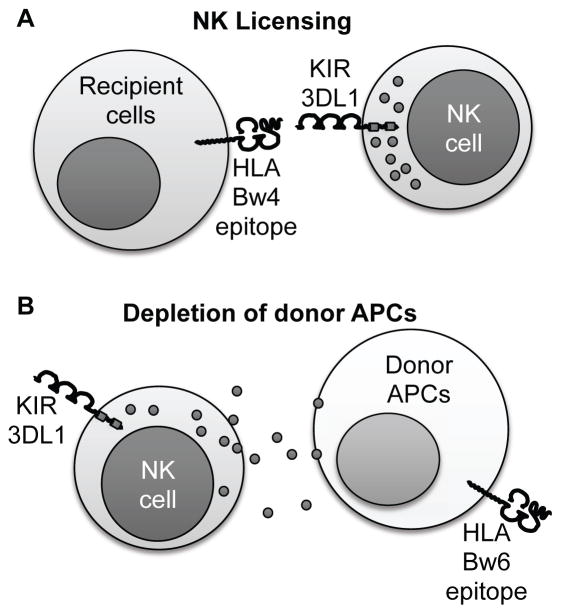
The significance of HVG NK cell activation in human lung allograft recipients has not been assessed. Thus, we tested the hypothesis that KIR3DL1 HVG-capable subjects, as defined by a licensing Bw4 ligand in the recipient but missing in the allograft (Figure 1), have improved CLAD-free survival following lung transplantation.
Figure 1. Proposed mechanism for NK cell mediated protection from rejection.
(A) NK cell licensing can occur following stimulation of recipient KIR3DL1 inhibitory receptors by recipient HLA antigens bearing the Bw4 epitope. (B) Bw4-licensed NK cells will kill cells that do not express HLA-Bw4, such as the depicted Bw6/Bw6 donor APC. Thus, host-versus-graft depletion of donor APC would be expected to result in improved graft survival for KIR3DL1+Bw4+ recipients who receive Bw6/Bw6 organs.
Methods
These studies were conducted with the approval of the University of California, San Francisco (UCSF) Internal Review Board, under protocol numbers 13-11512, 13-10738 and 16-19103.
UCSF cohort
Recipients of lung allografts who provided informed consent and for whom DNA samples were available were included. Bw4 and Bw6 epitopes were predicted from the clinical HLA typing of donor and recipient. Recipient KIR genotyping was performed using the Luminex-based rSSO method (One Lambda, Canoga Park, CA, USA). Probe 065 is specific for KIR3DL1*004, *005, *006, and some rare variants. These KIR3DL1 genotypes have been shown to have low or no expression of KIR3DL1 and recipients heterozygous or homozygous for this probe were considered to be low expressers (16, 17). Acute cellular rejection scores derived from pathological interpretation of large and small airway biopsies were abstracted from clinical records as previously described (18). We divided acute rejection data into early and late using a cutoff of 180 days post transplant. Serial spirometry was performed on recipients per clinical protocols. Time to CLAD was calculated as the time from transplant to a forced expiratory volume in 1 second (FEV1) or forced vital capacity (FVC) decline by 20% from the post-transplant baseline (3, 18). Survival time and clinical characteristics were obtained from a UCSF center-specific United Network for Organ Sharing (UNOS) dataset and UCSF electronic medical records.
Bronchoalveolar lavage (BAL) NK cell phenotyping was performed on a nested sub-cohort of lung transplant recipients. This was a convenience sample of subjects who underwent clinical bronchoscopy between February 2014 and September 2016, for whom excess BAL fluid was available. BAL lymphocytes were prepared as previously described (13) and stained with PE anti-KIR2D (Miltenyi Biotec, San Diego, CA, USA #130-092-688), PE anti-KIR3D (Beckman Coulter, Brea, CA, USA #IM3292), PerCP anti-KIR3DL1 (Biolegend, San Diego, CA #312718), PECy7 anti-CD56 (Biolegend #318318), AL700 anti-CD45 (Biolegend #304024) and APCCy7 anti-CD3 (Biolegend #344818). NK cells were defined as CD56+ CD3− CD45+ lymphocytes. BAL fluid samples with fewer than 500 KIR2D/3D+ NK cells were excluded.
UNOS cohort
For a UNOS registry-based validation cohort, we included all lung allograft recipients transplanted between 1995 and 2011 with complete HLA typing data and follow-up reporting of BOS status. Because the presence of follow-up reporting excluded most subjects with early mortality, we left-truncated the data at 6 months. Follow-up data were included through June 2016. Bw4/Bw6 status was determined based on HLA-B typing as reported to UNOS. HLA Bw4-80I status was assigned for HLA-B*5, 17, 38, 49, 51, 52, 53, 57, 58, 59, 63, and 77; Bw4-80T status was assigned for HLA-B*13, 27, 37, 44 and 47. There was a discrepancy between Bw4/Bw6 status as calculated from HLA-B type and Bw4/Bw6 status as recorded in the UNOS dataset for 2.5% of subjects, and these subjects were excluded. Time to BOS was determined by iteration through follow up records using a Python script (version 2.7.12, available by request). The first date with a report of BOS not followed by a report without BOS was considered the date of BOS. Only the first transplant was included for subjects undergoing retransplantation.
Analysis
Differences in clinical characteristics between HVG-capable and non-HVG-capable subjects were assessed by Student’s t-test or chi-square test, as appropriate. Percentages of KIR3DL1 expressing cells as a total of KIR+ NK cells in the BAL fluid were compared using generalized estimating equation-adjusted linear models, with robust standard error estimates used for inference (13). Average rejection scores were compared by Student’s t-test. We performed Kaplan-Meier analysis of freedom from CLAD or death in the UCSF cohort, and overall survival, as well as freedom from BOS, death, or retransplantation in the UNOS cohort. Differences were assessed by log-rank test. Hazard ratios were determined using Cox proportional hazards modeling. Adjusted models included donor and recipient age, gender, and ethnicity; the type of transplant; indication for transplantation; and the number of HLA mismatches at A and B loci.
Statistics were computed in R version 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria) using the “gee” and “survival” packages (19, 20).
Results
To test whether KIR3DL1-mediated HVG NK cell activity plays a role in lung transplant outcomes, we established a cohort of KIR3DL1-expressing lung transplant recipients. Of 258 subjects genotyped, 6 (2.3%) were KIR3DL1 negative. Characteristics for the remaining 252 subjects are shown in Table 1. Bw4 recipients of Bw6 grafts, for which KIR3DL1-mediated HVG activity is expected, represented 26% of the total cohort. At least one low-expressing KIR3DL1 allele was present in 30% and 45% of the HVG group and non-HVG-capable group, respectively, with this difference approaching statistical significance (P = 0.05).
Table 1.
Subject characteristics
| Cohort | UCSF | UNOS | ||||
|---|---|---|---|---|---|---|
| Bw6 Donor / Bw4 Recipient | Yes | No | P-value | Yes | No | P-value |
| Subjects N | 66 | 187 | 2919 | 9926 | ||
| KIR3DL1 Low (%) | 30% | 45% | 0.05 | – | – | |
| Age, mean (SD) | ||||||
| Recipient | 54 (13) | 54 (12) | 0.99 | 50 (15) | 50 (15) | 0.11 |
| Donor | 30 (13) | 33 (14) | 0.06 | 32 (14) | 32 (14) | 0.69 |
| Male Gender % | ||||||
| Recipient | 42% | 59% | 0.03 | 54% | 54% | 0.92 |
| Donor | 61% | 60% | 1.00 | 60% | 60% | 0.81 |
| D/R Match | 61% | 63% | 0.89 | 68% | 68% | 0.88 |
| Recipient Race/Ethnicity | ||||||
| White | 82% | 74% | 0.13 | 87% | 86% | 0.16 |
| Black | 6% | 7% | 7% | 7% | ||
| Hispanic | 3% | 13% | 4% | 5% | ||
| Other | 9% | 6% | 2% | 2% | ||
| Donor Race/Ethnicity | ||||||
| White | 38% | 50% | 0.01 | 69.3% | 68.1% | <0.001 |
| Black | 9% | 12% | 12.9% | 16.5% | ||
| Hispanic | 48% | 27% | 14.8% | 12.3% | ||
| Other | 5% | 11% | 3.0% | 3.2% | ||
| Race/Ethnicity Matched | 33% | 47% | 0.07 | 65% | 63% | 0.20 |
| Indication Group | ||||||
| A - Obstructive | 17% | 26% | 0.48 | 43% | 43% | 0.96 |
| B - Pulmonary Vascular | 8% | 5% | 6% | 6% | ||
| C - CF & Autoimmune | 12% | 10% | 15% | 15% | ||
| D - Restrictive | 64% | 59% | 33% | 33% | ||
| Unclassified | 0% | 0% | 3% | 3% | ||
| Lung Transplant Type | ||||||
| Double | 85% | 89% | 0.37 | 56% | 56% | 0.79 |
| Single | 11% | 9% | 41% | 41% | ||
| Heart-Lung | 5% | 2% | 3% | 3% | ||
| A-locus mismatches | ||||||
| 0 | 12% | 9% | 0.66 | 8% | 7% | 0.57 |
| 1 | 29% | 35% | 43% | 42% | ||
| 2 | 59% | 56% | 49% | 50% | ||
| B-locus mismatches | ||||||
| 0 | 0% | 2% | 0.44 | 0% | 1% | <0.001 |
| 1 | 17% | 19% | 16% | 25% | ||
| 2 | 83% | 79% | 84% | 74% | ||
We identified subjects from the UNOS registry as a validation cohort. KIR typing was not available for this UNOS cohort, but given that the population expression frequency of KIR3DL1 is as high as 97% (8), we inferred that most of the Bw4+ recipients of Bw6/Bw6 grafts had the potential for KIR3DL1 HVG activity. The major statistically significant difference between the HVG and reference groups across both cohorts was in donor ethnicity, with donors for the HVG subset being more commonly Hispanic and less frequently Black, likely reflecting ethnic differences in HLA frequencies. HLA-B locus mismatches were more frequent in the HVG UNOS cohort, which is expected, as HLA-B locus mismatching is a part of the group definition. Comparing the two cohorts, subjects in the UCSF cohort were on average slightly older, more likely to receive a double lung transplant, and more likely to have interstitial lung disease as an indication. Both donors and recipients in the UCSF cohort were more ethnically diverse than in the UNOS cohort, consistent with the demographics of Northern California and surrounding areas.
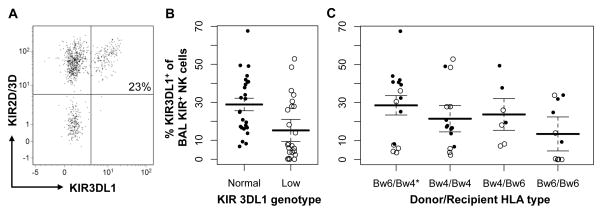
We investigated whether KIR3DL1 genotype and Bw4 epitope status affected the NK cell repertoire in BAL fluid. We evaluated the percentage of KIR3DL1-expressing cells from total KIR+ NK cells in 49 samples from a nested sub-cohort of 35 subjects with normal or low KIR3DL1 expression genotypes. Overall, KIR3DL1 expression (Figure 2A) was readily detectable in the BAL fluid of most subjects but was significantly lower in subjects with low-expressing genotypes (Figure 2B), with a 13.6% decrease in the mean percentage of KIR3DL1-expressing cells when compared with subjects with normal KIR3DL1 genotypes (P = 0.02). Notably, only 15% of the KIR3DL1 normal genotype subjects had <10% KIR3DL1-expressing NK cells, whereas 53% of the KIR3DL1-low genotype subjects had <10% KIR3DL1 expression (P = 0.006). We did not detect a statistically significant difference in KIR3DL1 expression based on donor Bw6/Bw6, recipient Bw4+ status, although this analysis was potentially limited by the small sample size (Figure 2C).
Figure 2. KIR3DL1 frequency in NK cells from post-lung transplant BAL fluid.
(A) KIR3DL1 expression was determined by flow cytometry in BAL fluid NK cells that express KIR2D and/or KIR3D for 49 BAL samples from 35 lung transplant recipients. (B) Subjects were stratified by recipient genotype, with “Low” indicating at least one low-expressing KIR3DL1 allele, or (C) by donor and recipient Bw4 typing. Lines indicate mean and standard error estimates from general estimating equation-adjusted generalized linear models. “○” indicates low genotype, while “●” indicates normal expression. KIR3DL1 expression of the total KIR+ NK cell population was less frequent in the low KIR3DL1 genotype by 13.5% (95% CI 1.9–25.0%, P = 0.02). Donor / recipient Bw4 status was not associated with KIR3DL1 expression.
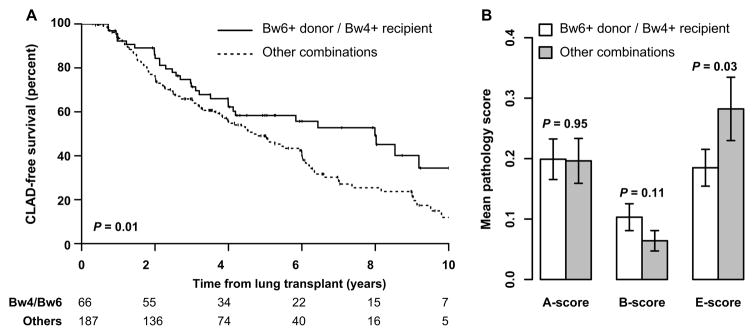
We analyzed CLAD-free survival as a function of HVG status (Table 2). As shown in Figure 3A, CLAD-free survival was higher in KIR3DL1+ Bw4+ recipients of Bw6/Bw6 grafts (P = 0.01 by log-rank test), with a 3.4-year difference in median survival. Using an unadjusted cox-proportional hazard model, this difference corresponded to a hazard ratio for CLAD or death of 0.62 (95% CI 0.41–0.92). Within the 148 subjects with normal predicted KIR3DL1 expression by genotype, the hazard ratio was 0.53 (95% CI 0.32–0.86). However, there was no significant association across the 105 subjects predicted to have low KIR3DL1 expression by genotype (HR 0.90, 95% CI 0.45–1.79). While decreased statistical power within the low KIR3DL1 genotype sub-group may explain some of the observed difference between these sub-groups, these data would also be consistent with a model where the frequency of KIR3DL1 receptors on recipient NK cells contributes to the HVG effect. Adjusting for donor and recipient age, gender, ethnicity, the type of transplant, indication for transplant, and the number of HLA mismatches at the A and B loci did not substantively change the findings (HR 0.59, 95% CI 0.38–0.92, P = 0.02).
Table 2.
CLAD-free survival by donor/recipient Bw4 status and KIR genotype in UCSF cohort
| KIR3DL1 genotype | HVG HLA mismatching1 | N | Median CLAD-free survival (years +/− 95% CI) | CLAD or death hazard ratio (+/− 95% CI) | P-value | ||
|---|---|---|---|---|---|---|---|
| Any | No | 187 | 4.56 | 3.89 – 5.60 | 1 | ||
| Yes | 66 | 7.99 | 3.99 - NA | 0.62 | 0.41 – 0.92 | 0.02* | |
| Normal | No | 102 | 4.65 | 3.82 – 6 | 1 | ||
| Yes | 46 | 7.99 | 4.65 - NA | 0.53 | 0.32 – 0.86 | 0.01* | |
| Low | No | 85 | 4.46 | 2.74 – 6.34 | 1 | ||
| Yes | 20 | 4.19 | 2.97 - NA | 0.90 | 0.45 – 1.79 | 0.76 | |
Donor Bw6/Bw6 and recipient Bw4+
Figure 3. CLAD-free survival and acute cellular rejection stratified by potential KIR3DL1-mediated host-versus graft NK cell activity in the UCSF cohort.
(A) CLAD-free survival in KIR3DL1+ Bw4+ recipients of Bw6+ lung allografts was compared with the remainder of the cohort using Kaplan-Meier analysis. Bw4+ recipients of Bw6+ lung allografts had improved CLAD-free survival by log-rank test (P = 0.01). The number of subjects at risk in each stratum is shown at 2-year intervals. (B) Average scores for perivascular (A-score, 946 biopsies on 233 subjects), bronchiolar (B-score, 937 biopsies on 233 subjects), and bronchial (E-score, 834 biopsies on 227 subjects) lymphocytic inflammation were quantified on large and small airway biopsies acquired within the first 6 months following transplant for 240 lung transplant recipients are shown stratified by Bw4+ recipients and Bw6+ donor status. Mean +/− 95% confidence intervals are shown, and differences between groups were determined by Student’s t-test.
We assessed acute cellular rejection for KIR3DL1+ recipients to determine if potential for 3DL1 activity was associated with reduced acute rejection (Figure 3B). In the first 6 months, we did not observe a difference in peri-vascular inflammation (A-score, P = 0.95) between groups. There was an increase in early large airway lymphocytic inflammation (E-score, P = 0.03) in subjects capable of HVG activity, but a trend towards decreased small airway inflammation (B-score, P = 0.11). No statistically significant differences were seen between groups in acute rejection scores after six months post-transplant (≥1033 biopsies on ≥225 subjects, P ≥ 0.65).
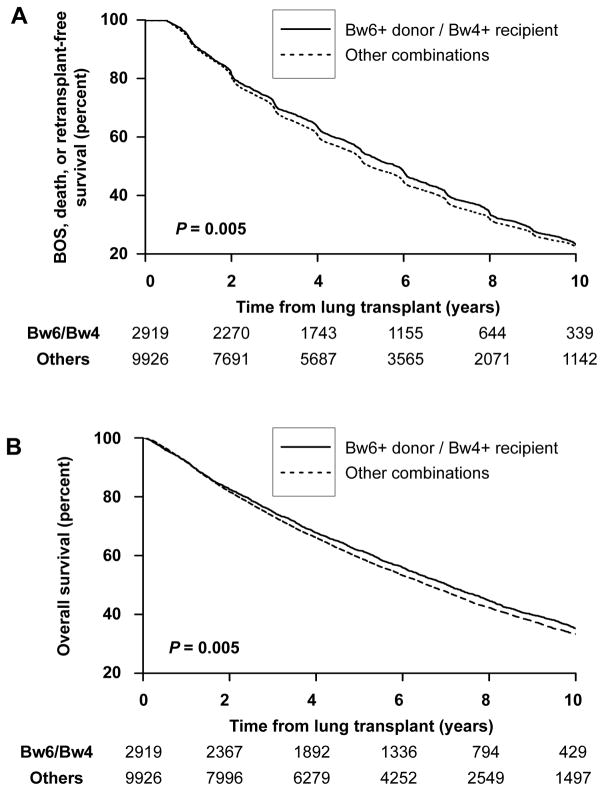
To test the validity of findings in the UCSF cohort, outcomes next were in HGV status-stratified subjects in the UNOS registry. Bw4+ recipients of Bw6/Bw6 grafts exhibited increased freedom from BOS, death, or retransplantation (Figure 4A, P = 0.005) and improved overall survival (Figure 4B, P = 0.005) compared with other combinations. The effect size was less than in the UCSF cohort. Adjusting for all the covariates in Table 1, the hazard ratio for BOS, death, or retransplantation was 0.94 (95% CI 0.90–0.99, P = 0.01). The adjusted hazard ratio for death alone was 0.93 (95% CI 0.88–0.98, P = 0.007). We also looked at the subset of White subjects receiving organs from White donors, where survival again was improved in the presence of HVG HLA mismatches (HR 0.93, 95% CI 0.87–0.99, P = 0.02).
Figure 4. Long-term outcomes by potential KIR3DL1-mediated host-versus graft NK cell activity in the UNOS cohort.
Freedom from BOS, death, or retransplantation (A) and overall survival (B) is shown for Bw4+ recipients of Bw6+ lung allografts compared other combinations was by Kaplan-Meier analysis. Bw4+ recipients of Bw6+ lung allografts had improved freedom from BOS, death, or retransplantation by log-rank test (A, P = 0.005) and improved overall survival (B, P = 0.005). The number of subjects at risk in each stratum is shown at 2-year intervals.
To test the impact of predicted KIR3DL1 ligand affinity and evaluate for an HLA-Bw4 gene dose effect, we subdivided subjects from the UNOS cohort by donor and recipient Bw4 type as well as by KIR3DL1 ligand interaction strength (Table 3). The strength of HLA and KIR interactions is determined the HLA ligand copy number and by the ligand affinity for the KIR receptor, with Bw4-80I ligands having greater affinity for KIR3DL1 than Bw4-80T ligands. Survival was higher for donor Bw6/Bw6 and recipient Bw4+ types than for any other combination, suggesting that both licensing and inhibition contribute to survival advantage. Having a higher-affinity Bw4-80I inhibitory ligand (HR per level increase 1.04, 95% CI 1.00–1.07, P = 0.03) and having multiple copies of HLA-Bw4 (HR per copy 1.06, 95% CI 1.02–1.11, P = 0.004) in the donor were both associated with worse survival, suggesting that the strength of donor NK cell inhibition is important. By contrast, lower ligand affinity and gene copies were associated with the largest improvements in survival, suggesting that low-level interactions are at least adequate for KIR licensing.
Table 3.
Overall survival by donor/recipient Bw4 status and Bw4 ligand strength in the UNOS cohort.
| Donor | Recipient | Inhibition | Licensing | N | Events | Median Survival (years +/− 95% CI) | Mortality hazard ratio (+/− 95% CI) | P-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Bw6/Bw6 | Bw4+ | − | + | 2919 | 1780 | 6.96 | 6.59 – 7.26 | 1 | ||
| Bw4+ | Bw4+ | + | + | 4891 | 3112 | 6.59 | 6.37 – 6.86 | 1.07 | 1.01 – 1.13 | 0.03* |
| Bw4+ | Bw6/Bw6 | + | − | 3045 | 1986 | 6.38 | 6.08 – 6.70 | 1.11 | 1.05 – 1.19 | <0.001*** |
| Bw6/Bw6 | Bw6/Bw6 | − | − | 1990 | 1273 | 6.49 | 6.14 – 6.82 | 1.09 | 1.01 – 1.17 | 0.02* |
| Donor | Recipient | Inhibition | Licensing | N | Median Survival (years +/− 95% CI) | Mortality hazard ratio (+/− 95% CI) | P-value | |||
| Bw6/Bw6 | Bw4+ | − | + | 2919 | 1780 | 6.96 | 6.59 – 7.26 | 1 | ||
| Bw4-80T+ | Bw4+ | + | + | 2272 | 1453 | 6.70 | 6.38 – 7.10 | 1.05 | 0.98 – 1.13 | 0.14 |
| Bw4-80I+ | Bw4+ | ++ | + | 2619 | 1659 | 6.48 | 6.16 – 6.84 | 1.08 | 1.01 – 1.15 | 0.03* |
| Bw6/Bw6 | Bw4+ | − | + | 2919 | 1780 | 6.96 | 6.59 – 7.26 | 1 | ||
| Bw4/Bw6 | Bw4+ | + | + | 3777 | 2385 | 6.69 | 6.47 – 7.04 | 1.05 | 0.98 – 1.11 | 0.15 |
| Bw4/Bw4 | Bw4+ | ++ | + | 1114 | 727 | 6.04 | 5.60 – 6.71 | 1.14 | 1.04 – 1.24 | 0.004** |
| Bw6/Bw6 | Bw6 | − | − | 1990 | 1273 | 6.49 | 6.14 – 6.82 | 1 | ||
| Bw6/Bw6 | Bw4-80T+ | − | + | 1540 | 939 | 7.16 | 6.69 – 7.63 | 0.89 | 0.82 – 0.97 | 0.007** |
| Bw6/Bw6 | Bw4-80I+ | − | ++ | 1379 | 841 | 6.73 | 6.17 – 7.13 | 0.96 | 0.88 – 1.04 | 0.32 |
| Bw6/Bw6 | Bw6/Bw6 | − | − | 1990 | 1273 | 6.49 | 6.14 – 6.82 | 1 | ||
| Bw6/Bw6 | Bw4/Bw6 | − | + | 2264 | 1377 | 7.00 | 6.60 – 7.38 | 0.91 | 0.84 – 0.98 | 0.02* |
| Bw6/Bw6 | Bw4/Bw4 | − | ++ | 655 | 403 | 6.70 | 6.13 – 7.41 | 0.95 | 0.85 –1.07 | 0.41 |
Discussion
In two cohorts of lung transplant recipients, we found improved CLAD-free survival in Bw4+ recipients who received lung allografts from Bw6/Bw6 donors as compared with other combinations, after adjusting for potential confounders. The improvement in CLAD-free survival was significant in the UCSF cohort with genotypes associated with normal, rather than low, KIR3DL1 expression, and associated with decreased inflammation of the large airways. In the UNOS cohort, donors with multiple copies of Bw4 ligands and with higher-affinity Bw4-80I ligands had the worst survival. NK cells expressing KIR3DL1 were present in the BAL fluid of most evaluated subjects, although low-expressing KIR3DL1 genotypes were associated with fewer KIR3DL1+ BAL NK cells and decreased effect of HVG mismatching.
Despite our hypothesis that decreased CLAD risk following HVG mismatching results from NK cell depletion of donor APC, we did not directly measure APC depletion, and a variety of other mechanisms could explain our observations. Indeed, the lack of effect on acute cellular rejection assessed by transbronchial biopsy is inconsistent with the murine lung transplant model, where NK cell depletion of APC resulted in decreased early acute cellular rejection. Alternatively, NK cell activation could modulate allo- or pathogen-specific T cell responses through cytokine release or contact-dependent interactions, or directly augment anti-fibrotic responses (21). Also, since HLA signal peptides are epitopes for HLA-E, HLA variations might result in the differential inhibition of NK cells through interactions with NKG2A rather than KIR3DL1 (22). Finally, despite multivariable adjustment, we cannot exclude unmeasured potential confounders. For example, these HLA-types could be in linkage disequilibrium with genes unrelated to NK cell activity that mediate the observed effect.
The most significant difference between the results from these two cohorts was the magnitude of the HVG mismatching effect, with a 3.4-year and a 7.4-month difference in the UCSF and UNOS cohorts, respectively. The larger number of subjects in the UNOS cohort might suggest this number more accurately reflects the true magnitude of the mismatching effect. Differences between these cohorts, such as the increased racial diversity of the UCSF cohort, might also explain some of the difference in effect size. Also, within the UNOS dataset, increased misclassification of subjects based on unknown KIR3DL1 status, inconsistent follow up reporting, or differences in HLA typing methods between centers may introduce bias towards the null, which could result in an underestimation of the effect size.
Increased survival in the UNOS cohort study was associated with decreased allograft inhibition as determined by gene copy and predicted KIR affinity. Interestingly, a similar effect of KIR interaction strength was not observed in the context of NK licensing. Although initial studies showed a linear relationship between the strength of NK cell licensing and NK cell activity (23), subsequent studies did not observe a requirement for NK cell licensing in protecting against viral infection (24) or in mediating antibody-dependent cell-mediated cytotoxicity (25). In the context of human lung transplantation, this analysis suggests that the presence but not the degree of NK cell licensing is important.
HLA-KIR interactions have been described involving HLA-A, B, and C molecules with other activating and inhibitory KIR receptors. We chose KIR3DL1/Bw4, because KIR3DL1 is highly prevalent and thus amenable to examination in the non-KIR typed UNOS cohort, however, HVG NK capability may be present in the absence of Bw6/Bw6 allograft to a Bw4+ recipient. These uninvestigated KIR-HLA relationships may have biased our findings towards the null. A more detailed dissection of other KIR ligand interactions might add to our understanding of the role of NK cells in lung transplant immune responses.
Given the relatively small effect observed attributable to KIR3DL1 ligand mismatching in the UNOS cohort, these data would not support modifying lung donor matching procedures. However, these findings provide a rationale for investigation of therapeutic strategies modulating NK cells or depleting donor APCs, such as targeting HLA class II-expressing cells during ex-vivo lung perfusion with cytotoxic antibodies (26) or blocking the interaction with HLA-Bw4 and KIR3DL1 in selected recipients.
Overall, we identified a sub-population of lung transplant recipients with improved survival associated with recipient NK cell licensing and absence of NK cell inhibition in the allograft. Prospective investigation is needed to determine if improved survival can be realized by enhancing NK cell control of allograft-specific immune responses.
Acknowledgments
Project funding came from Award Number IK2CX001034 from the Clinical Sciences Research & Development Service of the VA Office of Research and Development and the National Heart Lung and Blood Institute, National Institutes of Health, through the UCSF Career Development Program in Omics of Lung Diseases (grant number K12HL119997). The OPTN registry is supported in part by Health Resources and Services Administration contract 234-2005-370011C.
Abbreviations
- APC
antigen presenting cell
- BOS
bronchiolitis obliterans syndrome
- BAL
bronchoalveolar lavage
- CLAD
chronic lung allograft dysfunction
- CI
confidence interval
- FEV1
forced expiratory volume in 1 second
- FVC
forced vital capacity
- HR
hazard ratio
- HVG
host-versus-graft
- KIR
Killer cell immunoglobulin-like receptor
- MHC
Major Histocompatibility Complex
- NK
Natural Killer
- RAS
restrictive allograft syndrome
- UNOS
United Network for Organ Sharing
- UCSF
University of California, San Francisco
Footnotes
Disclaimer
The content is the responsibility of the authors alone and does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.HHS/HRSA. OPTN/SRTR 2012 Annual Data Report. 2014. [Google Scholar]
- 2.Yusen RD, Edwards LB, Kucheryavaya AY, Benden C, Dipchand AI, Dobbels F, et al. The registry of the International Society for Heart and Lung Transplantation: thirty-first adult lung and heart-lung transplant report--2014; focus theme: retransplantation. J Heart Lung Transplant. 2014;33(10):1009–24. doi: 10.1016/j.healun.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Verleden GM, Raghu G, Meyer KC, Glanville AR, Corris P. A new classification system for chronic lung allograft dysfunction. J Heart Lung Transplant. 2014;33(2):127–33. doi: 10.1016/j.healun.2013.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Todd JL, Palmer SM. Bronchiolitis obliterans syndrome: the final frontier for lung transplantation. Chest. 2011;140(2):502–8. doi: 10.1378/chest.10-2838. [DOI] [PubMed] [Google Scholar]
- 5.Rajalingam R. Polymorphic KIR-HLA System Regulates Natural Killer Cell Response. In: Ratcliffe M, editor. Encyclopedia of Immunology. Elsevier; 2016. pp. 369–80. [Google Scholar]
- 6.Anfossi N, Andre P, Guia S, Falk CS, Roetynck S, Stewart CA, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25(2):331–42. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–13. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 8.Du Z, Gjertson DW, Reed EF, Rajalingam R. Receptor-ligand analyses define minimal killer cell Ig-like receptor (KIR) in humans. Immunogenetics. 2007;59(1):1–15. doi: 10.1007/s00251-006-0168-4. [DOI] [PubMed] [Google Scholar]
- 9.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31(4):429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 10.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105(8):3053–8. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajalingam R. Variable interactions of recipient killer cell immunoglobulin-like receptors with self and allogenic human leukocyte antigen class I ligands may influence the outcome of solid organ transplants. Curr Opin Organ Transplant. 2008;13(4):430–7. doi: 10.1097/MOT.0b013e3283095248. [DOI] [PubMed] [Google Scholar]
- 12.Kwakkel-van Erp JM, van de Graaf EA, Paantjens AW, van Ginkel WG, Schellekens J, van Kessel DA, et al. The killer immunoglobulin-like receptor (KIR) group A haplotype is associated with bronchiolitis obliterans syndrome after lung transplantation. J Heart Lung Transplant. 2008;27(9):995–1001. doi: 10.1016/j.healun.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Greenland JR, Jewell NP, Gottschall M, Trivedi NN, Kukreja J, Hays SR, et al. Bronchoalveolar lavage cell immunophenotyping facilitates diagnosis of lung allograft rejection. Am J Transplant. 2014;14(4):831–40. doi: 10.1111/ajt.12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jungraithmayr W, Codarri L, Bouchaud G, Krieg C, Boyman O, Gyulveszi G, et al. Cytokine Complex-expanded Natural Killer Cells Improve Allogeneic Lung Transplant Function via Depletion of Donor Dendritic Cells. Am J Respir Crit Care Med. 2013;187(12):1349–59. doi: 10.1164/rccm.201209-1749OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ruggeri L, Capanni M, Urbani E, Perruccio K, Shlomchik WD, Tosti A, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 16.Pando MJ, Gardiner CM, Gleimer M, McQueen KL, Parham P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J Immunol. 2003;171(12):6640–9. doi: 10.4049/jimmunol.171.12.6640. [DOI] [PubMed] [Google Scholar]
- 17.Gardiner CM, Guethlein LA, Shilling HG, Pando M, Carr WH, Rajalingam R, et al. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J Immunol. 2001;166(5):2992–3001. doi: 10.4049/jimmunol.166.5.2992. [DOI] [PubMed] [Google Scholar]
- 18.Greenland JR, Jones KD, Hays SR, Golden JA, Urisman A, Jewell NP, et al. Association of large-airway lymphocytic bronchitis with bronchiolitis obliterans syndrome. Am J Respir Crit Care Med. 2013;187(4):417–23. doi: 10.1164/rccm.201206-1025OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Therneau TM, Lumley T. Survival Analysis. 2.38 ed. 2015. [Google Scholar]
- 20.Carey VJ, Lumley T, Ripley B. gee: Generalized Estimation Equation Solver. 2015. [Google Scholar]
- 21.Culley FJ. Natural killer cells in infection and inflammation of the lung. Immunology. 2009;128(2):151–63. doi: 10.1111/j.1365-2567.2009.03167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yunis EJ, Romero V, Diaz-Giffero F, Zuniga J, Koka P. Natural Killer Cell Receptor NKG2A/HLA-E Interaction Dependent Differential Thymopoiesis of Hematopoietic Progenitor Cells Influences the Outcome of HIV Infection. J Stem Cells. 2007;2(4):237–48. [PMC free article] [PubMed] [Google Scholar]
- 23.Brodin P, Karre K, Hoglund P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. 2009;30(4):143–9. doi: 10.1016/j.it.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Orr MT, Murphy WJ, Lanier LL. ‘Unlicensed’ natural killer cells dominate the response to cytomegalovirus infection. Nat Immunol. 2010;11(4):321–7. doi: 10.1038/ni.1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tarek N, Le Luduec JB, Gallagher MM, Zheng J, Venstrom JM, Chamberlain E, et al. Unlicensed NK cells target neuroblastoma following anti-GD2 antibody treatment. J Clin Invest. 2012;122(9):3260–70. doi: 10.1172/JCI62749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang CH, Sapra P, Vanama SS, Hansen HJ, Horak ID, Goldenberg DM. Effective therapy of human lymphoma xenografts with a novel recombinant ribonuclease/anti-CD74 humanized IgG4 antibody immunotoxin. Blood. 2005;106(13):4308–14. doi: 10.1182/blood-2005-03-1033. [DOI] [PubMed] [Google Scholar]